Summary
Neural Crest Cells (NCC) are a transient, embryonic cell population characterized by unusual migratory ability and developmental plasticity. To annotate and characterize cis-regulatory elements utilized by the human NCC we coupled a hESC differentiation model with genome-wide profiling of histone modifications, coactivator and transcription factor (TF) occupancy. Sequence analysis predicted major TFs binding at epigenomically annotated hNCC enhancers, including master NC regulator, TFAP2A, and nuclear receptors NR2F1 and NR2F2. Although many TF binding events occur outside enhancers, sites coinciding with enhancer chromatin signatures show significantly higher sequence constraint, nucleosomal depletion, correlation with gene expression and functional conservation in NCC isolated from chicken embryos. Simultaneous co-occupancy of TFAP2A and NR2F1/F2 is associated with permissive enhancer chromatin states, characterized by high levels of p300 and H3K27ac. Our results provide first global insights into human NC chromatin landscapes and a rich resource for studies of craniofacial development and disease.
Introduction
Transcriptional enhancers are the primary determinant of cell type-specific gene expression (Buecker and Wysocka, 2012; Bulger and Groudine, 2010, 2011). A central feature of enhancers is their ability to function as integrated TF binding platforms, recognized both by major lineage specifiers and DNA binding effectors of signaling pathways (Buecker and Wysocka, 2012; Mullen et al., 2011; Trompouki et al., 2011). Recent studies showed that epigenomic profiling of chromatin features commonly associated with enhancers, including occupancy of general transcriptional coactivators, hypersensitivity to nucleases and enrichment of certain histone marks at flanking nucleosomes, allows for identification of enhancers in a genome-wide, cell type-specific and conservation-independent manner (Heintzman et al., 2009; Rada-Iglesias et al., 2011; Visel et al., 2009).
We reasoned that hESC differentiation models combined with epigenomic enhancer annotation and sequence analysis of the underlying DNA, can be used as an unbiased approach to identify main TFs driving gene expression in transient cell types arising during human development. As a proof of concept, we focus here on the NCC, a vertebrate-specific transient embryonic cell group that is ectodermal in origin, but upon delamination from the neural tube acquires a remarkably broad differentiation potential and ability to migrate throughout the body to give rise to craniofacial bones and cartilages, peripheral nervous system, pigment cells, and certain cardiac structures (Gammill and Bronner-Fraser, 2003; Sauka-Spengler and Bronner-Fraser, 2008). Aberrant NC development is associated with a broad variety of congenital malformations, known as neurocristopathies, which due to a critical contribution of the NC to the head mesenchyme, often manifest in deafness, complex craniofacial defects, and include a large variety of syndromes, as well as non-syndromic manifestations, such as cleft lip and palate, one of the most common congenital defects (Birnbaum et al., 2009; Passos-Bueno et al., 2009).
Regulatory events that accompany NC formation occur at 3 to 6 weeks of human gestation and are largely inaccessible for studies in an embryonic context (Betters et al., 2010). To overcome this limitation we previously developed an in vitro hESC differentiation model, which recapitulates gene expression, migratory potential and differentiation characteristics of NCC (Bajpai et al., 2010). Here we use this model for genome-wide analyses of chromatin marking patterns, gene expression and TF occupancy in hNCC. Through these analyses we annotate hNCC enhancer regions and subsequently predict and confirm that they are commonly co-occupied by the NC lineage specifier TFAP2A and orphan nuclear receptors NR2F1/2. We further show that these TFs synergize to bring about active chromatin states and demonstrate the requirement for NR2F1 function in NC gene expression, enhancer activity and morphogenesis of the ectomesenchyme.
Results
Epigenomic profiling of hNCC
To obtain NCC for genomic analyses, we used an in vitro differentiation model in which hESC are first induced to form neuroectodermal spheres (hNEC) that subsequently give rise to migratory cells expressing early NC markers and recapitulating neuronal, mesenchymal and melanocytic differentiation potential of the NC (Bajpai et al., 2010). To examine chromatin patterns associated with NC regulatory regions we performed ChIP-seq analyses from hNCC population in which both premigratory and migratory anterior NC fates were represented, using antibodies recognizing p300, H3K4me1, H3K27ac, H3K4me3 and H3K27me3 (Figure S1A). We identified over 4300 genomic elements marked by the active enhancer signature (Heintzman et al., 2009; Rada-Iglesias et al., 2011), defined by the occupancy of p300, with simultaneous enrichment of H3K27ac and H3K4me1 at flanking regions and absence of H3K4me3 (listed in Supplementary Data 1). Typical of enhancers, identified elements were generally located within 1-200 kb away from the nearest transcription start site (TSS; Figure S1B), overlapped with FAIRE hypersensitive sites (Figure S1C) and were on average more evolutionary constrained than flanking non-coding regions (Figure S1D).
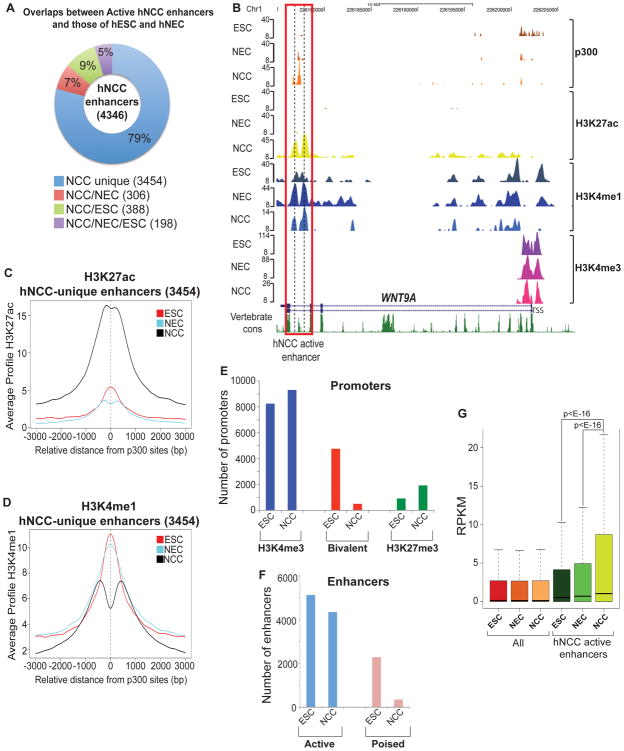
Comparisons of hNCC ChIP-seq results with the corresponding datasets from hESC and hNEC (Rada-Iglesias et al., 2011) revealed that 79% of the identified regions were marked by the active enhancer signature in hNCC, but not hESC or hNEC (Figure 1A, example shown in Figure 1B). H3K27ac, a histone mark tightly correlated with active enhancer states (Bonn et al., 2012; Cotney et al., 2012; Creyghton et al., 2010; Heintzman et al., 2009; Rada-Iglesias et al., 2011) was the most dynamic across cell types (Figure 1C, example shown in Figure 1B), whereas H3K4me1 was often present at the identified regions already in hESC (Figure 1D, example shown in Figure 1B), in agreement with its proposed role in enhancer priming (Bonn et al., 2012; Creyghton et al., 2010; Rada-Iglesias et al., 2011). In contrast to enhancers, considerably less cell type-specific variation was observed in chromatin marking patterns at proximal promoter regions (Figure S1E). Thus, enhancer utilization is highly dynamic even among the genetically matched and transient cell types examined in our studies.
Figure 1. Epigenomic mapping uncovers hNCC enhancer elements (see also Figure S1).
(A) Cell type-specificity of hNCC active enhancer regions. (B) p300, H3K27ac, H3K4me1 and H3K4me3 enrichment profiles in hESC, hNEC and hNCC at a representative hNCC enhancer (e.g. WNT9A). The peak height corresponds to normalized fold-enrichments calculated by QuEST. (C) Average hNCC H3K27ac and (D) H3K4me1 ChIP-seq signal profiles around the central position of p300 peaks at unique hNCC active enhancers. (E) Number of H3K4me3-only, bivalent (i.e. enriched in both H3K4me3 and H3K27me3) and H3K27me3-only promoters in hESC and hNCC. (F) Number of active and poised enhancers in hESC and hNCC. (G) Gene expression, measured as RPKMs, calculated for all human ENSEMBL genes and for those closest (within 100 Kb) to hNCC active enhancers. Expression levels presented as boxplots. P-values calculated using paired Wilcoxon tests.
hESC-associated chromatin features are not prevalent in hNCC
Due to their extreme developmental plasticity, human NCC are postulated to share many properties of hESC (d'Aquino et al., 2011; Kaltschmidt et al., 2011). Among prevalent features of hESC chromatin are so-called bivalent promoters (marked by both H3K4me3 and H3K27me3) (Zhao et al., 2007) and poised developmental enhancers characterized by the presence of H3K27me3 instead of H3K27ac (Rada-Iglesias et al., 2011). We observed these features to be much less abundant in hNCC, whereas the number of enhancers and promoters marked by active signatures is comparable between the two cell types (Figure 1 E-F). This result is not an artifact of ineffective H3K27me3 ChIP-seq in hNCC, as the number of H3K27me3-only marked promoters is higher in hNCC than in hESC. In addition, we cannot exclude the possibility that a portion of the observed hNCC bivalent promoters and poised enhancers arises from heterogeneity in the hNCC population, rather than true bivalency. Regardless, our results suggest that hNCC did not broadly coopt typical hESC chromatin features, albeit it remains a possibility that rare cells within our population do exhibit hESC-like properties.
Functional annotation of hNCC enhancers reveals association with craniofacial development and disease
To test whether presence of the identified putative enhancer regions positively correlates with expression of nearby genes, we performed polyA transcriptome analysis of hNCC by RNA-seq and validated the results by RT-qPCR analysis of selected transcripts (Supplementary Data 2, Figure S1F). Transcriptome profiling confirmed our earlier observations that hNCC express well-characterized NC markers (e.g. TWIST1, SNAI1, SNAI2, SOX9, SOX10, SOX11, FOXD3, NGFR (p75), TFAP2A) and showed that these cells are HOX-negative and DLX1/2 -positive, a gene expression pattern consistent with the dorsal-anterior cranial NCC, which preferentially differentiate into ectomesenchymal derivatives and give rise to the majority of craniofacial bones and cartilages (Minoux and Rijli, 2010). We next examined expression of transcripts arising from the TSS closest to hNCC enhancers and observed a strong positive correlation between identified regions and genes highly expressed and upregulated in hNCC (Figure 1G).
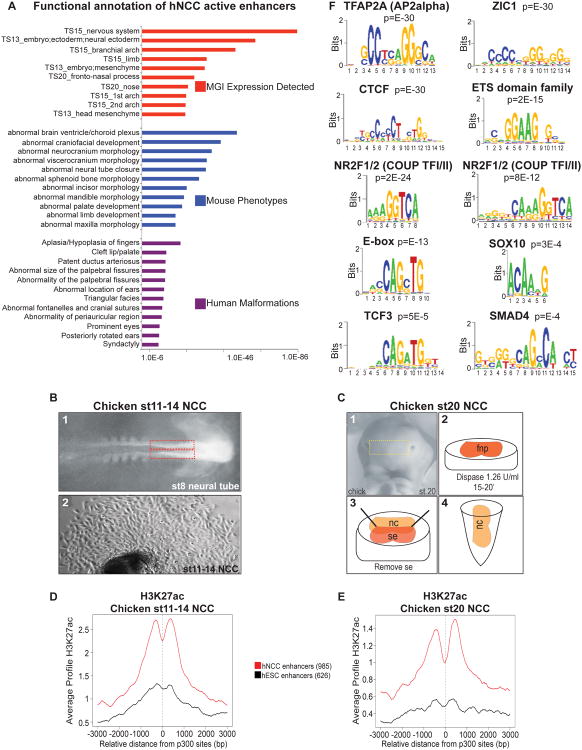
Functional annotation of hNCC enhancers using GREAT (McLean et al., 2010) showed association with genes expressed in neurectoderm, branchial arches and head mesenchyme consistent with the dorsal-anterior pre- and postmigratory NC identity of the analyzed cell population (Figure 2A, top panel). Strikingly, when we considered mouse phenotypes associated with deletion of genes linked to the identified enhancers, the most enriched categories included abnormalities of cranial bones, neural tube defects, and malformations of NC-derived facial structures, such as the mandible, maxilla and palate (Figure 2A, middle panel). Moreover, when associations with human malformations were considered, enriched categories included cleft lip/palate, abnormalities of palpebral fissures and ear structures and other craniofacial anomalies, all consistent with defects of cranial NC-derived head mesenchyme (Passos-Bueno et al., 2009), as well as open (patent) ductus arteriosus, a specialized heart blood vessel derived largely from the cardiac NC (Ivey et al., 2008) (Figure 2A, bottom panel). Association with hand malformations was also detected, which is not unexpected, as limb anomalies are commonly associated with neurocristopathies (Tucker et al., 1999). A large number of genes previously implicated in NC development, Wnt signaling, cell migration and malformations including cleft lip/palate, frontonasal dysplasia and patent ductus arteriosus have at least one hNCC enhancer located in relative proximity (select functional categories and corresponding genes are shown in Supplementary Table 1). Taken together, our data strongly suggests that the identified elements represent bona fide active enhancers relevant for NC gene expression, human craniofacial development and neurocristopathogenesis.
Figure 2. Functional annotation and sequence analysis of active hNCC enhancers (see also Figure S2).
(A) Annotation of hNCC active enhancers. Mouse Genome Informatics (MGI) Expression Detected (red) contains information on tissue and developmental stage– specific expression in mouse; Mouse Phenotypes (blue) ontology contains data on mouse genotype-phenotype associations; Human Malformations (purple) ontology contains data on human genotype - phenotype associations. The x-axis values correspond to Binomial raw (uncorrected) p-values. (B) Dorsal anterior neural folds from st8 chicken embryos were excised (red line) and cultured ex vivo for 72 hours, resulting in emigration of NCC estimated to correspond to chicken st11-14 cranial NCC. (C) Chicken st20 NCC were isolated in vivo from frontonasal prominences (FNP). FNPs were incubated with dispase in order to remove the surface ectoderm and forebrain neuroectoderm, which resulted in FNPs solely comprised of NCC. (D-E) Average ChIP-seq H3K27ac profiles from chicken NCC at st11-14 (D) or st20 (E) generated around central positions of hNCC enhancers conserved in the chicken genome. (F) Select overrepresented DNA sequence motifs enriched at hNCC active enhancers. Statistical significance (p-value) of the motifs over-representation is shown.
Conservation of enhancer H3K27ac patterns in chicken embryo NCC
Regulatory regions commonly show lower evolutionary constraint than coding sequences (Bulger and Groudine, 2010, 2011). Nonetheless, for 23% of the hNCC enhancer regions, corresponding conserved sequences can be identified in the chicken genome. Therefore, to further support biological relevance of the identified human enhancers, we examined conservation of H3K27ac, a chromatin mark closely associated with enhancer activity, at these corresponding regions in chicken NCC. To obtain chicken NCC we dissected the dorsal anterior neural folds from embryos prior to the onset of NC migration (st8) and cultured them ex vivo for 72 hours (approximately st11-14), allowing NCC to emigrate out (Figure 2B). In parallel, we dissected the frontonasal prominences of st20 embryos and separated NCC away from the epithelium (Figure 2C) (Brugmann et al., 2010). We chose the frontonasal prominence over other facial prominences because it is solely comprised of NC mesenchyme. These early and late chicken NCC populations were then analyzed by H3K27ac ChIP-seq, average H3K27ac enrichment profiles were generated over chicken genomic regions corresponding to hNCC enhancers or, to control for specificity, those corresponding to hESC enhancers. Remarkably, regions corresponding to hNCC enhancers showed high enrichment of H3K27ac in chicken NCC (both early and late) with a characteristic central dip in enrichment profile, indicative of the presence of nucleosomal depletion/hypermobility regions typical of enhancers (Figure 2D-E). As exemplified by the conserved enhancer acetylation patterns at select NC gene loci, corresponding chicken enhancers were acetylated either in both chicken NCC populations, or preferentially in one population (e.g. compare PLXND1 and ETS1 loci with MYCN locus) (Figure S2A). Taken together, these results validate biological relevance of our analysis and uncover conservation of H3K27ac patterns at NCC enhancers between species.
To further substantiate functional conservation of identified regulatory regions, we selected three human elements and tested their activity in vivo in the zebrafish transgenic enhancer reporter assay (Figure S2B). The selected enhancers (named A-C) were proximal to, respectively, WNT1, CCND1 and BMP7 genes and corresponded to uncharacterized genomic sequences, with exception of the WNT1 proximal enhancer, which falls within a broader region homologous to that commonly used as WNT1 NC-specific driver in mouse conditional knockout experiments (Echelard et al., 1994). Importantly, all three tested enhancers showed GFP expression in cranial NCC migrating to anterior branchial arches, as demonstrated by colocalization with Sox10 reporter-labeled cells (in red) (Figure S2C). In addition, enhancers proximal to WNT1 and BMP7 showed activity in the dorsal neural tube, where delaminating NCC could be observed (Figure S2C).
Widespread association of hNCC enhancers with the NC master regulator TFAP2A
We hypothesized that interrogation of DNA sequence motifs most recurrent at hNCC enhancer regions will reveal identity of major TFs driving NC gene expression. The most highly overrepresented motif corresponds to the recognition sequence for TFAP2A (a.k.a. AP2α), a TF implicated in NC induction, specification and differentiation (Figure 2F) (de Croze et al., 2011). TFAP2A deletion in mice results in severe dysmorphogenesis of the face, skull, sensory organs and cranial ganglia (Schorle et al., 1996), whereas in humans mutations in TFAP2A cause autosomal dominant branchiooculofacial syndrome (BOFS), characterized by malformations of craniofacial structures of NC origin (Milunsky et al., 2008).
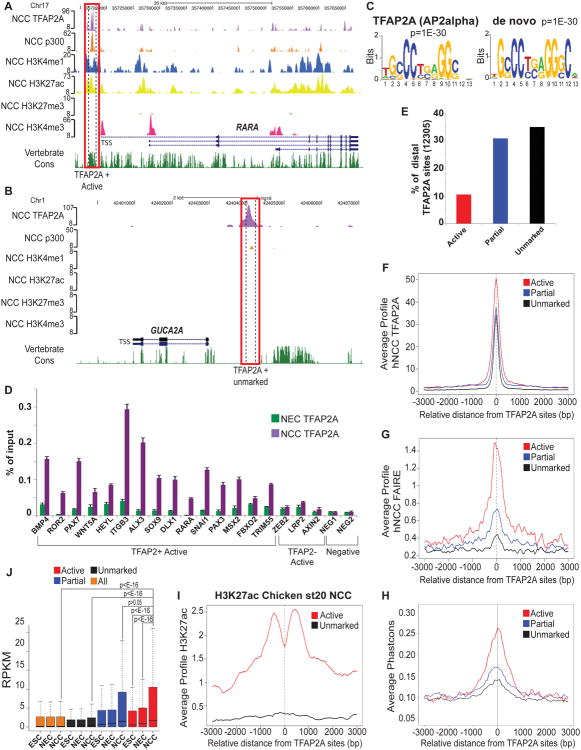
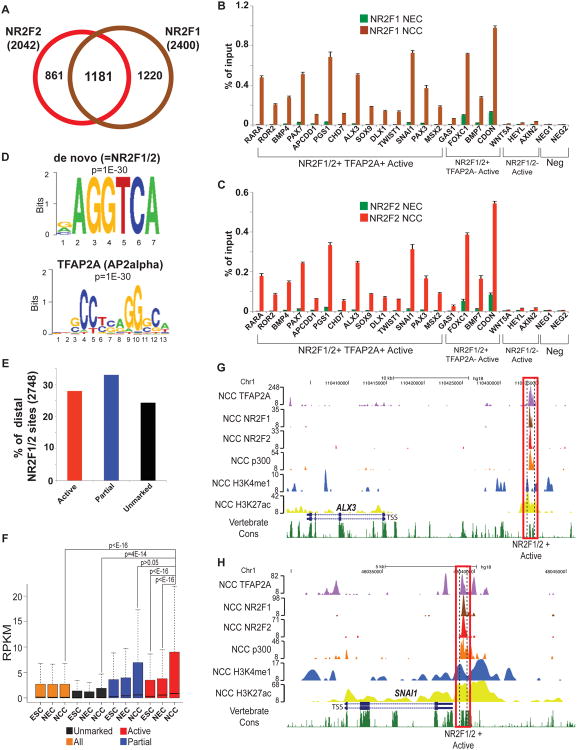
Despite the critical role of TFAP2A, its genomic location in the context of NC remains unknown. To test whether TFAP2A is broadly associated with hNCC enhancers we performed TFAP2A ChIP-seq analysis, which identified 16,851 high-confidence binding sites, of which 12,305 (73%) were promoter-distal (Figure 3A-B). As expected, the TFAP2A consensus binding sequence was the most overrepresented motif among bound regions, even in a de novo analysis (Figure 3C) with ∼90% of bound regions containing at least one strong TFAP2A binding site. Importantly, 30% of all hNCC enhancer regions were bound by TFAP2A (a representative locus is shown in Figure 3A), indicating extensive occupancy of TFAP2A at hNCC enhancers. To further validate our data we performed ChIP-qPCR analyses from cells corresponding to two developmental time points: (i) neuroectodermal rosettes (hNEC) just prior to NC migration and, (ii) hNCC after migration from rosettes. Low/no TFAP2A binding was detected at any of the interrogated regions in hNEC, but significant TFAP2A enrichment was observed at enhancers bound by TFAP2A according to ChIP-seq in hNCC (Figure 3D).
Figure 3. Occupancy of TFAP2A at active hNCC enhancers (see also Figures S3 and S9).
(A-B) ChIP-seq enrichment profiles in hNCC for two representative TFAP2A bound loci, either (A) overlapping an active hNCC enhancer or (B) occurring within unmarked chromatin. The peak height corresponds to normalized fold-enrichments calculated by QuEST. (C) Top overrepresented motifs enriched at distal TFAP2A-bound regions based on matches to known transcription factors (left) or through de novo motif analysis (right). (D) TFAP2A ChIP-qPCR analyses of select distal TFAP2A sites identified by ChIP-seq performed from chromatin isolated from hNCC or neurectodermal spheres (hNEC), just prior to attachment and emigration of hNCC. Error bars represent standard deviation from three technical replicates. (E) Percentage of distal TFAP2A sites that overlapped active hNCC enhancers (red), regions with a partial active enhancer signature (blue) or unmarked chromatin (black). (F-H) Average TFAP2A ChIP-seq (F), FAIRE-seq (G) and vertebrate Phastcons (H) signal profiles around the central position of distal TFAP2A regions overlapping active hNCC enhancers (red), or displaying a partial active enhancer signature (blue) or occurring within unmarked chromatin (black). (I) Average chicken st20 NCC H3K27ac ChIP-seq signal profiles around the central position of TFAP2A bound regions conserved in the chicken genome and overlapping active hNCC enhancers (red) or occurring within unmarked chromatin (black). (J) Gene expression, measured as RPKMs, was calculated for all human ENSEMBL genes and for those closest (within 100Kb) to distal TFAP2A bound regions overlapping active hNCC enhancers (red), displaying a partial active enhancer signature (blue) or occurring within unmarked chromatin (black). Expression levels presented as boxplots. P-values calculated using Wilcoxon tests.
Chromatin signatures facilitate identification of relevant TFAP2A binding sites
Although nearly a third of hNCC enhancer regions is occupied by TFAP2A, surprisingly, only 10% of distal TFAP2A sites coincide with the presence of the active enhancer signature, with an additional 30% of sites occurring within regions marked by a ‘partial’ signature (that is only p300 or only enhancer-associated histone modifications) (Figure 3E). To examine whether the presence or absence of enhancer signatures at TFAP2A sites reflects biologically meaningful differences we compared genomic properties of TFAP2A sites with or without enhancer chromatin signatures. On average, TFAP2A enrichment levels at regions overlapping active signatures were less than 1.5 fold higher than over unmarked regions (Figure 3F). Indeed, robust TFAP2A binding at sites occurring within unmarked chromatin was evident upon examination of individual loci (for a representative example, see Figure 3B), indicating that unmarked sites do not simply represent weak affinity associations. Strikingly, both nucleosomal depletion/hypermobility measured by FAIRE, as well as DNA sequence conservation, were substantially stronger at TFAP2A sites overlapping active signatures than at those unmarked, with sites overlapping ‘partial’ signatures falling in between (Figure 3G-H). Although, on average, TFAP2A-bound regions overlapping active signatures were more evolutionary constrained in sequence, we could still identify many unmarked TFAP2A sites conserved in the chicken genome. Nonetheless, when we compared H3K27ac patterns over these corresponding regions in chicken NCC, we observed that conserved TFAP2A sites that coincided with active signatures in hNCC were also enriched for H2K27ac in chicken NCC, while conserved unmarked TFAP2A sites showed no such enrichment (Figure 3I, Figure S3), supporting functional conservation of TFAP2A binding events.
Importantly, TFAP2A sites overlapping active or partial enhancer signatures were linked to genes highly expressed and upregulated in hNCC, but unmarked TFAP2A sites showed no correlation with elevated expression in any of the examined cell types (Figure 3J). Similarly, functional annotation of TFAP2A sites overlapping active enhancers linked them to NC and craniofacial genes, whereas unmarked TFAP2A sites lacked clear association with NC or other tissues (data not shown). Taken together, these results suggest that chromatin signatures can facilitate identification of TFAP2A sites relevant for NC gene expression (and perhaps more generally, identification of productively engaged TF sites, see Discussion) and indicate that TFAP2A binding is not sufficient for the assembly of the productive enhanceosome complexes.
Sequence analysis of hNCC enhancers detects recognition motifs of major NC TFs and signaling effectors
We hypothesized that to activate NC gene expression, TFAP2A cooperates with additional sequence-specific TFs binding at hNCC enhancer elements. We reasoned that such factors should be well expressed in NC and their recognition motifs overrepresented at hNCC enhancers. We therefore reexamined enriched hNCC enhancer sequence motifs, which in addition to the TFAP2A motif, included those for zinc finger protein ZIC1, the insulator binding protein CTCF, two highly related orphan nuclear receptors NR2F1 and NR2F2 (a.k.a. COUP TF1 and COUP TF2), ETS-domain TFs, and Helix-loop-Helix TFs recognizing the E-box (Figure 2F). Also detectable, albeit with lower statistical significance, were recognition motifs for the WNT-signaling effector TCF3, the BMP-signaling effector SMAD4 and the SOXE family TFs, such as SOX10 (Figure 2F). These results are in close agreement with genetic and embryological NC studies in model organisms, which implicated Zic1 (Sato et al., 2005), Sox10 (Honore et al., 2003), Ets-1 (Theveneau et al., 2007), and E-box binding proteins c-Myc, Twist, Snail1/2 as critical regulators of NC development (Aybar et al., 2003; Bellmeyer et al., 2003; Soo et al., 2002; Taneyhill et al., 2007). Moreover, Wnt and Bmp signaling plays a major role in NC formation (Garcia-Castro et al., 2002; Liem et al., 1995) and it is not unexpected that effectors of these pathways, such as TCF3 and SMAD4 converge on hNCC enhancers. Thus, our analysis identified majority of the known NC TFs and signaling effectors, with a notable exception of FoxD3 (Teng et al., 2008). However, FoxD3 has been shown to predominantly act as a repressor (Pohl and Knochel, 2001), and it is conceivable that it binds genomic sites distinct from active enhancer regions. Regardless, our data underscore validity of the epigenomic approach and suggest that humans utilize the same major NC TFs as other vertebrates.
Nuclear receptor NR2F1 regulates NC gene expression
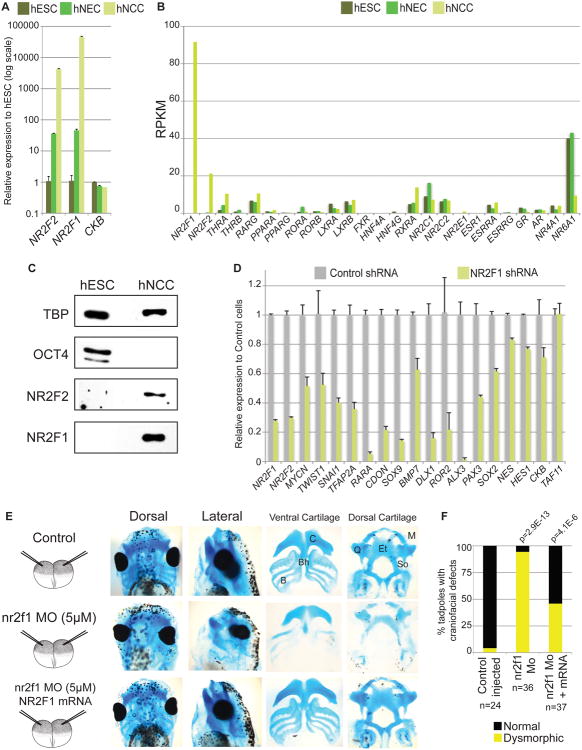
Interestingly, two of the highly enriched motifs represent recognition sites for TFs previously not implicated in NC development, CTCF and NR2F1/2. In contrast to the ubiquitously expressed CTCF, expression of NR2F1 and NR2F2 mRNA was dramatically upregulated during hESC differentiation to hNCC (over 40000-fold and 4000-fold, respectively; Figure 4A). Indeed, NR2F1 and NR2F2 were the most highly expressed and most highly upregulated in hNCC of all interrogated nuclear receptors (Figure 4B). This upregulation was accompanied by extensive chromatin remodeling at the NR2F1 and NR2F2 gene loci (Figure S4A), as has been reported for other master developmental regulators undergoing activation (Cotney et al., 2012), and by the induction of respective protein products (Figure 4C). We reasoned that NR2F1 and NR2F2 represent candidates for new NC TFs that may synergize with TFAP2A at hNCC enhancers. Consistently with such possibility, NR2F1/2 recognition motif is overrepresented not only at hNCC enhancer regions, but also at TFAP2A distal sites (Figure S4B), with a strong preference for those overlapping enhancer signatures.
Figure 4. NR2F1/2 are induced in hNCC, regulate expression of NC genes in vitro and craniofacial development in vivo (see also Figures S4 and S5).
(A) Expression of NR2F1 and NR2F2 was measured in hESC, hNEC and hNCC using RT-qPCR. Expression normalized to that of the housekeeping gene EEF2 and presented relative to hESC in a logarithmic scale is shown. Error bars represent standard deviation from three technical replicates. (B) Expression of select nuclear receptors, including NR2F1 and NR2F2 as measured by RNA-seq (RPKMs). (C) Immunoblot analyses of TBP, OCT4, NR2F2 and NR2F1 in hESC and hNCC. (D) Expression of indicated genes was measured by RT-qPCR in hNCC expressing shRNA against NR2F1 (green). For all genes, expression is normalized to that of the housekeeping gene EEF2 and presented relative to hNCC expressing control-scrambled shRNA (gray). Error bars represent standard deviation from three technical replicates. (E) Craniofacial defects in tadpoles raised from 2-cell stage embryos injected into both blastomeres with nr2f1 MO, control-injected or co-injected with nr2f1 MO and human NR2F1 mRNA. Embryos were stained with alcian blue beyond stage 40-45 to visualize craniofacial cartilages. For each set of injected embryos, dorsal and lateral views of the head are shown, as well as dissected ventral (i.e. C: ceratohyal, Bh: basihyal, B: branchial) and dorsal cartilages (M: Meckel, Q: quadrate, Et: ethmoid, So: subocular). (F) Penetrance of craniofacial dysmorphisms in analyzed tadpoles. P-values were calculated using hypergeometric tests and represent the statistical significance of the nr2f1 MO induced defects (vs control) and of the NR2F1 mRNA rescues (vs nr2f1 MO), respectively.
To establish whether NR2F1 function is important for transcription of NC genes we induced expression of a NR2F1-targeting shRNA during differentiation of hESC to hNCC. In several independent experiments, we observed approximately a 50-70% reduction in NR2F1 mRNA compared to control cells expressing a scrambled shRNA (Figure 4D, Figure S4C). Importantly, while genes associated with neural induction (e.g. NES, HES1) or housekeeping genes (e.g. TAF11, CKB) were only modestly affected, multiple genes involved in NC and/or craniofacial development (e.g. SNAI1, DLX1, ALX3, MYCN, SOX9, CDON, RARA, ROR2), as well as TFAP2A and NR2F2, were significantly downregulated upon NR2F1 knockdown (Figure 4D, Figure S4C), suggesting a major role for NR2F1 in NC gene regulation.
NR2F1 downregulation perturbs craniofacial development in Xenopus
Due to the fundamental contribution of the NC into the head skeleton and face, craniofacial defects are a hallmark of human neurocristopathies and perturbations of NC regulators in animal models. NR2F1/2 are highly conserved across vertebrates and we turned to the Xenopus laevis embryo model to examine functional consequences of NR2F1/2 depletion for craniofacial morphogenesis. RNA in situs for Xenopus nr2f1 and nr2f2 demonstrated that both genes are expressed in similar structures during Xenopus laevis embryogenesis, including brain, eye primordia, heart field and early somites, in accordance with earlier reports (Figure S5A-B). With respect to NC, no expression was detected at the neural plate border/premigratory crest at the early neurula stage, but by the late neurula stage nr2f2 expression was apparent in the streams of migrating cranial NCC (Figure S5A-B; see arrows; of note, this was less evident for nr2f1, but the nr2f2 probe generally gave stronger signal). At the tailbud stage strong expression of both nr2f1 and nr2f2 was observed in the first, second and third branchial arch (Figure S5A-B), in agreement with a potential role of these receptors in the migratory cranial NCC and ectomesenchyme.
To examine functional consequences of nr2f1 knockdown on NC-derived cranial cartilage, both blastomeres of 2-cell stage embryos were injected with morpholino oligonucleotide (MO) targeting translation of nr2f1 (Figure 4E). Resultant morphants exhibited severe and penetrant craniofacial dysmorphisms and defects of NC derived cartilages (p=2.9×E-13), which were absent in control embryos and partially rescued (p=4.1×E-6) in embryos co-injected with MO and mRNA of the corresponding human factor (Figure 4E-F). We also performed experiments with spatially restricted knockdowns, in which nr2f1 MO was co-injected with a fluorescent lineage tracer into a single dorsal-animal (DA) blastomere of 8-cell stage embryos. Such targeted injection restricts the MO to the neural tube and DA structures on the injected side of the embryo (overlapping with the majority of the cranial NC and its derivatives), while leaving the uninjected side as a control for relative comparisons (Figure S5C). Phenotypic analysis of the DA blastomere-injected nr2f1 morphants revealed craniofacial dysmorphisms and cranial cartilage defects on the injected side; these malformations were rescued by co-injection of the corresponding human mRNA (Figure S5D-E). In sum, our results demonstrate that nr2f1 downregulation is associated with ectomesenchyme defects typical of perturbations in NC regulators. Certainly, nr2f1 morphants show additional abnormalities consistent with reported roles of nr2f1 in other tissues. In addition, we cannot exclude the possibility of non-cell autonomous contribution to observed ectomesenchyme defects. Nonetheless, considering in vivo phenotype and autonomous requirement for NR2F1 in NC gene expression in vitro, our cumulative observations are in agreement with the direct role of NR2F1 in cranial NC regulation.
NR2F1 and NR2F2 occupy enhancers located in proximity of critical NC genes
To investigate NR2F1 and NR2F2 genomic occupancy in hNCC we performed ChIP-seq analyses, which identified 2400 and 2042 high confidence sites, respectively, with a pronounced overlap in binding patterns of the two factors, consistently with similar DNA binding specificity and NR2F1-NR2F2 heterodimerization (Cooney et al., 1992) (Figure 5A). NR2F1 sites identified as NR2F2-negative with our stringent calling cutoffs, had nonetheless subthreshold NR2F2 enrichment and vice versa (Figure S6A-B). Furthermore, average enrichment profiles at NR2F1/2 co-occupied, NR2F1-only or NR2F2-only occupied sites revealed binding of both nuclear receptors in all three categories (Figure S6C-E), suggesting that our peak calling cut-offs underestimated the true degree of NR2F1 and NR2F2 binding overlap. We therefore considered a combined NR2F1/2 binding dataset (comprised of 3262 genomic regions, Figure 5A, Supplementary Data 3) for further analyses. ChIP-qPCR from an independently derived hNEC and hNCC samples confirmed hNCC-specific association of NR2F1 and NR2F2 with regions identified by ChIP-seq, as well as enrichment of NR2F2 at NR2F1-only sites (e.g. FOXC1, PAX7) and of NR2F1 at NR2F2-only sites (e.g. PGS1, APCDD1) (Figure 5B-C).
Figure 5. Genome-wide analysis of NR2F1/2 occupancy in hNCC (see also Figure S6).
(A) Overlap between NR2F1 (brown) and NR2F2 (red) binding sites in hNCC. (B-C) Chromatin isolated from hNCC and neurectodermal spheres (hNEC), just prior to attachment and emigration of hNCC was used for NR2F1 (B) and NR2F2 (C) ChIP-qPCRs. Error bars represent standard deviation from three technical replicates. (D) Top overrepresented motifs enriched at distal NR2F1/2-bound regions based on either de novo motif analysis (top, which matches NR2F1/2 motif) or matches to known transcription factors (bottom). (E) Percentage of distal NR2F1/2 sites that overlapped active hNCC enhancers (red), regions with a partial active enhancer signature (blue) or that occurred within unmarked chromatin (black). (F) Expression levels, measured as RPKMs, were calculated for all human ENSEMBL genes and for those closest (within 100 Kb) to distal NR2F1/2 bound regions overlapping active hNCC enhancers (red), or displaying a partial active enhancer signature (blue) or occurring within unmarked chromatin (black). Expression levels presented as boxplots. P-values calculated using Wilcoxon tests. (G-H) ChIP-seq enrichment profiles in hNCC for two representative NR2F1/2-bound loci (proximal to ALX3 (G) and SNAI1 (H)) overlapping active hNCC enhancers.
A strong overrepresentation of the NR2F1/2 and TFAP2A motifs was detected at NR2F1/2 bound sites (Figure 5D). Moreover, the majority of identified NR2F1/2 regions coincided with active (28%) or partial (33%) enhancer chromatin signatures (Figure 5E). As observed for TFAP2A, NR2F1/2 sites overlapping active signatures displayed higher nucleosomal depletion and sequence conservation than unmarked sites (Figure S6F-G) and showed much stronger association with genes upregulated in hNCC (Figure 5F), further generalizing the usefulness of combining TF binding data with epigenomic profiles for identification of relevant TF sites. Inspection of individual loci revealed NR2F1/2 binding in proximity of NC genes downregulated upon knockdown of NR2F1, including SNAI1, TFAP2A, ALX3, CDON, MYCN, RARA, ROR2, DLX1, SOX9 and NR2F2 (examples shown in Figure 5G-H).
NR2F1/2 binding sites are required for activity of select hNCC enhancers
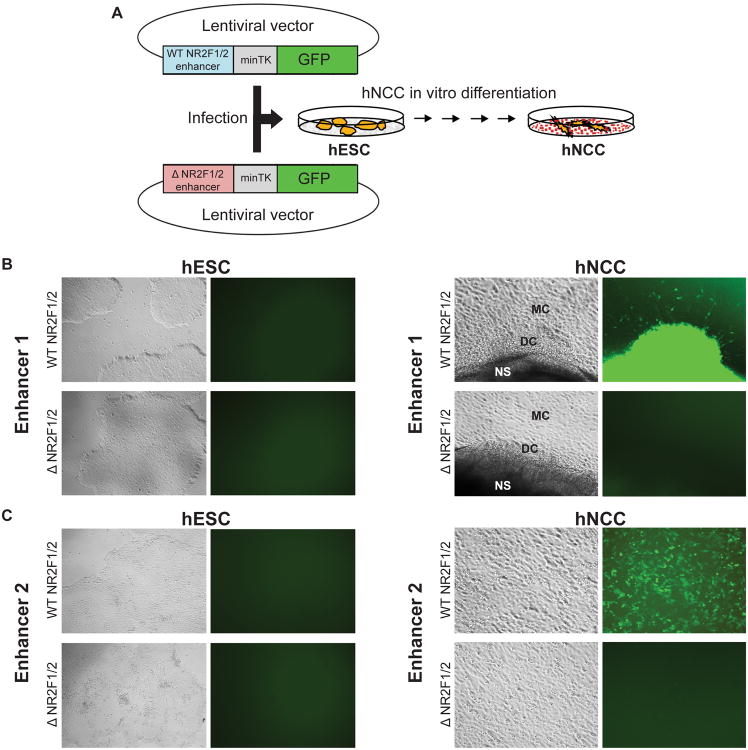
Association of NR2F1 binding sites with active enhancer regions located in proximity of genes induced in the NC, as well as downregulation of NC genes upon NR2F1 knockdown, suggest that in the context of hNCC NR2F1 acts primarily as an activator. However, in other contexts NR2F1/2 were shown to function as repressors (Lin et al., 2011; Tsai and Tsai, 1997). To address whether presence of the NR2F1/2 DNA recognition sequence contributes to the enhancer activity, we selected two NR2F1/2-bound hNCC enhancers, each containing a single NR2F1/2 motif. These regions were proximal to, respectively, BMP7 (Enhancer 1) and CDON (Enhancer 2) genes, whose expression was induced during differentiation of hESC to hNCC, and downregulated upon NR2F1 knockdown in hNCC (Figure 4D). hESC were transduced with GFP reporters containing either wild-type (WT) enhancer, or a mutant version with deletion of the NR2F1/2 binding motif (ΔNR2F1/2) (Figure 6A). In hESC, neither WT nor ΔNR2F1/2 reporters were active, but both WT reporters were induced during differentiation to hNCC, with the first reporter being active most strongly in the delaminating hNCC and attached rosettes, with some activity in migratory hNCC (Figure 6B), and the second reporter showing activity specific to a fraction of the migratory hNCC (Figure 6C). In support of the NR2F1/2 activating role, for both reporters deletion of the NR2F1/2 binding site strongly diminished enhancer activity (Figure 6B-C, quantified in Figure S7).
Figure 6. In vitro reporter assays demonstrate the importance of NR2F1/2 sites for hNCC enhancer activity (see also Figure S7).
(A) Schematic representation of in vitro reporter assays. Select enhancers with intact (WT) or deleted (Δ) NR2F1/2 sites were cloned in front of a minimal promoter driving GFP expression. The resulting lentiviral vectors were used to infect hESC and GFP expression was subsequently followed during hNCC differentiation. (B-C) Bright field and GFP images for two select enhancers in either hESC (left panels) or hNCC (right panels). In both cases, representative images obtained with intact (WT) enhancers or enhancers with deleted (Δ) NR2F1/2 sites are shown. NS: neurosphere, DC: delaminating neural crest cells, MC: migratory NCC.
Simultaneous binding and cooperation of NR2F1/2 and TFAP2A at hNCC enhancers
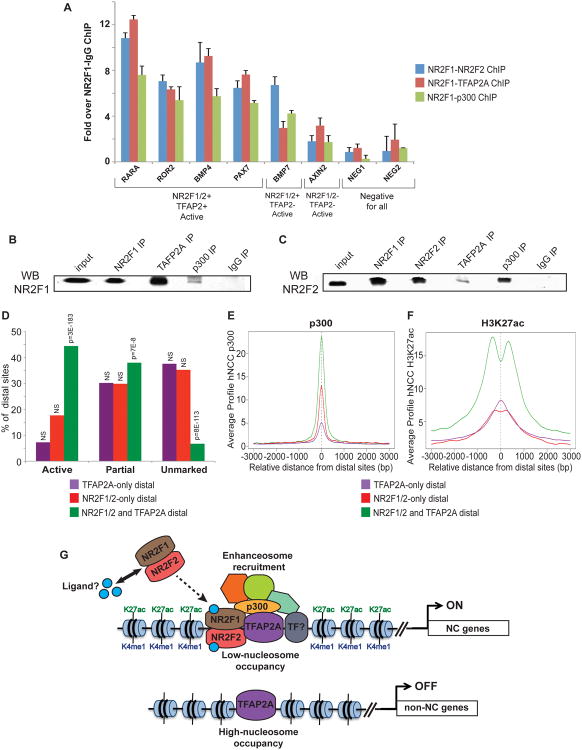
Next we proceeded to examine a potential interplay between NR2F1/2 and TFAP2A. Comparisons of binding datasets showed that a substantial fraction (38%) of NR2F1/2 distal sites was also bound by TFAP2A, with co-occupied sites characterized by a very strong association with cranial NC-derived structures, skull formation and craniofacial anomalies, but not with neural or neural tube ontologies (Figure S8A). Many TFAP2A and NR2F1/2 co-occupied sites are located in proximity of genes encoding NC transcriptional regulators (e.g. SOX9, TFAP2A, SNAI1), signaling mediators involved in NC formation and differentiation (e.g. BMP7, BMP4, NOTCH1, EFNB1), and genes associated with craniofacial anomalies in humans (e.g. ALX3, CHD7, TCOF1) (genomic coordinates of all co-occupied distal regions are listed in Supplementary Data 4, along with names of proximal genes). To directly demonstrate that NR2F1/2 and TFAP2A co-bound hNCC enhancers are indeed simultaneously occupied by these TFs, we performed sequential ChIP assays, which showed enrichment of NR2F1-NR2F2, NR2F1-TFAP2A and NR2F1-p300 signals over NR2F1-IgG signal at hNCC enhancers, but not at control regions (Figure 7A). Furthermore, co-immunoprecipitation assays revealed association of the four proteins (i.e. NR2F1, NR2F2, TFAP2A and p300) in hNCC nuclear extracts (Figure 7B-C). Time-course ChIP-qPCR experiments showed that NR2F1 and TFAP2A bind hNCC enhancer regions at similar time points during differentiation, coinciding with the onset of hNCC migration and gain of H3K27ac (Figure S8B-E).
Figure 7. TFAP2A, NR2F1/2 simultaneously bind and cooperate at hNCC enhancers (see also Figure S8).
(A) Sequential ChIPs were performed using NR2F1 as the first antibody, followed by NR2F2 (blue), TFAP2A (red) or p300 (green) antibodies. Resulting material was analyzed by qPCR analyses of select hNCC enhancers. Y-axis shows fold enrichment over a NR2F1-IgG sequential ChIP. Error bars represent standard deviation from three technical replicates. (B-C) NR2F1 (B) and NR2F2 (C) immunoblot analyses of NR2F1, NR2F2, TFAP2A, p300 or IgG immunoprecipitates from hNCC nuclear extracts. (D) Percentage of distal sites bound only by TFAP2A (purple), bound only by NR2F1/2 (red) or co-bound by both TFAP2A and NR2F1/2 (green) and overlapping active hNCC enhancers, or regions with a partial active enhancer signature or occurring within unmarked chromatin. P-values were calculated using hypergeometric tests, based on comparisons with all TFAP2A and NR2F1/2 combined bound regions (n=13990). NS=non-significant (p>0.05). (E-F) Average p300 (E) and H3K27ac (F) ChIP-seq signal profiles generated around the central position of distal regions bound only by TFAP2A (purple), bound only by NR2F1/2 (red) or co-bound by both TFAP2A and NR2F1/2 (green). (G) Proposed model of hNCC enhancer activation, whereby a master lineage specifier, TFAP2A, and effectors of ligand-dependent signaling, NR2F1/2, simultaneously bind enhancer sequences and synergistically lead to the establishment of transcriptionally permissive enhancer chromatin states and gene activation. Additional TFs (depicted as ‘TFs?’), including NC specifiers and effectors of major signaling pathways (e.g. BMP, WNT) are also likely involved in the combinatorial recruitment of productive enhanceosomes
Synergy between TFAP2A and NR2F1/2 is suggested by the tendency of the co-occupied sites to occur within regions marked by the active chromatin signatures (Figure 7D). Nonetheless, direct regulation of TFAP2A and NR2F2 expression by NR2F1 makes dissecting the contribution of each factor to the enhanceosome complex formation difficult in knockdown experiments. Instead, we examined whether genomic elements bound by both TFs, compared to regions bound by a single TF, display higher binding signals, coactivator occupancy and more pronounced H3K27ac levels. On average, co-bound regions had modestly elevated levels of TF binding, substantially increased levels of p300, and much higher and broader enrichment of H3K27ac (Figure S8F-H, Figure 7E-F). These results argue for a synergistic function of TFAP2A and NR2F1/2 in establishing permissive chromatin states at hNCC enhancers.
Discussion
To the best of our knowledge our study represents the first genome-wide mapping of chromatin states and TF occupancy in NCC of any organism and illustrates how coupling of hESC differentiation models with epigenomic profiling holds predictive power in the analysis of major TFs driving gene expression in developmentally transient cell states. Below we discuss broad implications of our work for studies of human development and disease and for understanding general mechanisms governing cell type-specific transcriptional regulation during embryogenesis.
Integration of lineage and signaling information at NC enhancers
Our results suggest that sequence information encoded by hNCC enhancers is simultaneously read by a master lineage specifier, TFAP2A, and by nuclear receptors, leading in turn to the establishment of the transcriptionally permissive enhancer chromatin states (Figure 7G). Nuclear receptors are effectors of ligand-dependent signaling, and although the physiological ligand of NR2F1/2 has not been identified, a structural study convincingly argues that these TFs indeed function as ligand-induced activators (Kruse et al., 2008). Beyond providing a direct link between the signaling environment and the transcriptional machinery controlling NC function, the importance of identifying such ligand in the future is underscored by observations that human craniofacial development is particularly sensitive to environmental changes resulting from fetal exposure (Lammer et al., 1985).
Whereas our work identified TFAP2A and NR2F1/2 as major hNCC enhancer binding proteins, analysis of motifs overrepresented at hNCC enhancers suggests that other TFs, such as ETS, E-box and SoxE factors, as well as transcriptional effectors of signaling pathways, including Wnt and BMP, also assemble at hNCC enhancers (Betancur et al., 2010; Sauka-Spengler and Bronner-Fraser, 2008). Future genome-wide studies will be needed to clarify the binding pattern overlap and spatio-temporal relationships among these factors. Nonetheless our results suggest that, as has been reported in other systems (Mullen et al., 2011; Trompouki et al., 2011), synergistic function of lineage specifiers and signaling effectors converges at NC enhancer elements.
Chromatin signatures facilitate identification of TF sites engaged in productive enhanceosomes
TFAP2A and NR2F1/2 binding sites that coincide with the presence of active signatures display much higher levels of nucleosomal depletion/hypermobility, association with gene expression and functional conservation than TF sites not overlapping with enhancer signatures. A similar correlation may hold true for other master regulators in different cell types. For example, only 16% of OCT4 binding sites coincide with the presence of the active signature in hESC, but those that do, show a strong correlation with genes expressed in hESC, whereas unmarked OCT4 sites exhibit no such association (Figure S9). These observations suggest that considering chromatin signatures may facilitate identification of TF binding events engaged in productive enhanceosomes, and thus relevant for gene expression within the analyzed cell type. Certainly, TF binding events occurring outside of the chromatin signatures may also be biologically meaningful, for example by priming future enhancer sites (Zaret and Carroll, 2011), or by functioning in other tissues. Nevertheless, decreased sequence conservation at TF sites lacking active signatures suggests that at least some of these sites may represent irrelevant binding events (Biggin, 2011).
A role for nuclear receptors NR2F1/2 in NC gene regulation
NR2F1/2 have been implicated in various biological processes including brain, eye and vascular development (reviewed in (Lin et al., 2011; Tsai and Tsai, 1997)), but their role in the NC has not been previously examined. In mouse models, substantial redundancy between the two receptors was reported, with many phenotypes only revealed upon NR2F1/2 double knockdown/knockout in the relevant cell types (Naka et al., 2008; Tang et al., 2010). Interestingly, although our expression and genomic occupancy data are consistent with the potential redundancy between the two receptors, knockdown of NR2F1 is sufficient to perturb NC gene expression in hNCC and morphogenesis of the ectomesenchyme in the frog. In contrast, Nr2f1 knockout mice undergo relatively unaltered embryogenesis, albeit cranial ganglia anomalies (consistent with NC defects) were reported in these animals (Qiu et al., 1997). Distinct requirements for individual NR2F receptors in different species may be related to the observation that in hNCC the NR2F2 locus is under direct control of NR2F1, and thus expression of both receptors is affected by NR2F1 knockdown. Regardless, our results strongly suggest that NR2F1 and NR2F2 genes should be systematically screened for mutations in human craniofacial disorders of unknown etiology. In support of this notion, individual cases of patients with a deletion of a single NR2F1 allele or with a microdeletion of a larger chromosomal region spanning NR2F2 gene reported numerous craniofacial abnormalities typically observed in human neurocristopathies (Brown et al., 2009; Poot et al., 2007).
Resource for studies of NC development and disease
The enhancer repertoire reported here provides a rich resource to study NC development. First, many identified regulatory elements are associated with genes that were previously shown to be critical for the NC function and craniofacial development, but whose regulatory regions were heretofore unknown. Second, a substantial subset of the identified enhancers is linked to genes that were not previously implicated in the NC, but which encode signaling regulators, TFs and cell migration factors. Thus, our study identified a wealth of new candidates that can be examined for their role in NC specification and migration. Third, our work uncovered direct genomic targets of the critical NC lineage specifier TFAP2A, which can be followed up on a locus-specific basis. Lastly, enhancer regions reported here are useful for development of reporter tools for further characterization of NCC populations in vitro and in vivo.
It is now well documented that enhancer mutations are linked to various human pathologies, including neurocristopathies (Amiel et al., 2010; Sakabe et al., 2012), but their role has not been systematically examined, largely due to the scarcity of information on the relevant genomic sequences. Over 500 Mendelian human congenital disorders involve craniofacial malformations with likely NC involvement. Although chromosomal locations linked to many craniofacial disorders have been identified, these regions are often large, and disease-associated mutations outside of the coding sequences are difficult to pinpoint within broad non-coding territories (Jugessur et al., 2009). With datasets provided here, hNCC enhancers located within specific disease-associated chromosomal regions can be readily detected and their sequences characterized for recurrent mutations in patients. Moreover, our work will aid understanding of complex multi-genic malformations, such as the cleft lip/palate (Birnbaum et al., 2009; Jugessur et al., 2009), as SNPs located in proximity of the hNCC enhancers and TF binding sites could be easily incorporated into genome-wide association study (GWAS) pipelines.
Experimental procedures
hNCC derivation
hESC (H9 line) were differentiated into hNCC as previously described (Bajpai et al., 2010). Briefly, hESC were incubated with 2mg/ml collagenase. Once detached, clusters of 100-200 cells were plated in hNCC differentiation medium: 1:1 Neurobasal medium/D-MEM F-12 medium (Invitrogen), 0.5× B-27 supplement with Vitamin A (50× stock, Invitrogen), 0.5× N-2 supplement (100× stock, Invitrogen), 20 ng/ml bFGF (Peprotech), 20 ng/ml EGF (Sigma-Aldrich), 5 μg/ml bovine insulin (Sigma-Aldrich) and 1× Glutamax-I supplement (100× stock, Invitrogen). Medium was changed every other day. After six-seven days of differentiation, resultant neuroepithelial spheres attached and gave rise to migratory hNCC. Three-four days after the appearance of the first hNCC, cells were harvested for subsequent experiments.
Chromatin immunuprecipitation (ChIP), sequential ChIP, FAIRE and antibodies
ChIP and FAIRE assays were performed from approximately 107 hNCC cells per experiment (Rada-Iglesias et al., 2011). Sequential ChIPs were performed as previously described with slight modifications (Furlan-Magaril et al., 2009). All antibodies used in this study have been previously reported as suitable for ChIP and/or ChIP-seq: p300 (sc-585, Santa Cruz Biotechnology), H3K4me1 (ab8895, Abcam), H3K27ac (ab4729, Abcam), H3K4me3 (39159, Active Motif), H3K27me3 (39536, Active Motif), NR2F1 (PP-H8132-00, Perseus Proteomics), NR2F2 (PP-H7147-00, Perseus Proteomics), TFAP2A (sc-184, Santa Cruz Biotechnology).
ChIP-seq
DNA libraries were prepared from hNCC p300 ChIP, hNCC FAIRE, hNCC H3K4me3 ChIP, hNCC H3K4me1 ChIP, hNCC H3K27me3 ChIP, hNCC H3K27ac ChIP, hNCC NR2F1 ChIP, hNCC NR2F2 ChIP, hNCC TFAP2A ChIP, hNCC input DNA, St11-14 Chicken H3K27ac ChIP, St20 Chicken H3K27ac ChIP, St11-14 Chicken input DNA, St20 Chicken input DNA ChIP-seq, FAIRE-seq and input libraries were prepared according to Illumina protocol and sequenced using Illumina Genome Analyzer. All sequences were mapped by ELAND software (Illumina Inc) and analyzed by QuEST 2.4 software (Valouev et al., 2008). Aforementioned human and chicken ChIP-seq datasets have been deposited into GEO repository under accession numbers GSE28876 and GSE38066, respectively. Histone modification and p300 ChIP-seq datasets from hESC and hNEC have been previously reported and deposited into GEO database (accession number GSE24447) (Rada-Iglesias et al., 2011).
Enhancer classification
Distal p300 enriched elements were defined as those that were not enriched in H3K4me3. Subsequently, those distal p300 elements in hESC, hNEC or hNCC were classified in the following groups (Supplementary Data 1):
Active enhancers: distal p300 regions located within 2 Kb of regions enriched in H3K4me1 and H3K27ac.
Poised enhancers: distal p300 regions located within 2 Kb of regions enriched in H3K27me3 and more than 2 Kb away from regions enriched in H3K27ac.
In addition, when comparing the binding profiles of transcription factors (NR2F1/2, TFAP2A) with the location of hNCC enhancers, we considered two additional categories.
Partial active enhancers: distal transcription factor bound regions that overlapped p300-bound regions not corresponding to active enhancers or distal regions not enriched in p300 but enriched in H3K4me1 and H3K27ac.
Unmarked: distal transcription factor bound regions located more than 2 Kb away from regions enriched in p300, H3K27ac, H3K4me1 or H3K27me3.
shRNA-mediated NR2F1 knockdown
shRNA against NR2F1 was purchased from Open biosystems(sequence:CGCCCACCCAGCAGAAATACAATAGTGAAGCCACAGATGTATTGTATTT CTGCTGGGTGGGCTT, Oligo ID: V2LHS-239287). The NR2F1 shRNA-encoding sequence was cloned in the pTRIPZ vector, in which shRNA expression is doxycycline inducible. pTRIPZ vectors expressing either NR2F1 shRNA or control-scrambled shRNA were used to transduce hESC. Infected hESC were then expanded in media containing puromycin (1 μg/μl) for at least two passages. hESC were then differentiated into hNCC, having both puromycin (1 μg/μl) and doxycycline (1 μg/μl) in the corresponding media. Three days after the appearance of the first hNCC, total RNA was isolated and cDNA generated as described above. RT-qPCR analysis of gene expression was performed using two independent biological replicates for both NR2F1 and control shRNA expressing cells and performing each qPCR reaction in triplicate. Fold changes in gene expression were calculated using the delta-delta Ct method and the EEF2 gene as a loading control. For all genes, expression is normalized to that of the housekeeping gene EEF2 and presented relative to hNCC expressing control-scrambled shRNA.
In vitro enhancer reporter assays
Representative active hNCC enhancers bound by NR2F1/2 and with a uniquely strong match to the NR2F1/2 binding motif were selected. Wild-type enhancer sequences or enhancer sequences with deleted NR2F1/2 binding motif (Supplementary data 5) were cloned into a lentiviral vector (Sin-minTK-eGFP) in front of a minimal TK promoter driving GFP expression. hESC were transduced with the corresponding lentiviruses and subsequently expanded for at least two passages in Neomycin (0.2 mg/ml) containing media. Infected hESC were then differentiated into hNCC and GFP fluorescence levels were monitored. For quantification of the reporter assays, hNCCs were separated into single cell suspension by trypsin treatment. Cell GFP fluorescence was analyzed on CS&T calibrated BD FACS Aria II SORP flow cytometer on 488nm laser line. Percent of GFP positive cells were determined by histogram interval gate set on GFP negative control hNCC cells such that the gate encompasses <0.2% control population.
Supplementary Material
Highlights.
Chromatin profiling identifies thousands of human NC enhancers
Many enhancers are functionally conserved in the avian embryo NC
Chromatin signatures predict novel NC transcriptional regulators
TFAP2A and NR2F1/2 cooperate to promote permissive enhancer chromatin states
Acknowledgments
We thank G. Crump for insightful comments on this work, R. Srinivasan for assistance and A. K. Roos, E. Calo-Velazquez and K. Murata for manuscript reading; Z. Weng and Stanford Sequencing Service Center for Illumina sequencing. This work was supported by RO1 GM095555, CIRM RN1 00579-1 and W.M. Keck Foundation grants to J.W. A.R-I. was supported by an EMBO long-term fellowship and Siebel Foundation.
Footnotes
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amiel J, Benko S, Gordon CT, Lyonnet S. Disruption of long-distance highly conserved noncoding elements in neurocristopathies. Ann N Y Acad Sci. 2010;1214:34–46. doi: 10.1111/j.1749-6632.2010.05878.x. [DOI] [PubMed] [Google Scholar]
- Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130:483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- Bajpai R, Chen DA, Rada-Iglesias A, Zhang J, Xiong Y, Helms J, Chang CP, Zhao Y, Swigut T, Wysocka J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–962. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellmeyer A, Krase J, Lindgren J, LaBonne C. The protooncogene c-myc is an essential regulator of neural crest formation in xenopus. Dev Cell. 2003;4:827–839. doi: 10.1016/s1534-5807(03)00160-6. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betters E, Liu Y, Kjaeldgaard A, Sundstrom E, Garcia-Castro MI. Analysis of early human neural crest development. Dev Biol. 2010;344:578–592. doi: 10.1016/j.ydbio.2010.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, Baluardo C, Ferrian M, Almeida de Assis N, Alblas MA, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41:473–477. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet. 2012;44:148–156. doi: 10.1038/ng.1064. [DOI] [PubMed] [Google Scholar]
- Brown KK, Alkuraya FS, Matos M, Robertson RL, Kimonis VE, Morton CC. NR2F1 deletion in a patient with a de novo paracentric inversion, inv(5)(q15q33.2), and syndromic deafness. Am J Med Genet A. 2009;149A:931–938. doi: 10.1002/ajmg.a.32764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugmann SA, Powder KE, Young NM, Goodnough LH, Hahn SM, James AW, Helms JA, Lovett M. Comparative gene expression analysis of avian embryonic facial structures reveals new candidates for human craniofacial disorders. Hum Mol Genet. 2010;19:920–930. doi: 10.1093/hmg/ddp559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012 doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339:250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney AJ, Tsai SY, O'Malley BW, Tsai MJ. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol Cell Biol. 1992;12:4153–4163. doi: 10.1128/mcb.12.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Oh S, Demare LE, Reilly SK, Gerstein MB, Noonan JP. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012 doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Aquino R, Tirino V, Desiderio V, Studer M, De Angelis GC, Laino L, De Rosa A, Di Nucci D, Martino S, Paino F, et al. Human neural crest-derived postnatal cells exhibit remarkable embryonic attributes either in vitro or in vivo. Eur Cell Mater. 2011;21:304–316. doi: 10.22203/ecm.v021a23. [DOI] [PubMed] [Google Scholar]
- de Croze N, Maczkowiak F, Monsoro-Burq AH. Reiterative AP2a activity controls sequential steps in the neural crest gene regulatory network. Proc Natl Acad Sci U S A. 2011;108:155–160. doi: 10.1073/pnas.1010740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y, Vassileva G, McMahon AP. Cis-acting regulatory sequences governing Wnt-1 expression in the developing mouse CNS. Development. 1994;120:2213–2224. doi: 10.1242/dev.120.8.2213. [DOI] [PubMed] [Google Scholar]
- Furlan-Magaril M, Rincon-Arano H, Recillas-Targa F. Sequential chromatin immunoprecipitation protocol: ChIP-reChIP. Methods Mol Biol. 2009;543:253–266. doi: 10.1007/978-1-60327-015-1_17. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Bronner-Fraser M. Neural crest specification: migrating into genomics. Nat Rev Neurosci. 2003;4:795–805. doi: 10.1038/nrn1219. [DOI] [PubMed] [Google Scholar]
- Garcia-Castro MI, Marcelle C, Bronner-Fraser M. Ectodermal Wnt function as a neural crest inducer. Science. 2002;297:848–851. doi: 10.1126/science.1070824. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honore SM, Aybar MJ, Mayor R. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev Biol. 2003;260:79–96. doi: 10.1016/s0012-1606(03)00247-1. [DOI] [PubMed] [Google Scholar]
- Ivey KN, Sutcliffe D, Richardson J, Clyman RI, Garcia JA, Srivastava D. Transcriptional regulation during development of the ductus arteriosus. Circ Res. 2008;103:388–395. doi: 10.1161/CIRCRESAHA.108.180661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 2009;15:437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt B, Kaltschmidt C, Widera D. Adult Craniofacial Stem Cells: Sources and Relation to the Neural Crest. Stem Cell Rev. 2011 doi: 10.1007/s12015-011-9340-9. [DOI] [PubMed] [Google Scholar]
- Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, et al. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6:e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer EJ, Chen DT, Hoar RM, Agnish ND, Benke PJ, Braun JT, Curry CJ, Fernhoff PM, Grix AW, Jr, Lott IT, et al. Retinoic acid embryopathy. N Engl J Med. 1985;313:837–841. doi: 10.1056/NEJM198510033131401. [DOI] [PubMed] [Google Scholar]
- Liem KF, Jr, Tremml G, Roelink H, Jessell TM. Dorsal differentiation of neural plate cells induced by BMP-mediated signals from epidermal ectoderm. Cell. 1995;82:969–979. doi: 10.1016/0092-8674(95)90276-7. [DOI] [PubMed] [Google Scholar]
- Lin FJ, Qin J, Tang K, Tsai SY, Tsai MJ. Coup d'Etat: an orphan takes control. Endocr Rev. 2011;32:404–421. doi: 10.1210/er.2010-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milunsky JM, Maher TA, Zhao G, Roberts AE, Stalker HJ, Zori RT, Burch MN, Clemens M, Mulliken JB, Smith R, et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am J Hum Genet. 2008;82:1171–1177. doi: 10.1016/j.ajhg.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka H, Nakamura S, Shimazaki T, Okano H. Requirement for COUP-TFI and II in the temporal specification of neural stem cells in CNS development. Nat Neurosci. 2008;11:1014–1023. doi: 10.1038/nn.2168. [DOI] [PubMed] [Google Scholar]
- Passos-Bueno MR, Ornelas CC, Fanganiello RD. Syndromes of the first and second pharyngeal arches: A review. Am J Med Genet A. 2009;149A:1853–1859. doi: 10.1002/ajmg.a.32950. [DOI] [PubMed] [Google Scholar]
- Pohl BS, Knochel W. Overexpression of the transcriptional repressor FoxD3 prevents neural crest formation in Xenopus embryos. Mech Dev. 2001;103:93–106. doi: 10.1016/s0925-4773(01)00334-3. [DOI] [PubMed] [Google Scholar]
- Poot M, Eleveld MJ, van 't Slot R, van Genderen MM, Verrijn Stuart AA, Hochstenbach R, Beemer FA. Proportional growth failure and oculocutaneous albinism in a girl with a 6.87 Mb deletion of region 15q26.2-->qter. Eur J Med Genet. 2007;50:432–440. doi: 10.1016/j.ejmg.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Pereira FA, DeMayo FJ, Lydon JP, Tsai SY, Tsai MJ. Null mutation of mCOUP-TFI results in defects in morphogenesis of the glossopharyngeal ganglion, axonal projection, and arborization. Genes Dev. 1997;11:1925–1937. doi: 10.1101/gad.11.15.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakabe NJ, Savic D, Nobrega MA. Transcriptional enhancers in development and disease. Genome Biol. 2012;13:238. doi: 10.1186/gb-2012-13-1-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sasai N, Sasai Y. Neural crest determination by co-activation of Pax3 and Zic1 genes in Xenopus ectoderm. Development. 2005;132:2355–2363. doi: 10.1242/dev.01823. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Schorle H, Meier P, Buchert M, Jaenisch R, Mitchell PJ. Transcription factor AP-2 essential for cranial closure and craniofacial development. Nature. 1996;381:235–238. doi: 10.1038/381235a0. [DOI] [PubMed] [Google Scholar]
- Soo K, O'Rourke MP, Khoo PL, Steiner KA, Wong N, Behringer RR, Tam PP. Twist function is required for the morphogenesis of the cephalic neural tube and the differentiation of the cranial neural crest cells in the mouse embryo. Dev Biol. 2002;247:251–270. doi: 10.1006/dbio.2002.0699. [DOI] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 2007;134:1481–1490. doi: 10.1242/dev.02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K, Xie X, Park JI, Jamrich M, Tsai S, Tsai MJ. COUP-TFs regulate eye development by controlling factors essential for optic vesicle morphogenesis. Development. 2010;137:725–734. doi: 10.1242/dev.040568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng L, Mundell NA, Frist AY, Wang Q, Labosky PA. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development. 2008;135:1615–1624. doi: 10.1242/dev.012179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theveneau E, Duband JL, Altabef M. Ets-1 confers cranial features on neural crest delamination. PLoS One. 2007;2:e1142. doi: 10.1371/journal.pone.0001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell. 2011;147:577–589. doi: 10.1016/j.cell.2011.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Tsai MJ. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Ferguson CA, Bach I, Rosenfeld MG, Sharpe PT. Conserved regulation of mesenchymal gene expression by Fgf-8 in face and limb development. Development. 1999;126:221–228. doi: 10.1242/dev.126.2.221. [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, Carroll JS. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 2011;25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.