Abstract
The LspA1 and LspA2 proteins of Haemophilus ducreyi 35000 are two very large macromolecules that can be detected in concentrated culture supernatant fluid. Both of these proteins exhibit homology with the N-terminal region of the Bordetella pertussis filamentous hemagglutinin (FHA), which is involved in secretion of the latter macromolecule. The lspA2 open reading frame is flanked upstream by a gene, lspB, that encodes a predicted protein with homology to the B. pertussis FhaC outer membrane protein that is involved in secretion of FHA across the outer membrane. The H. ducreyi lspB gene encodes a protein with a predicted molecular mass of 66,573 Da. Reverse transcription-PCR analysis suggested that the lspB gene was transcribed together with the lspA2 gene on a single mRNA transcript. Polyclonal H. ducreyi LspB antiserum reacted with a 64-kDa antigen present in the Sarkosyl-insoluble cell envelope fraction of H. ducreyi 35000, which indicated that the LspB protein is likely an outer membrane protein. Concentrated culture supernatant fluids from H. ducreyi lspB and lspA1 lspB mutants did not contain detectable LspA1 and detectable LspA2, respectively. However, complementation of the lspB mutant with the wild-type lspB gene on a plasmid restored LspB protein expression and resulted in release of detectable amounts of the LspA1 protein into culture supernatant fluid. When evaluated in the temperature-dependent rabbit model of infection, the lspB mutant was attenuated in the ability to cause lesions and was never recovered in a viable form from lesions. These results indicated that the H. ducreyi LspB protein is involved in secretion of the LspA1 and LspA2 proteins across the outer membrane.
Haemophilus ducreyi is an unencapsulated, gram-negative bacillus and is the etiologic agent of the sexually transmitted genital ulcer disease known as chancroid (2, 62). Chancroid, although rarely seen in the United States, is common in some developing countries (9). Relatively little is known about the bacterial gene products essential for virulence expression by H. ducreyi (52). However, several possible virulence factors of H. ducreyi have been identified to date. These include a number of gene products localized to the outer membrane, including both proteins (19, 20, 53, 56) and lipooligosaccharide (11-13, 21, 23, 59). In addition, H. ducreyi has been shown to produce at least two toxins (4, 16, 42, 43) and a copper-zinc superoxide dismutase (50, 51). Functionally, the ability of H. ducreyi to attach to (3, 15) and invade (61) human cells in vitro, the ability to attach to extracellular matrix components (8), and the ability to resist phagocytosis (1, 69) have been suggested as possible virulence mechanisms. However, when organisms were tested in the human challenge model for experimental chancroid (7, 54), only H. ducreyi mutants lacking expression of the peptidoglycan-associated lipoprotein (22), the hemoglobin-binding outer membrane protein HgbA (6), and the DsrA outer membrane protein (10) were shown to exhibit reduced virulence.
It has been reported that the H. ducreyi 35000 chromosome contains two very large, unlinked, paralogous open reading frames (ORFs), lspA1 and lspA2, which encode proteins with moderate similarity to the filamentous hemagglutinin (FHA) of Bordetella pertussis (67). The LspA1 and LspA2 proteins expressed by H. ducreyi 35000 can be detected as soluble proteins, have apparent molecular weights of approximately 260,000, and are present in culture supernatant fluid. The level of expression of LspA2 is much lower than that of LspA1, so it is often difficult to detect LspA2 in culture supernatant fluid by Western blot analysis (66). Inactivation of the lspA1 gene results in significantly increased levels of LspA2 in culture supernatant fluid (66), suggesting that expression of these two proteins may be linked through a complex regulatory network. Moreover, an H. ducreyi lspA1 lspA2 double mutant is significantly less virulent than its wild-type parent strain in the temperature-dependent rabbit model of infection (66) and is unable to inhibit the phagocytic activity of certain cell lines in vitro (63).
The H. ducreyi 35000 lspA2 ORF is flanked immediately upstream by lspB, a gene encoding an ortholog of the FhaC outer membrane protein involved in the secretion of FHA by B. pertussis (29, 31, 68). H. ducreyi LspA1, LspA2, and LspB have been proposed to be components of a two-partner secretion system (31, 33) in which the LspB protein is likely the sole accessory protein involved in secretion of the LspA1 and LspA2 proteins across the outer membrane of H. ducreyi. In this paper, we report that the LspB protein of H. ducreyi is required for secretion of both LspA1 and LspA2 across the outer membrane.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The H. ducreyi strains and plasmids used in this study are listed in Table 1. Wild-type H. ducreyi strains were routinely cultivated on chocolate agar (CA) plates at 33°C in a humidified atmosphere containing 95% air and 5% CO2 as described previously (47). Mutant or plasmid-containing H. ducreyi strains were grown either on CA plates containing chloramphenicol (0.5 μg/ml) or on GC-heme agar plates (55) containing both chloramphenicol (0.5 μg/ml) and kanamycin (30 μg/ml) as necessary. For some experiments, H. ducreyi strains were grown at 33 to 34°C in a gyratory water bath at 90 rpm in a modified version of a Columbia broth-based medium (sCB) previously described for growing Haemophilus somnus (28, 67). sCB consisted of 35 g of Columbia broth (Difco Laboratories, Detroit, Mich.) per liter, 0.1% (wt/vol) Trizma base (Sigma Chemical Co., St. Louis, Mo.), equine hemin (25 μg/ml; Sigma), 1% (vol/vol) IsoVitaleX (Becton Dickinson Microbiology Systems, Cockeysville, Md.), and 2.5% (vol/vol) heat-inactivated fetal bovine serum (JRH BioSciences, Lenexa, Kans.) (67). H. ducreyi concentrated culture supernatant fluid (CCS) was prepared as described previously (67).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| H. ducreyi strains | ||
| 35000 | Wild-type strain isolated in Winnipeg, Manitoba, Canada | 26 |
| 35000.1 | Isogenic lspA1 mutant; Kanr | 66 |
| 35000.88 | Isogenic lspB mutant; Chlr | This study |
| 35000.188 | Isogenic lspA1 lspB mutant; Kanr Chlr | This study |
| A77 | Serum-sensitive strain | 7 |
| Plasmids | ||
| pACYC184 | Cloning vector able to replicate in both E. coli and H. ducreyi; Chlr | 14 |
| pWKS30 | Low-copy cloning vector for E. coli | 64 |
| pCW156 | pRSETB (Invitrogen) with a 0.22-kb PCR product encoding amino acids 27 to 100 of LspB ligated into the BamHI and EcoRI sites; Ampr | This study |
| pCW158 | pWKS30 with the 2.88-kb PCR product containing the entire lspB gene ligated into the PstI and BamHI sites; Ampr | This study |
| pCW159 | pCW158 with the 0.7-kb SmaI fragment of pCWnpCAT1 (containing a nonpolar chloramphenicol resistance cartridge) ligated into the HpaI site; Chlr Ampr | This study |
| pCW177 | 1.95-kb PvuII-NruI fragment of pACYC184 ligated to the 1.2-kb MamI-EcoRV fragment (kanamycin resistance gene) of pLS88; Kanr Chlr | This study |
| pCW173 | pACYC184 with the 2.65-kb SalI-BamHI fragment containing the lspB gene ligated into the SalI and BamHI sites; Chlr | This study |
| pCW225 | 4.3-kb PvuII-NruI fragment of pCW173 ligated to the 1.2-kb MamI-EcoRV fragment (kanamycin resistance gene) of pLS88; Kanr | This study |
| pCWnpCAT1 | 0.7-kb PCR product containing a promoterless chloramphenicol resistance cartridge from pACYC184 ligated into the SmaI site of pBluescript II KS(+) | This study |
Kan, kanamycin; Chl, chloramphenicol; Amp, ampicillin.
Escherichia coli XL1-Blue (Stratagene Corp., La Jolla, Calif.) was used as the host for general cloning manipulations. E. coli strains were grown in Luria-Bertani medium (49) supplemented with ampicillin (100 μg/ml), kanamycin (50 μg/ml), or chloramphenicol (30 μg/ml) when appropriate for maintenance of plasmids. For protein expression, E. coli was grown in 2× YT medium (49). Plasmid constructs used to construct isogenic H. ducreyi mutants were transformed into and isolated from E. coli HB101 before they were linearized and electroporated into H. ducreyi.
DNA methods.
Plasmid purification, phenol-chloroform extraction, restriction enzyme digestion, fill-in reactions with the Klenow fragment of DNA polymerase I and deoxynucleoside triphosphates, agarose gel electrophoresis, ligation, transformation of chemically competent E. coli strains, and Southern blot analysis were performed as described previously (49). Nucleotide sequence analysis was performed by using Big Dye terminator chemistry and a model 373A automated DNA sequencer (Applied Biosystems, Inc., Foster City, Calif.). Southern blot hybridization analysis with a DNA probe labeled with [α-32P]dCTP by the random-primer method was performed as described previously (16). The probe was a 1.37-kb portion of the lspB gene produced by PCR amplification with oligonucleotide primers 5′-CCCGATTGAGAATTGGTTGTC-3′ and 5′-TTGGATCCCGGTTATTCGGAGCAATCG-3′.
Production of a polyclonal H. ducreyi 35000 LspB-specific antiserum.
A 226-bp region from lspB encoding amino acids 27 to 100 of the LspB protein was amplified by PCR from H. ducreyi 35000 genomic DNA by using primers 5′-TTGGATCCCGGTTATTCGGAGCAATCG-3′ (BamHI site underlined) and 5′-TTGAATTCCGTTGATTTTTGTGCCGTATTTTG-3′ (EcoRI site underlined). The gel-purified PCR product was digested with BamHI and EcoRI and ligated into the polyhistidine (six-His) fusion protein vector pRSETB (Invitrogen Inc., Carlsbad, Calif.) to produce pCW156. Nucleotide sequence analysis of the DNA insert in pCW156 confirmed the absence of polymerase-induced base changes. E. coli BL21(DE3)(pLysS)(pCW156) was grown in 2× YT medium; the six-His LspB fusion protein was induced by addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and was subsequently purified by chromatography by using Talon metal affinity resin (Clontech Laboratories, Palo Alto, Calif.). Mice were immunized by the intraperitoneal route with 50 μg of the purified fusion protein emulsified in 50% (vol/vol) Freund's complete adjuvant (Difco) and then boosted with 25 μg of this protein emulsified in 50% (vol/vol) incomplete Freund's adjuvant (Difco) 1 month later. Two weeks after the booster injection, blood was drawn from these mice and pooled, and the resultant serum was used in a Western blot analysis as described below.
RT-PCR analysis.
H. ducreyi total RNA was isolated from broth cultures, treated with DNase, and subjected to multiplex reverse transcription (RT)-PCR analysis by using the Titan one-tube RT-PCR system (Roche Molecular Biochemicals, Indianapolis, Ind.) as previously described (67). RT-PCR was used to reverse transcribe and amplify transcripts derived from the lspB gene, from a region spanning the lspB-lspA2 intergenic region, and from the H. ducreyi pal gene (53). The two primers described above for amplification of the 226-bp region from lspB encoding amino acids 27 to 100 of the LspB protein yielded a 226-bp lspB-specific product. Primers 5′-TGGAATACCGCTTAAAGGTTTTG-3′ and 5′-TCAACGATTGCATTAGATGAGTCTG-3′ yielded a 431-bp product that spanned the lspB-lspA2 intergenic region. A previously described primer pair that yielded a 354-bp pal-specific product (67) was included as a positive amplification control.
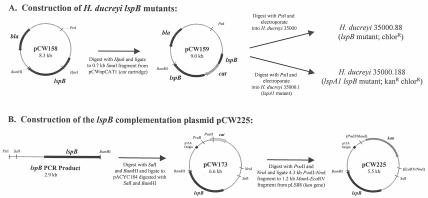
Construction of H. ducreyi mutants. (i) Construction of an lspB mutant.
A 2.88-kb portion of the H. ducreyi lspB gene (nucleotides 797 to 3679 in the sequence deposited in the GenBank database under accession number AF289079) was amplified by PCR by using primers 5′-TTCTGCAGTTAAAAACTGCACCCCCG-3′ (PstI site underlined) and 5′-TTGGATCCGTAATTTTGGTTAAAAACTGATTG-3′ (BamHI site underlined), digested with PstI and BamHI, and ligated into the low-copy-number vector pWKS30 (64) that had been digested with PstI and BamHI. The resultant plasmid, pCW158 (Fig. 1A), was linearized by digestion with HpaI and ligated to the 0.7-kb SmaI fragment of pCWnpCAT1 containing a nonpolar promoterless chloramphenicol resistance cartridge (cat) to produce plasmid pCW159. Plasmid pCWnpCAT1 (the source of the nonpolar promoterless cat cassette) was constructed by using SmaI to digest the product obtained by PCR-based amplification of the cat ORF from pACYC184 (14) with forward primer 5′-TTTCCCGGGTGACTAACTAGAGGAAGCTAAAATGGAGAAAAAAATCACTG-3′ and reverse primer 5′-TTTCCCGGGTCCATTATCCTTCCAGAAATTACGCCCCG CCCTGCC-3′ (SmaI sites underlined) and ligating the product into pBluescript KS(+). These primers were essentially the same as those used by Lukomski and colleagues to generate the nonpolar promoterless cat cassette contained in pSL1 (38). Plasmid pCW159 was linearized by digestion with PstI and used to electroporate H. ducreyi 35000 as previously described (27). H. ducreyi transformants were selected on CA containing chloramphenicol. One mutant, 35000.88, was randomly selected and used for further analysis.
FIG. 1.
Construction of the mutants and recombinant plasmid pCW225 used in this study. (A) Construction of the H. ducreyi lspB and lspA1 lspB mutants; (B) construction of recombinant plasmid pCW225. Restriction sites in parentheses are not present in pCW225 and reflect cloning junctions.
(ii) Construction of an lspA1 lspB mutant.
Plasmid pCW159 (Fig. 1A) was linearized by digestion with PstI and was used to electroporate the H. ducreyi lspA1 mutant 35000.1 (66). H. ducreyi transformants were selected on CA containing chloramphenicol. One mutant, 35000.188, was randomly selected and used for further analysis.
Complementation of the H. ducreyi lspB mutant.
A 2.88-kb DNA fragment containing the H. ducreyi lspB gene was amplified by PCR with Pfu DNA polymerase (Stratagene) and primers 5′-TTCTGCAGTTAAAAACTGCACCCCCG-3′ (PstI site underlined) and 5′-TTGGATCCGTAATTTTGGTTAAAAACTGATTG-3′ (BamHI site underlined). After digestion with SalI (the SalI site was present in the amplified fragment) and BamHI, the resultant 2.65-kb DNA fragment was ligated into pACYC184, and the ligation reaction mixture was used to electroporate H. ducreyi strain A77 to obtain pCW173 (Fig. 1B). Plasmid pCW173 was digested with PvuII and NruI to excise the chloramphenicol resistance gene, and the 4.3-kb fragment from the digest was ligated to the 1.2-kb MamI-EcoRV fragment (containing the kanamycin resistance gene) of pLS88 to produce plasmid pCW225. Plasmid pCW225 was transformed into the lspB mutant 35000.88, and the resulting transformants were selected on GC-heme agar containing kanamycin and chloramphenicol.
Isolation and fractionation of cell envelopes.
The Sarkosyl-insoluble cell envelope fraction was isolated as previously described (36) from H. ducreyi strains grown overnight in sCB.
SDS-PAGE and Western blot analysis.
To detect LspA1 and LspA2, samples containing H. ducreyi CCS were heated at 100°C for 5 min in sample buffer (45), resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by using a 7.5% (wt/vol) polyacrylamide separating gel, and transferred to nitrocellulose as described previously (67). The membranes were blocked with phosphate-buffered saline (PBS) containing 0.05% (vol/vol) Tween 20 and 3% (wt/vol) skim milk and incubated with monoclonal antibodies (MAbs) in the form of hybridoma supernatants. The MAbs used in this study have been described previously (67). MAb 40A4 is LspA1 specific, MAb 1H9 is LspA2 specific, and MAb 11B7 recognizes both LspA1 and LspA2. MAbs bound to proteins on nitrocellulose membranes were detected by using 125I-labeled goat anti-mouse immunoglobulin G (25) followed by autoradiography.
To detect LspB, Sarkosyl-insoluble H. ducreyi cell envelope fractions (20 μg of protein/lane) or H. ducreyi whole-cell lysates (37) were heated in sample buffer (45) containing 5% (vol/vol) 2-mercaptoethanol, resolved by SDS-PAGE with a 10% (wt/vol) polyacrylamide separating gel, and transferred to nitrocellulose. For Western blot analysis, the membranes were blocked and processed as described above except that a 1:2,000 dilution of the polyclonal mouse LspB antiserum was used as the source of primary antibody.
Serum bactericidal assay.
The serum bactericidal assay was performed as previously described (65), except that normal human serum was used as the source of complement.
Virulence testing.
The temperature-dependent rabbit model for experimental chancroid (47) was used to evaluate the virulence of the H. ducreyi strains described in this study. Lesions were scored on days 2, 4, and 7 postinfection by using the following scoring system: 0, no change; 1, erythema; 2, induration; 3, nodule; 4, pustule or necrosis. A statistical analysis of lesion scores was performed as described previously (5, 56). On day 7 postinfection, the animals were euthanized, and the lesions which had been initially inoculated with 105 CFU were excised from each rabbit, bisected with a sterile scalpel blade, and rinsed with PBS to recover pustular material. PBS washes were spread onto CA to recover viable H. ducreyi.
Nucleotide sequence accession number.
The nucleotide sequence of the H. ducreyi 35000 lspB locus has been deposited in the GenBank database under accession number AF289079.
RESULTS
Determination of the nucleotide sequence of the H. ducreyi lspB gene.
It was previously determined that the lspA2 gene of H. ducreyi 35000 was flanked upstream by an ORF (designated lspB) that encoded a protein with homology to the protein encoded by the B. pertussis fhaC gene (67, 68). Because only the 3′ portion of the H. ducreyi lspB gene had been sequenced previously (67), we determined the complete nucleotide sequence of the H. ducreyi 35000 lspB gene, as well as that of the upstream flanking DNA. Approximately 2 kb of DNA upstream of the available sequence of the partial H. ducreyi lspB gene was amplified by PCR by using a previously described method (40) and was sequenced. The resultant nucleotide sequence was confirmed by subsequent sequence analysis of a PCR product produced independently by amplifying this region from the H. ducreyi 35000 chromosome with different oligonucleotide primers and the high-fidelity thermostable DNA polymerase Pfu (data not shown).
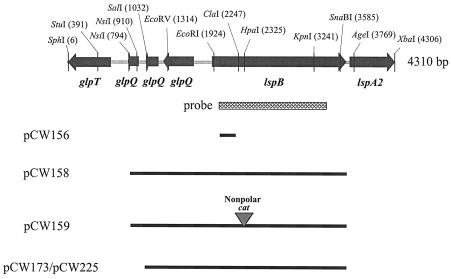
Nucleotide sequence analysis revealed that the complete lspB ORF contained 1,770 bases (nucleotides 1900 to 3669 in Fig. 2) and encoded a protein with a predicted molecular mass of 66,573 Da. Immediately upstream of the lspB ORF were three small putative ORFs which encoded predicted polypeptides with homology to different regions of the glycerophosphodiester phosphodiesterase GlpQ (Fig. 2). The predicted LspB protein contained a putative 26-amino-acid signal peptide sequence and had a carboxy-terminal phenylalanine residue; the latter fact was consistent with the possibility that this protein might be associated with the outer membrane of H. ducreyi (58). PHI-BLAST analysis revealed significant similarity of the predicted H. ducreyi LspB protein to the H. somnus IbpB protein (GenBank accession no. BAC78648; Expect = e-148), as well as to the FhaC proteins of Bordetella bronchiseptica (GenBank accession no. AAF21946; Expect = e-64) and B. pertussis (GenBank accession no. NP_880575; Expect = e-62). An alignment of the predicted H. ducreyi LspB protein with the IbpB protein of H. somnus and the FhaC protein of B. pertussis is shown in Fig. 3.
FIG. 2.
Partial restriction endonuclease map of the H. ducreyi 35000 lspB locus and locations of DNA inserts of recombinant plasmids used in this study. The lspB gene was flanked upstream by three small putative ORFs with homology to regions of the glycerophosphodiester phosphodiesterase glpQ gene and downstream by the lspA2 gene. Only a portion of the lspA2 ORF is shown. The region amplified by PCR (nucleotides 1977 to 3342) and used as a probe for Southern blot analysis (see Fig. 4A) is indicated by the cross-hatched box.
FIG. 3.
Alignment of the amino acid sequences of H. ducreyi LspB (Hd LspB), H. somnus IbpB (Hs IbpB), and B. pertussis FhaC (Bp FhaC). Identical amino acids are indicated by asterisks, and conserved amino acids are indicated by periods.
Detection of lspB sequences in H. ducreyi strains.
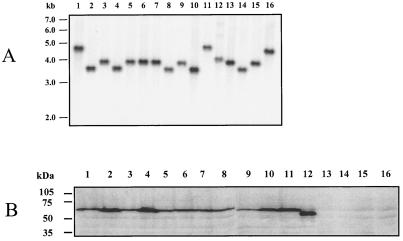
To determine the prevalence of the lspB gene in H. ducreyi strains, we performed a Southern blot analysis with genomic DNA from 16 H. ducreyi strains (67) isolated in diverse geographic regions (Fig. 4A). These 16 strains included 4 strains (A77, 6V, E1673, and 78226) (Fig. 4A, lanes 13 to 16) whose CCS were previously shown to not contain detectable LspA1 and LspA2 (67) and which were found to be essentially avirulent in an animal model of experimental chancroid (5). An lspB-specific probe (see Fig. 2 for location) hybridized to a single band in all strains examined, indicating that the lspB gene was conserved among these strains and was present as a single copy. The lspB probe was observed to hybridize to restriction fragments that were approximately five different sizes in the H. ducreyi strains examined: 4.8 kb (Fig. 4A, lanes 1 and 11), 4.6 kb (Fig. 4A, lane 16), 4.0 kb (Fig. 4A, lane 12), 3.9 kb (Fig. 4A, lanes 3, 5 to 7, 9, 13, and 15), and 3.6 kb (Fig. 4A, lanes 2, 4, 8, 10, and 14). This indicated that there was some genetic diversity among these strains in this region of the chromosome.
FIG. 4.
Southern and Western blot analyses of H. ducreyi strains to detect lspB genes and LspB protein expression. (A) For Southern blot analysis, chromosomal DNA from each strain was digested with EcoRV, electrophoresed through a 0.8% (wt/vol) agarose gel, transferred to nitrocellulose, and probed with an α-32P-labeled 1.37-kb DNA region of the H. ducreyi lspB ORF. The positions of DNA size markers are indicated on the left. (B) Whole-cell lysates of H. ducreyi strains were subjected to SDS-PAGE and Western blot analysis by using a polyclonal LspB antiserum as described in Materials and Methods. The positions of molecular size markers are indicated on the left. Lane 1, 35000; lane 2, RO18; lane 3, 181; lane 4, CA173; lane 5, WPB506; lane 6, BG411; lane 7, 041; lane 8, 1145; lane 9, 1151; lane 10, Cha-I; lane 11, Hd12; lane 12, CIP 542; lane 13, A77; lane 14, 6V; lane 15, E1673; lane 16, 78226.
The H. ducreyi 35000 lspB and lspA2 genes are present on a single transcript.
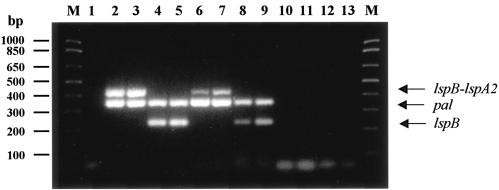
Because the translation initiation codon of the lspA2 ORF was separated from the translational stop codon of the lspB ORF by only 46 nucleotides, we hypothesized that the lspB and lspA2 ORFs could be cotranscribed, yielding a single mRNA. Attempts to determine the size of the transcript(s) containing the lspB and lspA2 sequences by Northern blot analysis were not successful (data not shown), probably because of the extremely large size (>17,000 nucleotides) of the predicted transcript. Therefore, we sought to localize the lspB and lspA2 sequences to the same RNA transcript by RT-PCR analysis of RNA isolated from broth-grown H. ducreyi. Using primers that spanned the lspB-lspA2 junction, we amplified a 431-bp product from both strain 35000 (Fig. 5, lane 6) and an H. ducreyi lspA1 mutant (Fig. 5, lane 7) that is known to express the LspA2 protein at readily detectable levels (66). A 226-bp lspB-specific product was also obtained from H. ducreyi 35000 (Fig. 5, lane 8) and the lspA1 mutant (Fig. 5, lane 9). No PCR products were obtained from RNA samples not subjected to RT prior to PCR amplification (Fig. 5, lanes 10 to 13), indicating that the PCR products shown in lanes 6 to 9 of Fig. 5 were not a result of DNA contamination of the RNA samples. Primers to reverse transcribe and amplify a 354-bp region of the H. ducreyi pal gene transcript (53) were included as an internal positive control. These results indicated that the 3′ region of the lspB transcript and the 5′ region of the lspA2 transcript were present on the same mRNA molecule.
FIG. 5.
Multiplex RT-PCR analysis of H. ducreyi lspB-containing transcripts. The following templates were included in the reaction mixtures: lane 1, no template (negative control); lanes 2 and 4, 100 ng of H. ducreyi wild-type strain 35000 genomic DNA (positive control); lanes 3 and 5, 100 ng of H. ducreyi lspA1 mutant genomic DNA (positive control); lanes 6, 8, 10, and 12, 1 μg of H. ducreyi wild-type strain 35000 total RNA; lanes 7, 9, 11, and 13, 1 μg of H. ducreyi lspA1 mutant total RNA. The primer sets included in the reaction mixtures were as follows: lanes 1 to 3, 6, 7, 10, and 11 contained both pal-specific (354-bp product) and lspB-lspA2 (431-bp product) primers; lanes 4, 5, 8, 9, 12, and 13 contained both pal-specific (354-bp product) and lspB-specific (226-bp product) primers. The reaction mixtures loaded in lanes 10 to 13 were not subjected to the RT step of the RT-PCR procedure and served as controls to detect DNA contamination of RNA preparations. Lane M contained DNA size markers.
Detection of LspB protein expression by H. ducreyi strains.
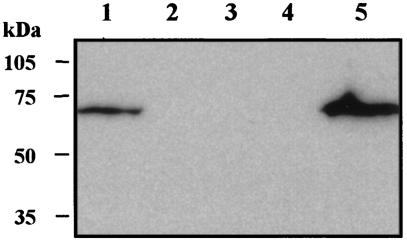
To detect and localize the expression of the LspB protein by various H. ducreyi strains, we produced a polyclonal LspB antiserum by immunizing mice with a six-His-LspB fusion protein (described in Materials and Methods). This antiserum was used in a Western blot analysis to probe whole-cell lysates prepared from the same strains that were analyzed by Southern blotting as described above. This polyclonal LspB antiserum reacted with an antigen having an apparent molecular weight of approximately 64,000 from 11 H. ducreyi strains (Fig. 4B, lanes 1 to 11), including strain 35000. The immunoreactive antigen from strain CIP 542 (Fig. 4B, lane 12) was slightly smaller and had an apparent molecular weight of approximately 58,000. The functional significance of the size difference, if any, is not apparent, and CIP 542 has previously been shown to release LspA1, but not LspA2, into CCS (67). This polyclonal LspB antiserum also reacted with an antigen of the same apparent size that was present in the Sarkosyl-insoluble cell envelope fraction of H. ducreyi 35000 (Fig. 6, lane 1), a result which indicated that the LspB protein is likely present in the outer membrane of H. ducreyi.
FIG. 6.
Western blot analysis of the Sarkosyl-insoluble cell envelope fraction from wild-type, mutant, and complemented H. ducreyi strains with a polyclonal LspB antiserum. Lane 1, wild-type strain 35000; lane 2, lspB mutant 35000.88; lane 3, lspA1 lspB mutant 35000.188; lane 4, 35000.88(pCW177) (vector-only control); lane 5, 35000.88(pCW225). The positions of molecular mass markers are indicated on the left.
The same antiserum did not react with the four avirulent strains (A77, 6V, E1673, and 78226) (Fig. 4B, lanes 13 to 16) whose CCS lacked detectable LspA1 or LspA2 (5, 59). Interestingly, when we PCR amplified and sequenced the lspB gene from strain A77, we found that the lspB ORF in this strain contained a 7-nucleotide (5′-CTTATAT-3′) insertion 894 nucleotides downstream from the ATG start codon. This insertion altered the reading frame, resulting in a premature translational stop codon in this gene (data not shown). This mutation is likely responsible for the lack of LspB protein expression in strain A77 (Fig. 4B, lane 13).
Construction of isogenic H. ducreyi lspB and lspA1 lspB mutants.
To investigate the role of the H. ducreyi LspB protein in the secretion of the LspA1 and LspA2 proteins, we constructed two isogenic mutants as described in Materials and Methods (Fig. 1A). An lspB mutant (35000.88) was constructed to investigate the role of the LspB protein in the secretion of the LspA1 protein. Similarly, an lspA1 lspB mutant (35000.188) was constructed to investigate the role of the LspB protein in the secretion of the LspA2 protein (because an lspA1 mutant expresses readily detectable levels of LspA2 in CCS [66]). A nonpolar promoterless chloramphenicol resistance cassette (38) was used to construct the lspB mutations in order to eliminate potential polar effects on expression of the downstream lspA2 gene. Mutants were initially identified from pools of antibiotic-resistant transformants by PCR analysis and were subsequently confirmed by Southern blot analysis to contain the desired lspB mutations (data not shown).
Characterization of membrane proteins and CCS from wild-type and mutant H. ducreyi strains.
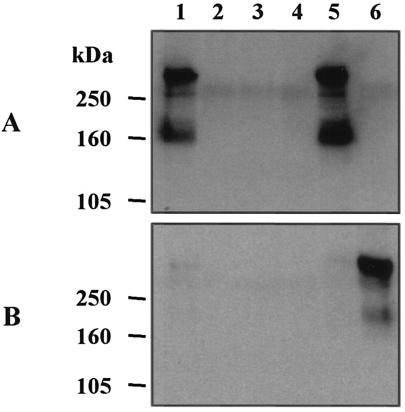
Western blot analysis of whole-cell lysates (data not shown) and Sarkosyl-insoluble cell envelope fractions of the 35000.88 (lspB) and 35000.188 (lspA1 lspB) mutants with the polyclonal LspB antiserum confirmed that these two mutants did not produce the LspB protein (Fig. 6, lanes 2 and 3, respectively). Western blot analysis was also performed with CCS prepared from wild-type and lspB mutant strains; these CCS were probed with LspA1- and LspA2-specific MAbs to evaluate the effect of this mutation on the secretion of the LspA1 and LspA2 proteins by H. ducreyi (Fig. 7). CCS from the lspB mutant 35000.88 (Fig. 7A, lane 2) did not contain detectable LspA1. Similarly, CCS from the lspA1 lspB mutant 35000.188 (Fig. 7B, lane 3) did not contain detectable LspA2 protein, whereas the CCS from the lspA1 mutant 35000.1 contained readily detectable levels of LspA2 (Fig. 7B, lane 6). RT-PCR analysis of total RNA from the lspA1 lspB mutant revealed the presence of a transcript derived from lspA2 (data not shown), indicating that insertion of the nonpolar promoterless cat cartridge into the lspB gene did not eliminate transcription of the downstream lspA2 gene. In addition, Western blot analysis with MAb 11B7 showed that whole-cell lysates of both the lspB and lspA1 lspB mutants contained immunoreactive LspA proteins (data not shown), a result which suggested that the LspA protein(s) accumulated inside these mutants. Taken together, these results indicated that the H. ducreyi LspB protein is probably involved in the secretion of both the LspA1 and LspA2 proteins.
FIG. 7.
Western blot analysis of CCS from wild-type, mutant, and complemented H. ducreyi strains with LspA1-specific MAb 40A4 (A) and LspA2-specific MAb 1H9 (B). Lane 1, wild-type strain 35000; lane 2, lspB mutant 35000.88; lane 3, lspA1 lspB mutant 35000.188; lane 4, 35000.88(pCW177) (vector-only control); lane 5, 35000.88(pCW225); lane 6, lspA1 mutant 35000.1. lspA1 mutant 35000.1 expressed readily detectable levels of LspA2 (66) and was used as a control to detect LspA2 expression. The positions of molecular mass markers are indicated on the left.
The growth of the lspB mutant 35000.88 and the lspA1 lspB mutant 35000.188 in sCB was no different from the growth of the wild-type strain 35000 (data not shown), indicating that the lspB mutation did not affect the ability of these strains to grow in vitro. We also evaluated the bactericidal activity of normal human serum against the lspB mutant and the lspA1 lspB mutant and found that these two mutants were as serum resistant as wild-type strain 35000 (data not shown). Western blot analysis confirmed that both of these mutants expressed the DsrA protein (data not shown) that is responsible for the expression of serum resistance by H. ducreyi (20).
Complementation of the H. ducreyi lspB mutant.
Complementation of the lspB mutation in strain 35000.88 with the H. ducreyi lspB gene provided in trans on plasmid pCW225 (Fig. 1B) was performed to confirm that the lspB gene was responsible for the phenotypic effects described above. Strain 35000.88(pCW225) (Fig. 6, lane 5), but not the vector-only control 35000.88(pCW177) (Fig. 6, lane 4), expressed LspB protein that was detectable in the Sarkosyl-insoluble cell envelope fraction, and it also was able to secrete the LspA1 protein (Fig. 7A, lanes 5 and 4, respectively). These results confirmed that an undetected secondary mutation was not responsible for the lack of detectable LspA1 in CCS from the lspB mutant strain 35000.88. It should also be noted that the apparent level of expression of LspB in the complemented mutant 35000.88(pCW225) (Fig. 6, lane 5) was greater than that in the wild-type parent strain (Fig. 6, lane 1).
Virulence analysis of wild-type, mutant, and complemented H. ducreyi strains.
The lspB mutant 35000.88 was evaluated in the temperature-dependent rabbit model to determine this mutant's ability to produce dermal lesions relative to that of wild-type parent strain 35000. In one experiment, the lspB mutant 35000.88 produced significantly lower lesion scores (P < 0.0001) than wild-type strain 35000 produced for two different inoculum sizes (Table 2, experiment 1). Furthermore, viable lspB mutants were not recovered from any of the lesions selected from any rabbit in this experiment, whereas wild-type strain 35000 was recovered from all seven rabbits in this experiment (data not shown). These data indicated that the lspB mutant was substantially attenuated in this animal model.
TABLE 2.
Lesion formation by wild-type and mutant H. ducreyi strains in the temperature-dependent rabbit modela
| Expt | Strain | Inoculum size (CFU) | Lesion score (mean ± SD) on day:
|
P valueb | ||
|---|---|---|---|---|---|---|
| 2 | 4 | 7 | ||||
| 1 | 35000 | 105 | 4.00 ± 0.00 | 4.00 ± 0.00 | 4.00 ± 0.00 | |
| 35000.88 (lspB mutant) | 105 | 3.00 ± 0.00 | 2.75 ± 0.49 | 2.71 ± 0.49 | 0.0001c | |
| 35000 | 104 | 3.63 ± 0.53 | 3.63 ± 0.53 | 3.71 ± 0.49 | ||
| 35000.88 (lspB mutant) | 104 | 3.00 ± 0.00 | 2.25 ± 0.49 | 1.86 ± 1.07 | ||
| 2 | 35000 | 105 | 4.00 ± 0.00 | 3.88 ± 0.35 | 3.88 ± 0.35 | |
| 35000.88(pCW177) | 105 | 3.00 ± 0.00 | 2.88 ± 0.35 | 2.63 ± 0.74 | 0.0003c | |
| 35000.88(pCW225) | 105 | 3.13 ± 0.35 | 3.13 ± 0.35 | 3.25 ± 0.46 | 0.0001 (0.0088)c,d | |
| 35000 | 104 | 3.13 ± 0.35 | 3.13 ± 0.64 | 3.00 ± 0.93 | ||
| 35000.88(pCW177) | 104 | 2.88 ± 0.35 | 2.00 ± 0.53 | 1.63 ± 0.52 | ||
| 35000.88(pCW225) | 104 | 3.00 ± 0.00 | 2.25 ± 0.46 | 2.38 ± 0.74 | ||
Seven rabbits were used in experiment 1, and eight rabbits were used in experiment 2.
P value calculated for the difference between wild-type and test strain lesion scores for the three scoring days by using both inoculum-sizes.
Significant difference.
The P value of 0.0088 refers to the difference between the 35000.88(pCW177) and 35000.88(pCW225) strains.
We also evaluated the complemented mutant strain 35000.88(pCW225) and its vector-only control, strain 35000.88(pCW177), in this animal model (Table 2, experiment 2). The presence of the lspB gene in trans did not completely restore the virulence of the lspB mutant to wild-type levels (P < 0.0001). However, the lesion scores obtained with complemented lspB mutant 35000.88(pCW225) were significantly different (P < 0.0088) from the lesion scores obtained with the vector-only control, strain 35000.88(pCW177). Viable H. ducreyi organisms were recovered from all eight rabbits infected with wild-type 35000 strain and from one rabbit inoculated with the complemented mutant 35000.88(pCW225), whereas no viable organisms were recovered from any rabbit inoculated with the vector-only control 35000.88(pCW177) (data not shown). The lesion score differences suggested that, although the presence of the lspB gene on a plasmid did not confer wild-type virulence on lspB mutant strain 35000.88 (Table 2, experiment 2), it did restore partial virulence compared to the virulence of the lspB mutant harboring only the plasmid vector.
DISCUSSION
The predicted protein product of the H. ducreyi lspB gene exhibits homology (42% similarity) to B. pertussis FhaC, which is the sole outer membrane accessory protein involved in the secretion of FHA (encoded by fhaB) (29, 31, 68). B. pertussis FhaB and FhaC are the prototypic components of the recently described two-partner secretion system involved in the export of large (100- to 500-kDa) virulence factors (33). This specialized secretion system is characterized by a channel-forming β-barrel outer membrane protein (TpsB) and a cognate large exoprotein (TpsA) synthesized as a preprotein that undergoes extensive proteolytic processing during secretion. The large exoprotein is probably transported across the cytoplasmic membrane in a Sec-dependent manner and then guided by the conserved N-proximal secretion domain comprised of approximately 110 amino acids across the periplasm in an extended conformation (with the C-terminal region of the TpsA preprotein acting as an intramolecular chaperone) directly to the transporter such that translocation across both membranes is coupled (33).
Despite the high levels of homology among the TpsA exoprotein members in the secretion domain, there is relatively limited overall identity at the primary amino acid sequence level. The presence of a large number of repeated β-strands that fold into amphipathic β-helices is common among proposed TpsA members, including H. ducreyi LspA1 and LspA2 (34), suggesting that a specialized secretion system is required for large proteins rich in β-structure. The genes for the secreted TpsA exoprotein and its cognate TpsB outer membrane transporter are typically present in the same operon (33, 34). The TpsB exporter proteins are typically approximately 60-kDa proteins, contain several transmembrane β-strands, including an amphipathic C-terminal 10-amino-acid region, and are predicted to form an integral transmembrane β-barrel channel in the outer membrane through which they translocate the cognate exoprotein (31). Each TpsB protein appears to be specific for secreting only its cognate TpsA exoprotein (30), although the FhaC exporter proteins of B. pertussis and B. bronchiseptica appear to be functionally interchangeable for the secretion of B. pertussis FHA (32), probably because of the high level of primary amino acid sequence identity between these two proteins.
PHI-BLAST analysis revealed that orthologs of the H. ducreyi LspB protein are encoded in a large number of bacterial genomes, including many genomes that have been recently sequenced but have yet to have their TpsAB systems functionally characterized (data not shown). Members of the TpsAB family that have been characterized previously include systems that produce and export the Ca2+-independent cytolysins of Serratia marcescens, Proteus mirabilis, and H. ducreyi, the HxuA heme:hemopexin-binding protein of Haemophilus influenzae, and several adhesions, including the HMW1 and HMW2 proteins of H. influenzae and the FHA proteins of B. pertussis and B. bronchiseptica (reviewed in references 33 and 34). Genes that encode members of the TpsAB family have also been identified in the genomes of H. somnus (GenBank accession no. BAC78648), Pasteurella multocida (39), E. coli (46), Pseudomonas aeruginosa (57), Neisseria meningitidis (60), Fusobacterium nucleatum (35), Xanthomonas campestris (17), Yersinia pestis (44), Photorhabdus luminescens (18), and Ralstonia solanacearum (48), but they have yet to be functionally characterized. In addition, several of these genomes, including those of H. ducreyi, B. pertussis, P. multocida, P. luminescens, and R. solanacearum, encode two or more TpsAB systems.
The H. ducreyi lspB gene is located directly upstream of the lspA2 ORF and encodes a protein which, consistent with the characteristics of TpsB transporters (33, 34), is present in the Sarkosyl-insoluble cell envelope fraction of H. ducreyi and has a predicted molecular mass of 66,573, an extensive β-sheet conformation, and a carboxy-terminal phenylalanine residue typical of outer membrane proteins. The majority of the identity between the H. ducreyi LspB protein and the well-studied B. pertussis FhaC transporter was found in the β-strand regions and in loops 1 and 8 (L1 and L8) (24). Southern blot analysis with an lspB-specific DNA probe resulted in identification of a single hybridizing band in the 16 H. ducreyi strains included in this study, indicating that a single lspB gene was conserved among strains of this pathogen. Among the bacterial proteins with homology to LspB, the IbpB outer membrane transporter protein of H. somnus (GenBank accession no. BAC78648) exhibited the highest degree of similarity (61%). Interestingly, the H. ducreyi hhdB gene product, a putative TpsB exporter involved in the secretion of the HhdA hemolysin (cytolysin) (43), exhibited only 30% similarity to the LspB protein.
It is interesting that H. ducreyi strain A77, which has an lspB gene but which does not secrete the LspA1 or LspA2 protein (67), contains a 7-nucleotide insertion in the lspB ORF that results in a premature translational stop codon. Strain A77 has been reported previously to be serum sensitive (20, 41), to be deficient in adherence to human foreskin fibroblasts (3, 5), to be deficient in microcolony formation (5), to lack a galactose residue in the N-acetyllactosamine portion of its lipooligosaccharide (59), and to be avirulent in the temperature-dependent rabbit model (5). In light of the reports of other phenotypic changes accumulated by A77, the discovery of the lspB mutation in this strain raises the possibility that A77 has a hypermutator phenotype that has rendered this strain avirulent.
Northern blot analysis of H. ducreyi total RNA with lspB- and lspA2-specific probes was not successful in determining the size of the transcript(s) derived from these two genes (data not shown), likely because of the extremely large size predicted for this transcript (>17,000 nucleotides) if the genes were cotranscribed. The lack of a discrete hybridizing band on a Northern blot probed with lspB suggested that the lspB gene was not transcribed as part of a monocistronic operon but likely was cotranscribed with lspA2. Therefore, we confirmed by RT-PCR analysis that the H. ducreyi lspB gene was, in fact, cotranscribed with the lspA2 gene. This finding was notable considering our observation that LspA2 is very difficult (66) and sometimes impossible (67) to detect in CCS from wild-type strain 35000, whereas LspB can be readily detected by Western blot analysis (Fig. 6). Why LspA2 is present at barely detectable levels in CCS from wild-type strain 35000 is not apparent, but it could be due to some type of posttranscriptional regulation or posttranslational processing.
We constructed two independent mutants to investigate the role of the LspB protein in the secretion of the LspA1 and LspA2 proteins. An lspB mutant was constructed to address the role of LspB in the secretion of the LspA1 protein. Similarly, an lspA1 lspB double mutant was constructed to address the role of LspB in the secretion of the LspA2 protein because LspA2 can be readily detected in CCS from an lspA1 mutant (66). CCS from the lspB mutant and the lspA1 lspB mutant did not contain detectable LspA1 and LspA2, respectively, indicating that the single LspB protein encoded by the H. ducreyi genome is involved in the secretion of both of these proteins across the outer membrane. Furthermore, the lspB mutant was significantly less virulent in the temperature-dependent rabbit model than the wild-type parent strain 35000, suggesting that the ability to secrete the LspA1 and LspA2 proteins was required for the full expression of virulence by H. ducreyi in this animal model. These results confirm that these two proteins are involved in virulence expression by this pathogen, which was first demonstrated by the finding that an lspA1 lspA2 double mutant of H. ducreyi is substantially attenuated in the temperature-dependent rabbit model (66).
Complementation of the lspB mutation in strain 35000.88 with the wild-type lspB gene on a plasmid restored the ability of this mutant to secrete LspA1 in vitro (Fig. 7). We were unable to perform a similar complementation analysis of lspA1 lspB mutant 35000.188 because this mutant and the plasmid containing the wild-type lspB gene both possessed a kanamycin resistance gene, a condition that precluded stable maintenance of the plasmid. Nonetheless, RT-PCR analysis confirmed that the lspA2 gene was transcribed in the lspA1 lspB mutant, indicating that the presence of the nonpolar promoterless cat cartridge in the lspB gene did not prevent transcription of the downstream lspA2 gene.
Complementation with the wild-type lspB gene in the lspB mutant 35000.88 could only partially restore the defect in virulence expression in the animal model (Table 2). However, the complemented mutant [35000.88(pCW225)] did yield higher lesion scores than the vector-only control strain 35000.88(pCW177), and the difference was significant. The failure of complementation with the wild-type lspB gene to fully restore virulence to this mutant may involve the relative level of expression of the LspB protein by this strain. It appeared that LspB was overexpressed by this complemented mutant (Fig. 6, lane 5), and the increased abundance of LspB may have had a detrimental effect on the structure or function of the outer membrane in this strain. Moreover, we were never able to successfully clone the full-length lspB gene on the higher-copy-number plasmid pLS88 in H. ducreyi (data not shown), a result which suggested that the LspB protein was toxic when it was overexpressed from this multicopy plasmid in H. ducreyi. Therefore, it seemed possible that the 35000.88(pCW225) strain was under stress and would not exhibit complete restoration of virulence in vivo since it did not truly represent the wild-type state with respect to LspB expression.
Collectively, the data obtained in the present study indicate that H. ducreyi contains a single lspB gene whose protein product is involved in secretion of both the LspA1 and LspA2 proteins across the outer membrane. It will be interesting to determine the exact mechanism of secretion of H. ducreyi LspA1 and LspA2, to characterize the manner in which these large proteins interact with the LspB transporter in the outer membrane, and to compare this mechanism with that described for the secretion of FHA across the outer membrane of B. pertussis.
Acknowledgments
This study was supported by U.S. Public Health Service grant AI32011 to E.J.H. and by National Research Service award F32-AI09845 to C.K.W.
We are grateful to Michelle Alfa for providing H. ducreyi A77, and we thank Christopher Elkins for his generous gift of DsrA antibody. We appreciate the assistance of Nikki Wagner with the nucleotide sequence analysis and the assistance of Robert Blick and Joseph Nika with the rabbit model.
Editor: B. B. Finlay
REFERENCES
- 1.Ahmed, H. J., C. Johansson, L. A. Svensson, K. Ahlman, M. Verdrengh, and T. Lagergard. 2002. In vitro and in vivo interactions of Haemophilus ducreyi with host phagocytes. Infect. Immun. 70:899-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albritton, W. L. 1989. Biology of Haemophilus ducreyi. Microbiol. Rev. 53:377-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alfa, M. J., P. Degagne, and T. Hollyer. 1993. Haemophilus ducreyi adheres to but does not invade cultured human foreskin cells. Infect. Immun. 61:1735-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alfa, M. J., P. Degagne, and P. A. Totten. 1996. Haemophilus ducreyi hemolysin acts as a contact cytotoxin and damages human foreskin fibroblasts in cell culture. Infect. Immun. 64:2349-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfa, M. J., M. K. Stevens, P. Degagne, J. Klesney-Tait, J. D. Radolf, and E. J. Hansen. 1995. Use of tissue culture and animal models to identify virulence-associated traits of Haemophilus ducreyi. Infect. Immun. 63:1754-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al Tawfiq, J. A., K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. [DOI] [PubMed] [Google Scholar]
- 7.Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 178:1684-1687. [DOI] [PubMed] [Google Scholar]
- 8.Bauer, M. E., and S. M. Spinola. 1999. Binding of Haemophilus ducreyi to extracellular matrix proteins. Infect. Immun. 67:2649-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bong, C. T., M. E. Bauer, and S. M. Spinola. 2002. Haemophilus ducreyi: clinical features, epidemiology, and prospects for disease control. Microbes Infect. 4:1141-1148. [DOI] [PubMed] [Google Scholar]
- 10.Bong, C. T., R. E. Throm, K. R. Fortney, B. P. Katz, A. F. Hood, C. Elkins, and S. M. Spinola. 2001. DsrA-deficient mutant of Haemophilus ducreyi is impaired in its ability to infect human volunteers. Infect. Immun. 69:1488-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozue, J. A., M. V. Tullius, J. Wang, B. W. Gibson, and R. S. Munson, Jr. 1999. Haemophilus ducreyi produces a novel sialyltransferase. Identification of the sialyltransferase gene and construction of mutants deficient in the production of the sialic acid-containing glycoform of the lipooligosaccharide. J. Biol. Chem. 274:4106-4114. [DOI] [PubMed] [Google Scholar]
- 12.Campagnari, A. A., R. Karalus, M. A. Apicella, W. Melaugh, A. J. Lesse, and B. W. Gibson. 1994. Use of pyocin to select a Haemophilus ducreyi variant defective in lipooligosaccharide biosynthesis. Infect. Immun. 62:2379-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campagnari, A. A., L. M. Wild, G. E. Griffiths, R. J. Karalus, M. A. Wirth, and S. M. Spinola. 1991. Role of lipopolysaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect. Immun. 59:2601-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole, L. E., T. H. Kawula, K. L. Toffer, and C. Elkins. 2002. The Haemophilus ducreyi serum resistance antigen DsrA confers attachment to human keratinocytes. Infect. Immun. 70:6158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cope, L. D., S. R. Lumbley, J. L. Latimer, J. Klesney-Tait, M. K. Stevens, L. S. Johnson, M. Purven, R. S. Munson, Jr., T. Lagergard, J. D. Radolf, and E. J. Hansen. 1997. A diffusible cytotoxin of Haemophilus ducreyi. Proc. Natl. Acad. Sci. 94:4056-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 18.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J. F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Medigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 19.Elkins, C., C.-J. Chen, and C. E. Thomas. 1995. Characterization of the hgbA locus encoding a hemoglobin receptor from Haemophilus ducreyi. Infect. Immun. 63:2194-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elkins, C., K. J. Morrow, Jr., and B. Olsen. 2000. Serum resistance in Haemophilus ducreyi requires outer membrane protein DsrA. Infect. Immun. 68:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filiatrault, M. J., R. S. Munson, Jr., and A. A. Campagnari. 2001. Genetic analysis of a pyocin-resistant lipooligosaccharide (LOS) mutant of Haemophilus ducreyi: restoration of full-length LOS restores pyocin sensitivity. J. Bacteriol. 183:5756-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fortney, K. R., R. S. Young, M. E. Bauer, B. P. Katz, A. F. Hood, R. S. Munson, Jr., and S. M. Spinola. 2000. Expression of peptidoglycan-associated lipoprotein is required for virulence in the human model of Haemophilus ducreyi infection. Infect. Immun. 68:6441-6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson, B. W., A. A. Campagnari, W. Melaugh, N. J. Phillips, M. A. Apicella, S. Grass, J. Wang, K. L. Palmer, and R. S. Munson, Jr. 1997. Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J. Bacteriol. 179:5062-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guedin, S., E. Willery, J. Tommassen, E. Fort, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2000. Novel topological features of FhaC, the outer membrane transporter involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 275:30202-30210. [DOI] [PubMed] [Google Scholar]
- 25.Gulig, P. A., G. H. McCracken, Jr., C. F. Frisch, K. H. Johnston, and E. J. Hansen. 1982. Antibody response of infants to cell surface-exposed outer membrane proteins of Haemophilus influenzae type b after systemic Haemophilus disease. Infect. Immun. 37:82-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammond, G. W., C. J. Lian, J. C. Wilt, and A. R. Ronald. 1978. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob. Agents Chemother. 13:608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen, E. J., J. L. Latimer, S. E. Thomas, M. Helminen, W. L. Albritton, and J. D. Radolf. 1992. Use of electroporation to construct isogenic mutants of Haemophilus ducreyi. J. Bacteriol. 174:5442-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Inzana, T. J., and L. B. Corbeil. 1987. Development of a defined medium for Haemophilus somnus isolated from cattle. Am. J. Vet. Res. 48:366-369. [PubMed] [Google Scholar]
- 29.Jacob-Dubuisson, F., C. Buisine, N. Mielcarek, E. Clement, F. D. Menozzi, and C. Locht. 1996. Amino-terminal maturation of the Bordetella pertussis filamentous haemagglutinin. Mol. Microbiol. 19:65-78. [DOI] [PubMed] [Google Scholar]
- 30.Jacob-Dubuisson, F., C. Buisine, E. Willery, G. Renauld-Mongenie, and C. Locht. 1997. Lack of functional complementation between Bordetella pertussis filamentous hemagglutinin and Proteus mirabilis HpmA hemolysin secretion machineries. J. Bacteriol. 179:775-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacob-Dubuisson, F., C. El Hamel, N. Saint, S. Guedin, E. Willery, G. Molle, and C. Locht. 1999. Channel formation by FhaC, the outer membrane protein involved in the secretion of the Bordetella pertussis filamentous hemagglutinin. J. Biol. Chem. 274:37731-37735. [DOI] [PubMed] [Google Scholar]
- 32.Jacob-Dubuisson, F., B. Kehoe, E. Willery, N. Reveneau, C. Locht, and D. A. Relman. 2000. Molecular characterization of Bordetella bronchiseptica filamentous haemagglutinin and its secretion machinery. Microbiology 146:1211-1221. [DOI] [PubMed] [Google Scholar]
- 33.Jacob-Dubuisson, F., C. Locht, and R. Antoine. 2001. Two-partner secretion in Gram-negative bacteria: a thrifty, specific pathway for large virulence proteins. Mol. Microbiol. 40:306-313. [DOI] [PubMed] [Google Scholar]
- 34.Kajava, A. V., N. Cheng, R. Cleaver, M. Kessel, M. N. Simon, E. Willery, F. Jacob-Dubuisson, C. Locht, and A. C. Steven. 2001. Beta-helix model for the filamentous haemagglutinin adhesin of Bordetella pertussis and related bacterial secretory proteins. Mol. Microbiol. 42:279-292. [DOI] [PubMed] [Google Scholar]
- 35.Kapatral, V., I. Anderson, N. Ivanova, G. Reznik, T. Los, A. Lykidis, A. Bhattacharyya, A. Bartman, W. Gardner, G. Grechkin, L. Zhu, O. Vasieva, L. Chu, Y. Kogan, O. Chaga, E. Goltsman, A. Bernal, N. Larsen, M. D'Souza, T. Walunas, G. Pusch, R. Haselkorn, M. Fonstein, N. Kyrpides, and R. Overbeek. 2002. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 184:2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klesney-Tait, J., T. J. Hiltke, I. Maciver, S. M. Spinola, J. D. Radolf, and E. J. Hansen. 1997. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J. Bacteriol. 179:1764-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis, D. A., M. K. Stevens, J. L. Latimer, C. K. Ward, K. Deng, R. Blick, S. R. Lumbley, C. A. Ison, and E. J. Hansen. 2001. Characterization of Haemophilus ducreyi cdtA, cdtB, and cdtC mutants in in vitro and in vivo systems. Infect. Immun. 69:5626-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukomski, S., R. A. Hull, and S. I. Hull. 1996. Identification of the O antigen polymerase (rfc) gene in Escherichia coli O4 by insertional mutagenesis using a nonpolar chloramphenicol resistance cassette. J. Bacteriol. 178:240-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.May, B. J., Q. Zhang, L. L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2001. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ochman, H., F. J. Ayala, and D. L. Hartl. 1993. Use of polymerase chain reaction to amplify segments outside boundaries of known sequences. Methods Enzymol. 218:309-321. [DOI] [PubMed] [Google Scholar]
- 41.Odumeru, J. A., G. M. Wiseman, and A. R. Ronald. 1985. Role of lipopolysaccharide and complement in susceptibility of Haemophilus ducreyi to human serum. Infect. Immun. 50:495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer, K. L., W. E. Goldman, and R. S. Munson, Jr. 1996. An isogenic haemolysin-deficient mutant of Haemophilus ducreyi lacks the ability to produce cytopathic effects on human foreskin fibroblasts. Mol. Microbiol. 21:13-19. [DOI] [PubMed] [Google Scholar]
- 43.Palmer, K. L., and R. S. Munson, Jr. 1995. Cloning and characterization of the genes encoding the haemolysin of Haemophilus ducreyi. Mol. Microbiol. 18:821-830. [DOI] [PubMed] [Google Scholar]
- 44.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 45.Patrick, C. C., S. E. Pelzel, P. A. Gulig, C. J. McCracken, J. D. Radolf, and E. J. Hansen. 1989. Antigenic evidence for the synthesis of two different lipooligosaccharides by some strains of Haemophilus influenzae type b. Infect. Immun. 57:1971-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 47.Purcell, B. K., J. A. Richardson, J. D. Radolf, and E. J. Hansen. 1991. A temperature-dependent rabbit model for production of dermal lesions by Haemophilus ducreyi. J. Infect. Dis. 164:359-367. [DOI] [PubMed] [Google Scholar]
- 48.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.San Mateo, L. R., M. M. Hobbs, and T. H. Kawula. 1998. Periplasmic copper-zinc superoxide dismutase protects Haemophilus ducreyi from exogenous superoxide. Mol. Microbiol. 27:391-404. [DOI] [PubMed] [Google Scholar]
- 51.San Mateo, L. R., K. L. Toffer, P. E. Orndorff, and T. H. Kawula. 1999. Neutropenia restores virulence to an attenuated Cu,Zn superoxide dismutase-deficient Haemophilus ducreyi strain in the swine model of chancroid. Infect. Immun. 67:5345-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spinola, S. M., M. E. Bauer, and R. S. Munson, Jr. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 70:1667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spinola, S. M., T. J. Hiltke, K. R. Fortney, and K. L. Shanks. 1996. The conserved 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi has homology to PAL. Infect. Immun. 64:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spinola, S. M., L. M. Wild, M. A. Apicella, A. A. Gaspari, and A. A. Campagnari. 1994. Experimental human infection with Haemophilus ducreyi. J. Infect. Dis. 169:1146-1150. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, M. K., L. D. Cope, J. D. Radolf, and E. J. Hansen. 1995. A system for generalized mutagenesis of Haemophilus ducreyi. Infect. Immun. 63:2976-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stevens, M. K., S. Porcella, J. Klesney-Tait, S. R. Lumbley, S. E. Thomas, M. V. Norgard, J. D. Radolf, and E. J. Hansen. 1996. A hemoglobin-binding outer membrane protein is involved in virulence expression by Haemophilus ducreyi in an animal model. Infect. Immun. 64:1724-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 58.Stuyve, M., M. Moons, and J. Tommassen. 1991. Carboxyterminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 59.Sun, S., B. Schilling, L. Tarantino, M. V. Tullius, B. W. Gibson, and R. S. Munson, Jr. 2000. Cloning and characterization of the lipooligosaccharide galactosyltransferase II gene of Haemophilus ducreyi. J. Bacteriol. 182:2292-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 61.Totten, P. A., J. C. Lara, D. V. Norn, and W. E. Stamm. 1994. Haemophilus ducreyi attaches to and invades human epithelial cells in vitro. Infect. Immun. 62:5632-5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vakevainen, M., S. Greenberg, and E. J. Hansen. 2003. Inhibition of phagocytosis by Haemophilus ducreyi requires expression of the LspA1 and LspA2 proteins. Infect. Immun. 71:5994-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 65.Ward, C. K., and T. J. Inzana. 1994. Resistance of Actinobacillus pleuropneumoniae to bactericidal antibody and complement is mediated by capsular polysaccharide and blocking antibody specific for lipopolysaccharide. J. Immunol. 153:2110-2121. [PubMed] [Google Scholar]
- 66.Ward, C. K., J. L. Latimer, J. R. Nika, M. Vakevainen, J. R. Mock, K. Deng, R. J. Blick, and E. H. Hansen. 2003. Mutations in the lspA1 and lspA2 genes of Haemophilus ducreyi affect the virulence of this pathogen in an animal model system. Infect. Immun. 71:2478-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ward, C. K., S. R. Lumbley, J. L. Latimer, L. D. Cope, and E. J. Hansen. 1998. Haemophilus ducreyi secretes a filamentous hemagglutinin-like protein. J. Bacteriol. 180:6013-6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Willems, R. J., C. Geuijen, H. G. Van Der Heide, G. Renauld, P. Bertin, W. M. van den Akker, C. Locht, and F. R. Mooi. 1994. Mutational analysis of the Bordetella pertussis fim/fha gene cluster: identification of a gene with sequence similarities to haemolysin accessory genes involved in export of FHA. Mol. Microbiol. 11:337-347. [DOI] [PubMed] [Google Scholar]
- 69.Wood, G. E., S. M. Dutro, and P. A. Totten. 2001. Haemophilus ducreyi inhibits phagocytosis by U-937 cells, a human macrophage-like cell line. Infect. Immun. 69:4726-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]