Abstract
The immune events that take place in the central nervous system (CNS) during cryptococcal infection are incompletely understood. We used competitive reverse transcription-PCR to delineate the time course of the local expression of mRNAs encoding a variety of cytokines and inducible nitric oxide synthase (iNOS) during progressive murine cryptococcal meningoencephalitis and assessed the CNS inflammatory response using immunohistochemistry. Interleukin 18 (IL-18), transforming growth factor β1, and IL-12p40 mRNAs were constitutively expressed in the brains of infected and uninfected mice; IL-2 mRNA was not detected at any time. Increased levels of transcripts corresponding to IL-1α, tumor necrosis factor alpha (TNF-α), and iNOS were detected as early as day 1 postinfection, with TNF-α rising by ∼30-fold and iNOS increasing by ∼5-fold by day 7. Each remained at these levels thereafter. IL-4, IL-6, and gamma interferon transcripts were detected on day 5, and IL-1β and IL-10 transcripts were detected beginning on day 7. Once detected, each remained at a relatively constant level through 28 days of infection. This cytokine profile does not suggest a polarized Th1 or Th2 response. Immunohistochemistry did not reveal inflammatory infiltrates before day 7, despite the presence of cryptococci. Intraparenchymal abscesses with inflammatory cells in their peripheries were found beginning on day 10. The infiltrates were comprised primarily of cells expressing CD4, CD8, or CD11b; low numbers of cells expressing CD45R/B220 were also present. The persistence of Cryptococcus observed in the CNS may result from an ineffective immune response, perhaps owing to an insufficient anticryptococcal effector function of endogenous glial cells resulting from competing pro- and anti-inflammatory cytokines. These data detail the immune response in the brain and could be important for the future design of specific immunomodulatory therapies for this important opportunistic infection.
The ubiquitous yeast Cryptococcus neoformans is an opportunistic pathogen that causes life-threatening disease, predominantly in patients with impaired cell-mediated immunity, such as those with AIDS. If the host is unable to effectively clear the primary pulmonary infection, widespread hematogenous dissemination may occur. Cryptococcus appears to have a predilection for establishing meningoencephalitis, as also occurs in murine models. The immunopathogenesis of central nervous system (CNS) infection remains incompletely understood (10, 36). Previous studies have suggested that phagocytic effector cells in the brain (e.g., microglia and astrocytes), as well as cell-mediated immunity (CMI) and cytokine release, all play roles in brain-specific immune response (1, 11, 17, 23, 32, 40).
Most studies of cryptococcal infection in murine models have focused on pulmonary infection, in which the cytokines tumor necrosis factor alpha (TNF-α), interleukin 1β (IL-1β), IL-12, IL-18, and gamma interferon (IFN-γ) are produced during the first week of C. neoformans infection. These cytokines appear to stimulate protective CMI (23, 28). Alveolar macrophages are likely the major source of TNF-α and IL-1β early in pulmonary infection, whereas the cellular sources of early IL-12 and IL-18 have not been identified. CD4+ and CD8+ T cells and NK cells are potential sources of IFN-γ early in the course of infection (23, 24). Cryptococcal virulence factors may also modulate early signaling molecules of the host response. For example, the polysaccharide capsule from C. neoformans decreases TNF-α and IL-1β production by alveolar macrophages in vitro and induces production of IL-10 (an anti-inflammatory inhibitor of Th1 immune response), thereby providing mechanisms by which the capsule may down-regulate CMI in lung or brain (2, 52, 53).
Assessment of mRNA levels is a useful surrogate to measure the expression of numerous biologically important molecules, particularly when small amounts of these molecules are produced. Analysis of mRNA using reverse transcription (RT)-PCR provides a sensitive tool for studying cytokine regulation. To quantify this assay, competitive PCR, which employs the simultaneous amplification of a DNA fragment of interest and of a competitor template of known concentration within the same PCR, may be used. The competitor DNA contains primer-binding sites identical to those of the target and is presumed to undergo amplification with an efficiency equal to that of the target. The competitor is designed to yield a final PCR product that is slightly larger or smaller than that of the target, allowing the subsequent resolution and identification of their respective products by using standard agarose gel electrophoresis (18, 29, 44, 58).
To further characterize host defense mechanisms during cerebral cryptococcal infection, we used competitive RT-PCR to examine the temporal mRNA expression of a variety of cytokines and inducible nitric oxide synthase (iNOS) during the course of experimental murine cryptococcal meningoencephalitis. We concurrently analyzed the inflammatory-cell types involved in the brain (including adherent meninges) response by using immunohistochemistry.
MATERIALS AND METHODS
Mice.
Four- to 5-week-old male BALB/c mice were obtained from Charles River Laboratories (Portage, Mich.). The animals were housed three to five per cage under conventional conditions and provided sterilized food and acidified water ad libitum. All experiments were performed with the approval of the Institutional Animal Care and Use Committee of the California Institute for Medical Research under the guidelines set forth by the Office of Laboratory Animal Welfare of the National Institutes of Health.
C. neoformans.
An encapsulated serotype A strain of C. neoformans (CDC9759) was used as described previously (12-14, 37). To enhance virulence, two consecutives passages in mice were made by intravenous infection and recovery of yeasts from brain tissue. A single encapsulated colony was picked and inoculated into 5 ml of synthetic amino acid medium fungal broth (21) and incubated at 35°C for 72 h on a gyratory shaker at 140 rpm. A 100-μl sample of this culture was inoculated into 5 ml of fresh synthetic amino acid medium fungal broth and incubated for 48 h (log phase) on a gyratory shaker at 35°C. The yeast cells were harvested and washed twice with sterile pyrogen-free saline by low-speed centrifugation (1,000 × g), counted with a hemacytometer, and diluted in saline to 1.6 × 105 cells/ml (inoculum). Serial dilutions of this suspension were plated on Sabouraud dextrose agar, and the resulting colony counts showed an inoculum viability of 78%.
Intravenous inoculation.
Mice were infected by intravenous injection in a lateral tail vein with 0.25 ml of the inoculum (4 × 104 cryptococcal yeast cells per mouse). Uninfected control mice were inoculated with 0.25 ml of sterile pyrogen-free saline.
Quantification of C. neoformans in organs.
The course of infection was determined on days 1, 3, 5, 7, 10, 15, 21, and 28 postinfection by quantitative plating of organ homogenates. In brief, five infected mice were euthanatized by CO2 asphyxiation at each time point. The brain and spleen of each mouse were aseptically removed and disrupted using a mechanical homogenizer (TekMar, Cincinnati, Ohio) as described previously (12-14, 37). The number of viable C. neoformans organisms was determined by quantitative plating of serially diluted homogenates onto Sabouraud dextrose agar plates containing chloramphenicol (50 mg/liter). The plates were incubated at 35°C for 3 days, and the numbers of CFU per entire organ were determined.
RNA extraction and tissue immunohistochemistry.
Mice were perfused to remove peripheral blood cells from the brain and spleen. Three infected mice at each time point and three uninfected mice on days 5 and 28 were deeply anesthetized by inhalation of methoxyflurane (Metofane; Schering-Plough Animal Health, Union, N.J.) vapors. The right atrium of the heart was nicked, and perfusion was done by inserting a 22-gauge needle into the left ventricle, followed by slow injection of 25 ml of 10°C phosphate-buffered saline (PBS). The brain was removed, and the two hemispheres were separated along the medial plane; the spleen was also removed. The tissues (half brains or entire spleens) were put into individually marked polypropylene tubes and immediately frozen in a dry-ice- acetone bath. All tissues were stored at −80°C for subsequent RNA extraction. The remaining half brain was put into a disposable base mold, embedded in Tissue-Tek OCT (Sakura Finetek, Inc., Torrance, Calif.), frozen, and stored at −80°C for subsequent sectioning and immunohistochemical analysis.
Total RNA was extracted by homogenizing frozen perfused brain or spleen tissue in 5 or 3 ml of TRIzol reagent (GibcoBRL, Life Technologies, Rockville, Md.), respectively, using a mechanical homogenizer at room temperature. The method used was essentially that supplied by the manufacturer. After homogenization, chloroform (1.0 or 0.6 ml, respectively) was added, and the tubes were mixed vigorously and then centrifuged at 10,000 × g and 4°C for 20 min. Total RNA was precipitated from the aqueous phase with an equal volume of isopropanol and pelleted by centrifugation at 13,000 × g for 10 min at room temperature. Pellets containing RNA were washed once with 75% ethanol and resuspended in 200 μl of diethylpyrocarbonate (DEPC)-treated water. The concentrations and purities of the RNA preparations were determined by spectrophotometry using absorbance at 260 and 280 nm. The A260/A280 ratio of the samples was ≥1.8. To confirm the integrity of the RNA and to assess possible DNA contamination, 10 μg of each total-RNA sample was electrophoresed and bands were visualized by ethidium bromide staining.
To remove possible traces of genomic DNA not visualized by the electrophoresis, 10 μg of each RNA sample was treated with 10 U of RNase-free DNase (RQ1; Promega, Madison, Wis.) at 37°C for 30 min according to the manufacturer's instructions. The DNase was subsequently inactivated by incubation at 65°C for 10 min.
RT.
Each DNase-treated RNA sample (5 μg) was reverse transcribed using 1 μl (0.5 μg) of oligo(dT)12-18 primer (GibcoBRL), 1 μl (200 U) of SuperScript II reverse transcriptase (GibcoBRL), 1 μl (40 U) of RNasin (Promega), 5 μl of 5 mM (each) deoxynucleoside triphosphate (Sigma, St. Louis, Mo.), 5 μl of 0.1 M dithiothreitol (GibcoBRL), 10 μl of 5× enzyme buffer (GibcoBRL), and 22 μl of DEPC-treated water (total reaction volume, 50 μl). Negative controls were performed using all components but without added reverse transcriptase. Total RNA from mouse brain or spleen (Ambion Inc., Austin, Tex.) was used for positive controls and for establishing reaction conditions. RNA samples, water, and the primer were initially mixed and incubated at 70°C for 8 min, followed by a quick chilling on ice. The other reagents were added, and the final reaction mixture was incubated at 42°C for 1 h. The resultant cDNA was stored frozen at −20°C until it was needed.
PCR.
The following description applies only to PCR done in experiments 2 and 3. The amplification mixture for each sample was made to a total volume of 50 μl. It contained 0.5 μl (1 μg/μl) each of a 3′ and a 5′ gene-specific primer (Table 1) (Operon, Alameda, Calif.), 0.5 μl (5 U/μl) of Taq DNA polymerase (Promega), 2.5 μl of cDNA, 21 μl of DEPC-treated water, and 25 μl of the corresponding premix tube of FailSafe PCR (Epicentre Technologies, Madison, Wis.). The PCR conditions for each primer pair were optimized using cDNA samples prepared from spleen mRNA and the FailSafe PCR buffers. When possible for a primer pair, the plasmid pPQRS, a gift from R. M. Locksley (University of California—San Francisco) (44), was amplified as a positive control. The plasmid contains a polycompetitor insert for IL-2, IL-4, IL-5, IL-10, IL-12p40, IFN-γ, TNF-α, transforming growth factor β1 (TGF-β1), iNOS, and hypoxanthine phosphoribosyl transferase (HPRT) arranged so that the products amplified from the plasmid template by each specific primer pair differ slightly in size from those amplified from the cellular target cDNA (44). Negative controls were samples in which (i) the reverse transcriptase was omitted in the RT step to test for DNA contamination and (ii) Taq polymerase was not added. The PCR was carried out in a thermal cycler (Gene Mate; Intermountain Scientific) using a hot start at 65°C for 2 min and cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 2 min (repeated for 35 cycles), with a final end extension of 10 min at 72°C for all targets tested. Preliminary experiments demonstrated that under these conditions the samples had not yet reached an amplification plateau. Ten microliters of each PCR mixture was electrophoresed through 1.5% agarose gels, stained with 0.5 μg of ethidium bromide per ml, visualized with a UV transilluminator, and photographed. The sizes of the PCR products were verified by comparison with a 100-bp DNA ladder run in parallel on the same gel. The specificities of the transcript-derived bands were determined in gels run with negative controls.
TABLE 1.
Sequences of oligonucleotide primers used for PCR amplification of cytokine, iNOS, and HPRT mRNAs and product sizes predicted for sample cDNA and competitor DNA (pPQRS)
| Gene | Primer sequencea | Product cDNA size (bp) | Product pPQRS size (approx) (bp) |
|---|---|---|---|
| IL-1α | 5′ ATGGCCAAAGTTCCTGACTTGTTT | 615 | |
| 5′ CCTTCAGCAACACGGGCTGGTC | |||
| IL-1β | 5′ ATGGCAACTGTTCCTGAACTCAACT | 527 | |
| 5′ CAGGACAGGTATAGATTCTTTCCTTT | |||
| IL-2 | 5′ TCCACTTCAAGCTCTACAG | 247 | 320 |
| 5′ GAGTCAAATCCAGAACATGCC | |||
| IL-4 | 5′ CATCGGCATTTTGAACGAGGTCA | 240 | 360 |
| 5′ CTTATCGATGAATCCAGGCATCG | |||
| IL-6 | 5′ ATGAAGTTCCTCTCTGCAAGAGACT | 642 | |
| 5′ CACTAGGTTTGCCGAGTAGATCTC | |||
| IL-10 | 5′ CCAGTTTTACCTGGTAGAAGTGATG | 324 | 440 |
| 5′ TGTCTAGGTCCTGGAGTCCAGCAGACTCAA | |||
| IL-12p40 | 5′ AACTGGCGTTGGAAGCACGG | 368 | |
| 5′ GAACACATGCCCACTTGCTG | |||
| IL-12p35 | 5′ ACCTCAGTTTGGCCAGGGTC | 387 | |
| 5′ CAAGGCACAGGGTCATCATC | |||
| IL-18 | 5′ ACTGTACAACCGCAGTAATACGG | 434 | |
| 5′ AGTGAACATTACAGATTTATCCC | |||
| IFN-γ | 5′ CATTGAAAGCCTAGAAAGTCTG | 267 | 320 |
| 5′ CTCATGAATGCATCCTTTTTCG | |||
| TFN-α | 5′ GTTCTATGGCCCAGACCCTCACA | 383 | 600 |
| 5′ TCCCAGGTATATGGGCTCATACC | |||
| TGF-β1 | 5′ TGGACCGCAACAACGCCATCTATGAGAAAACC | 525 | |
| 5′ TGGAGCTGAAGCAATAGTTGGTATCCAGGGCT | |||
| iNOS | 5′ TGGGAATGGAGACTGTCCCAG | 306 | 390 |
| 5′ GGGATCTGAATGTGATGTTTG | |||
| HPRT | 5′ GTTGGATACAGGCCAGACTTTGTTG | 352 | 450 |
| 5′ GAGGGTAGGCTGGCCTATAGGCT | |||
| β-Actin | 5′ GTGGGCCGCTCTAGGCACCAA | 550 | |
| 5′ CTCTTTGATGTCACGCACGATTTC |
All primer sequences are listed as 5′ to 3′. The top primer sequence is the forward primer, and the bottom sequence is the reverse primer for each pair. The primer pair sequences used for IL-2, IL-4, IL-10, and iNOS were derived from reference 44. The forward primer sequences for TNF-α, IFN-γ, and HPRT were derived from reference 44, and the reverse primer sequences for these pairs were provided by E. Wakil, D. B. Corry, and L. Stowring (personal communications). IL-1α, IL-1β, IL-6, β-actin, IL-12p35, and IL-12p40 primer pair sequences were provided by C. A. Hunter (personal communication). Primer pair sequences for IL-18 and TGF-β1 were derived from reference 28.
Determination of cytokine mRNA concentration.
Samples from infected mice yielding products of the expected size following PCR with specific cytokine or iNOS primers (IL-4, IL-10, IFN-γ, TNF-α, and iNOS), where no detectable product was found from uninfected control mice, were subjected to quantification using competitive PCR analysis as described previously (18, 29, 44, 58). Cytokines for which a competitor was not available on the pPQRS plasmid or that were constitutively produced were not further quantified. Briefly, six serial PCRs were set up in which a constant volume of experimental cDNA was added to serial fivefold dilutions of linearized pPQRS plasmid, also in a constant volume. Primers for IL-4, IL-10, IFN-γ, TNF-α, or iNOS cDNA were used for amplification. PCR products were electrophoresed through an agarose gel and stained with ethidium bromide. The dilution showing the point of equivalence in staining intensity of the PCR products from competitor (pPQRS) and from the tissue samples was used to determine the concentration of the mRNA derived from a particular sample. To control for possible variation in the efficiencies of the RT step among different experimental samples, HPRT mRNA concentrations (a “housekeeping” gene, presumed to be expressed at constant amounts) were also calculated for each sample.
Immunohistochemistry.
Direct and indirect immunohistochemistry was performed on acetone-fixed 8-μm-thick cryostat sections of brain (42). Endogenous peroxides were inhibited, and the nonspecific protein-binding sites were blocked by incubating sections with 1% bovine serum albumin (Sigma) in PBS plus 0.0001 M sodium azide for 30 min at room temperature in a humidified chamber. The following rat anti-mouse monoclonal antibodies (BD Pharmingen, San Diego, Calif.) were used: biotinylated anti-CD4, 0.5-mg/ml stock diluted 1:100; biotinylated anti-CD8, 0.5-mg/ml stock diluted 1:100; biotinylated anti-CD11b, 0.5-mg/ml stock diluted 1:100; and purified anti-CD45R/B220, 0.5-mg/ml stock diluted 1:200. The antibodies (0.2 ml) were added to cover the tissue sections on the slides, and the slides were incubated for 1 h at room temperature. For CD4, CD8, and CD11b, peroxidase-conjugated streptavidin (1.0-mg/ml stock diluted 1:500; Jackson ImmunoResearch, West Grove, Pa.) was added and the slides were incubated for 30 min at room temperature. An indirect method was used to detect CD45R/B220, adding biotinylated rabbit anti-rat immunoglobulin G (IgG) (0.5-mg/ml diluted 1:100; Vector Laboratories, Inc., Burlingame, Calif.) as the secondary antibody. Reactions were developed using the VECTASTAIN ABC detection system (Vector) for 30 min at room temperature. The reactions were developed using diaminobenzidine nickel concentrate (Pierce, Rockford, Ill.) diluted 1:10 in stable peroxide substrate buffer as a chromogen. The slides were washed three times in PBS for 5 min each time after each binding step, using gentle agitation. Sections were counterstained with hematoxylin and mounted. For each antibody, sections of murine spleen were used as a positive control and nonspecific isotype-matched (purified rat IgG2a and purified rat IgG2b) antibodies were used as negative controls. Specificity was confirmed by comparing the observed distribution of label with the anatomically expected distribution of positive cells in the spleen. The CD4/CD8 ratio was estimated by counting the CD4-labeled cells in five fields at ×400 magnification in one section and counting labeled cells in five fields of another serial section stained for CD8.
Statistical analysis.
The yeast burden in tissues is presented as log10 geometric mean CFU per organ ± 95% confidence interval (n = 5 mice per time point). Quantification of cytokine transcripts is presented as median log10 DNA copies (n = 3 mice per time point). Statistical analyses were done using the open-access program GMC (http://www.forp.usp.br/restauradora/gmc/gmc.html). A nonparametric Kruskal-Wallis analysis of variance followed by a test for multiple comparisons (22) was used to test for differences in yeast burdens in tissues for the various times sampled. Levels of cytokine mRNA transcripts in the brain on the different days sampled were compared in the same way (22). A P value of <0.05 was considered significant. Spearman's rank correlation test was used for correlation analysis. A correlation with an r value of ≥0.60 at a level of 1% (α = 0.01) was considered significant.
RESULTS
Fungal burden.
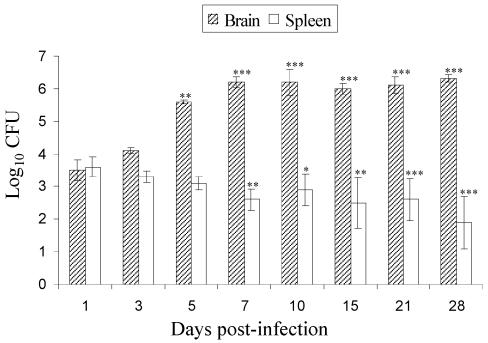
The recovery of CFU from the various samples from the brain and spleen is presented in Fig. 1. In the early phase of infection (days 1 and 3), the yeast burdens in the brain and spleen were similar. By days 5 and 7, the brain CFU had increased compared to day 1 CFU (P < 0.01). From day 5 through day 28, brain CFU remained at ∼106/organ, whereas the spleen CFU decreased significantly with time (P < 0.05 to 0.01). These data demonstrate clearance of the organisms from the spleen compared to proliferation in the brain. In addition, these data delineate the temporal severity of infection, allowing comparison with the cytokine and histological responses.
FIG. 1.
Yeast burdens in brains and spleens of BALB/c mice after intravenous infection with 4 × 104 cells of C. neoformans (isolate 9759). The data are expressed as log10 geometric mean CFU per organ ± 95% confidence interval (n = 5 mice/time). The differences between brain and spleen at each time compared to day 1 postinfection were statistically significant as indicated (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Cytokine profiles.
An initial study of BALB/c mice infected systemically with C. neoformans was done to determine the feasibility of following mRNA expression for eight cytokines in the brain (Table 2, experiment 1). The brains from groups of four infected and two uninfected mice on days 3 and 10 of infection and from two infected and one uninfected mouse on day 21 were examined. mRNA transcripts were followed by semiquantitative RT-PCR methodology as described for studies of murine toxoplasmosis (27). In this initial experiment, β-actin was used as the control housekeeping gene, and in most cases the primer pairs used (27) were different from those listed in Table 1. IL-1α, IL-12p35, and TGF-β1 were constitutively expressed by infected and uninfected mice on all days. IL-2 transcripts were not detected on any day in any mouse. IFN-γ, IL-6, and IL-10 were detected in no animals on day 3 of infection but were found on day 10 and day 21 only in infected mice (Table 2, experiment 1). No quantification was done during these studies, which led us to perform the studies described below.
TABLE 2.
Expression of cytokine and iNOS mRNAs during the course of experimental meningoencephalitis due to C. neoformans, analyzed by RT-PCR
| Cytokine or control | Expt no. | Expressiona
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Infected mice
|
Uninfected mice
|
||||||||||||
| 1b | 3 | 5 | 7 | 10 | 15 | 21 | 28 | 3 | 5 | 10 | 28 | ||
| Proinflammatory | |||||||||||||
| IL-1α | 1 | ++++ | ++++ | ++ | ++ | ++ | |||||||
| 2 | −−− | +++ | +++ | +++ | ++− | +++ | +++ | +++ | −−− | −−− | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | ±±− | ±−− | |||
| IL-1β | 1 | ND | ND | ND | ND | ND | |||||||
| 2 | −−− | −−− | −−− | +++ | +++ | +++ | +++ | +++ | −−− | −−− | |||
| 3 | −−− | −−− | −−− | +++ | ++++ | +++ | +++ | +++ | −−− | −−− | |||
| TNF-α | 1 | ND | ND | ND | ND | ND | |||||||
| 2 | +−− | ++− | +++ | ++− | ++− | +−− | ++− | ++− | −−− | −−− | |||
| 3 | +++ | +++ | ### | ### | #### | ### | ### | ### | −−− | +−− | |||
| IL-6 | 1 | −−−− | ++++ | ++ | −− | −− | |||||||
| 2 | −−− | −−− | +−− | +−− | +++ | +++ | +++ | +++ | −−− | −−− | |||
| 3 | −−− | −−− | +++ | +++ | ++++ | +++ | +++ | +++ | −−− | −−− | |||
| T-helper 2 | |||||||||||||
| IL-4 | 1 | ND | ND | ND | ND | ND | |||||||
| 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| 3 | −−− | −−− | +++ | ### | #### | ### | ### | ### | −−− | −−− | |||
| IL-10 | 1 | −−−− | −+++ | ++ | −− | −− | |||||||
| 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| 3 | −−− | −−− | −−− | ### | #### | ### | ### | ### | −−− | −−− | |||
| T-helper 1 | |||||||||||||
| IFN-γ | 1 | −−−− | ++++ | ++ | −− | −− | |||||||
| 2 | −−− | −−− | +++ | +++ | +++ | ++− | +++ | +++ | −−− | −−− | |||
| 3 | −−− | −−− | ### | ### | #### | ### | ### | ### | −−− | −−− | |||
| IL-2 | 1 | −−−− | −−−− | −− | −− | −− | |||||||
| 2 | −−− | −−− | −−− | −−− | −−− | −−− | −−− | −−− | −−− | −−− | |||
| 3 | −−− | −−− | −−− | −−− | −−−− | −−− | −−− | −−− | −−− | −−− | |||
| Inducible enzyme | |||||||||||||
| iNOS | 1 | ND | ND | ND | ND | ND | |||||||
| 2 | −−+ | +++ | ++− | +++ | +++ | ++− | +−− | −−− | −−− | −−− | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | ### | ### | ### | −−− | −−− | |||
| Constitutive | |||||||||||||
| IL-12p40 | 1 | ND | ND | ND | ND | ND | |||||||
| 2 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | +++ | +++ | |||
| IL-18 | 1 | ND | ND | ND | ND | ND | |||||||
| 2 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | +++ | +++ | |||
| TGF-β1 | 1 | ++++ | ++++ | ++ | ++ | ++ | |||||||
| 2 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | +++ | +++ | |||
| IL12p35 | 1 | ++++ | ++++ | ++ | ++ | ++ | |||||||
| 2 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | +++ | +++ | |||
| Internal controls | |||||||||||||
| HPRT | 1 | ND | ND | ND | ND | ND | |||||||
| 2 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | +++ | +++ | |||
| β-actin | 1 | ++++ | ++++ | ++ | ++ | ++ | |||||||
| 2 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | |||
| 3 | +++ | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | +++ | +++ | |||
RNA samples were obtained from two to four mice per time point. The symbols indicate whether mRNA for a specific gene was detected after PCR amplification for each animal (i.e., one symbol represents one animal). ND, RT-PCR was not done for that time point; −, no mRNA was detected for that gene at the lower limit of sensitivity of the assay; +, mRNA expression was detected for that gene; ±, mRNA expression was at the limit of detection; #, mRNA expression was detected for that gene and, as measured by competitive PCR, was at a significantly higher level than mRNA expression for the gene on days 1 and 3 postinfection (P < 0.05 to 0.001) (not used for products when quantification was not performed by RT-PCR; those are indicated as mRNA detected only [+]).
Day postinfection.
Using the information obtained from the initial study, we designed a more detailed experiment examining earlier time points, a broader repertoire of cytokines, and quantitation of some cytokine transcripts to more clearly define the temporal sequence of expression. We studied the expression of various cytokines during the 28-day course of cryptococcal meningoencephalitis. Two separate infection experiments were performed (Table 2, experiments 2 and 3). In the initial experiment (Table 2, experiment 2), we sought to optimize the RT-PCR method and to examine the expression of the various mRNA transcripts. In the second, replicate experiment (Table 2, experiment 3), the optimized methods were used for the PCR and mRNA expression was quantified by the competitive-PCR method. Overall, the levels of expression and times of appearance of the cytokine mRNAs were very similar for each experiment. After optimization of the PCR conditions for each cytokine (Table 2, experiment 3), we detected mRNAs for 11 cytokines (IL-1α, IL-1β, IL-4, IL-6, IL-10, IL-12p40, IL-12p35, IL-18, IFN-γ, TNF-α, TGF-β1) and for iNOS, as well as the constitutively expressed genes for HPRT and β-actin, in the brains of infected mice (Table 2, experiment 3). Transcripts encoding IL-2 in the brain, if present at all, were at levels below the sensitivity afforded by this assay. However, mRNA for IL-2 was readily demonstrated in control experiments using RNA isolated from the spleens of the same animals (data not shown). The reproducibility of the appearance of mRNA transcripts from individual mice at a given time point is demonstrated in Table 2 and Fig. 2. In addition, cDNA was made from the RNA derived from each tissue sample for each animal on three separate occasions. Replicate PCRs using these different cDNA preparations showed them to give identical PCR results.
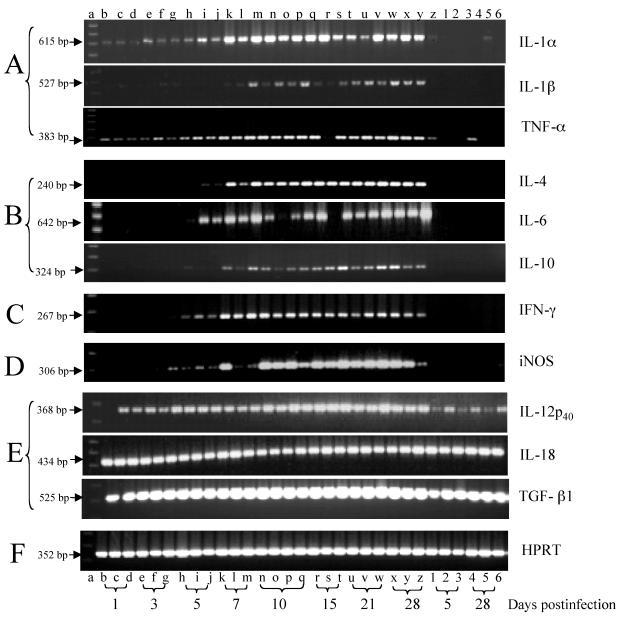
FIG. 2.
Temporal expression of cytokine, iNOS, and HPRT mRNAs in brains of uninfected BALB/c mice (lanes 1 to 6) and BALB/c mice infected with 4 × 104 cells of C. neoformans (lanes b to z). Total RNA was extracted from half of each brain from uninfected mice on days 5 and 28 and from infected mice on days 1, 3, 5, 7, 10, 15, 21, and 28. The RNA samples were obtained from three or four mice per time point. RT-PCR was performed, and the PCR products were electrophoresed on 1.5% agarose gels containing 0.5 μg of ethidium bromide per ml and observed with a UV transilluminator. Lane a, molecular size marker (100-bp ladder). (A) Proinflammatory cytokines; (B) Th2 cytokines; (C) Th1 cytokines; (D) inducible enzyme; (E) constitutive cytokines; (F) internal control.
(i) Constitutive cytokines.
mRNAs encoding HPRT, IL-12p40, IL-18, and TGF-β1 were detected at all sampling times and at similar levels in the brains, regardless of the presence or absence of infection (Fig. 2); the same was true for β-actin and IL-12p35 (data not shown). Quantification by competitive PCR could not be done for two mRNAs because the primer pair utilized to amplify IL-12p40 mRNA has a different sequence than that used for the IL-12p40 insert of pPQRS and IL-18 was not included in the pPQRS plasmid. Thus, we examined potential differences by densitometry scans of ethidium-stained bands.
Densitometry analysis suggested comparable levels of transcripts in all animals and throughout the time course for each of these genes (data not shown). These results indicate that mRNAs encoding IL-12p35, IL-12p40, IL-18, and TGF-β1 are constitutively expressed in the brain. One possible exception was observed for IL-12p40, for which the transcript concentration increased 40% compared to the baseline beginning on day 10 of infection.
(ii) Inducible cytokines.
The levels of IL-1α, IL-1β, IL-4, IL-6, IL-10, IFN-γ, TNF-α, and iNOS mRNAs were induced or elevated in the brain during the course of infection (Fig. 2). Cytokine mRNAs for IL-1β, IL-4, IL-6, IL-10, and IFN-γ were not detectable in uninfected controls. TNF-α was detected in one uninfected mouse (Fig. 2, lane 4), but this animal was not included in further analyses. IL-1α was detected in repeated amplifications in some controls, but not others, as a barely visible band and may be expressed on a constitutive basis at very low levels. In infected animals, these cytokine mRNAs showed differences in their times of initial appearance postinfection. IL-1α, TNF-α, and iNOS were detectable 1 day postinfection, whereas IL-4, IL-6, and IFN-γ were detected beginning on day 5 and IL-1β and IL-10 were detected beginning on day 7 postinfection (Fig. 2).
(iii) Cytokine mRNA concentration.
The method of competitive PCR used to quantify the number of transcripts is illustrated in Fig. 3. For IL-4, there was a significant increase in transcript abundance on days 7, 15 and 21 (P < 0.01) and on day 28 (P < 0.001) in comparison with the levels at the first detectable time point, day 5. However, maximal induction had occurred by day 7 postinfection and remained constant thereafter (Fig. 4).
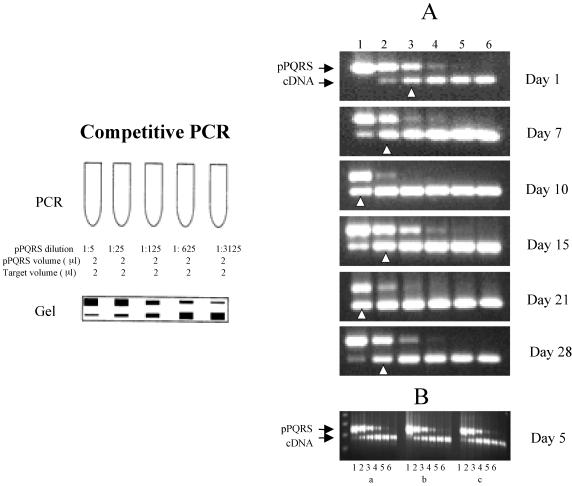
FIG. 3.
Competitive PCR. (Left panel) Schematic of a typical PCR experiment (described in Materials and Methods). (A) Titration of iNOS mRNA transcripts in the brains of infected BALB/c mice sampled at different times postinfection. A constant amount of experimental cDNA was titrated against sequential dilutions of linear pPQRS plasmid competitor during the PCR amplification. Lane 1, 3.8 × 105 copies of plasmid DNA/μl; lane 2, 7.7 × 104 copies of plasmid DNA/μl; lane 3, 1.5 × 104 copies of plasmid DNA/μl; lane 4, 3.1 × 103 copies of plasmid DNA/μl; lane 5, 6.2 × 102 copies of plasmid DNA/μl; lane 6, 1.2 × 102 copies of plasmid DNA/μl. The arrowheads indicate the points of equivalence in copy numbers of the two different PCR products. The sizes of the PCR products were 390 bp for pPQRS iNOS and 306 bp for iNOS. (B) Example of the reproducibility of the pPQRS methodology for the expression of iNOS transcripts in three mice (a, b, and c) at the same time postinfection. The lanes are the same as for panel A.
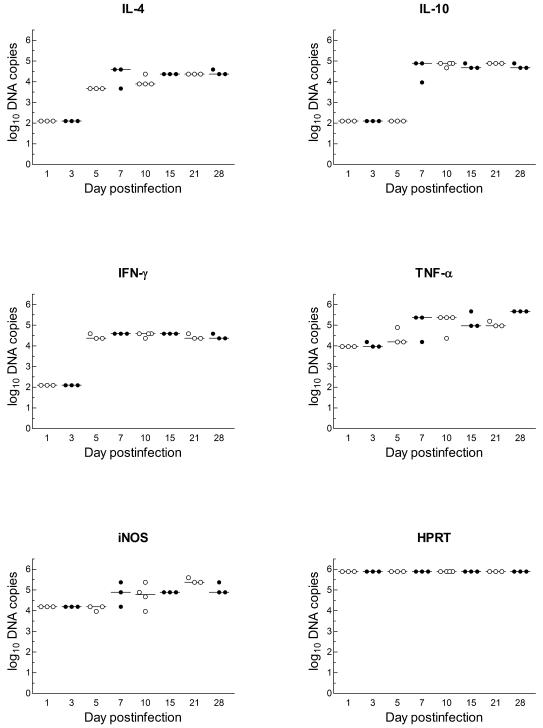
FIG. 4.
Quantification of DNA copies of cytokines (IFN-γ, TNF-α, IL-4, and IL-10), iNOS, and HPRT during the course of cerebral cryptococcal infection in BALB/c mice, determined by competitive PCR as described in Materials and Methods. The symbols represent individual mice, and the bars represent the medians for the time points examined. On days 1 and 3, IFN-γ, IL-4, and IL-10 were not detected, and IL-10 was not detected on day 5. The statistical significance of differences during the evolution of infection and the correlation among the cytokines are discussed in the text. Values of 2 indicate that no product was detected, and the lower limit of sensitivity of the assay was ∼100 copies.
The induction of measurable quantities of TNF-α mRNA began on day 1 and persisted during the entire course of infection. The numbers of copies (log10) of TNF-α mRNA were significantly greater on days 7, 10, 15, 21, and 28 than on day 1 or 3 (P < 0.01 to 0.001). These levels remained high and were higher on day 28 postinfection (P < 0.05 to 0.01) than at any preceding time point (Fig. 4).
Figure 4 also shows that the results for iNOS were similar to those for TNF-α (days 7 through 28 compared to day 5; P < 0.05 to 0.01). After day 5 or 7, there was no significant change in the levels of mRNA transcripts for IL-4 (day 7), IFN-γ (day 5), and IL-10 (day 7) (Table 2 and Fig. 4).
Correlation analysis of the transcript levels of induced genes that had been quantified was done by the Spearman rank correlation test. These results showed direct correlation at the 1% level between the expression levels of IL-4 and iNOS (r = 0.73). No other significant correlations in gene expression were found.
Inflammatory-cell response in brain.
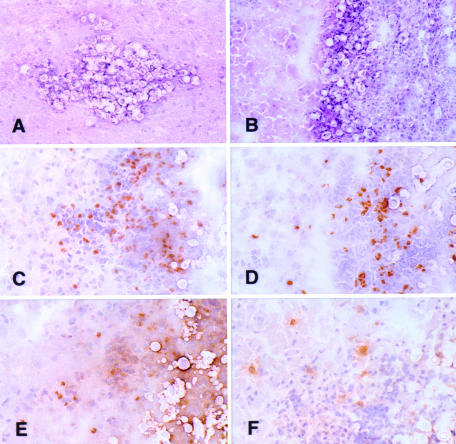
To determine whether the observed variations in cytokine levels were associated with particular cell types recruited to the site of the infection, immunohistochemical staining for T cells, B cells, and monocytes was performed on frozen sections of brain obtained at various times during the course of infection. Small cryptococcal pseudocysts containing densely packed accumulations of encapsulated organisms were observed by day 5 postinfection. The first inflammatory cells associated with these lesions were detected on day 7; these cells were CD11b+. Since few neutrophils were identified in any of the infiltrates, the CD11b+ cells were probably predominantly macrophages or monocytes (Table 3). On day 10, acute meningitis and intraparenchymal abscesses with many encapsulated cryptococci were evident. The inflammatory cells consisted of CD4+ and CD8+ T lymphocytes, CD11b+ cells, and a few CD45R/B220+ B lymphocytes. The CD4/CD8 ratio was estimated to be 2.5:1. The same pattern of inflammation was seen on days 10, 15, 21, and 28 postinfection (Fig. 5).
TABLE 3.
Presence of T lymphocytes (CD4+ and CD8+), B lymphocytes (CD45R/B220+), and monocytes (CD11b+) in the brain during cryptococcal meningoencephalitis
| Surface marker | Presence on daya:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 5 | 7 | 10 | 15 | 21 | 28 | |
| CD4 | − − − | − − − | − − −* | −* − −* | −* − +* +* | +* −* −* | −* +* +* | +* +* +* |
| CD8 | − − − | − − − | − − −* | −* − −* | −* − +* +* | +* −* −* | −* +* +* | +* +* +* |
| CD11b | − − − | − − − | − − −* | +* − +* | +* − +* +* | +* +* +* | −* +* +* | +* +* +* |
| CD45R/B220 | − − − | − − − | − − −* | −* − −* | −* − +* +* | +* −* −* | −* +* +* | +* −* +* |
−, absence of labeled cells; +, presence of labeled cells; *, Cryptococcus in tissue. CD4, CD8, and CD11b were detected directly using biotinylated rat anti-mouse antibodies, and CD45R/B220 was detected indirectly using purified rat anti-mouse antibody followed by biotinylated rabbit anti-rat immunoglobulin G. All were observed after immunoperoxidase staining. Three or four mice were evaluated at each time point as shown by the number of symbols.
FIG. 5.
(A and B) Cryosections of typical intraparenchymal cryptococcal abscesses from day 10 (A) and day 21 (B) postinfection (hematoxylin and eosin staining). (C to F) Adjacent serial cryosections (immunoperoxidase with hematoxylin counterstain) demonstrating the inflammatory response in a cryptococcal abscess from day 28 postinfection. Intact brain parenchyma is on the left side of the field in each panel. (C) Anti-CD4; (D) anti-CD8; (E) anti-CD11b; (F) anti-CD45R/B220. Numerous CD4+ and CD8+ lymphoid cells and CD11b+ cells are shown in the abscess. There are fewer B cells in the infiltrate in panel F. Magnifications: ×206 (A), ×171 (B), and ×294 (C to F).
In general, abscesses increased in size with time, and they were more numerous, containing more thickly encapsulated budding yeast. The inflammatory-cell response and the numbers of T cells, B cells, and monocytes visualized in the lesions generally correlated with the numbers of yeast cells present in the abscesses. No necrosis, vasculitis, granulomatous foci, or multinucleated or epithelioid cells were observed at any time.
DISCUSSION
Numerous reports have described the immune interactions that take place in the lungs following infection with C. neoformans (19, 20, 23, 24, 40, 43). Less is known about the host-pathogen interplay in the brain, the organ most frequently involved in extrapulmonary infection and in infections with a fatal outcome.
Although the CNS has traditionally been regarded as an immunologically privileged site, activated T cells can traffic across the blood-brain barrier into the CNS for immune surveillance. In this way, when subjected to an injury or infection, the CNS can mobilize and develop an immune response involving infiltrating CD4+ and CD8+ T cells, B cells, macrophages and neutrophils, and activated resident cells (e.g., microglia, astrocytes, and endothelial cells) (6, 7, 10, 17, 31, 57).
When they are activated, endogenous CNS cells express major histocompatibility complex class I and class II molecules and may therefore act as antigen-presenting cells. They also express complement receptors; produce cytokines, chemokines, and molecules with bactericidal activity, such as nitric oxide (NO); and are capable of phagocytosis (1, 7, 10, 25, 26, 30, 32, 57). Microglia, acting as antigen-presenting cells, stimulate T-cell proliferation and cytokine secretion, which in turn stimulates these semiprofessional phagocytes to ingest and more effectively kill invading organisms (1, 57).
Whether an immune response within the CNS is initiated, amplified, or suppressed depends on a number of factors, including (i) the activation state of microglia, (ii) cytokine and cytokine receptor levels in glial and immune cells, (iii) relative expression of immune-enhancing and immune-suppressing cytokines, (iv) the locations of these cytokines within the CNS, and (v) the temporal sequence in which a particular cell is exposed to various cytokines (57). The actions of cytokines on the vasculature in the brain also may be of pathophysiological relevance. There is increasing evidence that IL-1 activates the intercellular adhesion molecules, such as intercellular adhesion molecule 1, vascular cell adhesion molecules, E-selectin, and complement regulatory proteins, and it may also cause platelet adhesion (34).
Previous studies of brain cytokine expression in cryptococcosis have used mice infected via the natural route of infection (the respiratory tract) or mice preimmunized with cryptococcal antigen (soluble or heat-killed yeast) and subsequently challenged with C. neoformans via the intracerebral route. In both situations, the immune response in the brain is a secondary immune response (20, 23, 25). The intravenous inoculation of naive mice with C. neoformans, as in the present study, results in direct and preferential dissemination of the yeast to the brain, allowing the primary immune response in the brain to be followed. In most patients with cryptococcal CNS infection, there is an inapparent preceding event which may be brief and initiate a minimal immune response. The model of CNS infection after intravenous inoculation used in the present study may approximate this situation. The model used in our studies also provides a consistent level of fungal burden in the CNS from mouse to mouse that is not afforded by the pulmonary model.
Using RT-PCR analysis, we found that mRNAs for cytokines, such as TGF-β1 and IL-18, were expressed constitutively and stayed at constant levels in infected and uninfected animals. This was also true of IL-12p35. A similar observation about TGF-β1 has been made in patients with cerebral malaria (9). This finding is consistent with the idea that TGF-β1, produced by endogenous glial cells, acts as an anti-inflammatory and neuroprotective molecule and contributes to normal functioning of the CNS (57). The expression of IL-1α at barely detectable levels in some uninfected controls, but not in others, is in accord with previous reports of whether this inflammatory cytokine is constitutively produced in the brain (55). However, the increase in expression during progressive infection observed during our studies is similar to the pattern of expression in the brain reported in studies of Toxoplasma gondii (27).
Normally, IL-12 is produced by monocytes and B cells (and activated microglial cells in the CNS) and induces the production of IFN-γ by natural killer (NK) and T cells and the development of a Th1 cellular immune response against infection (15, 19). Neutralization of either IL-12 or IFN-γ blocks the development of protective CMI (23, 43). IL-12 has a positive effect on host immune response, and the cytokine has been shown to improve the treatment of cryptococcosis in a murine model (13). Some investigators (16, 51) have detected IL-12p40 by immunohistochemistry and/or mRNA assay in normal neural tissue; other reports indicate that IL-12p40 mRNA production in the brain is not constitutive (48, 56). In two experiments with three animals each, we showed constitutive expression of mRNA for the IL-12p40 subunit.
We also demonstrated the following. First, the transcripts for the proinflammatory cytokines IL-1α, IL-1β, IL-6, and TNF-α; the Th1 cytokine IFN-γ; the Th2 cytokines IL-4 and IL-10; and iNOS were induced above baseline levels, if any, in the brain during the course of cryptococcal infection. Second, the expression of IL-1α, TNF-α, and iNOS mRNAs showed similar kinetics, with early detection on day 1 and increased levels at later time points. Third, the expression of IL-4, IL-6, and IFN-γ mRNAs was detected on day 5, and expression of IL-1β and IL-10 was detected on day 7. Fourth, the expression of mRNA for IL-4, IL-10, and IFN-γ remained constant in later stages of infection. Similar results were obtained in the murine model of cerebral candidiasis (3), in AIDS patients with meningeal cryptococcosis, and in experimental murine cryptococcal meningoencephalitis (36).
The initial expression of IL-12, IL-18, TGF-β1, IL-1α, TNF-α, and iNOS transcripts prior to the appearance of observable infiltrating inflammatory cells suggests that these mRNAs were produced by resident cells in the CNS. In contrast, IL-1β, IL-4, IL-6, IL-10, and IFN-γ transcripts were detected only after day 5 or 7 postinfection. Rises in the levels of these transcripts corresponded with the initial arrival of infiltrating cells (CD4+ T cells, CD8+ T cells, CD11b+ macrophages, and CD45R/B220+ B cells) as observed by immunohistochemistry, which requires significant numbers of cells to be present before they can be detected in tissue sections. However, the precise cellular origins of each cytokine remain to be determined. Despite the infiltration of T cells, however, IL-2 was not detected in the brains, suggesting that T-cell proliferation may not occur.
TNF-α has effects similar to those of IL-1, and early production of TNF-α is required to prevent the establishment of cryptococcal foci in the CNS (23). Our data indicate that TNF-α and iNOS are produced in direct response, and in proportion, to the magnitude of the infectious burden seen in the brain. A similar result was observed during the development of cerebral candidiasis (3).
IFN-γ is considered to play a critical role in the activation of macrophages through induction of NO, a principal mediator for macrophage killing activity in mice. In addition, IFN-γ enhances the antigen-presenting activity of macrophages and acts as a stimulus for IL-12 production, causing the expansion of a Th1 cell population. Protection observed in immunized mice was mediated via IFN-γ due to the activation of effector cells already present at the site of infection or after their recruitment to the site (1, 10). Treatment with IFN-γ results in a reduction in the cryptococcal burden in the CNS in infected mice (49), and based on these data, it has been possible to use IFN-γ in adjuvant immunotherapy (14, 37).
Three different forms of nitric oxide synthase that can affect brain function have been identified in the brain: type I, neuronal nitric oxide synthase; type II, iNOS; and type III, endothelial nitric oxide synthase (33). iNOS is normally not expressed in the brain but is active for 4 to 8 h synthesizing NO in nanomolar concentrations when induced (33, 50). NO acts in part as a neurotransmitter and as a hormone but is cytotoxic at high concentrations and might be involved in neural degeneration. In immune cells, NO is converted to products like peroxynitrite, which are highly toxic to microorganisms (33). IFN-γ, TNF-α, IL-2, and IL-1β mediate the induction of expression of iNOS in microglia and astrocytes, whereas TGF-β1, IL-4, and IL-10 inhibit iNOS induction (33). NO suppresses the functions of proinflammatory cytokines and eliminates inflammatory cells in the CNS, possibly through an apoptotic mechanism (34, 35, 57). This phenomenon has been described in Paracoccidioides brasiliensis pulmonary infection; in the early stage, NO is important for killing the organisms, but persistent NO production likely contributes to the immunosuppression observed during infection (8).
In view of our results with respect to the expression of IL-12, TNF-α, IFN-γ, and the infiltrating cells in the brain, we might have expected a resolution of infection or at least a reduction in the numbers of fungi in the brain. However, the concomitant expression of TGF-β1, IL-4, and IL-10 might have acted as immunosuppressive cytokines, allowing the continuation of the infectious process. In addition, although in the early stages of infection NO contributes to the killing of yeasts, the expression of iNOS by endogenous cells may have been modulated by the immunosuppressive cytokines or NO may cause immunosuppression itself, thereby permitting progression of the infection and death. The former possibility is supported by the correlation observed between levels of IL-4 and iNOS mRNAs. The paradoxical depression of iNOS may be observed in the brain, but not in the lungs, as the result of the neuroprotective action of microglia, which express suppressive cytokines, such as TGF-β1, to a greater degree than the proinflammatory cytokines IL-1β, IL-6, IL-12, IFN-γ, and TNF-α under natural conditions (34, 35, 57).
C. neoformans virulence factors, such as the polysaccharide capsule or melanin, can modulate phagocytosis, antigen processing, cytokine production, lymphocyte proliferation, and fungicidal activity (4, 38, 39, 45, 46, 52-54) and therefore hinder the evolution of an effective immune response (2, 11). Our finding of the absence of IL-2 production is consistent with the impairment of local T-cell proliferation. In addition, in the presence of specific antibody to the polysaccharide capsule, microglial cells are potent effector cells against C. neoformans, resulting in prolonged survival and reduced organ tissue fungal burden, as well as earlier and better-organized granuloma formation in infected mice.
The fact that an avirulent strain, unlike a virulent strain, induces the host to mount an effective Th1 response (5) that subsequently protects against challenge with a virulent strain demonstrates the importance of the pathogen in determining the immune response. For example, mannoprotein from acapsular strains and nonmelanogenic C. neoformans strains promotes Th1 responses that coincide with increased antifungal activity of effector cells (5, 41, 52). Experimental studies with different host genetic backgrounds showed that this also influences T helper cell differentiation (20, 47). CBA/J and CB-17 mice normally develop a Th1-type response to C. neoformans, while C57BL/6 mice develop Th2-type responses during pulmonary cryptococcal infection (19, 20, 25). A logical next step will be to study the cytokine messages in the brains of different mouse strains, infected with different Cryptococcus strains (differing in virulence and serotype), to characterize differences in their immune responses in cerebral cryptococcal infection. The activities of cytokines may not directly correlate with mRNA or protein levels, as their function may be further modulated at the level of posttranslational modification, processing multimer assembly or release from the cell of origin, or presentation or sequestration by specific carrier molecules and as a result of varying receptor types on neighboring target cells. It will be interesting to correlate the mRNA levels shown here with the results of future studies measuring cytokine protein expression to determine whether they correlate in the brain similarly to the correlations described for expression in the lungs during murine cryptococcosis (28).
In summary, we present new detailed information on the timing of the mRNA expression of specific immune modulatory molecules in intracranial cryptococcal infection in the murine experimental system. This should prove useful to other investigators for studying the genetic bases of host resistance or point to new interventions via therapy with cytokines (or by elimination of cytokines with antibody or other inhibitors) to promote more efficient elimination of the fungus from the CNS.
Acknowledgments
We thank Christopher A. Hunter and Roberto Macina for instruction and advice. We thank M. Martinez, M. Homola, P. Chu, L. Ellis, and K. Gabriel for their assistance. We also thank Luiz de Souza for his assistance with the statistical analyses.
Claudia M. L. Maffei was supported by a scholarship from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP).
Editor: T. R. Kozel
REFERENCES
- 1.Aguirre, K., E. A. Havell, G. W. Gibson, and L. L. Johnson. 1995. Role of tumor necrosis factor and gamma interferon in acquired resistance to Cryptococcus neoformans in the central nervous system of mice. Infect. Immun. 63:1725-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almeida, G. M., R. M. Andrade, and C. A. Bento. 2001. The capsular polysaccharides of Cryptococcus neoformans activate normal CD4(+) T cells in a dominant Th2 pattern. J. Immunol. 167:5845-5851. [DOI] [PubMed] [Google Scholar]
- 3.Ashman, R. B., E. M. Bolitho, and A. Fulurija. 1995. Cytokine mRNA in brain tissue from mice that show strain-dependent differences in the severity of lesions induced by systemic infection with Candida albicans yeast. J. Infect. Dis. 172:823-830. [DOI] [PubMed] [Google Scholar]
- 4.Barluzzi, R., A. Brozzetti, D. Delfino, F. Bistoni, and E. Blasi. 1998. Role of the capsule in microglial cell-Cryptococcus neoformans interaction: impairment of antifungal activity but not of secretory functions. Med. Mycol. 36:189-197. [PubMed] [Google Scholar]
- 5.Barluzzi, R., A. Brozzetti, G. Mariucci, M. Tantucci, R. G. Neglia, F. Bistoni, and E. Blasi. 2000. Establishment of protective immunity against cerebral cryptococcosis by means of an avirulent, nonmelanogenic Cryptococcus neoformans strain. J. Neuroimmunol. 109:75-86. [DOI] [PubMed] [Google Scholar]
- 6.Blasi, E., R. Barluzzi, R. Mazzolla, P. Mosci, and F. Bistoni. 1992. Experimental model of intracerebral infection with Cryptococcus neoformans: roles of phagocytes and opsonization. Infect. Immun. 60:3682-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasi, E., R. Barluzzi, R. Mazzolla, L. Pitzurra, M. Puliti, S. Saleppico, and F. Bistoni. 1995. Biomolecular events involved in anticryptococcal resistance in the brain. Infect. Immun. 63:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bocca, A. L., E. E. Hayashi, A. G. Pinheiro, A. B. Furlanetto, A. P. Campanelli, F. Q. Cunha, and F. Figueiredo. 1998. Treatment of Paracoccidioides brasiliensis-infected mice with a nitric oxide inhibitor prevents the failure of cell-mediated immune response. J. Immunol. 161:3056-3063. [PubMed] [Google Scholar]
- 9.Brown, H., G. Turner, S. Rogerson, M. Tembo, J. Mwenechanya, M. Molyneux, and T. Taylor. 1999. Cytokine expression in the brain in human cerebral malaria. J. Infect. Dis. 180:1742-1746. [DOI] [PubMed] [Google Scholar]
- 10.Buchanan, K. L., and H. A. Doyle. 2000. Requirement for CD4+ T lymphocytes in host resistance against Cryptococcus neoformans in the central nervous system of immunized mice. Infect. Immun. 68:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchanan, K. L., and J. W. Murphy. 1998. What makes Cryptococcus neoformans a pathogen? Emerg. Infect. Dis. 4:71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemons, K. V., R. Azzi, and D. A. Stevens. 1996. Experimental systemic cryptococcosis in SCID mice. J. Med. Vet. Mycol. 34:331-335. [DOI] [PubMed] [Google Scholar]
- 13.Clemons, K. V., E. Brummer, and D. A. Stevens. 1994. Cytokine treatment of central nervous system infection: efficacy of interleukin-12 alone and synergy with conventional antifungal therapy in experimental cryptococcosis. Antimicrob. Agents Chemother. 38:460-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemons, K. V., J. E. Lutz, and D. A. Stevens. 2001. Efficacy of recombinant gamma interferon for treatment of systemic cryptococcosis in SCID mice. Antimicrob. Agents Chemother. 45:686-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Decken, K., G. Kohler, K. Palmer-Lehmann, A. Wunderlin, F. Mattner, J. Magram, M. K. Gately, and G. Alber. 1998. Interleukin-12 is essential for a protective Th1 response in mice infected with Cryptococcus neoformans. Infect. Immun. 66:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deckert, M., S. Soltek, G. Geginat, S. Lutjen, M. Montesinos-Rongen, H. Hof, and D. Schluter. 2001. Endogenous interleukin-10 is required for prevention of a hyperinflammatory intracerebral immune response in Listeria monocytogenes meningoencephalitis. Infect. Immun. 69:4561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobrick, P., K. Miksits, and H. Hahn. 1995. L3T4(CD4)-, Lyt-2(CD8)- and Mac-1(CD11b)-phenotypic leukocytes in murine cryptococcal meningoencephalitis. Mycopathologia 131:159-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilliland, G., S. Perrin, K. Blanchard, and H. F. Bunn. 1990. Analysis of cytokine mRNA and DNA: detection and quantitation by competitive polymerase chain reaction. Proc. Natl. Acad. Sci. USA 87:2725-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoag, K. A., M. F. Lipscomb, A. A. Izzo, and N. E. Street. 1997. IL-12 and IFN-gamma are required for initiating the protective Th1 response to pulmonary cryptococcosis in resistant C.B-17 mice. Am. J. Respir. Cell Mol. Biol. 17:733-739. [DOI] [PubMed] [Google Scholar]
- 20.Hoag, K. A., N. E. Street, G. B. Huffnagle, and M. F. Lipscomb. 1995. Early cytokine production in pulmonary Cryptococcus neoformans infections distinguishes susceptible and resistant mice. Am. J. Respir. Cell Mol. Biol. 13:487-495. [DOI] [PubMed] [Google Scholar]
- 21.Hoeprich, P., and P. Finn. 1972. Obfuscation of the activity of antifungal antimicrobics by culture media. J. Infect. Dis. 126:353-361. [DOI] [PubMed] [Google Scholar]
- 22.Hollander, M., and D. A. Wolfe. 1973. Nonparametric statistical methods. Wiley, New York, N.Y.
- 23.Huffnagle, G. B., and M. F. Lipscomb. 1998. Cells and cytokines in pulmonary cryptococcosis. Res. Immunol. 149:387-396. [DOI] [PubMed] [Google Scholar]
- 24.Huffnagle, G. B., M. F. Lipscomb, J. A. Lovchik, K. A. Hoag, and N. E. Street. 1994. The role of CD4+ and CD8+ T cells in the protective inflammatory response to a pulmonary cryptococcal infection. J. Leukoc. Biol. 55:35-42. [DOI] [PubMed] [Google Scholar]
- 25.Huffnagle, G. B., and L. K. McNeil. 1999. Dissemination of C. neoformans to the central nervous system: role of chemokines, Th1 immunity and leukocyte recruitment. J. Neurovirol. 5:76-81. [DOI] [PubMed] [Google Scholar]
- 26.Huffnagle, G. B., R. M. Strieter, L. K. McNeil, R. A. McDonald, M. D. Burdick, S. L. Kunkel, and G. B. Toews. 1997. Macrophage inflammatory protein-1α (MIP-1α) is required for the efferent phase of pulmonary cell-mediated immunity to a Cryptococcus neoformans infection. J. Immunol. 159:318-327. [PubMed] [Google Scholar]
- 27.Hunter, C. A., J. S. Abrams, M. H. Beaman, and J. S. Remington. 1993. Cytokine mRNA in the central nervous system of SCID mice infected with Toxoplasma gondii: importance of T-cell-independent regulation of resistance to T. gondii. Infect. Immun. 61:4038-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawakami, K., M. Tohyama, X. Qifeng, and A. Saito. 1997. Expression of cytokines and inducible nitric oxide synthase mRNA in the lungs of mice infected with Cryptococcus neoformans: effects of interleukin-12. Infect. Immun. 65:1307-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein, S. A., O. G. Ottmann, K. Ballas, T. S. Dobmeyer, M. Pape, E. Weidmann, D. Hoelzer, and U. Kalina. 1999. Quantification of human interleukin 18 mRNA expression by competitive reverse transcriptase polymerase chain reaction. Cytokine 11:451-458. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. C., D. W. Dickson, C. F. Brosnan, and A. Casadevall. 1994. Human astrocytes inhibit Cryptococcus neoformans growth by a nitric oxide-mediated mechanism. J. Exp. Med. 180:365-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, S. C., Y. Kress, M. L. Zhao, D. W. Dickson, and A. Casadevall. 1995. Cryptococcus neoformans survive and replicate in human microglia. Lab. Investig. 73:871-879. [PubMed] [Google Scholar]
- 32.Lee, S. C., W. Liu, D. W. Dickson, C. F. Brosnan, and J. W. Berman. 1993. Cytokine production by human fetal microglia and astrocytes. Differential induction by lipopolysaccharide and IL-1 beta. J. Immunol. 150:2659-2667. [PubMed] [Google Scholar]
- 33.Licinio, J., P. Prolo, S. M. McCann, and M. L. Wong. 1999. Brain iNOS: current understanding and clinical implications. Mol. Med. Today 5:225-232. [DOI] [PubMed] [Google Scholar]
- 34.Licinio, J., and M. L. Wong. 1997. Pathways and mechanisms for cytokine signaling of the central nervous system. J. Clin. Investig. 100:2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loddick, S. A., M. L. Wong, P. B. Bongiorno, P. W. Gold, J. Licinio, and N. J. Rothwell. 1997. Endogenous interleukin-1 receptor antagonist is neuroprotective. Biochem. Biophys. Res. Commun. 234:211-215. [DOI] [PubMed] [Google Scholar]
- 36.Lortholary, O., L. Improvisi, N. Rayhane, F. Gray, C. Fitting, J. M. Cavaillon, and F. Dromer. 1999. Cytokine profiles of AIDS patients are similar to those of mice with disseminated Cryptococcus neoformans infection. Infect. Immun. 67:6314-6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lutz, J. E., K. V. Clemons, and D. A. Stevens. 2000. Enhancement of antifungal chemotherapy by interferon-gamma in experimental systemic cryptococcosis. J. Antimicrob. Chemother. 46:437-442. [DOI] [PubMed] [Google Scholar]
- 38.Monari, C., T. R. Kozel, A. Casadevall, D. Pietrella, B. Palazzetti, and A. Vecchiarelli. 1999. B7 costimulatory ligand regulates development of the T-cell response to Cryptococcus neoformans. Immunology 98:27-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monari, C., C. Retini, B. Palazzetti, F. Bistoni, and A. Vecchiarelli. 1997. Regulatory role of exogenous IL-10 in the development of immune response versus Cryptococcus neoformans. Clin. Exp. Immunol. 109:242-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, J. W. 1998. Protective cell-mediated immunity against Cryptococcus neoformans. Res. Immunol. 149:373-386. [DOI] [PubMed] [Google Scholar]
- 41.Pietrella, D., R. Cherniak, C. Strappini, S. Perito, P. Mosci, F. Bistoni, and A. Vecchiarelli. 2001. Role of mannoprotein in induction and regulation of immunity to Cryptococcus neoformans. Infect. Immun. 69:2808-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polak, J., and S. V. Noorden. 1997. Introduction to immunocytochemistry, 2nd ed. Springer-Verlag, Inc., New York, N.Y.
- 43.Qureshi, M. H., T. Zhang, Y. Koguchi, K. Nakashima, H. Okamura, M. Kurimoto, and K. Kawakami. 1999. Combined effects of IL-12 and IL-18 on the clinical course and local cytokine production in murine pulmonary infection with Cryptococcus neoformans. Eur. J. Immunol. 29:643-649. [DOI] [PubMed] [Google Scholar]
- 44.Reiner, S. L., S. Zheng, D. B. Corry, and R. M. Locksley. 1993. Constructing polycompetitor cDNAs for quantitative PCR. J. Immunol. Methods 165:37-46. [DOI] [PubMed] [Google Scholar]
- 45.Retini, C., A. Vecchiarelli, C. Monari, F. Bistoni, and T. R. Kozel. 1998. Encapsulation of Cryptococcus neoformans with glucuronoxylomannan inhibits the antigen-presenting capacity of monocytes. Infect. Immun. 66:664-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Retini, C., A. Vecchiarelli, C. Monari, C. Tascini, F. Bistoni, and T. R. Kozel. 1996. Capsular polysaccharide of Cryptococcus neoformans induces proinflammatory cytokine release by human neutrophils. Infect. Immun. 64:2897-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Romagnani, S. 1996. Th1 and Th2 in human diseases. Clin. Immunol. Immunopathol. 80:225-235. [DOI] [PubMed] [Google Scholar]
- 48.Stalder, A. K., A. Pagenstecher, N. C. Yu, C. Kincaid, C. S. Chiang, M. V. Hobbs, F. E. Bloom, and I. L. Campbell. 1997. Lipopolysaccharide-induced IL-12 expression in the central nervous system and cultured astrocytes and microglia. J. Immunol. 159:1344-1351. [PubMed] [Google Scholar]
- 49.Stevens, D. A., T. J. Walsh, F. Bistoni, E. Cenci, K. V. Clemons, G. Del Sero, C. Fe d'Ostiani, B. J. Kullberg, A. Mencacci, E. Roilides, and L. Romani. 1998. Cytokines and mycoses. Med. Mycol. 36(Suppl. 1):174-182. [PubMed] [Google Scholar]
- 50.Suzuki, Y., S. Fujii, T. Tominaga, T. Yoshimoto, T. Akaike, H. Maeda, and T. Yoshimura. 1999. Direct evidence of in vivo nitric oxide production and inducible nitric oxide synthase mRNA expression in the brain of living rat during experimental meningitis. J. Cereb. Blood Flow Metab. 19:1175-1178. [DOI] [PubMed] [Google Scholar]
- 51.Turka, L. A., R. E. Goodman, J. L. Rutkowski, A. A. Sima, A. Merry, R. S. Mitra, T. Wrone-Smith, G. Toews, R. M. Strieter, and B. J. Nickoloff. 1995. Interleukin 12: a potential link between nerve cells and the immune response in inflammatory disorders. Mol. Med. 1:690-699. [PMC free article] [PubMed] [Google Scholar]
- 52.Vecchiarelli, A. 2000. Immunoregulation by capsular components of Cryptococcus neoformans. Med. Mycol. 38:407-417. [DOI] [PubMed] [Google Scholar]
- 53.Vecchiarelli, A., C. Retini, C. Monari, and A. Casadevall. 1998. Specific antibody to Cryptococcus neoformans alters human leukocyte cytokine synthesis and promotes T-cell proliferation. Infect. Immun. 66:1244-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vecchiarelli, A., C. Retini, C. Monari, C. Tascini, F. Bistoni, and T. R. Kozel. 1996. Purified capsular polysaccharide of Cryptococcus neoformans induces interleukin-10 secretion by human monocytes. Infect. Immun. 64:2846-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vitkovic, L., J. Bockaert, and C. Jacque. 2000. “Inflammatory” cytokines: neuromodulators in normal brain? J. Neurochem. 74:457-471. [DOI] [PubMed] [Google Scholar]
- 56.Wolf, S. F., D. Sieburth, and J. Sypek. 1994. Interleukin 12: a key modulator of immune function. Stem Cells 12:154-168. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, B. G., and H. Link. 1998. Immune regulation within the central nervous system. J. Neurol. Sci. 157:1-12. [DOI] [PubMed] [Google Scholar]
- 58.Zhou, N. M., P. Matthys, C. Polacek, P. Fiten, A. Sato, A. Billiau, and G. Froyen. 1997. A competitive RT-PCR method for the quantitative analysis of cytokine mRNAs in mouse tissues. Cytokine 9:212-218. [DOI] [PubMed] [Google Scholar]