Abstract
The outer membrane protein CD (OMPCD) of Moraxella catarrhalis is an outer membrane protein with several attributes of a potential vaccine antigen. We isolated four transposon mutants of strain O35E on the basis of their reduced binding to A549 human lung cells in microcolony formation assays, and we determined that they contain a transposon in ompCD. We also found that these transposon insertions had pleiotropic effects: mutants grew slower, became serum sensitive, bound ∼10-fold less to A549 cells, and appeared transparent when grown on solid medium. We confirmed that these various phenotypes could be attributed solely to disruption of ompCD by constructing the isogenic strain O35E.CD1. O35E-ompCD was cloned, and recombinant Escherichia coli bacteria expressing the gene product exhibited a 10-fold increase in adherence to A549 cells. This is the first report of M. catarrhalis ompCD mutants, and our findings demonstrate that this gene product is an adhesin for human lung cells.
The gram-negative bacterium Moraxella catarrhalis is a pathogen of the human respiratory tract that causes otitis media in young children (17, 20, 33, 44) and lower respiratory tract infections in adults with chronic obstructive pulmonary disease (44, 62, 63). Patients with underlying conditions appear to be particularly susceptible, as illustrated by the increasing number of cases of M. catarrhalis-caused wound infections, bronchitis, conjunctivitis, sinusitis, bacteremia, pneumonia, meningitis, pericarditis, and endocarditis (8, 14, 17, 33, 44, 51, 65, 68, 70, 71).
Little is known about pathogenesis by M. catarrhalis. Most research has thus far focused on the identification and characterization of a few outer membrane proteins for vaccine development purposes. These include the adhesins UspA1 (30, 36, 37, 40, 41), UspA2H (36, 41), Hag (21, 22, 31, 54), McaP (69), and MID (23, 24, 26, 43, 52), the serum resistance factor UspA2 (3, 4, 15, 18, 36, 40, 41), the iron acquisition proteins CopB (2, 5, 11, 28, 29, 64), LbpA/LbpB (11, 13, 19, 74), TbpA/TbpB (13, 16, 50, 74), and OmpB1 (12, 13, 38, 39), and the highly conserved proteins outer membrane protein E (OMPE) (9, 10, 45) and OMPCD (32, 46, 47, 61). Although no specific biological function has been attributed to OMPCD, this molecule is predicted to be structurally similar to bacterial porins (47) and binds middle ear mucin (59). Thus, OMPCD may be involved in nutrient acquisition and/or adherence to mucosal surfaces.
The present study describes the isolation and characterization of ompCD mutants of the M. catarrhalis wild-type strain O35E, and the data demonstrate that OMPCD is an adhesin for A549 human lung epithelial cells.
MATERIALS AND METHODS
Strains, plasmids, tissue culture cell lines, and growth conditions.
Strains and plasmids are described in Table 1. M. catarrhalis strains were grown at 37°C in Todd Hewitt (TH) broth (Difco) or on TH agar plates in an atmosphere of 92.5% air-7.5% CO2. M. catarrhalis transposon mutants were selected with 20 μg of kanamycin (KAN)/ml. Escherichia coli strains were grown in Luria-Bertani (LB) broth (Difco) or on LB agar plates. For the selection of recombinant E. coli clones, the LB medium was supplemented with either 100 μg of ampicillin/ml, 50 μg of KAN/ml, or 15 μg of chloramphenicol/ml. For adherence and serum bactericidal assays with recombinant E. coli cells, 5-ml cultures were grown overnight at 37°C with shaking (200 rpm). These overnight cultures were diluted into 20 ml of fresh broth supplemented with 0.25 ml of 1000X CopyControl induction solution (Epicentre) and grown at 37°C for 2 h with vigorous shaking (300 rpm). Chang (conjunctival epithelium; ATCC CCL20.2), A549 (type II alveolar lung epithelium; ATCC CCL85), and human middle ear epithelial cells (HMEE) were cultured as described elsewhere (31).
TABLE 1.
Plasmids and strains
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| M. catarrhalis strains | ||
| O35E | Wild-type isolate | 4 |
| O35E.1 | uspA1 isogenic mutant of strain O35E; adherence negative for Chang cells | 3 |
| O35E.2 | uspA2 isogenic mutant of strain O35E; serum sensitive | 3, 4 |
| O35E.TN2 | hag transposon mutant of strain O35E; adherence negative for A549 and HMEE cells | 31 |
| O35E.TN52 | ompCD transposon mutant of strain O35E | This study |
| O35E.TN313 | ompCD transposon mutant of strain O35E | This study |
| O35E.TN593 | ompCD transposon mutant of strain O35E | This study |
| O35E.TN649 | ompCD transposon mutant of strain O35E | This study |
| O35E.CD1 | Isogenic ompCD mutant of strain O35E | This study |
| E. coli strain | ||
| EPI300 | Cloning strain | Epicentre |
| Plasmids | ||
| pCC1 | Cloning vector | Epicentre |
| pUC19 | Cloning vector | New England Biolabs |
| pUC4K | Source of the kanr cassette | AmershamBiosciences |
| pCC1.3 | pCC1 into which the control insert provided by the manufacturer was cloned; adherence negative, serum sensitive | This study |
| pELCD | Truncated O35E-ompCD gene cloned into pUC19 | This study |
| pELCDKAN | pELCD containing the kanr cassette of pUC4K in the middle of the truncated O35E-ompCD gene | This study |
| pMHCD1.2 | Complete O35E-ompCD ORF cloned into pCC1; adheres to A549 cells | This study |
Recombinant DNA techniques.
Standard molecular biology methods were performed as described previously (60). M. catarrhalis genomic DNA was prepared with the Invitrogen Easy-DNA kit. Plasmid DNA was purified with the QIAprep Spin Miniprep system (Qiagen). The North2South chemiluminescent nucleic acid hybridization and detection system (Pierce) was used to perform Southern blotting experiments. A 1.2-kb DNA fragment containing a kanr cartridge was obtained from the plasmid pUC4K and used as a probe in some of these experiments. The 1.2-kb ompCD-specific DNA probe was obtained by PCR using the oligonucleotide primers P1 and P2 (see below).
PCR and cloning.
Amplification of DNA fragments was performed with the Platinum Pfx DNA polymerase (Invitrogen) unless indicated otherwise. The ompCD-specific oligonucleotide primers P1 (5′-GTGACAGTCAGCCCACTA-3′) and P2 (5′-TTGCTACCAGTGATTACTGC-3′) were used to amplify a 1.2-kb DNA fragment from strain O35E that corresponds to a truncated open reading frame (ORF). The primers P3 (5′-GGATCGCTATGCTAAAATAGGTGC-3′) and P4 (5′-TCAAAAGCTAAGAAAACCGCT-3′) were used to generate a 1.6-kb amplicon from O35E containing the complete ompCD ORF and which was utilized as template in sequencing reactions as well as in cloning experiments with the Epicentre CopyControl PCR cloning system. Taq DNA polymerase (Invitrogen) was used in other PCR-based experiments. The plasmid pCC1.3 corresponds to the Epicentre CopyControl vector pCC1, into which the manufacturer's control DNA insert was cloned.
Transposon mutagenesis and adherence assays.
M. catarrhalis O35E transposon mutants were obtained using the EZ::TN <KAN-2> transposome (Epicentre), and mutants were screened in microcolony formation assays to identify those substantially reduced in their adherence to A549 cells, as we have previously reported (31). The method used to quantitatively measure the adherence of M. catarrhalis to human tissue culture cell lines has been described elsewhere and involves a 3-h incubation prior to washing unbound bacteria (31). Adherence assays with E. coli recombinant cells involved a 5-min incubation prior to washing unbound bacteria.
Construction of isogenic mutants.
An amplicon of 1.2 kb containing a truncated ompCD ORF from strain O35E was generated with the primers P1 and P2 (see above) and was ligated into the vector pUC19, yielding the recombinant plasmid pELCD. The latter was linearized with DraIII, treated with Pfu DNA polymerase (Stratagene) to render the restricted ends blunt, and ligated with a 1.2-kb SmaI DNA fragment containing the kanr cassette from the plasmid pUC4K. This ligation mixture was introduced into E. coli TOP10, and transformants were selected for resistance to KAN, thereby yielding the plasmid pELCDKAN. A 2.4-kb amplicon, which corresponds to a truncated O35E-ompCD gene interrupted by the kanr cartridge in the middle of the ORF, was generated from pELCDKAN using the primers P1 and P2. This PCR product was then electroporated into M. catarrhalis strain O35E. The resulting kanr colonies were screened by PCR with primers P1 and P2 to identify potential isogenic ompCD mutants (data not shown). Southern blotting experiments were performed to confirm that proper allelic exchange had occurred in the isogenic mutant O35E.CD1 (data not shown).
MAbs and characterization of selected protein antigens.
The UspA1- and UspA2-specific monoclonal antibody (MAb) 17C7 (4), the UspA1-specific MAb 24B5 (18), and the Hag-specific MAb 5D2 (54) have been described elsewhere. The OMPCD MAbs 1D3 and 3.9H have been reported previously (46). Outer membrane vesicles were prepared as described by others (49, 53). Whole-cell lysates of M. catarrhalis strains and E. coli recombinant cells were prepared as previously reported (18, 53). These preparations were heated at 100°C for 15 min, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and either stained with Coomassie blue or electrophoretically transferred to polyvinylidene difluoride membranes (Millipore) for Western blot analysis as previously described (31).
Serum resistance assays.
Serum bactericidal assays were performed as reported by Aebi et al. (3). Results are expressed as the percentage (± standard deviation) of bacteria surviving incubation with serum. These assays were performed on at least three separate occasions.
RNA purification and QRT-PCR.
The methods, primers, and probes used for RNA purification and quantitative real-time PCR (QRT-PCR) in these experiments have been described elsewhere (31).
Nucleotide sequence analysis.
The nucleotide sequence data were analyzed as previously reported (31).
Statistical methods.
All statistical analyses were performed using a Mann-Whitney test and GraphPad Prism 2.01 software. P values of <0.05 were considered statistically significant.
Nucleotide sequence accession number.
The nucleotide sequence of the M. catarrhalis O35E ompCD gene has been deposited in GenBank under the accession number AY493741.
RESULTS
ompCD mutations have pleiotropic effects.
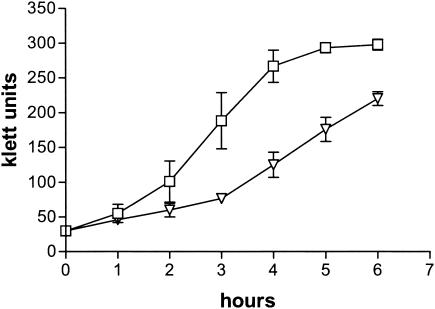
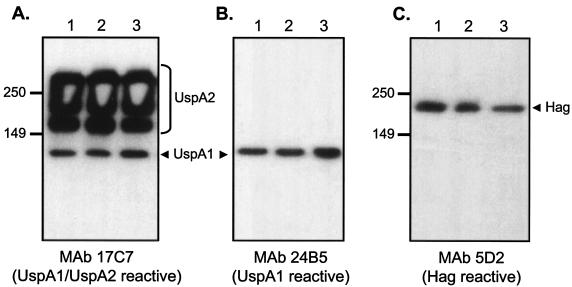
We recently described a mutagenesis and screening approach which identified nine adherence mutants in M. catarrhalis strain O35E (31). These mutants showed substantially lower binding to A549 human lung cells in microcolony formation assays, and all contained a transposon in the hag gene. This screening approach also yielded four adherence mutants that were not described in our original report because their growth was significantly impaired. Figure 1 illustrates this defect for the representative mutant O35E.TN52. We also found that these four slower-growing mutants expressed wild-type levels of Hag, UspA1, and UspA2 (Fig. 2). Since Hag appears to be the main adhesin for A549 and HMEE cells (23, 31), we reasoned that the apparent adherence defect exhibited by the four slow-growing mutants in microcolony assays might simply result from their poor growth.
FIG. 1.
Growth of M. catarrhalis strains in broth cultures. Symbols: squares, O35E; inverted triangles, O35E.TN52.
FIG. 2.
Western blot analysis of M. catarrhalis strains. Proteins present in outer membrane vesicles prepared from the wild-type strain O35E (lane 1), the transposon mutant O35E.TN52 (lane 2), and the isogenic mutant O35E.CD1 (lane 3) were resolved by SDS-PAGE and analyzed by Western blotting with the UspA1- and UspA2-specific MAb 17C7 (A), the UspA1-specific MAb 24B5 (B), and the Hag-specific MAb 5D2 (C). Note that samples were heated at 100°C for 15 min prior to Western blot analysis. Under these conditions, UspA1 migrates as a 125-kDa protein and UspA2 migrates as a high-molecular-weight aggregate that is expressed in large amounts (bracket on the right side of panel A). Positions of molecular mass markers are shown to the left in kilodaltons. The arrowheads indicate selected antigens.
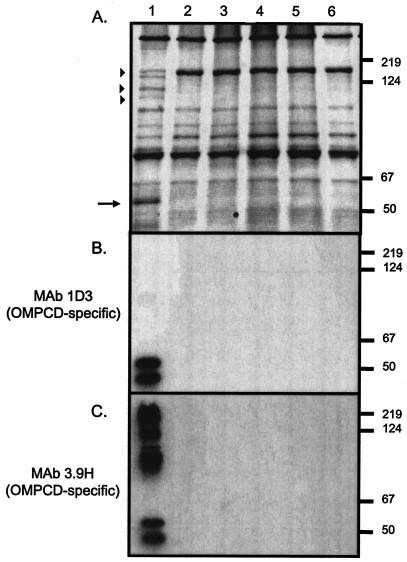
Even though Southern blotting experiments suggested that these four isolates are not siblings (data not shown), their colonies all appeared transparent in contrast to the opaque morphology of the parent strain O35E (Fig. 3). We also observed that all four isolates lacked expression of a major OMP of 55 kDa (Fig. 4A). Several larger antigens were also absent in the mutants (Fig. 4A).
FIG. 3.
Appearance of M. catarrhalis strains on agar plates. The wild-type strain O35E (bottom left) and the transposon mutant O35E.TN52 (top right) were streaked onto TH agar plates and photographed with lighting from above against a black background. Plates were incubated for 24 h.
FIG. 4.
Western blot analysis of M. catarrhalis strains. Proteins present in outer membrane vesicles prepared from the wild-type strain O35E (lane 1) as well as the transposon mutants O35E.TN52 (lane 2), O35E.TN313 (lane3), O35E.TN593 (lane 4), and O35E.TN649 (lane 5) and from the isogenic mutant O35E.CD1 (lane 6) were resolved by SDS-PAGE and stained with Coomassie blue (A) or analyzed by Western blotting with the OMPCD-specific MAbs 1D3 (B) and 3.9H (C). Positions of molecular mass markers are shown to the right in kilodaltons. The arrow as well as arrowheads indicate antigens that are missing in the transposon as well as the O35E.CD1 mutants.
It was previously shown that OMPCD is a heat-modifiable protein that migrates with a molecular mass of approximately 55 kDa (61). We therefore tested by Western blotting whether the major OMP missing from the transparent mutants was OMPCD. Lanes 2 to 5 in Fig. 4B demonstrate that the mutants no longer detectably expressed OMPCD. Western blotting experiments with the OMPCD-specific MAb 3.9H (Fig. 4C) also suggested that the higher-molecular-mass antigens missing from outer membrane vesicles of the mutants (Fig. 4A) were multimers of OMPCD. When we analyzed the mutants by PCR, the ompCD-specific primers P1 and P2 yielded an amplicon of 1.2 kb in the parent strain O35E and one of 2.4 kb in the transposon mutants (data not shown). These results are consistent with the 1,221-bp EZ::TN <KAN-2> transposon having inserted into the ompCD gene of all four independently isolated transparent mutants. This was confirmed by Southern blot analysis with ompCD- and transposon-specific probes (data not shown).
We constructed the isogenic ompCD strain O35E.CD1 to test whether the various phenotypes of our four transposon mutants could be attributed solely to disruption of the ompCD gene. We found that O35E.CD1 is transparent (data not shown), has an OMP profile indistinguishable from that of the transposon mutants (Fig. 4A, lane 6), lacks expression of OMPCD (Fig. 4B and C, lane 6), expresses wild-type levels of Hag, UspA1, and UspA2 (Fig. 2), and exhibits a growth defect similar to that of the transposon mutants (data not shown). Furthermore, we observed that O35E.TN52 (4.4% ± 8.2% survival) and O35E.CD1 (1.8% ± 2.5% survival) were sensitive to 10% normal human serum, whereas O35E was resistant (117.8% ± 15.4% survival). The uspA2 mutant O35E.2 (2.3% ± 2.4% survival) was used as a serum-sensitive control (3, 36), and heat inactivation of the serum abolished its ability to kill mutants (data not shown).
Taken together, these results demonstrate that ompCD mutations affect the growth, colony morphology, and serum resistance of M. catarrhalis O35E, but they do not affect expression of the adhesins UspA1 and Hag or that of the serum resistance factor UspA2.
The transposon insertion in O35E-ompCD does not significantly affect expression of the gene located directly downstream.
BLAST searches of the patented M. catarrhalis genome through NCBI databases identified the ompCD ORF (nucleotides 79050 to 77770 of AX067466) as well as the ORF located directly downstream (nucleotides 77514 to 75670 of AX067466). This ORF is predicted to encode a protein with 70% identity (83% similarity) to Salmonella enterica serovar Typhimurium GTP-binding elongation factor family protein BipA (NP_462889.1). To demonstrate that the various phenotypes of ompCD mutants were not due to a polar effect on expression of the gene downstream of ompCD, we used QRT-PCR to measure the expression of M. catarrhalis bipA. We used expression of the unlinked M. catarrhalis gene purH, which we recently reported to be located downstream of the hag gene (31), as a normalization control. No significant change in bipA expression was observed between the wild-type strain O35E (relative expression ± standard deviation, 1.0 ± 0.7), the O35E.TN52 transposon mutant (1.73 ± 0.3), and the O35E.CD1 isogenic mutant (2.2 ± 0.9). Previous work and these data make it unlikely that the various phenotypes of the ompCD mutants are due to changes in transcription of bipA.
OMPCD expression affects the adherence of strain O35E to A549 human lung cells.
The apparently reduced binding of ompCD mutants to A549 cells in microcolony formation assays might simply reflect their slower growth rate. These assays entail a 40-h incubation with A549 cells (31). We therefore measured the adherence of our mutants after 3 h of incubation, as previously reported (31). Table 2 shows that the transparent mutants attached substantially less well to A549 cells, as did the isogenic ompCD strain O35E.CD1. It should be noted that the hag mutant O35E.TN2 (31) was used as an adherence negative control in the assays.
TABLE 2.
Adherence of M. catarrhalis strains to human cells in vitro
| Strain | Description | % Adherencea
|
||
|---|---|---|---|---|
| A549b | HMEEc | Changd | ||
| O35E | wild type, opaque colony morphology | 40.5 ± 4.5 | 27.9 ± 4.2 | 17.4 ± 2.7 |
| O35E.TN2 | Tn mutant, transposon in hag gene, opaque colony morphology | 1.3 ± 0.3*f | 0.7 ± 0.1* | NDe |
| O35E.1 | Isogenic uspA1 mutant, opaque colony morphology | ND | ND | 1.5 ± 0.4* |
| O35E.TN52g | Tn mutant, transposon in ompCD gene, transparent colony morphology | 2.6 ± 0.4* | 23.9 ± 4.4 | 14.9 ± 1.8 |
| O35E.TN313g | Tn mutant, transposon in ompCD gene, transparent colony morphology | 4.4 ± 1.5* | ND | ND |
| O35E.TN593g | Tn mutant, transposon in ompCD gene, transparent colony morphology | 1.3 ± 0.2* | ND | ND |
| O35E.TN649g | Tn mutant, transposon in ompCD gene, transparent colony morphology | 0.8 ± 0.2* | ND | ND |
| O35E.CD1 | Isogenic ompCD mutant, transparent colony morphology | 5.1 ± 0.9* | ND | 16.1 ± 2.9 |
Adherence is expressed as the mean (± standard error) percentage of bacteria binding to monolayers.
Strains were incubated for 3 h with A549 cells prior to washing unbound bacteria.
Strains were incubated for 15 min with HMEE cells prior to washing unbound bacteria.
Strains were incubated for 15 min with Chang cells prior to washing unbound bacteria.
ND, not determined.
*, the difference in the P value compared to the value for the wild-type strain O35E was found to be statistically significant using a Mann-Whitney test.
Transposon mutants were first identified due to their reduced binding to A549 cells in microcolony formation assays.
To test whether this was a general defect in adherence, we measured the binding of ompCD mutants to Chang monolayers and found that they attached at nearly wild-type levels to these conjunctival cells (Table 2). UspA1 has been reported to be the major adhesin for Chang cells (3, 36), and the isogenic uspA1 mutant O35E.1 was used as an adherence negative control in our assay. Thus, the ompCD mutants express a functional UspA1 adhesin. We also found that the lack of OMPCD expression did not adversely affect M. catarrhalis binding to HMEE cells (Table 2). We previously reported that Hag is the major adhesin for HMEE cells (31); thus, the hag mutant O35E.TN2 was used as an adherence negative control in these experiments. Taken together, our results demonstrated that OMPCD expression specifically affects adherence to A549 human lung cells.
Cloning and expression of O35E ompCD by recombinant E. coli cells.
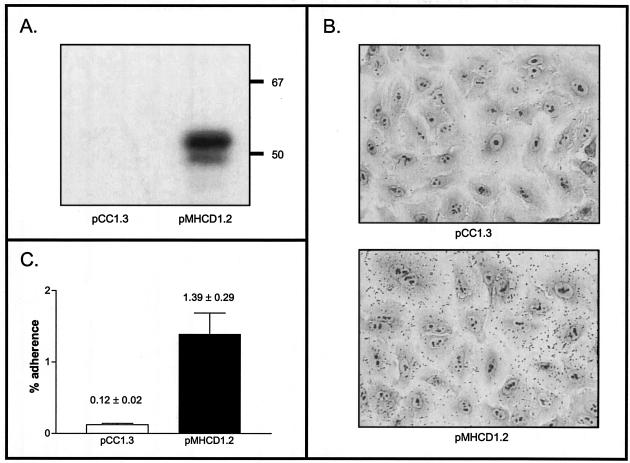
To determine whether OMPCD plays a direct role in serum resistance and adherence to A549 cells, we cloned and expressed this gene in E. coli strain EPI300 using Epicentre's CopyControl PCR cloning system. This system allowed the recombinant plasmid pMHCD1.2 to be maintained at a very low copy number. Under these conditions, OMPCD expression was not detectable (data not shown). Upon incubation at 37°C for 2 h and in the presence the CopyControl inducer solution, which boosts the plasmid copy number to 10 to 200 per cell, however, recombinant bacteria expressed detectable OMPCD (Fig. 5A). When we tested these induced cells in adherence assays, we found that OMPCD expression increased binding to A549 monolayers by 10-fold after only 5 min of incubation with these lung cells (Fig. 5B and C). Thus, OMPCD is an adhesin for A549 cells. OMPCD expression, however, did not confer serum resistance on E. coli (data not shown).
FIG. 5.
Western blot analysis of and adherence assays with E. coli recombinant bacteria. (A) Proteins present in whole-cell lysates of EPI300 pCC1.3 and EPI300 pMHCD1.2 were resolved by SDS-PAGE and then analyzed by Western blotting with the OMPCD-specific MAb 1D3. These lysates were prepared from cells grown in the presence of the Epicentre's CopyControl inducer solution for 2 h. Positions of molecular mass markers are shown to the right in kilodaltons. (B) Visual adherence assays. Recombinant bacteria were grown under the aforementioned conditions. Unbound bacteria were washed off A549 cells after a 5-min incubation, and the monolayers were stained with Giemsa. (C) Quantitative adherence assays. Results are expressed as the percentage (± standard error) of recombinant bacteria binding to A549 cells after 5 min of incubation.
The ompCD gene harbored by the plasmid pMHCD1.2 was sequenced to verify that no mutations were introduced by PCR. This O35E ompCD sequence was also found to be identical to that of ATCC 25240 ompCD (accession number L10755) and was deposited in the GenBank database.
DISCUSSION
The M. catarrhalis OMPCD protein exhibits numerous properties of a promising vaccine candidate. This antigen is surface exposed and is expressed by virtually all M. catarrhalis isolates tested to date (32, 47, 61). Immunization with recombinant OMPCD confers protective immunity in a mouse pulmonary clearance test (48), and the protein is an important target of the immune response in chronic obstructive pulmonary disease patients with M. catarrhalis infections (46). In addition, the predicted amino acid sequence of OMPCD is highly conserved among clinical isolates (32, 47), and our results extend these data. We found that the nucleotide and predicted amino acid sequences of O35E ompCD were identical to those of ATCC 25240 ompCD previously reported by Murphy and colleagues (47). The biological function(s) of OMPCD, however, has not been determined. Sequence analysis indicated that it is related to porins and that the most closely related gene product is the Pseudomonas aeruginosa porin OprF (47). Porins form a large family of OMPs that are involved in numerous biological functions, including nutrient acquisition (1, 34, 35). Since our data show that ompCD mutants have slower growth rates, OMPCD may therefore be involved in passage of a nutrient(s) across the M. catarrhalis outer membrane. Our isogenic ompCD mutants will facilitate the testing of this hypothesis.
Reddy and coworkers previously reported that OMPCD binds mucin glycoproteins, suggesting that it may be involved in adherence to mucosal surfaces (59). We found that ompCD mutants showed reduced binding to A549 human lung cells, supporting this hypothesis. We also recently showed that the M. catarrhalis O35E Hag protein is an adhesin for A549 cells (23, 31). Thus, one possible explanation for the decreased adherence of ompCD mutants is that the lack of OMPCD in the outer membrane affects the proper surface display of Hag, which in turn reduces adherence to A549 cells. Our results, however, argue against this possibility. We have previously shown that Hag is a major adhesin for middle ear cells (31). The ompCD mutant O35E.TN52 binds at near-wild-type levels to HMEE cells, whereas the hag mutant O35E.TN2 no longer attaches (Table 2). The ompCD mutant therefore expresses a functional Hag adhesin. Furthermore, OMPCD expression by recombinant E. coli bacteria increased adherence to A549 cells by 10-fold (Fig. 5). These data provide direct proof that OMPCD is an adhesin. Interestingly, P. aeruginosa OprF was recently shown to be an adhesin for A549 cells (7). Our results suggest that both Hag and OMPCD are involved in the binding of M. catarrhalis to A549 cells. Bacterial adherence is multifactorial and generally involves several steps, such as initial contact (from a distance) and close (tight) binding (27, 42, 66, 67). Since Pearson et al. demonstrated that Hag forms extended projections that cover M. catarrhalis O35E cells (54), Hag may initially contact A549 cells, whereas OMPCD is necessary for a closer interaction. Thus, both Hag and OMPCD may cooperate in specifically conferring adherence to A549 cells. We are currently investigating this hypothesis.
Our data also show that ompCD mutants are serum sensitive. This effect, however, might be indirect, since expression of recombinant OMPCD does not confer E. coli with the ability to resist the bactericidal activity of human complement. The lack of OMPCD expression in the outer membrane may thus affect proper surface display of a serum resistance factor, which in turn renders bacteria sensitive to complement killing. Candidate serum resistance factors include UspA2 (3, 36), CopB (29), fur-regulated genes (25), OmpE (45), and LOS (75). The transparent appearance of ompCD mutants may also be indirectly linked to the lack of OMPCD expression. For instance, it has been demonstrated for gram-negative pathogens such as Haemophilus influenzae (73) and Neisseria meningitidis (6) that changes in LOS structure and/or levels of expression cause differences in colony opacity.
Alternatively, recombinant E. coli bacteria may not express enough OMPCD to confer serum resistance, or the protein requires posttranslational modification that is not achieved in this heterologous genetic background. It is interesting that searches using the NCBI Conserved Domain Search tool indicate that OMPCD is related to the OmpA OMP and related peptidoglycan-associated (lipo)proteins family (COG2885; E value of 4e-24). E. coli K1 OmpA has been shown to mediate invasion of human brain microvascular endothelial cells (55, 57, 58) and thus is almost certainly involved in physical interactions with mammalian cells. Furthermore, E. coli OmpA plays a role in serum resistance. An ompA mutant showed greater sensitivity to normal human serum (72), and Prasadarao and colleagues recently demonstrated that OmpA contributes to serum resistance by binding to the complement C4b binding protein (56). Thus, M. catarrhalis OMPCD may very well play direct roles in resistance to complement killing and colony opacity.
In summary, this study reports the characterization of M. catarrhalis ompCD mutants isolated by their reduced adherence properties. Phenotypic analyses indicate that the lack of OMPCD expression leads to pleiotropic effects. The availability of isogenic strains as well as recombinant clones expressing the protein will facilitate establishing whether OMPCD is directly involved in nutrient acquisition, resistance to complement killing, and/or conferring the opaque appearance of M. catarrhalis wild-type strains. Our finding that OMPCD is an adhesin for human lung cells also suggests that a vaccine containing this protein (or portions thereof) may interfere with adherence, which is an important step in bacterial pathogenesis. Identifying an M. catarrhalis OMPCD epitope(s) involved in adherence and evaluating its vaccinogenic potential could significantly contribute to the development of a vaccine for this important human pathogen.
Acknowledgments
This study was supported in part by institutional start-up funds from the Medical College of Ohio, a grant from the Thrasher Research Fund (award number 02816-6), and a grant from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (AI051477) to E.R.L.
We thank Tim Murphy at the University of Buffalo and Eric Hansen at the University of Texas Southwestern Medical Center in Dallas for providing M. catarrhalis strains and antibodies. We also thank Thomas DeMaria at Ohio State University for providing cultures of human middle ear cells. We also thank Tim Murphy, Eric Hansen, Robert Blumenthal, and Mark Wooten for their helpful comments on the manuscript.
Editor: V. J. DiRita
REFERENCES
- 1.Achouak, W., T. Heulin, and J. M. Pages. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Aebi, C., L. D. Cope, J. L. Latimer, S. E. Thomas, C. A. Slaughter, G. H. McCracken, Jr., and E. J. Hansen. 1998. Mapping of a protective epitope of the CopB outer membrane protein of Moraxella catarrhalis. Infect. Immun. 66:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. L. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis 035E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aebi, C., B. Stone, M. Beucher, L. D. Cope, I. Maciver, S. E. Thomas, G. H. McCracken, Jr., P. F. Sparling, and E. J. Hansen. 1996. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect. Immun. 64:2024-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albiger, B., L. Johansson, and A. B. Jonsson. 2003. Lipooligosaccharide-deficient Neisseria meningitidis shows altered pilus-associated characteristics. Infect. Immun. 71:155-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Azghani, A. O., S. Idell, M. Bains, and R. E. Hancock. 2002. Pseudomonas aeruginosa outer membrane protein F is an adhesin in bacterial binding to lung epithelial cells in culture. Microb. Pathog. 33:109-114. [DOI] [PubMed] [Google Scholar]
- 8.Berner, R., R. F. Schumacher, M. Brandis, and J. Forster. 1996. Colonization and infection with Moraxella catarrhalis in childhood. Eur. J. Clin. Microbiol. Infect. Dis. 15:506-509. [DOI] [PubMed] [Google Scholar]
- 9.Bhushan, R., R. Craigie, and T. F. Murphy. 1994. Molecular cloning and characterization of outer membrane protein E of Moraxella (Branhamella) catarrhalis. J. Bacteriol. 176:6636-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhushan, R., C. Kirkham, S. Sethi, and T. F. Murphy. 1997. Antigenic characterization and analysis of the human immune response to outer membrane protein E of Branhamella catarrhalis. Infect. Immun. 65:2668-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonnah, R. A., R. H. Yu, H. Wong, and A. B. Schryvers. 1998. Biochemical and immunological properties of lactoferrin binding proteins from Moraxella (Branhamella) catarrhalis. Microb. Pathog. 24:89-100. [DOI] [PubMed] [Google Scholar]
- 12.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campagnari, A. A., K. L. Shanks, and D. W. Dyer. 1994. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect. Immun. 62:4909-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catlin, B. W. 1990. Branhamella catarrhalis: an organism gaining respect as a pathogen. Clin. Microbiol. Rev. 3:293-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, D., V. Barniak, K. R. VanDerMeid, and J. C. McMichael. 1999. The levels and bactericidal capacity of antibodies directed against the UspA1 and UspA2 outer membrane proteins of Moraxella (Branhamella) catarrhalis in adults and children. Infect. Immun 67:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen, D., J. C. McMichael, K. R. VanDerMeid, A. W. Masi, E. Bortell, J. D. Caplan, D. N. Chakravarti, and V. L. Barniak. 1999. Evaluation of a 74-kDa transferrin-binding protein from Moraxella (Branhamella) catarrhalis as a vaccine candidate. Vaccine 18:109-118. [DOI] [PubMed] [Google Scholar]
- 17.Christensen, J. J. 1999. Moraxella (Branhamella) catarrhalis: clinical, microbiological and immunological features in lower respiratory tract infections. APMIS Suppl. 88:1-36. [PubMed] [Google Scholar]
- 18.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, G. H. McCracken, Jr., and E. J. Hansen. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du, R. P., Q. Wang, Y. P. Yang, A. B. Schryvers, P. Chong, M. H. Klein, and S. M. Loosmore. 1998. Cloning and expression of the Moraxella catarrhalis lactoferrin receptor genes. Infect. Immun. 66:3656-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faden, H. 2001. The microbiologic and immunologic basis for recurrent otitis media in children. Eur. J. Pediatr. 160:407-413. [DOI] [PubMed] [Google Scholar]
- 21.Fitzgerald, M., R. Mulcahy, S. Murphy, C. Keane, D. Coakley, and T. Scott. 1997. A 200 kDa protein is associated with haemagglutinating isolates of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 18:209-216. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald, M., R. Mulcahy, S. Murphy, C. Keane, D. Coakley, and T. Scott. 1999. Transmission electron microscopy studies of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 23:57-66. [DOI] [PubMed] [Google Scholar]
- 23.Forsgren, A., M. Brant, M. Karamehmedovic, and K. Riesbeck. 2003. The immunoglobulin D-binding protein MID from Moraxella catarrhalis is also an adhesin. Infect. Immun. 71:3302-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forsgren, A., M. Brant, A. Mollenkvist, A. Muyombwe, H. Janson, N. Woin, and K. Riesbeck. 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J. Immunol. 167:2112-2120. [DOI] [PubMed] [Google Scholar]
- 25.Furano, K., and A. A. Campagnari. 2003. Inactivation of the Moraxella catarrhalis 7169 ferric uptake regulator increases susceptibility to the bactericidal activity of normal human sera. Infect. Immun. 71:1843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gjorloff Wingren, A., R. Hadzic, A. Forsgren, and K. Riesbeck. 2002. The novel IgD binding protein from Moraxella catarrhalis induces human B lymphocyte activation and Ig secretion in the presence of Th2 cytokines. J. Immunol. 168:5582-5588. [DOI] [PubMed] [Google Scholar]
- 27.Hardy, G. G., S. M. Tudor, and J. W. St. Geme III. 2003. The pathogenesis of disease due to nontypeable Haemophilus influenzae. Methods Mol. Med. 71:1-28. [DOI] [PubMed] [Google Scholar]
- 28.Helminen, M. E., I. Maciver, J. L. Latimer, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect. Immun. 61:2003-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helminen, M. E., I. Maciver, M. Paris, J. L. Latimer, S. L. Lumbley, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival in vivo. J. Infect. Dis. 168:1194-1201. [DOI] [PubMed] [Google Scholar]
- 30.Hill, D. J., and M. Virji. 2003. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol. Microbiol. 48:117-129. [DOI] [PubMed] [Google Scholar]
- 31.Holm, M. M., S. L. Vanlerberg, D. D. Sledjeski, and E. R. Lafontaine. 2003. The Hag protein of Moraxella catarrhalis strain O35E is associated with adherence to human lung and middle ear cells. Infect. Immun. 71:4977-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao, C. B., S. Sethi, and T. F. Murphy. 1995. Outer membrane protein CD of Branhamella catarrhalis: sequence conservation in strains recovered from the human respiratory tract. Microb. Pathog. 19:215-225. [DOI] [PubMed] [Google Scholar]
- 33.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 34.Klebba, P. E., and S. M. Newton. 1998. Mechanisms of solute transport through outer membrane porins: burning down the house. Curr. Opin. Microbiol. 1:238-247. [DOI] [PubMed] [Google Scholar]
- 35.Koebnik, R., K. P. Locher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol. 37:239-253. [DOI] [PubMed] [Google Scholar]
- 36.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luke, N. R., T. A. Russo, N. Luther, and A. A. Campagnari. 1999. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect. Immun. 67:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McMichael, J. C., M. J. Fiske, R. A. Fredenburg, D. N. Chakravarti, K. R. VanDerMeid, V. Barniak, J. Caplan, E. Bortell, S. Baker, R. Arumugham, and D. Chen. 1998. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect. Immun. 66:4374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meier, P. S., R. Troller, I. N. Grivea, G. A. Syrogiannopoulos, and C. Aebi. 2002. The outer membrane proteins UspA1 and UspA2 of Moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20:1754-1760. [DOI] [PubMed] [Google Scholar]
- 42.Merz, A. J., and M. So. 2000. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16:423-457. [DOI] [PubMed] [Google Scholar]
- 43.Mollenkvist, A., T. Nordstrom, C. Hallden, J. J. Christensen, A. Forsgren, and K. Riesbeck. 2003. The Moraxella catarrhalis immunoglobulin D-binding protein MID has conserved sequences and is regulated by a mechanism corresponding to phase variation. J. Bacteriol. 185:2285-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murphy, T. F., A. L. Brauer, N. Yuskiw, and T. J. Hiltke. 2000. Antigenic structure of outer membrane protein E of Moraxella catarrhalis and construction and characterization of mutants. Infect. Immun. 68:6250-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy, T. F., C. Kirkham, E. DeNardin, and S. Sethi. 1999. Analysis of antigenic structure and human immune response to outer membrane protein CD of Moraxella catarrhalis. Infect. Immun. 67:4578-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy, T. F., C. Kirkham, and A. J. Lesse. 1993. The major heat-modifiable outer membrane protein CD is highly conserved among strains of Branhamella catarrhalis. Mol. Microbiol. 10:87-97. [DOI] [PubMed] [Google Scholar]
- 48.Murphy, T. F., J. M. Kyd, A. John, C. Kirkham, and A. W. Cripps. 1998. Enhancement of pulmonary clearance of Moraxella (Branhamella) catarrhalis following immunization with outer membrane protein CD in a mouse model. J. Infect. Dis. 178:1667-1675. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, T. F., and M. R. Loeb. 1989. Isolation of the outer membrane of Branhamella catarrhalis. Microb. Pathog. 6:159-174. [DOI] [PubMed] [Google Scholar]
- 50.Myers, L. E., Y. P. Yang, R. P. Du, Q. Wang, R. E. Harkness, A. B. Schryvers, M. H. Klein, and S. M. Loosmore. 1998. The transferrin binding protein B of Moraxella catarrhalis elicits bactericidal antibodies and is a potential vaccine antigen. Infect. Immun. 66:4183-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neumayer, U., H. K. Schmidt, K. P. Mellwig, and G. Kleikamp. 1999. Moraxella catarrhalis endocarditis: report of a case and literature review. J. Heart Valve Dis. 8:114-117. [PubMed] [Google Scholar]
- 52.Nordstrom, T., A. Forsgren, and K. Riesbeck. 2002. The immunoglobulin D-binding part of the outer membrane protein MID from Moraxella catarrhalis comprises 238 amino acids and a tetrameric structure. J. Biol. Chem. 277:34692-34699. [DOI] [PubMed] [Google Scholar]
- 53.Patrick, C. C., A. Kimura, M. A. Jackson, L. Hermanstorfer, A. Hood, G. H. McCracken, Jr., and E. J. Hansen. 1987. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypeable Haemophilus influenzae. Infect. Immun. 55:2902-2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pearson, M. M., E. R. Lafontaine, N. J. Wagner, J. W. St. Geme III, and E. J. Hansen. 2002. A hag mutant of Moraxella catarrhalis strain O35E is deficient in hemagglutination, autoagglutination, and immunoglobulin D-binding activities. Infect. Immun. 70:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prasadarao, N. V. 2002. Identification of Escherichia coli outer membrane protein A receptor on human brain microvascular endothelial cells. Infect. Immun. 70:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prasadarao, N. V., A. M. Blom, B. O. Villoutreix, and L. C. Linsangan. 2002. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J. Immunol. 169:6352-6360. [DOI] [PubMed] [Google Scholar]
- 57.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1996. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect. Immun. 64:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reddy, M. S., T. F. Murphy, H. S. Faden, and J. M. Bernstein. 1997. Middle ear mucin glycoprotein: purification and interaction with nontypable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116:175-180. [DOI] [PubMed] [Google Scholar]
- 60.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 61.Sarwar, J., A. A. Campagnari, C. Kirkham, and T. F. Murphy. 1992. Characterization of an antigenically conserved heat-modifiable major outer membrane protein of Branhamella catarrhalis. Infect. Immun. 60:804-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sethi, S., N. Evans, B. J. Grant, and T. F. Murphy. 2002. New strains of bacteria and exacerbations of chronic obstructive pulmonary disease. N. Engl. J. Med. 347:465-471. [DOI] [PubMed] [Google Scholar]
- 63.Sethi, S., and T. F. Murphy. 2001. Bacterial infection in chronic obstructive pulmonary disease in 2000: a state-of-the-art review. Clin. Microbiol. Rev. 14:336-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sethi, S., J. M. Surface, and T. F. Murphy. 1997. Antigenic heterogeneity and molecular analysis of CopB of Moraxella (Branhamella) catarrhalis. Infect. Immun. 65:3666-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stefanou, J., A. V. Agelopoulou, N. V. Sipsas, N. Smilakou, and A. Avlami. 2000. Moraxella catarrhalis endocarditis: case report and review of the literature. Scand. J. Infect. Dis. 32:217-218. [DOI] [PubMed] [Google Scholar]
- 66.St. Geme, J. W., III. 1997. Bacterial adhesins: determinants of microbial colonization and pathogenicity. Adv. Pediatr. 44:43-72. [PubMed] [Google Scholar]
- 67.St. Geme, J. W., III. 2002. Molecular and cellular determinants of non-typeable Haemophilus influenzae adherence and invasion. Cell. Microbiol. 4:191-200. [DOI] [PubMed] [Google Scholar]
- 68.Thorsson, B., V. Haraldsdottir, and M. Kristjansson. 1998. Moraxella catarrhalis bacteraemia. A report on 3 cases and a review of the literature. Scand. J. Infect. Dis. 30:105-109. [DOI] [PubMed] [Google Scholar]
- 69.Timpe, J. M., M. M. Holm, S. L. Vanlerberg, V. Basrur, and E. R. Lafontaine. 2003. Identification of a Moraxella catarrhalis outer membrane protein exhibiting both adhesin and lipolytic activities. Infect. Immun. 71:4341-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Turner, H. R., M. R. Taylor, and W. R. Lockwood. 1985. Branhamella catarrhalis endocarditis in a patient receiving hemodialysis. South. Med. J. 78:1021-1022. [DOI] [PubMed] [Google Scholar]
- 71.Utsunomiya, T., K. Nakahara, M. Kuramochi, K. Hashiba, Y. Uzuka, and K. Matsumoto. 1984. Branhamella (Neisseria) catarrhalis endocarditis after insertion of a mitral prosthesis: a case report. Nippon Naika Gakkai Zasshi 73:1506-1511. [DOI] [PubMed] [Google Scholar]
- 72.Weiser, J. N., and E. C. Gotschlich. 1991. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]
- 74.Yu, R. H., R. A. Bonnah, S. Ainsworth, and A. B. Schryvers. 1999. Analysis of the immunological responses to transferrin and lactoferrin receptor proteins from Moraxella catarrhalis. Infect. Immun. 67:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zaleski, A., N. K. Scheffler, P. Densen, F. K. Lee, A. A. Campagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide Pk (Galα1-4Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]