Abstract
Background
hiPSCs are generated through epigenetic reprogramming of somatic tissue. Genomic imprinting is an epigenetic phenomenon through which monoallelic gene expression is regulated in a parent-of-origin-specific manner. Reprogramming relies on the successful erasure of marks of differentiation while maintaining those required for genomic imprinting. Loss of imprinting (LOI), which occurs in many types of malignant tumors, would hinder the clinical application of hiPSCs.
Results
We examined the imprinting status, expression levels and DNA methylation status of eight imprinted genes in five independently generated hiPSCs. We found a low frequency of LOI in some lines. Where LOI was identified in an early passage cell line, we found that this was maintained through subsequent passages of the cells. Just as normal imprints are maintained in long-term culture, this work suggests that abnormal imprints are also stable in culture.
Conclusions
Analysis of genomic imprints in hiPSCs is a necessary safety step in regenerative medicine, with relevance both to the differentiation potential of these stem cells and also their potential tumorigenic properties.
Keywords: Genomic imprinting, Loss of imprinting (LOI), DNA methylation, Histone modification, Human induced pluripotent cells
Background
Human induced pluripotent stem cells (hiPSCs) represent a promising therapeutic tool for many diseases, and might be useful for regenerating aged tissues and organs at high risk of failure [1,2]. However, the intrinsic self-renewal and pluripotency of hiPSCs potentially make them tumorigenic, hindering their clinical application [3-5]. hiPSCs are generated through epigenetic reprogramming of somatic tissue. It was initially thought that hiPSCs and human embryonic stem cells (hESCs) shared a high degree of epigenetic similarity [6,7]. However, recent reports have indicated that substantial differences exist between hiPSCs and hESCs with regard to gene expression, miRNA expression and DNA methylation [8-10]. Cell-of–origin-specific genetic and epigenetic differences exist in hiPSCs [11] and some of these stem cell lines spontaneously differentiate during serial passage [12]. Extensive evaluation of hiPSCs is consequently an essential component of the process required for their safe use in regenerative medicine.
Many types of malignant tumors are characterized by complex genetic and epigenetic alterations, including loss of heterozygosity (LOH) and loss of imprinting (LOI) [13,14]. Such alterations are presumed to represent the second hit, according to Knudson’s two-hit hypothesis (OMIM #167000) [15]. However, alterations in DNA methylation can also occur as the first hit during human carcinogenesis [16]. Alterations in the expression of imprinted genes represent one of the most common changes seen in cancer [17,18]. Some imprinted genes, including H19[19], GTL2[20], PEG1, PEG3[21], LIT1 (KCNQ1OT1) [22] and ZAC[23] are known to act, or are strongly implicated to act, as tumor suppressor genes (TSGs). Furthermore, imprinted genes play key roles in regulating growth and differentiation [24]. Thus the aberrant expression of imprinted genes may contribute to tumorigenesis or alter the differentiation potential of stem cells.
The monoallelic expression of imprinted genes is reliant on epigenetic mechanisms, most notably DNA methylation, which is established in the male and female germlines at discrete locations termed germline or gametic differentially methylated regions (gDMRs) [25]. Imprinted domains generally contain several genes displaying allele-specific expression and gDMRs within these domains act as imprinting centers or imprint control regions for the domain [26]. The majority of imprinted genes reside within these complex domains [27]. Although gametic DMRs are maintained throughout the life of the organism, genes within the domain can be imprinted in tissue- and developmentally specific manners [28].
In a recent paper, we demonstrated that hiPSCs exhibit epigenetic patterns distinct from hESCs [29]. After continuous passaging of the hiPSCs, these differences diminished such that over time the hiPSCs more closely resembled hESCs. However, we found that the imprinted DMRs showing abnormal methylation in early passage hiPSCs did not resolve during passaging. In this study we focused on the expression of imprinted genes in hiPSCs. Several reports on imprinted gene expression in hESCs demonstrate a substantial degree of instability [30]. Less is known regarding the stability of imprints in hiPSCs, although some work has begun [31]. We are particularly concerned with the stability of imprints in pluripotent stem cells during prolonged culture. Here, we examined the imprinting status and expression levels of eight imprinted genes and the methylation status of their DMRs in five independently derived hiPSCs. We found that the frequency LOI was very low in the early passaged lines. We also found that, in contrast, the epigenetic changes that took place at non-imprinted loci during prolonged culture for both normal and aberrant imprints were stably inherited despite prolonged passaging of the lines.
Results
Loss of heterozygosity (LOH) and loss of imprinting (LOI) in hiPSCs
We first determined whether hiPSCs showed LOH by comparing the restriction fragment length polymorphism (RFLP) patterns of the original tissue DNA with those of the hiPSC DNA samples. Samples in which RFLPs were present in the original DNA sample but absent or with an altered ratio in the hiPSC samples were considered to exhibit LOH. We found no evidence for LOH at the 8 loci tested (H19, IGF2, KCNQ1, LIT1, GTL2, PEG1, PEG3 and NDN).
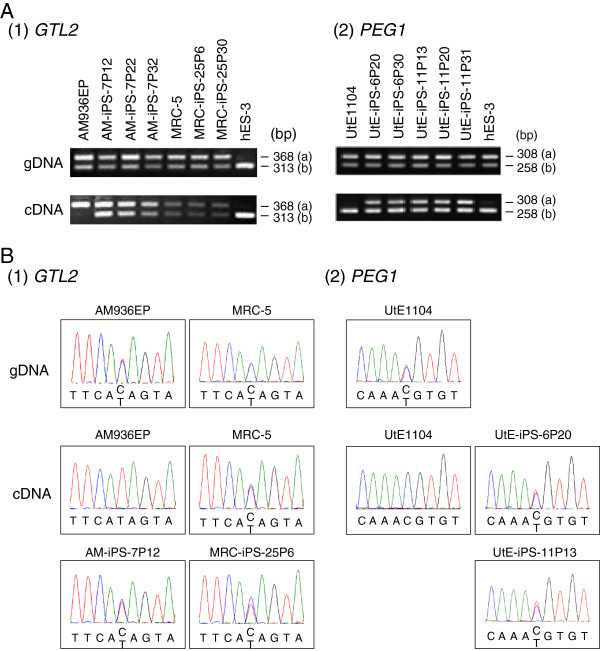
We next performed RT-PCR and RFLP analyses to identify samples that demonstrated loss of imprinting (LOI). Where expression of genes was low in undifferentiated cells, it was not possible to determine their imprinting status (H19 in MRC-iPS and UtE-iPS, IGF2 in PAE-iPS and GTL2 in Edom-iPS). Of the 16 informative loci, we identified LOI at three loci in hiPSCs, GTL2, PEG1 and PEG3, but we did not detect any LOI in hESCs (Table 1). Of particular interest, we observed loss of imprinting during the process of establishing the AM-iPSC (GTL2) and UtE-iPSC (PEG1) lines (Figure 1A). Where LOI was observed in early passage cells, this was maintained even after 30 or more passages (Figure 1).
Table 1.
LOI and MOI in hiPSCs
| |
|
H19 |
IGF2 |
PEG3 |
PEG1 |
GTL2 |
KCNQ1 |
NDN |
LIT1 |
||||||||
| |
|
(RsaI) |
(ApaI) |
(MnlI) |
(AflIII) |
(TaaI) |
(SmaI) |
(MboI) |
(RsaI) |
||||||||
| Cell |
Passage |
gDNA |
cDNA |
gDNA |
cDNA |
gDNA |
cDNA |
gDNA |
cDNA |
gDNA |
cDNA |
gDNA |
cDNA |
gDNA |
cDNA |
gDNA |
cDNA |
| AM936EP |
P9 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -2 |
P13 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -2 |
P19 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -2 |
P35 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -3 |
P9 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -3 |
P21 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -3 |
P29 |
a/b |
b |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -3 |
P36 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -7 |
P12 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a/b |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -7 |
P22 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a/b |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -7 |
P32 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a/b |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -8 |
P13 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -8 |
P20 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -8 |
P37 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -20 |
P8 |
N.T. |
a |
N.T. |
- |
N.T. |
- |
N.T. |
- |
N.T. |
a |
N.T. |
- |
N.T. |
- |
N.T. |
a |
| AM-iPS -20 |
P11 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -20 |
P14 |
a/b |
a |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -20 |
P16 |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| AM-iPS -20 |
P32 |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
a |
a |
- |
b |
- |
a/b |
a |
| PL551Ar |
P16 |
a/b |
a |
a/b |
a |
b |
- |
a/b |
N.D. |
b |
- |
b |
N.D. |
a |
- |
a |
- |
| PAE-iPS -05 |
P19 |
a/b |
a |
a/b |
N.D. |
b |
- |
a/b |
N.D. |
b |
- |
b |
- |
a |
- |
a |
- |
| PAE-iPS -05 |
P31 |
a/b |
a |
a/b |
N.D. |
b |
- |
a/b |
N.D. |
b |
- |
b |
- |
a |
- |
a |
- |
| PAE-iPS -11 |
P14 |
a/b |
a |
a/b |
N.D. |
b |
- |
a/b |
N.D. |
b |
- |
b |
- |
a |
- |
a |
- |
| PAE-iPS -11 |
P18 |
a/b |
a |
a/b |
N.D. |
b |
- |
a/b |
N.D. |
b |
- |
b |
- |
a |
- |
a |
- |
| PAE-iPS -11 |
P30 |
a/b |
a |
a/b |
N.D. |
b |
- |
a/b |
N.D. |
b |
- |
b |
- |
a |
- |
a |
- |
| MRC-5 |
- |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
a/b |
a/b |
N.D. |
a/b |
N.D. |
a |
- |
| MRC-iPS -16 |
P30 |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
ND |
a/b |
b |
a/b |
a |
a |
- |
| MRC-iPS -25 |
P6 |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
ND |
a/b |
b |
a/b |
a |
a |
- |
| MRC-iPS -25 |
P30 |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
ND |
a/b |
b |
a/b |
a |
a |
- |
| MRC-iPS -40 |
P11 |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
ND |
a/b |
b |
a/b |
a |
a |
- |
| MRC-iPS -40 |
P30 |
a/b |
N.D. |
b |
- |
a |
- |
b |
- |
a/b |
ND |
a/b |
b |
a/b |
a |
a |
- |
| UtE1104 |
P9 |
a/b |
N.D. |
a |
- |
a/b |
a/b |
a/b |
b |
a/b |
a/b |
b |
N.D. |
b |
- |
a |
- |
| UtE-iPS -6 |
P20 |
a/b |
N.D. |
a |
- |
a/b |
a/b |
a/b |
a/b |
a/b |
a/b |
b |
- |
b |
- |
a |
- |
| UtE-iPS -6 |
P31 |
a/b |
b |
a |
- |
a/b |
a/b |
a/b |
a/b |
a/b |
a/b |
b |
- |
b |
- |
a |
- |
| UtE-iPS -11 |
P13 |
a/b |
N.D. |
a |
- |
a/b |
N.D. |
a/b |
a/b |
a/b |
a |
b |
N.D. |
b |
- |
a |
- |
| UtE-iPS -11 |
P20 |
a/b |
N.D. |
a |
- |
a/b |
N.D. |
a/b |
a/b |
a/b |
a/b |
b |
N.D. |
b |
- |
a |
- |
| UtE-iPS -11 |
P30 |
a/b |
N.D. |
a |
- |
a/b |
N.D. |
a/b |
a/b |
a/b |
a |
b |
- |
b |
- |
a |
- |
| Edom22 |
P5 |
b |
- |
a/b |
a/b |
a |
- |
a/b |
b |
b |
- |
a/b |
a |
a |
- |
a |
- |
| Edom-iPS -1 |
P27 |
b |
- |
a/b |
N.D. |
a |
- |
a/b |
b |
b |
- |
a/b |
a |
a |
- |
a |
- |
| hES 3 |
P29 |
a/b |
a |
b |
- |
a/b |
a |
a/b |
b |
b |
- |
a |
- |
a/b |
b |
a |
- |
| SEES 1 |
P10 |
a/b |
a |
a/b |
a |
a |
- |
b |
- |
a/b |
b |
a |
- |
a |
- |
a |
- |
| SEES 4 | P9 | a/b | b | a/b | a | a | - | b | - | b | - | a | - | a/b | b | a | - |
A summary of LOI and MOI RFLP data for the 8 imprinted genes analyzed in 22 hiPSCs and 3 control hES cell lines. hiPSCs derived from extraembryonic amniotic membrane (AM-iPS), embryonic lung tissue (MRC-iPS), uterine endometrium (UtE-iPS), adult menstrual blood (Edom-iPS) and extraembryonic placental tissue (PAE-iPS). Samples were analyzed at the specified passage number. (-): not informative.
Figure 1.
Loss of imprinting in hiPSCs. Loss of allelic expression of GTL2 in cell line AM-iPS-7 (1) and PEG1 in cell lines UtE-iPS-6 and 11 (2). PCR products amplified from paired genomic DNA and cDNA samples were digested with the specified restriction enzymes (A). Results were confirmed by direct sequencing (B). AM936EP and UtE1104 are primary culture cells made from amniotic membrane and uterine endometrium cells, respectively. Band ‘a’: without a restriction site. Band ‘b’: with a restriction site.
Expression level of the imprinted genes in hiPSCs
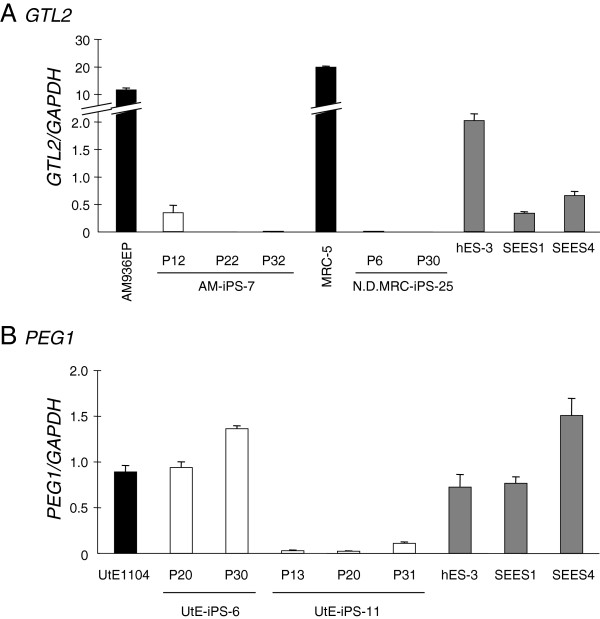
LOI can refer to silencing of an originally active allele or expression of a normally silent allele. Therefore, we compared the expression levels of the three genes that displayed LOI in hiPSCs and hESCs (Table 1). The expression of IGF2 and GTL2 was decreased in almost all the hiPSC lines in comparison with the hES cells (Additional files 1 and 2). GTL2 in cell line AM-iPS-7 and PEG1 in line UtE-iPS-11 showed apparent biallelic expression but their expression levels were relatively low in comparison to hESCs with reduced expression maintained stably through to late passages (Figure 2). In contrast, expression of PEG1 in cell line UtE-iPS-6 was not significantly different from that of hESCs. These results were in accordance with the DNA microarray analysis data we already reported [29]. Since, in two cases, LOI correlated with reduced gene expression, this has potential functional implications due to loss of function.
Figure 2.
Reduced expression of hiPSCs with LOI. Gene expression levels of GTL2 (A) and PEG1 (B) in the original and the passaged hiPSCs were compared to that of hESCs. The GAPDH ratio was calculated. The bars indicate the means ± standard deviation (SD) from two replicates. N.D.: not determined.
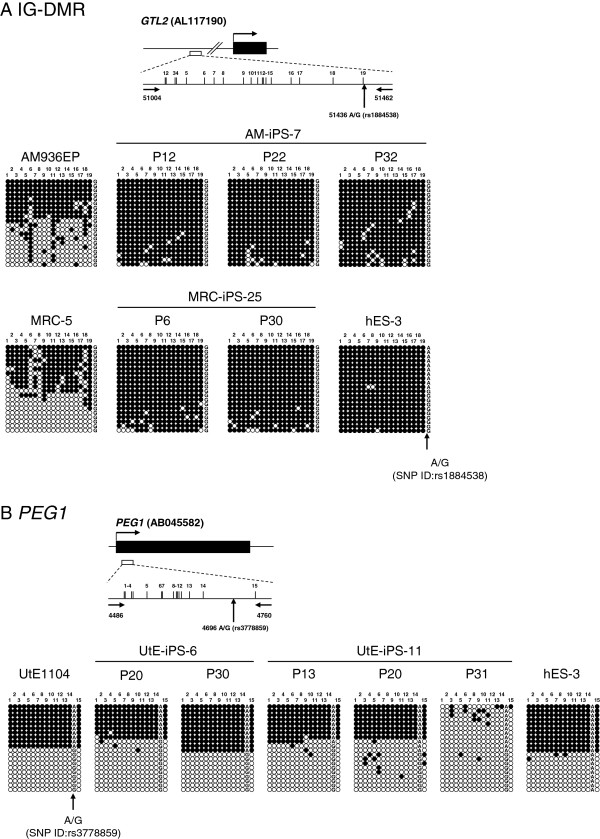
Analysis of the DNA methylation status and the histone modification of GTL2 and PEG1 DMRs in hiPSC lines
We determined the allele-specific methylation status of the GTL2 (IG-DMR) and PEG1 imprinted DMRs using polymorphic bisulfite-PCR sequencing (Figure 3). In cell line AM-iPS-7, which showed LOI and reduced expression of GTL2, we observed hypermethylation of IG-DMR, which was maintained during continuous passaging. IG-DMR methylation is normally present on the silent allele of GTL2[32], which suggests aberrant signaling between this DMR and GTL2 expression. In cell line UtE-iPS-11, in which there was LOI and reduced expression of PEG1, abnormal methylation was detected in passage 31 cells but not earlier passages. In cell line UtE-iPS-6, in which there was LOI but not reduced expression of PEG1, abnormal methylation was not detected. Allele-specific expression of some genes has been reported to be regulated by histone modification rather than direct DNA methylation [33-35]. We therefore analyzed histone modifications in the hiPS cell line by chromatin immunoprecipitation (ChIP) analyses using the following antibodies: dimethylated H3-Lys4 (H3K4me2), acetylated H3-Lys9 (H3K9ac), H3K9me2, and H3K27me3. H3K4me2 and H3K9ac mark active genes and H3K9me2 and H3K27me3 are repressive marks. In the GTL2 promoter region, H3K9me2 and H3K27me3 were enriched in AM-iPS-7 and MRC-iPS-25 cells (Figure 4D).
Figure 3.
Aberrant DNA methylation of hiPSCs with LOI. Bisulfite PCR sequencing methylation assay of genomic DNA prepared from AM-iPS-7 and MRC-iPS-25 at the IG-DMR (GTL2-DMR) (A) and UtE-iPS-6 and 11 at PEG1 (B). Each row represents a unique methylation profile within the pool of 20 clones sequenced. Closed and open circles represent methylated and unmethylated CpGs, respectively. The numbers represent the percentages of methylation by bisulfite sequencing. SNPs are shown by arrows.
Figure 4.
Reactivation of GTL2 expression by treatment with VPA. Reactivation of GTL2 expression by treatment with VPA (A). The expression level of GTL2 mRNA was restored by VPA treatment. Gene expression of the original cells and of the hiPSCs was compared to that of hESCs. The GAPDH ratio was calculated. The bars indicate the means ± SD from two replicates. The imprinting status of GTL2 was stable in response to VPA (B). Methylation status in bisulfite-PCR sequencing analyses of IG-DMR is unchanged (C). Histone modifications of the GTL2 promoter were changed by VPA (D). The immunoprecipitation/input ratio was calculated. The bars indicate the means ± SD from three replicates.
Reactivation of the imprinted genes by the HDACi treatment
Previous reports demonstrated that the GTL2 gene was aberrantly silenced in most mouse iPSC lines but that expression could be restored through HDACi treatment [36,37]. In our study, AM-iPS and MRC-iPS cells showed LOI of GTL2, with a reduction in gene expression and hypermethylation of the IG-DMR. To assess whether GTL2 expression could be restored, AM-iPS and MRC-iPS cells were treated with the HDAC inhibitor VPA (sodium valproate). VPA-treated cells did achieve a 3.0–5.8-fold increase in GTL2 expression levels (Figure 4A) and H3K4me2 and H3K9ac were enriched in its promoter region (Figure 4D). However, the DNA methylation pattern was stable under VPA treatment and the imprinting status of GTL2 was not changed, with cells maintaining biallelic expression of the gene (Figure 4B). These results suggested that the aberrant DNA methylation and imprinting that were established and maintained in early passages (Figure 4C) were not sustainably reversed by the treatment.
Discussion
Most hES and hiPS cell lines possess stable imprinted gene expression, at least in undifferentiated cells [30,31] and findings in this study]. This implies that imprints withstand the process of reprogramming and the rigors of growing in culture. In our study, we found that only three of the 22 hiPS cell lines we derived from a variety of somatic cell types showed LOI, and at only a few sites. The majority of cases had normal imprinting status. While LOI was rare in our hiPS cell lines, we found that it was maintained during prolonged passage, and resistant to VPA treatment. These abnormalities would preclude the use of these cell lines for therapeutic applications but might provide a mechanistic insight relevant to imprinting and reprogramming.
We previously reported that abnormal DNA methylation detected in early passage iPSCs diminished after continued passaging, such that these cells ultimately more resembled ESCs. However, abnormal DNA methylation at imprinted loci in ESCs occurs in response to continuous passaging [29]. Rugg-Gunn et al. suggested three possible explanations for LOI in hESCs [30]. First, the developmental onset of transcription might influence imprinted gene expression. Second, a particular imprinted gene’s expression might differ depending on whether it is regulated by maternally or paternally inherited methylation. Third, the pattern of imprinted gene expression might depend on whether the gene provides a growth advantage to hESCs. These possibilities might also apply to hiPSCs.
There are two caveats that apply to this work. First, we examined expression in undifferentiated cells. Consequently, we may have missed changes in imprinted gene expression where genes are expressed only in differentiated cells or where imprinting is tissue specific. Second, we examined total levels of expression and total methylation patterns of populations of cells. Therefore we cannot exclude the possibility that a small population within our samples could behave in a different manner from the general population. Nonetheless, our data are encouraging in suggesting that imprinting errors in iPSCs are derived from a variety of human somatic cell types.
One of the key advantages of iPSCs is that they can be derived from patients, supporting the further investigation of certain diseases, as well as the replacement of degenerated and damaged tissues. Careful analysis of imprinted genes should therefore be performed on all iPS cell lines since several published iPS cell lines that passed the necessary reprogramming criteria also showed aberrations in imprinted gene expression and DNA methylation of DMRs. This is particularly critical if these hiPSs are to be used for regenerative medicine since aberrations in imprinted genes could cause problems with cell differentiation and perhaps even cause tumors [38]. The analysis of imprinted genes is also essential for modeling of genetic diseases because abnormal imprinting can seriously confuse the disease phenotyping.
Recent advances in high-throughput technologies for gene expression analysis and DNA methylation analysis indicate the possibility that all newly generated stem cell lines can be characterized at the epigenetic level rapidly and precisely. However, our work and that of others suggest that certain imprinted loci may be more susceptible to LOI. This means that it might be possible to design targeted assays for specific loci as the first step in the characterization of newly generated cell lines, and also those that have been extensively passaged.
Conclusions
In conclusion, while imprinting errors may be rare in iPSCs, they are resistant to reversal strategies. The aberrant expression of imprinted genes in these lines is likely to hamper their use both for the understanding of certain pathologies and regenerative medicine.
Methods
Ethics statement
All experiments handling human cells and tissues were performed in line with the tenets of the Declaration of Helsinki. This study was approved by the Institutional Review Board of the National Institute for Child Health and Development and the Ethics Committee of Tohoku University School of Medicine.
DNA/RNA preparation of iPSCs
We generated 22 hiPSCs from extraembryonic amniotic membrane (AM-iPS), embryonic lung tissue (MRC-iPS), uterine endometrium (UtE-iPS), adult menstrual blood (Edom-iPS), and extraembryonic placental tissue (PAE-iPS) and characterized the pluripotent nature using culture methods described previously [39-41]. Prior to RNA and DNA preparation, feeder layers were removed from the undifferentiated cells by panning for 20 minutes.
Loss of heterozygosity (LOH) and loss of imprinting (LOI) analyses
PCR was performed on parental tissue and the genomic DNA of hiPSCs using the primer sequences summarized in Additional file 3. A PCR reaction mix containing 0.5 μM concentrations of each primer set, 200 μM dNTPs, 1× PCR buffer, and 1.25U of EX Taq Hot Start DNA Polymerase (Takara Bio, Tokyo, Japan) in a total volume of 20 μl was used. The following PCR program was used: 1 minute of denaturation at 94°C followed by 35 cycles of 30 seconds at 94°C, 30 seconds at 50-70°C, 30 seconds at 72°C and a final extension for 5 minutes at 72°C. PCR products were digested by unique polymorphic enzymes to identify samples that were heterozygous for a single nucleotide polymorphism (SNP). For samples found to be heterozygous for a SNP, RNA was prepared from matched hiPSCs, followed by reverse transcription-PCR (RT-PCR) and restriction digestion (Additional file 3) [42-49]. The digested PCR products were electrophoresed on 3% agarose gel.
Gene expression analysis
RNA expression levels of 8 imprinted genes were also analyzed by microarray and the real-time PCR. Microarray analysis was performed using an Agilent Whole Human Genome Microarray chip (G4112F, Agilent, Santa Clara, CA). Raw data were normalized and analyzed using GeneSpringGX11 software (Silicon Genetics, Redwood City, CA). The microarray data have been deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/). Real-time PCR reaction was done with SYBR Premix Ex Taq II (Takara Bio). In the case of PEG3 expression analysis, TaqMan Gene Expression Assay (Assay ID: Hs00300418-s1, Applied Biosystems, Foster City, CA) was carried out according to the manufacturer’s protocol using a StepOne Real-time PCR System (Applied Biosystems). The relative expression levels of the detected genes from these cells were estimated visually by comparing relative band intensities with the expression level of the housekeeping gene GAPDH.
Polymorphic bisulfite PCR methylation assay
We performed standard methylation assays using the SNPs and bisulfite sequencing [50]. The primary DMRs of eight imprinted genes (H19, GTL2, ZDBF2, PEG1, KCNQ1OT1, ZAC, PEG3 and SNRPN) were analyzed as described previously [50,51]. Each DNA sample was treated with sodium bisulfite using the EZ methylation kit (Zymo Research, Orange, CA) and amplified by PCR. PCR products were purified, cloned into pGEM-T (Promega, Medison, WI) and an average of 20 clones per individual were sequenced using reverse primer M13 and an automated ABI Prism 3130xl Genetic Analyzer (Applied Biosystems). To avoid any allelic bias, we used specific polymorphic sites. Sodium bisulfite modification treatments were carried out in duplicate for each DNA sample and at least three independent amplification experiments were performed for each individual examined.
Chromatin immunoprecipitation (ChIP) assay
ChIP analysis was performed using the Magna ChIP G Chromatin Immunoprecipitation Kit (Millipore, Temecula, CA) according to the manufacturer’s protocol. We used the following antibodies: dimethylated H3-Lys4, acetylated H3-Lys9, dimethylated H3-Lys9 and trimethylated H3-Lys27 (Millipore). The histone modifications were analyzed by real-time PCR. Real-time PCR reaction was done with SYBR Premix Ex Taq II (Takara Bio). The amount of precipitated DNA was determined as percentage relative to input DNA. Primers used are listed in Additional file 3.
Treatment of cells with sodium valproate
hiPSCs were plated at a density of 5 × 105 cells/60mm2 dish. Twenty-four hours later, they were treated with 1 mM sodium valproate (Wako, Tokyo, Japan) for the times stated. Total RNA was prepared and analyzed by the RT-PCR method. The methylation status of the DMRs was examined using the bisulfite PCR sequencing methylation assay described previously [51].
Abbreviations
DMR: Differentially methylated region; hESCs: Human embryonic stem cells; hiPSCs: Human induced pluripotent cells; LOH: Loss of heterozygosity; LOI: Loss of imprinting; MOI: Maintenance of imprinting; N.D: Not determined; N.T: Not tested; PCR: Polymerase chain reaction; RFLP: Restriction fragment length polymorphism; RT-PCR: Reverse transcription-PCR; SNP: Single nucleotide polymorphism; SD: Standard deviation.
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
AU and TA conceived and designed the study and wrote the manuscript. AU generated hiPSCs and hESCs. TA analyzed genomic imprinting. HH carried out the molecular study and data analysis and wrote the manuscript. H Okae, MS, NM and AS performed the molecular study and contributed to data analysis. MT, NK, H Okita, YM and HA prepared cell materials and contributed to data analysis. KN performed the transcriptome data analysis. All authors reviewed the results from the data analysis and approved the final manuscript.
Supplementary Material
Microarray analysis. Scatter plots of MRC-iPS-25P22 versus hES3 (A) and UtE-iPS-6P30 versus hES3 (B). Scatter plot comparing the spot intensities in hybridization with probes from hiPSCs (y axis) and hESCs (x axis). The magenta plots indicate the imprinted genes.
Gene expression analysis of the imprinted genes.H19 (A), IGF2 (B), PEG3 (C), PEG1 (D), GTL2 (E), KCNQ1 (F), NDN (G) and LIT1 (H). Gene expression of the original and hiPSCs was compared to that of hESCs. The GAPDH ratio was calculated. The bars indicate the means ± SD from two replicates.
PCR primers and conditions.
Contributor Information
Hitoshi Hiura, Email: h_hiura@med.tohoku.ac.jp.
Masashi Toyoda, Email: mtoyoda@tmig.or.jp.
Hiroaki Okae, Email: okaehiro@m.tohoku.ac.jp.
Masahiro Sakurai, Email: msaku913@affrc.go.jp.
Naoko Miyauchi, Email: naoko-k@med.tohoku.ac.jp.
Akiko Sato, Email: lukelab@oct-net.ne.jp.
Nobutaka Kiyokawa, Email: nkiyokawa@nch.go.jp.
Hajime Okita, Email: okita@nch.go.jp.
Yoshitaka Miyagawa, Email: miyayoshi0007@gmail.com.
Hidenori Akutsu, Email: akutsu-h@ncchd.go.jp.
Koichiro Nishino, Email: knishino@nch.go.jp.
Akihiro Umezawa, Email: umezawa@1985.jukuin.keio.ac.jp.
Takahiro Arima, Email: tarima@med.tohoku.ac.jp.
Acknowledgments
We would like to thank all the members of our laboratory for technical assistance and their support and valuable suggestions. In particular, we thank Dr. R. M. John and Mathew Van De Pette (Cardiff School of Biosciences) for comments on the manuscript. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI) (21028003, 23013003, and 23390385), the Takeda Foundation, Uehara Memorial Foundation and the Environment Research & Technology Development Fund (C1008) (TA).
References
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Blum B, Benvenisty N. The tumorigenicity of human embryonic stem cells. Adv Canc Res. 2008;100:133–158. doi: 10.1016/S0065-230X(08)00005-5. [DOI] [PubMed] [Google Scholar]
- Kiuru M, Boyer JL, O’Connor TP, Crystal RG. Genetic control of wayward pluripotent stem cells and their progeny after transplantation. Cell Stem Cell. 2009;4:289–300. doi: 10.1016/j.stem.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izpisua Belmonte JC, Ellis J, Hochedlinger K, Yamanaka S. Induced pluripotent stem cells and reprogramming: seeing the science through the hype. Nat Rev Genet. 2009;10:878–883. doi: 10.1038/nrg2700. [DOI] [PubMed] [Google Scholar]
- Maherali N, Ahfeldt T, Rigamonti A, Utikal J, Cowan C, Hochedlinger K. A high-efficiency system for the generation and study of human induced pluripotent stem cells. Cell Stem Cell. 2008;3:340–345. doi: 10.1016/j.stem.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, Miller J, Schlaeger T, Daley GQ, Feinberg AP. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TS, Gavina M, Chen CW, Sun B, Teng PN, Huard J, Deasy BM, Zimmerlin L, Peault B. Placental perivascular cells for human muscle regeneration. Stem Cells Dev. 2011;20:451–463. doi: 10.1089/scd.2010.0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A, Calo V, Bruno L, Rizzo S, Bazan V, Di Fede G. Hereditary ovarian cancer. Crit Rev Oncol Hematol. 2009;69:28–44. doi: 10.1016/j.critrevonc.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Balch C, Fang F, Matei DE, Huang TH, Nephew KP. Minireview: epigenetic changes in ovarian cancer. Endocrinology. 2009;150:4003–4011. doi: 10.1210/en.2009-0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG Jr. Genetics of human cancer. Annu Rev Genet. 1986;20:231–251. doi: 10.1146/annurev.ge.20.120186.001311. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Feinberg AP. DNA methylation, genomic imprinting and cancer. Curr Top Microbiol Immunol. 2000;249:87–99. doi: 10.1007/978-3-642-59696-4_6. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Schofield PN. Genomic imprinting and cancer. Mol Pathol. 1998;51:185–190. doi: 10.1136/mp.51.4.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. Imprinting of a genomic domain of 11p15 and loss of imprinting in cancer: an introduction. Canc Res. 1999;59(7 Suppl):1743s–1746s. [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi AK, Weber BL, Birrer MJ. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci USA. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda T, Asai A, Kuroiwa Y, Kobayashi S, Aisaka K, Nagashima G, Yoshida MC, Kondo Y, Kagiyama N, Kirino T, Kaneko-Ishino T, Ishino F. Tumour suppressor activity of human imprinted gene PEG3 in a glioma cell line. Genes Cells. 2001;6:237–247. doi: 10.1046/j.1365-2443.2001.00412.x. [DOI] [PubMed] [Google Scholar]
- Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, Kugoh H, Mukai T, Ikeguchi M, Oshimura M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Canc Sci. 2006;97:1147–1154. doi: 10.1111/j.1349-7006.2006.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamikihara T, Arima T, Kato K, Matsuda T, Kato H, Douchi T, Nagata Y, Nakao M, Wake N. Epigenetic silencing of the imprinted gene ZAC by DNA methylation is an early event in the progression of human ovarian cancer. Int J Canc. 2005;115:690–700. doi: 10.1002/ijc.20971. [DOI] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93:309–312. doi: 10.1016/S0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- Koerner MV, Barlow DP. Genomic imprinting-an epigenetic gene-regulatory model. Curr Opi Genet Dev. 2010;20:164–170. doi: 10.1016/j.gde.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opi Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet. 2008;9:129–140. doi: 10.1038/ni1560. [DOI] [PubMed] [Google Scholar]
- Nishino K, Toyoda M, Yamazaki-Inoue M, Fukawatase Y, Chikazawa E, Sakaguchi H, Akutsu H, Umezawa A. DNA methylation dynamics in human induced pluripotent stem cells over time. PLoS Genet. 2011;7:e1002085. doi: 10.1371/journal.pgen.1002085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg-Gunn PJ, Ferguson-Smith AC, Pedersen RA. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum Mol Genet. 2007;16 Spec No. 2:R243–R251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- Pick M, Stelzer Y, Bar-Nur O, Mayshar Y, Eden A, Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Monk D, Wagschal A, Arnaud P, Muller PS, Parker-Katiraee L, Bourc’his D, Scherer SW, Feil R, Stanier P, Moore GE. Comparative analysis of human chromosome 7q21 and mouse proximal chromosome 6 reveals a placental-specific imprinted gene, TFPI2/Tfpi2, which requires EHMT2 and EED for allelic-silencing. Genome Res. 2008;18:1270–1281. doi: 10.1101/gr.077115.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Ferrari F, Choi J, Walsh RM, Chen T, Ooi SS, Kim SY, Bestor TH, Shioda T, Park PJ, Hochedlinger K. Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet. 2012;44:398–405. doi: 10.1038/ng.1110. S391-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, Ko MS, Ohlsson R, Longo DL, Feinberg AP. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science. 2005;307:1976–1978. doi: 10.1126/science.1108080. [DOI] [PubMed] [Google Scholar]
- Nagata S, Toyoda M, Yamaguchi S, Hirano K, Makino H, Nishino K, Miyagawa Y, Okita H, Kiyokawa N, Nakagawa M, Yamanaka S, Akutsu H, Umezawa A, Tada T. Efficient reprogramming of human and mouse primary extra-embryonic cells to pluripotent stem cells. Genes Cells. 2009;14:1395–1404. doi: 10.1111/j.1365-2443.2009.01356.x. [DOI] [PubMed] [Google Scholar]
- Makino H, Toyoda M, Matsumoto K, Saito H, Nishino K, Fukawatase Y, Machida M, Akutsu H, Uyama T, Miyagawa Y, Okita H, Kiyokawa N, Fujino T, Ishikawa Y, Nakamura T, Umezawa A. Mesenchymal to embryonic incomplete transition of human cells by chimeric OCT4/3 (POU5F1) with physiological co-activator EWS. Exp Cell Res. 2009;315:2727–2740. doi: 10.1016/j.yexcr.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Cui CH, Uyama T, Miyado K, Terai M, Kyo S, Kiyono T, Umezawa A. Menstrual blood-derived cells confer human dystrophin expression in the murine model of Duchenne muscular dystrophy via cell fusion and myogenic transdifferentiation. Mol Biol Cell. 2007;18:1586–1594. doi: 10.1091/mbc.E06-09-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vambergue A, Fajardy I, Dufour P, Valat AS, Vandersippe M, Fontaine P, Danze PM, Rousseaux J. No loss of genomic imprinting of IGF-II and H19 in placentas of diabetic pregnancies with fetal macrosomia. Growth Horm IGF Res. 2007;17:130–136. doi: 10.1016/j.ghir.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10:1711–1718. doi: 10.1101/gr.161600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa O, Eccles MR, Szeto J, McNoe LA, Yun K, Maw MA, Smith PJ, Reeve AE. Relaxation of insulin-like growth factor II gene imprinting implicated in Wilms’ tumour. Nature. 1993;362:749–751. doi: 10.1038/362749a0. [DOI] [PubMed] [Google Scholar]
- Maegawa S, Yoshioka H, Itaba N, Kubota N, Nishihara S, Shirayoshi Y, Nanba E, Oshimura M. Epigenetic silencing of PEG3 gene expression in human glioma cell lines. Mol Carcinog. 2001;31:1–9. doi: 10.1002/mc.1034. [DOI] [PubMed] [Google Scholar]
- Pedersen IS, Dervan PA, Broderick D, Harrison M, Miller N, Delany E, O’Shea D, Costello P, McGoldrick A, Keating G, Tobin B, Gorey T, McCann A. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 1999;59:5449–5451. [PubMed] [Google Scholar]
- Higashimoto K, Soejima H, Yatsuki H, Katsuki T, Mukai T. An NsiI RFLP in the human long QT intronic transcript 1 (LIT1) J Hum Genet. 2000;45:96–97. doi: 10.1007/s100380050020. [DOI] [PubMed] [Google Scholar]
- Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, Yoshida MA, Niikawa N, Feinberg AP, Oshimura M. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet. 1999;8:1209–1217. doi: 10.1093/hmg/8.7.1209. [DOI] [PubMed] [Google Scholar]
- MacDonald HR, Wevrick R. The necdin gene is deleted in Prader-Willi syndrome and is imprinted in human and mouse. Hum Mol Genet. 1997;6:1873–1878. doi: 10.1093/hmg/6.11.1873. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato A, Otsu E, Hiura H, Tomatsu C, Utsunomiya T, Sasaki H, Yaegashi N, Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum Mol Genet. 2007;16:2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- Hiura H, Sugawara A, Ogawa H, John RM, Miyauchi N, Miyanari Y, Horiike T, Li Y, Yaegashi N, Sasaki H, Kono T, Arima T. A tripartite paternally methylated region within the Gpr1-Zdbf2 imprinted domain on mouse chromosome 1 identified by meDIP-on-chip. Nucleic Acids Res. 2010;38:4929–4945. doi: 10.1093/nar/gkq200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microarray analysis. Scatter plots of MRC-iPS-25P22 versus hES3 (A) and UtE-iPS-6P30 versus hES3 (B). Scatter plot comparing the spot intensities in hybridization with probes from hiPSCs (y axis) and hESCs (x axis). The magenta plots indicate the imprinted genes.
Gene expression analysis of the imprinted genes.H19 (A), IGF2 (B), PEG3 (C), PEG1 (D), GTL2 (E), KCNQ1 (F), NDN (G) and LIT1 (H). Gene expression of the original and hiPSCs was compared to that of hESCs. The GAPDH ratio was calculated. The bars indicate the means ± SD from two replicates.
PCR primers and conditions.