Abstract
Pyrococcus Furiosus (Pfu) is an excellent organism to generate reference samples for proteomics labs because of its moderately sized genome and very little sequence duplication within the genome. We demonstrated a stable and consistent method to prepare proteins in bulk that eliminates growth and preparation as a source of uncertainty in the standard. We performed several proteomic studies in different laboratories using each laboratory's specific workflow as well as separate and integrated data analysis. This study demonstrated that a Pfu whole cell lysate provides suitable protein sample complexity to not only validate proteomic methods, work flows, and benchmark new instruments, but also to facilitate comparison of experimental data generated over time and across instruments or labs.
Keywords: Pyrococcus furiosus (Pfu), Proteomics, Protein Complex Standard, MudPIT, OFFGEL electrophoresis, Directed LC-MS/MS
Introduction
Pyrococcus furiosus (Pfu) is an archaeon with 1,908 kilobases of DNA sequence and 2065 open reading frames (ORF)1. It is a hyperthermophilic anaerobe that was isolated from geothermally-heated marine sediments and thus it is a thermostable organism. It grows between 70 and 103 °C, with an optimum temperature of 100 °C and a doubling time of 37 minutes. Posttranslational modifications in archaea include glycosylation, phosphorylation, lipids and methylation. The proteome of Pfu has been analyzed by Lim et al and Holden et al2-3. Lee et al used peptide fractionation by immobilized pH gradients (IPG-IEF) followed by reversed-phase nano-HPLC and electrospray ionization tandem mass spectrometry (ESI-MS/MS) to identify a total of 900 proteins representing approximately 44% of the proteome were including many membrane proteins4. Pfu is a relatively simple organism with a moderately sized genome and very little sequence duplication within the genome or sequence conservation with more distant organisms such as human and mouse. As an organism with a relatively simple genome, Pfu should be an excellent organism for use as a standard in proteomics.
Proteomics is heavily driven by technologies to analyze and characterize proteins. Mass spectrometry has been a key technology for the analysis of proteins for 30 years and has become a vital part of proteomic strategies. Recent developments in proteomic workflows have encompassed all aspects of the process from sample preparation to separations to advances in mass spectrometry technology. A common process for analysis of complex protein mixtures is the use of “shotgun” proteomics where proteins are digested into mixtures of peptides and then analyzed by tandem mass spectrometry. The shotgun approach requires effective methods to digest the proteins and to separate the complex mixture of peptides. It also requires the use of tandem mass spectrometers to collect data, and software tools to quantify and identify the data. As workflows evolve, there is constant assessment and comparison of advances which requires the availability of protein mixtures with known content. The ready availability of consistent standards prevents “apples to oranges” comparisons when different or “home-brewed” standards are used to evaluate methods.
Early developments in proteomics were demonstrated using available protein mixtures such as yeast or E. coli cell lysates or synthetic mixtures such as SDS-PAGE standards5-7. While LC-MS/MS of synthetic mixtures yields the expected proteins, it also identified many additional proteins present in the mixtures as “contaminants”, suggesting improvements were needed in the preparation of these types of standards for use in proteomics. “Home brewed” synthetic standard mixtures of peptides and proteins were created by Purvine et al and Keller et al with more complexity than the SDS-PAGE standards as a way to test software approaches for protein identification8,9. More recently complicated standard protein mixtures for proteomic studies have been constructed which include mixtures of 20 or 48 reference proteins10, but standards of even greater complexity are needed so the use of cell lysates has been pursued.
By using cell lysates as more complicated “standard” samples, the tedious and difficult process of expressing, purifying and weighing out hundreds of proteins to create a mixture is circumvented. Complex standards based on yeast extracts11,12, T4 phage preparations13, and Shewanella oneidensis MR114 have been used. The difficulty with microorganisms as standards, however, is that they are often grown and prepared independently in the laboratories using them and thus how they are prepared creates a source of variability that complicates comparisons. For example, difference in growth media (e.g. rich media versus minimal media) will cause the expression of different sets of enzymes. While cell derived protein mixtures provide a challenging sample to test new methods, the fact that protein expression levels may change depending on growth conditions, lysis techniques, and strains can create variability when trying to compare methods between laboratories. Thus, to use a microorganism as a standard, a stable and consistent method to prepare proteins in bulk would be required to eliminate growth and preparation as a source of uncertainty in the standard.
The availability of a complex protein standard can challenge existing proteomic workflows and help develop and evaluate new strategies10. A standard cannot only be used to validate new methods and work flows, but can benchmark new instruments and methods. More importantly a complex standard can facilitate comparison of experimental data generated over time, and across instruments, methods, or labs. A standard should mimic a typical proteomic sample with a set of proteins of known sequences with diverse physical characteristics and minimal redundancy among proteins. If the sample is genetically distinct from human or mouse, it can also be used as a spike-in standard. Production of the standard should allow the creation of large amounts of the sample to provide year to year consistency and the proteins should be very stable at 4 °C and below for several years. In this joint study, we evaluated the suitability of Pyrococcus furiosus (Pfu) as complex protein standard for proteomics. Each of the three labs used different approaches to sample preparation and LC-MS analysis, however, a common data analysis step was performed on all the collected results to simplify comparison of the methodologies.
Experimental Methods
Preparation of the Complex Proteomic Standard (soluble extract from Pfu)
Pfu cell paste was purchased from the lab of Mike Adams (University of Georgia, Department of Biochemistry & Molecular Biology, Life Sciences Building, Green Street University of Georgia Athens, Georgia 30602-7229). P. furiosus (Pfu, DSM 3638) cells were produced by growth under anaerobic, reducing conditions at 90 °C in a 600-liter fermenter on maltose and peptides and harvested in the late log phase as described15.
While using clean-room precautions, 100g batches of frozen Pfu cell pellet were suspended in three volumes (w/v) of cold extraction buffer (10 mM Tris HCl, pH 8.0, 25 mM NaCl). The cell suspension was homogenized to a smooth consistency using a Kinematica Polytron PT 2100 for 3 minutes at 11,000 to 15,000 rpm. The suspension was then sonicated 5 times for 2 min at 50% duty Output 8 using Branson Sonifier 450 and the High-Gain 19mm horn, No: 101-147-031. Insoluble material was cleared from the lysate by centrifugation at 14,000 g for 60 minutes at 4 °C. The supernatant was transferred to a new bottle and centrifuged at 14,000 g for an additional 60 minutes at 4 °C. The supernatant was removed and combined with similarly processed supernatants from additional extractions to make one large uniform pool. The protein concentration of the pooled extract was determined by Bradford assay and then adjusted to 10 mg/ml with extraction buffer. Aliquots of 500 μg (50 μl/tube) per vial were prepared, frozen, and lyophilized.
Determination of reconstituted protein concentration: A vial of extract was reconstituted with 50 μl H2O by gentle pipetting. Aliquots of the reconstituted extract from 10 vials were diluted 30-fold and the protein concentration was measured in a Bradford assay. The average of triplicate measurements were then compared to a standard curve generated using dilutions of a protein of known concentration.
SDS-PAGE of proteins in extract: After vial reconstitution with 50 μl H2O by gentle pipetting, one μl (10 μg) of the sample was run on NuPAGE 4-12% or 12% Bis-Tris SDS-PAGE gel using MES buffer (Life Technologies). The gel was stained with Coomassie Brilliant Blue (Sigma B847-1EA).
Sample Preparation and LC-MS Analysis
(a) MudPIT and single phase LC/MS-MS analysis
Proteins were precipitated with trichloroacetic acid (TCA) using the following protocol: to 1 volume of sample solution (cold) add 1/3 volume of 100% (w/v) TCA (6.1N, Sigma), mix well to give a final TCA concentration of 25%, leave on ice for 3 hours. Spin for 30 minutes at 4 °C, aspirate the supernatant, leave 5 ∼ 10 μl in the tube so as not to disturb the pellet. Wash twice with ice-cold acetone (500 μl each). After each wash spin for 10 minutes. The protein pellet was dried either by air or by using a Speedvac for 1 ∼ 2 minutes.
Total proteins (100 μg) were dissolved in 8 μl 2% RapiGest (Waters) stock solution (less than 0.2% final conc.), 8 ul 5× Invitrosol (Invitrogen) and 50 uL 8 M urea (freshly prepared) (Sigma). The mixture was denatured with heat for 5 minutes at 60°C, and then sonicated for 2 hours in a water bath. Protein was then reduced by the addition of 500 mM tris(2-carboxyethyl)phosphine (TCEP) to a final concentration of 5 mM and incubated at room temperature for 20 minutes. Alkylation of reduced Cys residues was performed by adding 500 mM iodoacetamide to a final concentration of 10 mM and the solution was incubated at room temperature for 30 minutes in the dark. The solution was then diluted 2× to produce a urea concentration of 4 M by the addition of an equal volume of 100 mM Tris-HCl, pH 8.5. Trypsin (Promega) was added at ∼1:100 enzyme to substrate ratio (wt:wt) and incubated at 37°C for overnight in the dark. The digestion was terminated by adding 90% formic acid to a final concentration of 5% and the solution was stored at -20°C prior to analysis.
For multidimensional protein identification technology (MudPIT), total peptide mixtures were pressure-loaded onto a biphasic fused-silica capillary column (250 μm i.d.) consisting of 2.5 cm long strong cation exchange (5-μm partisphere, Whatman, Clifton, NJ) and 2.5 cm long reversed phase (Aqua C18, Phenomenex, Torrance, CA), which was prepared by slurry packing using an in-house high pressure vessel. The column was washed with buffer A (see below) for more than 10 column volumes (100 μm i.d.) with a through-hole union (Upchurch Scientific, Oak Harbor, WA). The analytical columns were pulled beforehand by a laser puller (Sutter Instrument Co., Novato, CA) with a 5 μm opening. It was packed with 3 μm reversed phase (Aqua C18, Phenomenex, Torrance, CA) to 15 cm long. The entire column setting (biphasic column-union-analytical column) was placed in line with an Agilent 1200 quaternary HPLC pump (Palo Alto, CA) for mass spectrometry analysis.
The digested proteins were analyzed using a 12-step MudPIT separation method as described previously.11,12 The buffer solutions used were as follows: water/acetonitrile/formic acid (95:5:0.1, v/v/v) as buffer A, water/acetonitrile/formic acid (20:80:0.1, v/v/v) as buffer B, 500 mM ammonium acetate with 5% acetonitrile and 0.1% formic acid as buffer C.
The elution gradient of step 1 was as follows: 10 min of 100% buffer A, a 5-min gradient from 0 to 15% buffer B, a 65-min gradient from 15 to 45% buffer B, a 15-min gradient from 45 to 100% buffer B, and 5 min of 100% buffer B. Step 2-11 had the following profile: 1 min of 100% buffer A, 4 min of X% buffer C, followed by the same gradient as step 1. The 4-min buffer C percentages (X) were 10, 20, 30, 40, 50, 60, 70, 80, 90, and 100%, respectively. The salt pulse for the final step (step 12) was 90% buffer C plus 10% buffer B (450 mM ammonium acetate in 12.5% acetonitrile).
For single phase LC-MS/MS analysis, 5 ug digested proteins were pressure-loaded onto a in-house pulled fused silica capillary (100 μm × 15 cm) packed with Aqua C18 material (RP, Phenomenex, Ventura, CA). A 90-minute linear gradient of buffer B from 0%-100% was applied.
Data-dependent tandem mass spectrometry (MS/MS) analysis was performed with a LTQ linear ion-trap and LTQ-Orbitrap hybrid mass spectrometer (ThermoFisher, San Jose, CA)16. Peptides eluted from the MudPIT column were directly electrosprayed into the mass spectrometer with the application of a distal 2.4-kV spray voltage12. A cycle of one full-scan MS spectrum (m/z 300-2000) was acquired followed by five or ten MS/MS events, sequentially generated on the first to the fifth or tenth most intense ions selected from the full MS spectrum for LTQ or LTQ-Orbitrap respectively. Resolution for Orbitrap analyzer was set at 60,000. All tandem mass spectra were collected using a normalized collision energy of 35% and an isolation window of 3 Da for both CID and ETD. Activation time for ETD was 100 ms. One micro scan was applied for all experiments in the Orbitrap or LTQ. Maximum ion injection times for full scan in Orbitrap and LTQ were 500 ms and 50 ms, respectively, and for MS/MS scan was 100 ms. AGC targets for LTQ and Orbitrap are 3e4 and 5e5 (full scan), 1e4 and 2e5 (MS/MS scan). The dynamic exclusion settings used were as follows: repeat count, 1; exclusion list size, 100; and exclusion duration, 180 second. MS scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFisher).
(b) Directed LC-MS/MS analysis
Both trichloroacetic acid (TCA) and acetone precipitation were done for sample preparation. The lyophilized sample containing 500 μg of total protein was dissolved in 200 μl of denaturation buffer (100 mM ammonium bicarbonate, 8 M urea, 0.1% RapiGest™(Waters)) and the protein solution split into two equal aliquots of 100 μl. 15 μl of a pure TCA solution and 400 μl of ice-cold acetone were added, respectively, to each aliquot and the solutions kept at 0°C (for TCA prep.) and -20°C (for acetone prep.) for 30 minutes. Both samples were centrifuged at 14 000 rpm at 4°C for 15 minutes, washed twice with ice-cold acetone (500 μl each) with spin for 10 minutes after each wash step. Finally, acetone was allowed to evaporate at room temperature.
The protein pellet (100 μg) was resuspended in 25 μl denaturation buffer (100 mM ammonium bicarbonate, 8 M urea, 0.1% RapiGest™(Waters)), sonicated for 5 min and spun down for 5 minutes at 20,000g at 4°C. A small aliquot of the supernatant was taken to determine the protein concentration using BCA assay (Pierce). Proteins in the supernatant were reduced with 5 mM tris(2-carboxyethyl)phosphine (TCEP) for 60 min at 37°C and alkylated with 10 mM iodoacetamide for 30 min in the dark before diluting the sample with 100 mM ammonium bicarbonate to a final urea concentration of 2 M. Proteins were digested by incubation with trypsin (1/50, w/w) over night at 37°C. The peptide solution was acidified with 2M HCl to a final concentration of 50 mM, mixed at 37°C for 30 minutes and the precipitated RapiGest removed by centrifugation at 14,000 rpm for 10 minutes. The peptides in the supernatant were extracted and desalted using C18 reversed-phase spin columns according to the manufacturer's instructions (Macrospin columns, Harvard Apparatus) and dried under vacuum.
For directed mass spectrometry, the setup of the μRPLC-MS system was as described previously17. The hybrid LTQ-FT-Ultra mass spectrometer was interfaced to a nanoelectrospray ion source (both Thermo Electron, Bremen, Germany) coupled online to a Tempo 1D-plus nanoLC (Applied Biosystems/MDS Sciex, Foster City, CA). Peptides were separated on a RP-LC column (75 μm × 15 cm) packed in-house with C18 resin (Magic C18 AQ 3 μm; Michrom BioResources, Auburn, CA, USA) using a linear gradient from 98% solvent A (98% water, 2% acetonitrile, 0.15% formic acid) and 2% solvent B (98% acetonitrile, 2% water, 0.15% formic acid) to 30% solvent B over 120 minutes at a flow rate of 0.3 μl/min. Each survey scan acquired in the ICR-cell at 100,000 FWHM was followed by MS/MS scans of the three most intense precursor ions in the linear ion trap with enabled dynamic exclusion for 30 seconds. Charge state screening was employed to select for ions with at least two charges and rejecting ions with undetermined charge state. The normalized collision energy was set to 32%, and one microscan was acquired for each spectrum.
For directed LC-MS/MS, all MS1 features were extracted from three data-dependent (DDA) LC-MS/MS runs of each sample, respectively, using the SuperHirn peak extraction and alignment algorithm18. After removing all features for which MS2-data were obtained and that could be detected in only one of the three DDA runs, the remaining 13 876 (TCA precipitation sample) and 10 553 (acetone precipitation sample) features were divided by their mass-to-charge (m/z) and charge (z) in three and two lists, respectively (see Supplement Table 2). Subsequently, the MS-parameters were adjusted to prevent triggering MS/MS-scans for precursors that do not fulfill the criteria of the features in the individual mass lists (z and m/z values). The scheduled mass lists were imported into the global mass list present in the MS-method file and activated. Directed MS-sequencing was carried out with the same settings as recently specified and optimized17.
In addition to the common data analysis described below, separate database searches and analyses of tandem mass spectra were performed on this data set. MS/MS spectra were searched against a decoy database (consisting of forward and reverse protein sequences) of the predicted proteome from Pyrococcus furiosus strain DSM 3638 protein NCBI database (NC_003413) containing a total of 4130 FASTA-formatted protein sequences using the SEQUEST search tool19. The search was performed with semi-tryptic cleavage specificity, mass tolerance of 25 ppm, methionine oxidation as variable modification and cysteine carbamidomethylation as a fixed modification. The database search results were post processed using the PeptideProphet20 and ProteinProphet21 program with enabled accurate mass (A) and decoy (-d) option. The peptide and protein probability score thresholds were set to 0.9 and 0.8, respectively, corresponding to a false discovery rate (FDR) below 1%. Detailed FDRs for all probability values calculated by Protein-/PeptideProphet are indicated in Supplement Table 2.
(c) Offline pI-based peptide fractionation
Prior to enzymatic digestion, the Pfu sample was desalted using a rapid chromatographic method using a gradient of 0.1% trifluoroacetic acid (TFA) in water (A) and 0.08%TFA in acetonitrile (B). The Pfu sample was dissolved in 6 M urea containing 1% acetic acid to completely denature the proteins, then 250 μg was loaded on a 4.6 × 100 mm mRP-C18 column (Agilent). After a 3 minute hold at 10%B, the gradient ramped to 70% B at 5 minutes, then 100%B at 6 minutes. After holding at 100%B for 2 minutes, the gradient returned to initial conditions. The fractions eluting between 6 and 9 minutes were combined and dried.
The dried, desalted sample (250 μg protein), was dissolved in 50% 2,2,2-trifluoroethanol (TFE, 99+ % Aldrich) in a 50 mM ammonium bicarbonate buffer with 4 mM DTT, then reduced at 95°C for 20 min. The reduced sample was subsequently alkylated with 16 mM iodoacetamide at room temperature for 1 hour. The unreacted iodoacetamide was quenched by the addition of DTT. After a 1:10 dilution with 25 mM ammonium bicarbonate buffer, trypsin (Agilent Technologies) was added at a 1:50 enzyme:substrate ratio and the sample was incubated overnight at 37°C. The digest was quenched by the addition of formic acid, then dried and stored at -20°C until fractionation.
For pI-based peptide separation, the OFFGEL Fractionator (Agilent Technologies) with a 24-well setup was used22. Fifteen minutes prior to sample loading, 24 cm long IPG gel strips (Immobiline DryStrips; GE Healthcare, Freiburg, Germany) with a linear pH gradient ranging from 3 to 10 were rehydrated in the assembled device with 40 μL of modified focusing buffer (no glycerol, 0.5 % carrier ampholyte mixture) per well. The digested sample (250 μg) was diluted in focusing buffer to a final volume of 3.6 ml and 150 μL of sample was loaded in each well. The sample was focused with a maximum current of 50 μA (typical voltages ranging from 500 V to 4000 V), until 50 kV was reached after 24 hours. The recovered fractions (volumes between 100 μL and 150 μL) were acidified with 5 μL formic acid then dried and stored at -20°C prior to LC-MS/MS analysis.
The OFFGEL electrophoresis fractions were dissolved in 20 μL of 3% acetonitrile in water with 0.1% formic acid. A 2 μL aliquot of each fraction was injected onto an LC-MS system consisting of a 1200 Series HPLC-Chip/MS system interfaced to a 6520 Q-TOF mass spectrometer (Agilent Technologies). The system was equipped with an HPLC-Chip (Agilent Technologies) interface that incorporated a 40 nL enrichment column and a 150 mm × 75 μm analytical column packed with Zorbax 300SB-C18 5 μm particles. Peptides were loaded onto the enrichment column with 97 % solvent A (water with 0.1 % formic acid) and 3 % B (90 % acetonitrile with 0.1 % formic acid) at 4 μL/min. The fractions were injected a total of 5 times. The gradient time was lengthened in two experiments to maximize identifications and those times are shown in parentheses. The elution gradient was as follows: load in 3% B, step to 5%B at 0.1 min, 10%B at 10 min, 45%B at 35 min (85 min or 155 min), then a 5-min gradient to 40%B, a 1-min step to 90%B and held for 4 min, then return to initial conditions and equilibrated for 14 min.
Data-dependent MS/MS acquisition was performed using an acquisition rate of 5 spectra/sec (m/z 300-3200) for MS and 3 spectra/sec (m/z 50-2000) for MS/MS. In each cycle, ten multiply-charged precursors were selected based on abundance and the precursors were excluded after 1 MS/MS for 0.3 min. In the four later experiments (B-D), a prototype data-dependent algorithm (subsequently released in MassHunter Acquisition version B.04) was used that significantly improved the precursor selection and subsequent MS/MS quality. Reference mass correction was done on-the-fly using a single reference mass at m/z 1221.9906.
Database Searching and Analysis of Tandem Mass Spectra
Mass spectra obtained from the various sample fractionation and data acquisition methods were analyzed using a common approach. Full MS and tandem mass spectra were extracted from raw files235 and the tandem mass spectra were searched against a Pyrococcus furiosus protein database containing 2,065 FASTA-formatted protein sequences. These protein sequences were obtained from the file AE009950.ffn found at ftp://ftp.ncbi.nih.gov/genbank/genomes/Bacteria/Pyrococcus_furiosus/ that was accessed from Genbank FTP site link on http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genome&Cmd=ShowDetailView&TermToSearch=228.
In order to accurately estimate peptide probabilities and false discovery rates, we used a double-decoy strategy: first, a Saccharomyces cerevisiae protein database containing 5,873 protein sequences, containing the translations of all systematically named ORFs, downloaded from the Saccharomyces Genome Database (database released on December 16, 2005), and 123 common contaminant proteins was added to the Pyrococcus furiosus database, for a total of 8,061 target database sequences. Second, a reverse decoy database containing the reverse sequences of all the 8,061 proteins was appended to the target database and the ProLuCID algorithm was used to find the best matching sequences from the combined database24,25.
ProLuCID searches were done on an Intel Xeon 80-processor cluster running under the Linux operating system. The peptide mass search tolerance was set to 3 Da for spectra acquired on the LTQ instrument, and 50 ppm for spectra acquired on the hybrid LTQ-Orbitrap and Q-TOF instruments. Since the peak selected for MS/MS analysis by the instrument control software is often not a monoisotopic ion, the search algorithm considers multiple isotopes, with a 50 ppm mass tolerance for each possible theoretical isotope peak. The mass of the amino acid Cysteine was statically modified by +57.02146 Da to take into account carboxyamidomethylation. No variable modifications were considered. No enzymatic cleavage conditions were imposed on the database search, so the search space included all candidate peptides whose theoretical mass fell within the mass tolerance window, regardless of their tryptic status. The data analysis method chosen did not make use of certain features available for high-resolution MS/MS data generated by the Q-TOF instrument, such as the presence of immonium ions. This compromise slightly reduced the number of proteins identified in the Q-TOF data but was necessary to enable a common analysis of all data sets.
The validity of peptide/spectrum matches (PSMs) was assessed8,9 using two search output parameters, the cross-correlation score (XCorr) and normalized difference in cross-correlation scores (DeltaCN)26. For Orbitrap and Q-TOF samples using the accurate precursor mass information, a third scoring parameter was included: DeltaMass, the absolute difference between the experimental precursor ion mass and the nearest theoretical isotope peak. The search results were grouped by charge state (+1, +2, +3, and greater than +3) and tryptic status (fully tryptic, half-tryptic, and non-tryptic), resulting in 12 distinct sub-groups. In each one of these sub-groups, the distribution of XCorr, DeltaCN, and DeltaMass values for (a) direct and (b) decoy database PSMs was obtained, then the direct and decoy subsets were separated by discriminant analysis. Full separation of the direct and decoy PSM subsets is not generally possible; therefore, peptide probabilities were calculated based on a non-parametric fit of the direct and decoy score distributions9. A peptide probability of 90% was set as the minimum threshold. The false discovery rate was calculated as the percentage of reverse decoy PSMs among all the PSMs that passed the 90% probability threshold, and further confirmed by the presence or absence of Saccharomyces cerevisiae PSMs. This procedure was independently performed on each data subset, resulting in a false discovery rate independent of tryptic status or charge state.
In addition, we required that every protein be supported by at least a unique peptide with probability greater than 99%. After this last filtering step, we estimate that both the protein and peptide false discovery rates were reduced to below 0.5%.
Results and Discussion
The goal of this study is to assess the use of Pyrococcus furiosus (Pfu) as a possible complex proteomics standard. The desired sample characteristics are: a) high stability to ensure a consistent supply of the material, b) the right degree of complexity and dynamic range, and c) high reproducibility – in order to provide a good metric for a wide variety of mass spectrometric approaches and different search engines. Pfu fully reconstitutes from its lyophilized state with the addition of water and its proteins were found to be stable during typical handling and storage (Supplementary material). A thermostable organism should be stable at low temperature making long term storage and use of materials possible.
To obtain complete and confident coverage of the Pfu proteome, we analyzed it with different LC-MS methods on three different platforms. They include: (1) a total of 20 MudPIT and single phase experiments, on two mass spectrometer platforms (LTQ and LTQ-Orbitrap), using two different fragmentation methods (CID and ETD); (2) directed LC-MS with two different protein precipitation protocols; and (3) LC-MS/MS analysis of pI-based peptide fractionation.
We identified a total of 1,517 unique Pyrococcus furiosus (Pfu) proteins, at a false discovery rate of less than 0.5% (Supplementary material), from all the experiments performed. For comparison, Table 1 – panel A for single experiments and B for replicate experiments - shows a summary of our results on different platforms and experimental setups. The MudPIT CID experimental setup identified, on average, 1,279 +/− 9 proteins per experiment. In order to obtain the most complete protein identification coverage of the Pfu sample, we repeated MudPIT CID experiments until we reached identification saturation (very few or no additional new proteins were identified with a new experiment)27,28. We performed a total of 15 MudPIT CID experiments in order to be certain of identification saturation – less than 3 additional proteins were identified in each of the last 8 experiments. Preparation of the Pfu sample excluded the insoluble fraction (e.g. membrane proteins) and the large number of proteins identified by each method represents a significant fraction of the soluble proteins. It was found that each method gave highly reproducible run-to-run identification of proteins and there was significant overlap in identifications between the different methods employed. This finding demonstrates that the test sample is complex enough to challenge analytic methods yet it is also simple enough to allow good reproducibility between analyses. Although a high percentage of the likely proteome under the growth conditions used to create the material was found, a challenge for further development is increasing the average sequence coverage of the identified proteins per analysis. Additionally, although we did not test it here, Pfu may make a good standard for the refinement of top-down techniques.
Table 1 (panels A, B).
Panel A: Average numbers of identified Pyrococcus furiosus proteins, peptides, and spectra in four categories of experimental setups and instruments. False discovery rates are estimated to be below 0.5%.
| Number of identified proteins | Number of identified peptides | Number of identified spectra | |
|---|---|---|---|
| Single phase-LTQ | 628 | 3,316 | 6,880 |
| MudPIT-Orbitrap-CID | 1,263 | 14,846 | 102,314 |
| Directed MS/MS (TCA-DDA)a | 932 | 6,085 | 9,880 |
| Directed MS/MS (acetone)a | 624 | 3,343 | 5,433 |
| aDetailed results of directed MS/MS combining the two protein precipitation approaches are listed in Table S2 and Figure S7.. | |||
| Panel B: Average numbers of identified Pyrococcus furiosus proteins, peptides, and spectra in three categories of experimental setups and instruments. In each case the results show averages and standard deviations (in parentheses) over three replicate experiments. False discovery rates are estimated to be below 0.5%. | |||
| Number of identified proteins | Number of identified peptides | Number of identified spectra | |
| MudPIT-LTQ | 1,279 (9) | 11,004 (522) | 105,215 (12,210) |
| MudPIT-Orbitrap-ETD | 1,015 (23) | 7,548 (1,611) | 52,992 (10,370) |
| QTOF-fractionation | 1,024 (47) | 9,328 (962) | 35,492 (7,205) |
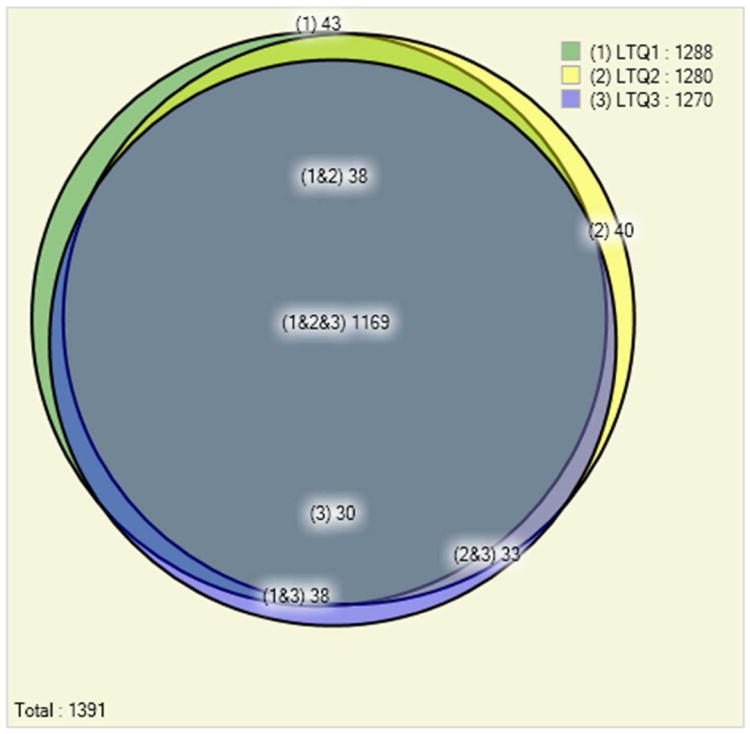
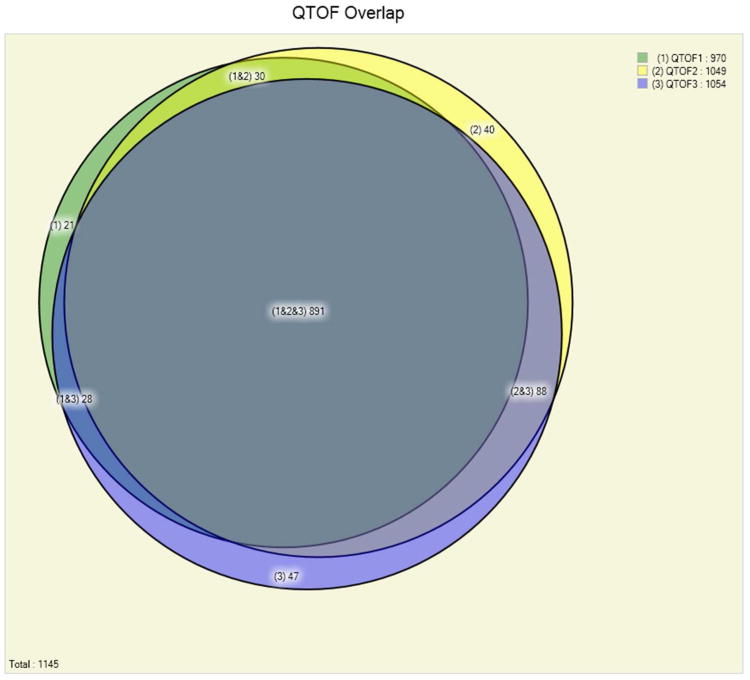
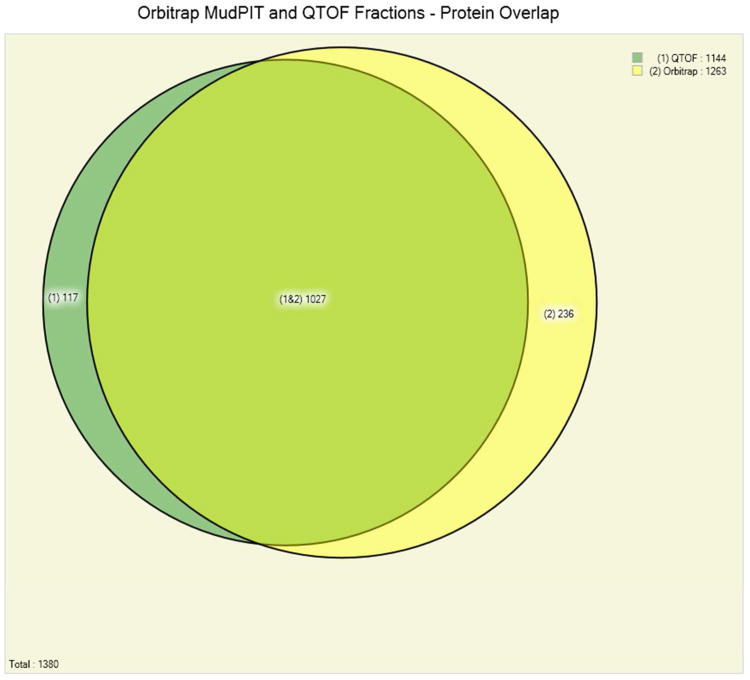
Figure 1 (panels A, B and C) shows the number of identified Pfu protein in three experiments from each of the methods employing extensive fractionation. It demonstrated very high reproducibility of the MudPIT experiments. In three replicate runs on the LTQ mass spectrometer 1,169 or 84% of the proteins are identified in all three replicates, while a further 109 (8%) are identified in at least two of the three replicates (Fig 1, panel A). This is a reflection of the large fraction of identified Pfu proteins in a single run: on average a MudPIT experiment results in the identification of 1,279 out of the total 1,517 Pfu proteins identified, therefore variability between replicate runs is likely to be small. A combination of IEF fractionation using the OFFGEL IEF system coupled with single dimension liquid chromatography and Q-TOF mass spectrometry produced a total of 891 or 78% of the proteins from three replicates. A further 146 (13%) are identified in at least two of the three replicates (Fig 1, panel B). A targeted mass spectrometry approach that avoided the use of extensive fractionation still produced good overlap in the proteins identified but these results consumed much less instrument time (Table 1). Overlap between the peptides observed with each of the methods was less extensive than for proteins (See Figure 2 in Supplementary – 3 panels from Figs 2, 6, 10). Employing electron transfer dissociation (ETD) in place of CID fragmentation resulted in few additional protein identifications (see Figure 3 in Supplementary – 2 panels from Figs 3, 4), although it was expected that ETD should identify more longer peptides. Proteins with post-translational or chemical modifications were found to be present at very low levels (less than 1%), that may due to no efforts were employed to enrich or identify modified peptides.
Figure 1 (Panels A,B,C).
Panel A. Comparison of the number of identified Pyrococcus furiosus proteins in three replicate 12-step MudPIT experiments on an LTQ mass spectrometer at a false discovery rate of below 0.5%. The replicate experiments show high reproducibility of the results: out of the total 1,391 proteins identified 84% (1,169) are identified in each one of the three experiments.
Panel B. Comparison of the number of identified Pyrococcus furiosus proteins in three replicate off-line fractionation experiments on a Q-TOF mass spectrometer at a false discovery rate of below 0.5%. The replicate experiments show high reproducibility of the results: out of the total 1,145 proteins identified 78% (891) are identified in each one of the three experiments.
Panel C. Comparison of the overall number of identified Pyrococcus furiosus proteins in 3 protein fractionation experiments on a Q-TOF mass spectrometer and a 12-step MudPIT experiment on an Orbitrap-LTQ mass spectrometer at a false discovery rate of below 0.5%. 1027 of the 1380 identified proteins (74%) are identified by both methods.
One striking advantage of the Pyrococcus furiosus (Pfu) protein standard is the simplicity of the data analysis. Pfu proteome has few peptides or proteins with high sequence similarity. In the absence of such similarities and redundancies, the protein parsimony analysis is greatly simplified: in a typical MudPIT experiment, out of the average 1,260 proteins with at least one identified peptide sequence, only an average of 4 proteins (less than 0.4%) are discarded due to sequence similarity with other proteins in the database. Furthermore, adding a decoy database from a different organism in the analysis does not introduce additional redundancies: out of the more than 12,000 Pfu peptides sequences identified in a MudPIT experiment, only three were shared sequences with Saccharomyces cerevisiae decoy proteins. Similar results were obtained for other decoy database choices (Homo sapiens, Escherichia coli). Table 2 compares the redundancy of the Pfu peptides to that seen in other common model organisms. Panel A compares the percentage of unique and shared peptides identified in a typical Pfu experiment versus a typical S. cerevisiae experiment. In the Pfu experiments, only 0.9% of the identified peptides are shared among two or more proteins. In contrast, in the S. cerevisiae experiment, 10.4% of the identified peptides are shared among multiple proteins. Panel B shows the number of decoy organism proteins (S. cerevisiae, E. coli and H. sapiens) that have common peptide sequences with identified Pfu proteins. The largely unique peptides derived from Pfu make it an ideal candidate for a proteomic standard. In instances of inter-experimental carryover the standard is not likely to be confused with materials originating from another sample.
Table 2 (panels A, B).
Panel A: Comparison of the percentage of shared peptides observed in a P. furiosus experiment and an S.cerevisiae experiment. Shared peptides are defined as identified peptides whose amino acid sequence is shared by two or more proteins in the database. The P. furiosus data set shows a level of redundancy that is an order of magnitude less than that of S. cerevisiae (0.9% vs. 10.4% shared peptides).
| Organism | Identified peptides | Unique peptides | Shared peptides | Percent shared |
|---|---|---|---|---|
| P. furiosus | 12,251 | 12,136 | 115 | 0.9% |
| S. cerevisiae | 7,786 | 6,977 | 809 | 10.4% |
| Panel B: Results of a P. furiosus proteomics experiment analyzed with a concatenated protein database formed by adding a decoy organism protein database to that of P. furiosus. Regardless of the decoy protein database added we identify 1,260 unique P. furiosus proteins and 4 redundant P. furiosus proteins. In addition a very small number of identified peptides (1 to 3, depending on the decoy database) have amino acid sequences shared with decoy organism proteins. | |||
| Decoy Organism | Unique P. furiosus peptides | Redundant P. furiosus peptides | Decoy organism proteins |
| S. cerevisiae | 1,260 | 4 | 3 |
| E. coil | 1,260 | 4 | 1 |
| H. sapiens | 1,260 | 4 | 1 |
Conclusion
We have identified a total of 1,517 proteins in the soluble fraction of Pyrococcus furiosus (Pfu) out of 2065 total ORFs, excluding membrane proteins, which are estimated to cover about 25% of the ORFs. Pfu can be readily grown in large quantities and potentially stored for a long period of time with minimal degradation given it's thermostability. The proteome is of moderate complexity with 2025 proteins predicted from the genome and there is very little duplication of sequence within the genome simplifying proteomic data analysis. These factors make the Pfu proteome an excellent resource to benchmark LC-MS platforms and proteomic workflows.
Supplementary Material
Acknowledgments
Catherine CL Wong and John R Yates acknowledge funding from NIH grants P41 RR011823/GM103533, R01 MH067880-09, P01 AG031097-03, R01 HL079442-07.
Footnotes
Notes: The authors declare no competing financial interest
References
- 1.Robb FT, Maeder DL, Brown JR, DiRuggiero J, Stump MD, Yeh RK, Weiss RB, Dunn DM. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 2001;330:134–57. doi: 10.1016/s0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- 2.Lim H, Eng J, Yates JR, 3rd, Tollaksen SL, Giometti CS, Holden JF, Adams MW, Reich CI, Olsen GJ, Hays LG. Identification of 2D-gel proteins: a comparison of MALDI/TOF peptide mass mapping to mu LC-ESI tandem mass spectrometry. J Am Soc Mass Spectrom. 2003;14:957–70. doi: 10.1016/s1044-0305(03)00144-2. [DOI] [PubMed] [Google Scholar]
- 3.Holden JF, Poole FL, Jr, Tollaksen SL, Giometti CS, Lim H, Yates JR, III, Adams MWW. Identification of membrane proteins in the hyperthermophilic archaeon Pyrococcus furiosus using proteomics and prediction programs. Comp Funct Genom. 2001;2:275–88. doi: 10.1002/cfg.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee AM, Sevinsky JR, Bundy JL, Grunden AM, Stephenson JL., Jr Proteomics of Pyrococcus furiosus, a hyperthermophilic archaeon refractory to traditional methods. J Proteome Res. 2009;8:3844–51. doi: 10.1021/pr801119h. [DOI] [PubMed] [Google Scholar]
- 5.McCormack AL, Eng JK, Yates IJR. Peptide sequence analysis on quadrupole mass spectrometers. Methods: A companion to Methods in Enzymology. 1994;6:274–273. [Google Scholar]
- 6.McCormack AL, Schieltz DM, Goode B, Yang S, Barnes G, Drubin D, Yates IJR. Direct analysis and identification of proteins in mixtures by LC-MS/MS and database searching at the low-femtomole level. Analytical Chemistry. 1997;69:767–776. doi: 10.1021/ac960799q. [DOI] [PubMed] [Google Scholar]
- 7.Eng J, McCormack A, Yates J. An Approach to Correlate Tandem Mass Spectral Data of Peptides with Amino Acid Sequences in a Protein Database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 8.Purvine S, Picone AF, Kolker E. Standard mixtures for proteome studies. OMICS. 2004;8:79–92. doi: 10.1089/153623104773547507. [DOI] [PubMed] [Google Scholar]
- 9.Keller A, Purvine S, Nesvizhskii AI, Stolyar S, Goodlett DR, Kolker E. Experimental protein mixture for validating tandem mass spectral analysis. OMICS. 2002;6:207–12. doi: 10.1089/153623102760092805. [DOI] [PubMed] [Google Scholar]
- 10.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJ. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods. 2009;6:423–30. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 12.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnology. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 13.Kurian Ponnamma, G DM, Suhocki Paul V, Gary Ledley. The use of bacteriophage T4 as a set of molecular weight and isoelectric point markers for two-dimensional electrophoresis. Electrophoresis. 2005;2:184–187. [Google Scholar]
- 14.Kolker E, Picone AF, Galperin MY, Romine MF, Higdon R, Makarova KS, Kolker N, Anderson GA, Qiu X, Auberry KJ, Babnigg G, Beliaev AS, Edlefsen P, Elias DA, Gorby YA, Holzman T, Klappenbach JA, Konstantinidis KT, Land ML, Lipton MS, McCue LA, Monroe M, Pasa-Tolic L, Pinchuk G, Purvine S, Serres MH, Tsapin S, Zakrajsek BA, Zhu W, Zhou J, Larimer FW, Lawrence CE, Riley M, Collart FR, Yates JR, 3rd, Smith RD, Giometti CS, Nealson KH, Fredrickson JK, Tiedje JM. Global profiling of Shewanella oneidensis MR-1: expression of hypothetical genes and improved functional annotations. Proc Natl Acad Sci U S A. 2005;102:2099–104. doi: 10.1073/pnas.0409111102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Verhagen MF, Menon AL, Schut GJ, Adams MW. Pyrococcus furiosus: large-scale cultivation and enzyme purification. Methods Enzymol. 2001;330:25–30. doi: 10.1016/s0076-6879(01)30368-3. [DOI] [PubMed] [Google Scholar]
- 16.Yates JR, Cociorva D, Liao L, Zabrouskov V. Performance of a linear ion trap-Orbitrap hybrid for peptide analysis. Anal Chem. 2006;78:493–500. doi: 10.1021/ac0514624. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt A, Gehlenborg N, Bodenmiller B, Mueller LN, Campbell D, Mueller M, Aebersold R, Domon B. An Integrated, Directed Mass Spectrometric Approach for In-depth Characterization of Complex Peptide Mixtures. Mol Cell Proteomics. 2008;7(11):2138–50. doi: 10.1074/mcp.M700498-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mueller LN, Rinner O, Schmidt A, Letarte S, Bodenmiller B, Brusniak MY, Vitek O, Aebersold R, Muller M. SuperHirn - a Novel Tool for High Resolution LC-MS-based Peptide/Protein Profiling. Proteomics. 2007;7(19):3470–80. doi: 10.1002/pmic.200700057. [DOI] [PubMed] [Google Scholar]
- 19.Yates JR, 3rd, Eng JK, McCormack AL, Schieltz D. Method to Correlate Tandem Mass Spectra of Modified Peptides to Amino Acid Sequences in the Protein Database. Anal Chem. 1995;67(8):1426–36. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- 20.Keller A, Nesvizhskii Al, Kolder E, Aebersold R. Empirical Statistical Model to Estimate the Accuracy of Peptide Identification Made by MS/MS and Database Search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 21.Nesvizhskii Al, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Specrometry. Anal Chem. 2002;75(17):4646–58. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 22.Hoerth P, Miller CA, Preckel T, Wenz C. Efficient Fractionation and Improved Protein Identification by Peptide OFFGEL Electrophoresis. Mol Cell Proteomics. 2006;5:1968–1974. doi: 10.1074/mcp.T600037-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., 3rd MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18:2162–8. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- 24.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 25.Xu T, V J, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR., III ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Molecular Cellular Proteomics. 2006;5:S174–S174. 671 Suppl. S. [Google Scholar]
- 26.Cociorva D, T DL, Yates JR., III Validation of Tandem Mass Spectrometry Database Search Results Using DTASelect. Current Protocols in Bioinformatics. 2007;Chapter 13 doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- 27.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 28.Durr E, Yu J, Krasinska KM, Carver LA, Yates JR, Testa JE, Oh P, Schnitzer JE. Direct proteomic mapping of the lung microvascular endothelial cell surface in vivo and in cell culture. Nat Biotechnol. 2004;22:985–92. doi: 10.1038/nbt993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.