Abstract
Stable isotope labeling via isobaric derivatization of peptides is a universally applicable approach that enables concurrent identification and quantification of proteins in different samples using tandem mass spectrometry. In this study, we evaluated the performance of amine-reactive isobaric Tandem Mass Tag (TMT), available as duplex and sixplex sets, with regard to their ability to elucidate protein expression changes. Using rat brain tissue from two different developmental time points, postnatal day 1 (p1) and 45 (p45), as a model system, we compared the protein expression ratios (p45/p1) observed using duplex TMT tags in triplicate measurements versus sixplex tag in a single LC-MS/MS analysis. A correlation of 0.79 in relative protein abundance was observed in the proteins quantified by these two sets of reagents. However, more proteins passed the criteria for significant fold change (-1.0 ≤ log2 ratio (p45/p1) ≥ +1.0 and p < 0.05) in the sixplex analysis. Nevertheless, in both methods most proteins showing significant fold change were identified by multiple spectra, increasing their quantification precision. Additionally, the fold change in p45 rats against p1, observed in TMT experiments, was corroborated by a metabolic labeling strategy where relative quantification of differentially expressed proteins was obtained using 15N-labeled p45 rats as an internal standard.
Keywords: tandem mass tag, TMT, isobaric tag, metabolic labeling, SILAM, mass spectrometry, quantitative proteomics, MudPIT
Introduction
Mass spectrometry (MS) – based protein quantification has emerged as a powerful technology for proteome wide quantitative profiling of differentially regulated proteins. One of the most popular methods for quantification is the isotopic labeling of biological samples prior to MS analysis. Stable isotope labeling enables multiple biological samples to be analyzed under the same chromatography and MS conditions. The samples can be labeled with isotopologues either at the protein level or peptide level and can be categorized into two main groups based on the method of quantification: mass-shift labels and isobaric labels. In mass-shift label-based quantification, isotopically labeled peptides show distinct peaks with a fixed mass difference in an MS1 spectra, and quantification can be achieved either by using the peak intensity of the precursor ions or area under the chromatographic peak to reflect the abundance of the peptide. This is the basis of quantification for several popular techniques, such as stable isotope labeling in mammals (SILAM)1, stable isotope labeling with amino acids in cell culture (SILAC),2, 3 isotope-coded affinity tags (ICAT),4 dimethyl labeling5 and others.6 On the other hand, in the isobaric labeling approach peptides labeled with isotopic variants of the labels have similar masses and appear as a single composite peak in an MS1 scan. Following fragmentation, the different tags produce unique reporter ions that can be detected and quantified in an MS/MS scan. Both peptide identification and quantification are derived from the same MS/MS spectrum. MS/MS-based quantitation is determined by the relative intensities of fragment peaks at fixed m/z values within an MS/MS spectrum. The isobaric labeling methods include family of reagents referred to as ‘isobaric mass tags’ such as isobaric tag for relative and absolute quantification (iTRAQ)7 and Tandem Mass Tag (TMT).8
All isobaric reagents contain three functional groups: a reporter ion group, a mass normalization group, and an amine-reactive group. The amine reactive group specifically reacts with N-terminal amine groups and ε-amine groups of lysine residues to attach the isobaric tags to peptides. The amine specificity of these reagents makes most peptides in the sample amenable to this labeling strategy; therefore nearly all peptides can be used for quantification. The mass normalization groups balance the mass difference among the reporter ion groups so that different isotopic variants of the tag have the same mass. After differential labeling, the labeled peptides are mixed and the resultant mixture gives rise to a set of single unresolved additive precursor ions in MS1 where the signal from the same peptide with different labels is summed, providing a moderate increase in sensitivity.9 During MS/MS analysis, the mass-balancing carbonyl moiety is released as a neutral loss, thereby liberating a singly charged reporter ion. The reporter ions from the TMT labels appear in the low mass range, distinct from peptide fragment peaks, while the remainder of the sequence informative b- and y-ions remains as additive isobaric signals. The summed intensity of b- and y- sequence ions aids sensitivity and the intensities of the reporter ion peaks are used as a surrogate measurement for the abundance of the peptide (and thus protein) between the different labeled samples. The peptide fragment peaks in the mass spectrum are matched to a database in order to identify the parent protein.
Isobaric tagging is a universally applicable approach, where proteins are independently isolated from samples of interest before labeling and pooling for downstream analysis. This allows direct ratiometric comparison of relative abundance of identified protein species among multiplexed samples. The isobaric tags are available as: duplex and sixplex in the case of TMT (Thermo Scientific) and fourplex and eightplex in the case of iTRAQ (AB Sciex). The duplex and sixplex TMT label reagents share chemical structure, but differ in the number of incorporated heavy isotopes.10 In TMT duplex labeling approach, derivatization by the two isobaric chemical labels enables a direct comparison of two samples in a single MS analysis. Labeling of a tryptic peptide that has a lysine at the C-terminus with TMT duplex leads to a mass shift of + 450.3 Th due to the incorporation of TMT labels at N-terminal α-amino group of the peptide and at the ε-amino group of the C-terminal lysine. The shift is + 458.3 for TMT sixplex. The sixplex labeling scheme allows up to six samples to be compared in a single MS run, thereby increasing experimental throughput for protein quantitation.
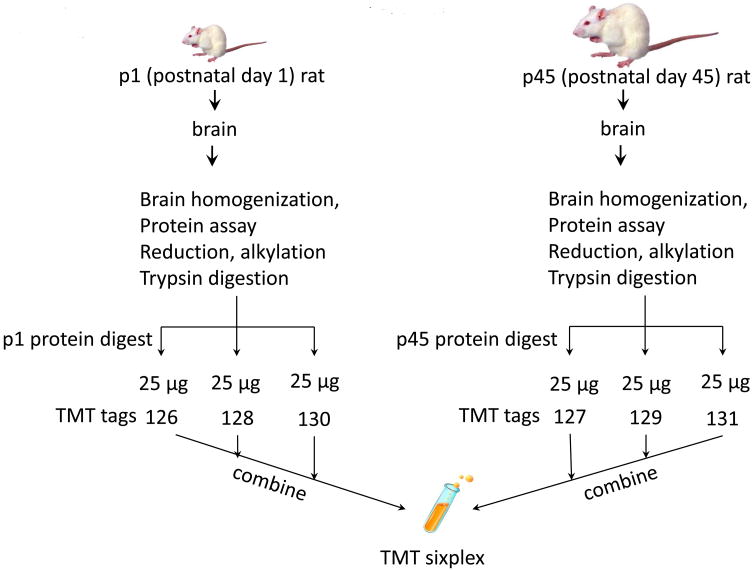
In this study we investigated ratiometric protein expression observed in a single sixplex versus a triplicate duplex TMT experiment. With the sixplex reagent triplicate measurements will be acquired within each tandem mass spectrum and with the duplex reagents three separate measurements are acquired. For the quantitative protein expression profiling by these two sets of TMT tags, we used protein extracts from rat brain at two different time points: postnatal day 1 (p1) and 45 (p45). The duplex and sixplex have nominal reporter ion masses at m/z 126-127 or 126-131. The measurements were carried out using the same p1 and p45 stock samples that were split after tryptic digestion. For the sixplex experiment, the p1 sample was split and equimolar amounts were labeled with three different channel reagent (m/z 126, 128, and 130) of sixplex reagents. Likewise, p45 was split in equal parts and labeled with the remaining three different channel reagent (m/z 127, 129, and 131) of sixplex reagents. Subsequently, all six channels of sixplex were mixed and analyzed on a LTQ Orbitrap Velos using the MudPIT method.11 For the duplex experiment, the p1 sample was labeled with one of the duplex tag (m/z 126) and p45 was labeled with the other tag (m/z 127). The derivatized p1 and p45 samples in duplex experiment were pooled and divided into four aliquots (each one containing a total of 100 micrograms of proteins) and three were used for MudPIT analysis, thus providing three technical replicates. The p45/p1 quantitative profile of proteins commonly observed in the sixplex and duplex experiment showed a modest correlation of R2 = 0.79 between them.
Experimental Section
Isolation of rat brains
Sprague-Dawley rats were used for this study. The rats were maintained in a temperature-controlled (23 °C) facility with a 12-h light/dark cycle and provided ad libitum food and water. On post-natal day 1 and 45, the pups were subjected to halothane by inhalation until unresponsive at the same time of day, and the brains were quickly removed and frozen with liquid nitrogen and stored at −80 °C. All methods involving animals were approved by the Institutional Animal Research Committee and accredited by the American Association for Accreditation of Laboratory Animal Care.
Sample preparation
The p1 and p45 rat brain cortices were homogenized in the ice-cold buffer (1 g of tissue/10 mL of buffer) containing 4 mM HEPES, pH 7.4, 0.32 M sucrose and Protease Inhibitor Cocktail tablet (Roche, Indianapolis, IN) in a Teflon hand-held dounce grinder. The protein concentration was determined with a BCA protein assay (Sigma-Aldrich, St. Louis, MO). Five hundred micrograms of proteins from each animal were methanol/chloroform/water precipitated. During TMT-based quantitation, equal amounts of proteins are usually aliquoted from parallel samples and subjected to reduction, alkylation and digestion steps prior to labeling. The precipitation of proteins becomes necessary to exchange the buffer used during cell or tissue lysis that may contain primary amines and hence incompatible with TMT labeling reaction. The resolubilization of precipitated proteins can be difficult and poor recovery of proteins might negatively affect MS based quantitation. Unlike TMT manual protocol where protein precipitation is performed after cysteine reduction and alkylation step and resolubilization of acetone-precipitated pellet in TEAB buffer, we performed protein precipitation prior to cysteine modification step and SDS was used to achieve efficient dissolution of the protein pellets. The precipitate from each tissue was dissolved in 0.1 M triethyl ammonium bicarbonate (TEAB) dissolution buffer and SDS, and subsequently denatured and alkylated. The precipitation procedures rarely yield 100% recoveries; however, methanol/chloroform precipitation method results in reduced protein loss with several advantages when compared with other precipitation methods.12 The sample was diluted with water prior to trypsin addition, yielding a final SDS concentration of ≤ 0.05% (wt/vol) during digestion (concentrations > 0.1% (w/v) will inhibit trypsin activity).13 A vial of trypsin (Promega, Madison, WI) was reconstituted in 40 μl Milli-Q water and added to each sample (1:25, w/w) and the trypsin digestion was carried out at 37 °C overnight.
TMT labeling
The TMT labeling was performed according to the manufacturer's instructions (Thermo Fisher Scientific, Rockford, IL) with some modifications. The TMT reagents (0.8 mg) were dissolved in 100 μl of anhydrous acetonitrile. For the sixplex experiment, each of the labeling reaction mixtures contained 25 μl of the TMT reagent and 75 μl (50 μg) of the tryptic digest in TEAB buffer to ensure that the organic (acetonitrile) content was between 25-30% (v/v) for the reagent's stability.14 Aliquots of the p1 sample tryptic digest were derivatized with sixplex chemical labels 126, 128 and 130 Th (Thomson), while labels 127, 129 and 131 Th were added to the p45 sample tryptic digest aliquoted in three different tubes. For duplex experiment, 200 μg of the tryptic digest each from p1 and p45 were reacted with 126 and 127 Th TMT reagents, respectively. Figure 1 illustrates schematically the workflow used to implement this strategy. After the labeling, reaction mixtures were incubated at room temperature for 1 h, 15 μl of 5% hydroxylamine solution in water was added to quench the labeling reaction. Each TMT-modified digest from sixplex was then combined into one sample and vacuum dried. Similarly, duplex samples were pooled into one sample and vacuum dried. The lyophilized TMT-labeled peptides were reconstituted with 1 mL of buffer A (0.1% formic acid (FA), 5% acetonitrile (ACN) in water) centrifuged at 14,000 rpm for 30 minutes to remove particulates prior to loading into the MudPIT trapping column. For sixplex, a single MS run was performed by loading 150 μg of the TMT-modified digest into the MudPIT column. Three MS runs, comprising three technical replicates, of 100 μg modified peptides (mixed in 1:1 ratio) were performed for the duplex experiment. With sixplex, the equivalent of three technical replicates (“triple- duplex”) was performed in one MS run since six TMT tags were used. From our experience we have observed that the total amount of protein loading on the biphasic MudPIT trap column does not affect the ratio of each TMT channel.
Figure 1.
Schematic diagram showing the design workflow used for the TMT experiments. The protein extracts from rat brain at two different postnatal developmental time points, p1 and p45, were digested using trypsin, and peptides were labeled with sixplex or duplex TMT reagents. Labeled peptides were combined and subjected to MudPIT analysis in LTQ Orbitrap Velos.
Multidimensional Protein Identification Technology (MudPIT) Analysis
MS analysis of TMT sixplex and duplex samples was performed using MudPIT technology. Capillary columns were prepared in-house from particle slurries in methanol. An analytical RPLC column was generated by pulling a 100 μm ID/360 μm OD capillary (Polymicro Technologies, Inc., Phoenix, AZ) to 3 μm ID tip. The pulled column was packed with reverse phase particles (Jupiter C18, 4 μm dia., 90 Å pores, Phenomenex, Torrance, CA) until 15 cm long. A MudPIT trapping column was prepared by creating a Kasil frit at one end of an undeactivated 250 μmID/360 μm OD capillary (Agilent Technologies, Inc., Santa Clara, CA), which was then successively packed with 2.5 cm strong cation exchange particles (Luna SCX, 5 μm dia., 100 Å pores, Phenomenex, Torrance, CA) and 2.5 cm reverse phase particles (Jupiter C18, 10 μm dia., 90 Å pores, Phenomenex, Torrance, CA). The MudPIT trapping column was equilibrated using buffer A prior to sample loading. After sample loading and prior to MS analysis, the resin-bound peptides were desalted with 1 mL of buffer A by letting it flow through the biphasic trap column. MudPIT and analytical columns were assembled using a zero-dead volume union (Upchurch Scientific, Oak Harbor, WA).
LC-MS/MS analysis was performed on LTQ Orbitrap Velos (Thermo Scientific, San Jose, CA, USA) interfaced at the front end with a quaternary HP 1100 series HPLC pump (Agilent Technology, Santa Clara, CA, USA) using an in-house built electrospray stage. Electrospray was performed directly from the analytical column by applying the ESI voltage at a tee (150 μm ID, Upchurch Scientific) directly downstream of a 1:1000 split flow used to reduce the flow rate to 250 nL/min through the columns. A fully automated 11-step MudPIT run was performed on each sample using a three mobile phase system consisting of buffer A (5% acetonitrile (ACN); 0.1% formic acid (FA) (Sigma Aldrich, San Louis, MO, USA), buffer B (80% ACN, 0.1% FA), and buffer C (500 mM ammonium acetate, 5% ACN, 0.1% FA). The first step was 60 min, whereas subsequent steps were 120 min each. Each MudPIT run includes steps with 0, 10, 20, 30, 40, 50, 60, 70, 80 and 100% buffer C run for 4 min at the beginning of the gradient, except step #10-11 which include salt bump of 90% buffer C with 10% buffer B for 4 min. A schematic representation of the gradient profile is reported in Supplemental Table1 (Supporting Information).
As peptides were eluted from the microcapillary column, they were electrosprayed directly into a mass spectrometer with the application of a distal 2.4 kV spray voltage. Peptides were analyzed using a Top-10 data-dependent acquisition method in which fragmentation spectra are acquired for the top ten peptide ions above a predetermined signal threshold. For each cycle, survey full–scan MS spectra (m/z 300-1600) were acquired in the Orbitrap with a mass resolution of 30,000 at m/z 400 with an automatic gain control (AGC) target of 1×106 ions and the maximal injection time of 250 ms. Each full scan was followed by the selection of the most intense ions, up to 10, for higher-energy collisional dissociation (HCD)-MS/MS analysis in the Orbitrap. The HCD dissociation mode enables simultaneous production of TMT reporter ions and fragment ions of the peptides. In all cases, one microscan was recorded. MS/MS scans were acquired in the Orbitrap with a mass resolution of 7500. The target value was 30,000 ions with injection time of 150 ms. Once analyzed, the selected peptide ions were dynamically excluded from further analysis for 120 s to allow for the selection of lower-abundance ions for subsequent fragmentation and detection using the setting for repeat count = 1, repeat duration = 30 ms and exclusion list size = 500. Ions with singly or unassigned charge states were rejected. The minimum MS signal for triggering MS/MS was set to 5000 and an activation time of 0.1 ms were used. The m/z isolation width for MS/MS fragmentation was set to 2 Th. For MS/MS, precursor ions were activated using 45% normalized collision energy.
Protein identification and Database searches
Tandem mass spectra were extracted from the Xcalibur data system format (raw) into MS2 format using RawXtract1.9.9.2. The MS/MS spectra were searched with the ProLuCID15 algorithm against the EBI rat IPI database (ftp://ftp.ebi.ac.uk/pub/databases/IPI/, version 3.71, release date March 24, 2010) that was concatenated to a decoy database in which the sequence for each entry in the original database was reversed. The search parameters include 10 ppm peptide precursor mass tolerance, 0.6 Da for the fragment mass tolerance, static cysteine modification of 57.02146 amu and static N-terminus and lysine modification of 229.1629 for sixplex and 225.1558 for duplex TMT labels. The search space also included all fully– and semi–tryptic peptide candidates of length of at least six amino acids. Maximum number of internal miscleavages was kept unlimited, thereby allowing all cleavage points for consideration. The ProLuCID outputs were assembled and filtered using the DTASelect2.016 program that groups related spectra by protein and removes those that do not pass basic data-quality criteria. DTASelect2.0 combines XCorr and DeltaCN measurements using a quadratic discriminant function to compute a confidence score to achieve a user-specified false discovery rate. The estimated false positive rate was kept at about 1% at the protein level.
Census,17 a software tool for quantitative proteomic analysis, was used to extract the relative intensities of reporter ions for each peptide from the identified tandem mass spectra for normalization. The mass tolerance and intensity threshold for the reporter ions in Census were set at 0.05 Da and 6000, respectively.
Data normalization
Similar to other high–throughput experiments, TMT experiments are also affected by numerous factors that can lead to unwanted, random or systematic (non-biological) variation. To obtain reliable results, the collected data have to be normalized and analyzed using proper statistical methods.
The proteins are normalized based on the assumption that total intensity remains the same for each of the six tags in sixplex or for either of the two tags in duplex experiments. The rationale for this choice is based on the assumption that the protein amount from six or two samples is the same. After normalization, the relative quantification for p45/p1 in sixplex experiment was derived from the ratio of the reporter ions over the average of the reporter ions (126, 128 and 130) corresponding to the p1 tryptic peptides. For the duplex experiment, the relative quantification was based on the ratio of the reporter ion (127) corresponding to the p45 tryptic peptides, over the ratio of the reporter ion (126) corresponding to the p1 tryptic peptides, in the same replicate experiments. For sixplex, proteins whose p45/p1 ratios were observed in at least two of the three technical replicates were included, whereas for duplex, proteins whose p45/p1 ratios were observed in at least two of the three replicate MudPIT runs are included in further statistical analysis of the data. Student's t-test was employed to evaluate the significance of observed protein changes. A significantly increased level of proteins in p45 sample requires p45/p1 ratio is ≥ 1.0 in log2 scale with p <0.05. The down-regulated proteins in p45 or alternatively, enriched proteins in p1 samples had p45/p1 ratio ≤ -1.0 in log2 scale (p < 0.05). The statistical computing and graphics were performed in R software environment.
Results
In this study, we evaluated strategies of using duplex TMT tags in triplicate measurements versus the use of sixplex tags to label a triplicate sample. In addition, we determined whether the choice of duplex or sixplex TMT tags influences the resulting protein abundance ratios observed in isobaric labeling experiments. Using rat brain homogenates from p1 and p45 rats, we performed one sixplex (each condition labeled with 3 distinct tags) and triplicate duplex (each condition labeled with one of the two tags) MudPIT runs in the LTQ Orbitrap Velos. The sixplex can be considered as triple-duplex experiment which allowed three analytical replicates to be run together in a mass spectrometer. In the sixplex experiment, the tryptic peptides from p1 rat brain sample were labeled in triplicate using m/z 126, 128 and 130 isobaric TMT tags, while the peptides from p45 rat brain sample were labeled in triplicate using m/z 127, 129 and 131 isobaric TMT tags. The six labeled peptide pools were mixed in a 1:1:1:1:1:1 ratio. In the duplex experiment, the tryptic peptides from a p1 sample were labeled with m/z 126 while the p45 tryptic digest was labeled with m/z 127 isobaric TMT tags. The schematic representation of the experimental design is depicted in Figure 1. After lysis and tryptic digestion, peptides were derivatized with TMT reagent. TMT reagents derivatize primary amine groups; hence they tag virtually all proteins/peptides except those lacking both lysine and reactive N-terminal amino acids. To determine protein quantification accurately, it is imperative that all the peptides should be fully labeled.9, 14 In order to test the labeling efficiency of TMT reagents for our samples, the database searching on ProLuCID was carried out by setting TMT tags as a variable modification, an approach that has been reported previously18, on either N-terminal amine or lysine residue. This is different from the standard database searching criteria for TMT-modified peptides, in which TMT tag is set as a fixed modification. With a variable modification during a search, both labeled and unlabeled peptides can be identified and used to calculate the labeling efficiency, which is defined as the percent of labeled peptides among all identified peptides. The TMT labeling efficiency was very high; about 99.0% of lysine and >96.0% of N–terminal amines were found modified, both for duplex and sixplex tags (Table 1).
Table 1.
The labeling efficiency of sixplex and duplex TMT tags at N-terminal α-amine group and ε-amine group of lysine in the tryptic peptides.
| Sample | Percent Labeling | Standard Deviation (3 repeat injections) | ||
|---|---|---|---|---|
| N-terminus modification | Lysine Modification | N-terminus modification | Lysine Modification | |
| Sixplex* | 96.6 | 99.2 | NA | NA |
| Duplex | 97.3 | 99.1 | 0.60 | 0.03 |
Only single LC-MS run
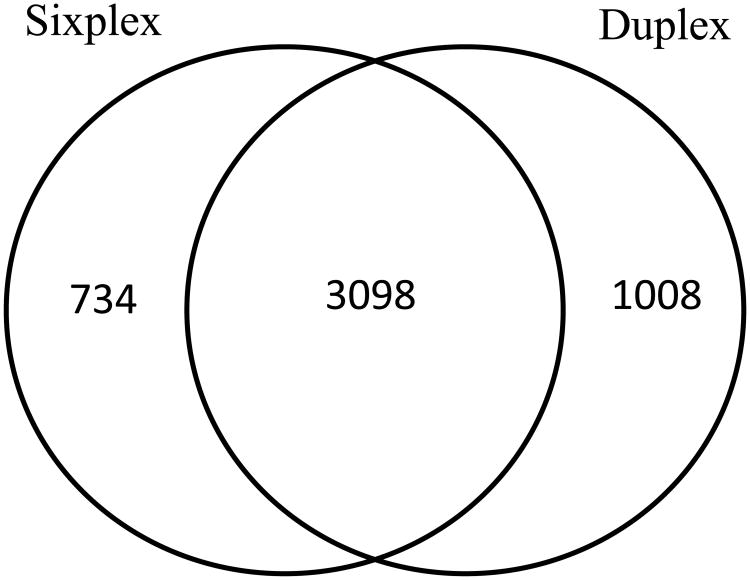
The relative intensities of the reporter ions depict the relative concentration of the peptides in the sample. In order to acquire quality spectra; we used an ion selection threshold of 5000 counts during data acquisition in MS/MS mode. Since, fluctuation of peptide ratios is greater in peptides with low reporter ion intensities,19, 20 to exclude low intensity TMT reporter ion measurements, the intensity threshold was set to 6000 during post-acquisition filtering of data in Census. The percentage of quantified proteins (out of total identified) obtained is very high both for sixplex and duplex TMT experiments, which makes sense since both quantification and identification information originates from the same MS/MS spectrum. Table 2 shows the number of identified proteins in sixplex and triplicate duplex runs along with the number and percentage of quantified proteins. The Venn diagram, presented in Figure 2, demonstrates the common and unique proteins quantified in the sixplex and duplex analyses. It shows that 3098 proteins were commonly identified in sixplex and duplex experiments. In addition, 734 and 1008 proteins were exclusively identified in sixplex and duplex experiments, respectively.
Table 2. Summary of the total proteins that were identified and quantified in sixplex and duplex TMT experiments.
| TMT experiments | Identified Proteins | Quantified Proteins | % of proteins quantified | Differentially expressed proteins (-1.0 ≤ log2 p45/p1 ratio ≥ +1.0, p < 0.05) |
|---|---|---|---|---|
| Sixplex | 3996 | 3832 | 96% | 886 |
| Duplex*: | ||||
| Replicate 1 | 4429 | |||
| Replicate 2 | 4092 | |||
| Replicate 3 | 4174 | |||
| Duplex Avg. | 4232 | 4106 | 97% | 562 |
3 MudPIT runs (twice as much total material loaded than Sixplex)
Figure 2.
Venn diagram depicting the overlapped and unique proteins quantified by the sixplex and duplex TMT labeling experiments.
A distribution of proteins in p1 or p45 sample, for sixplex experiment, in accordance to their reporter ion (128/126, 130/126, and 130/128 for p1 and 129/127, 131/127, and 131/129 for p45) ratiometric expression is depicted in Supplementary Figure 1a. The theoretical quantitative ratio of any two labeled p1 sample (126, 128 and 130) and p45 sample (127, 129 and 131) should be approx. 1, since they are analytical replicates. However, during sample handling and labeling variability are introduced, hence, not all relative protein ratios are likely to agree with the theoretical value. Specifically, for the mixing ratios of 1:1 for p1 or p45 samples, the observed ratios reveal that 93% of proteins are within ± 0.25 of theoretical ratio of 1:1 for p1 samples and 95% of proteins are within ± 0.25 of theoretical ratio of 1:1 for p45 samples in the sixplex TMT experiment. The boxplot in Supplementary Fig 1b shows the spread of technical replicates of the three p1/p1 and three p45/p45 ratios observed in the TMT sixplex experiment. Since the samples were split after tryptic digestion and before labeling, the narrow dispersion of these log2 ratios from 1:1 in case of p1:p1 and p45:p45 indicates the good reproducibility in sample handling, labeling and analysis. This also demonstrates the quantification accuracy since majority of p1/p1 or p45/p45 ratios are within ± 0.25 of 1.0 (ratio 0.80 to 1.25), suggesting that, in a comparison of p45 against p1, ratios outside these limits are likely to be due to biological, rather than technical, variation captured by TMT. Supplementary Figure 2a and b shows histograms of log2 transformed individual ratios of three p45/p1pairs (127/126, 129/128 and 131/130) labeled with sixplex and each of the three replicates labeled with duplex (127/126) experiments. Boxplot in Supplementary Fig 2c shows the degree of dispersion in the data. The median ratio for p45/p1 is 1.0 but it exhibited, as expected, much higher variability.
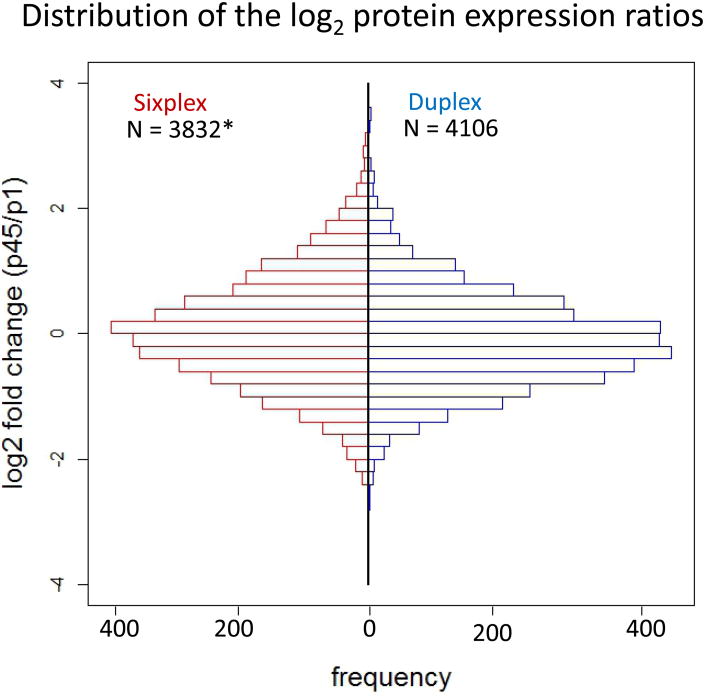
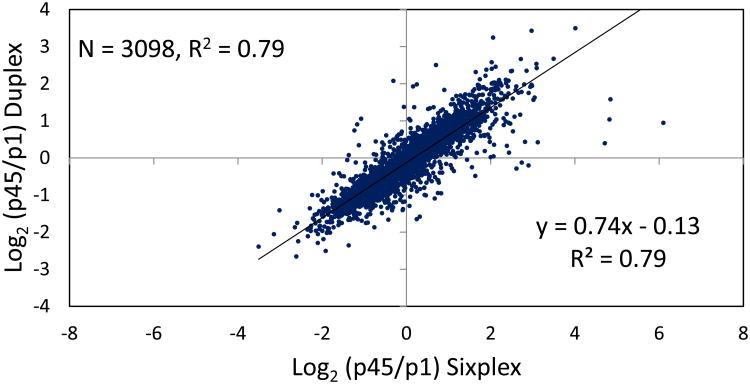
A distribution of the average relative abundance ratios of the p45/p1, corresponding to the proteins quantified in sixplex and duplex, in log2 scale is illustrated in Figure 3. The distribution of ratios is approximately normal (centered on 0 in log2 scale). This is necessary as it ensures that the normalization and statistics used during the analysis are appropriate.13 For the sixplex and duplex protein expression level correlation study, we compared the average p45/p1 protein ratios detected by the sixplex study with their corresponding ratios measured by the triplicate duplex analyses. The ratios observed by sixplex and duplex were positively correlated (correlation coefficient R2 = 0.79). The slope of the regression serves as an indication of the relative sensitivity of one quantification strategy to the other.21 A slope of 1.0 would indicate equal sensitivity between sixplex and duplex TMT experiments. Figure 4 shows the scatter plot of log2-transformed ratio of the common proteins in the two TMT experiments. A slope less than 1.00 indicates that the sensitivity of sixplex in the detection of relative protein expression is better than the duplex. Ninety six percent of the total quantified proteins (3832) in the sixplex fell within standard deviation (SD) of 0.5 or less with a median SD of 0.05 in the three p45/p1 ratios in the sixplex experiment. In duplex, about 94% of the total quantified proteins (4106) have SD of 0.5 or less with a median SD of 0.11 in the three replicate analyses. Histograms for the SD of sixplex and duplex TMT experiments are shown in Supplementary Figure 3. Since more than 90% of the quantifiable proteins in sixplex and duplex fell within SD of 0.5, we considered a 2-fold (≥ 1.0 or ≤ -1.0 in log2 scale) expression change as a meaningful cutoff, representing significant differences in the differential proteome in p45 and p1. Upon filtering the proteins with the relative abundance ratio (between p45 and p1) of at least ± 2-fold and p-value < 0.05, 886 proteins in sixplex and 562 proteins in duplex were mapped to be differentially expressed (Table 2). Proteins with SD value more than 0.5 are not included when deriving potentially significant proteins. A total of 357 proteins were observed together in sixplex and duplex experiments as meeting the significantly changing criteria, -1.0 ≤ log2 (p45/p1 ratio) ≥ 1.0 and p value of < 0.05, with a correlation coefficient (R2) of 0.96.
Figure 3.
Frequency histogram of the p45/p1 ratios depicting distribution of log2 protein expression ratios observed in the sixplex and duplex TMT experiments. The data were normalized as described under Experimental Procedures to compensate for differences in the sample preparation, handling and labeling. Positive values for the log2 ratios correspond to up-regulated proteins (proteins “enriched”) in p45 samples whereas negative values are for downregulated proteins in p45 (proteins “enriched” in p1 samples).
* 6 proteins less in the histogram for scaling of y-axis
Figure 4.
Scatterplot of correlation of postnatal day 45 versus postnatal day 1 (p45/p1) protein expression ratios determined by sixplex and duplex. The log2-transformed ratios of p45/p1 from the sixplex experiment were plotted against those from the duplex experiment.
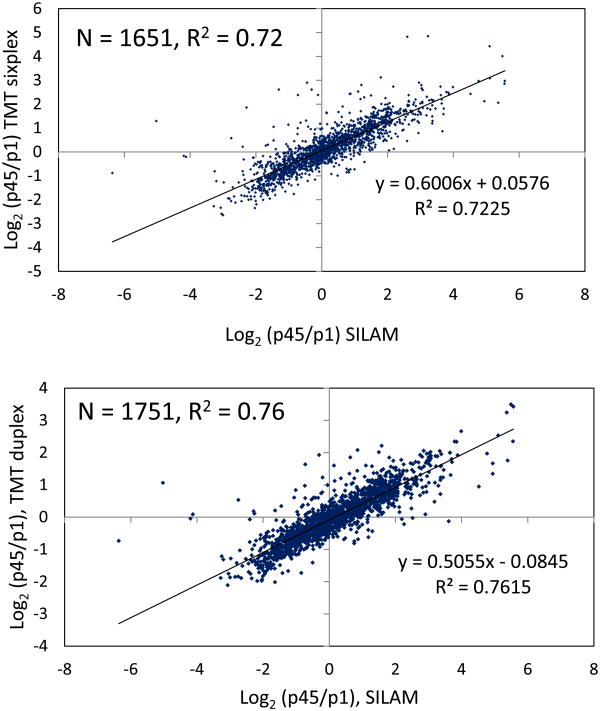
The SILAM strategy uses metabolic labeling of rodents with 15N, a heavy stable isotope of nitrogen, for quantitative proteomic analysis.1 The heavy isotope labeled tissues serve as internal standard for relative quantitation of differentially expressed proteins. We observed comparable quantification results when we examined the TMT data with SILAM-based p45 versus p1 quantification results. The details of SILAM based sample preparation and quantification data can be found elsewhere.22, 23 Most proteins showed the same tendency of change regardless of the labeling strategy. The correlation coefficient (R2) of p45/p1 ratios of commonly identified proteins in TMT and SILAM are 0.72 for sixplex vs SILAM and 0.76 for duplex vs SILAM (Figure 5). Supplementary Figure 4 and 5 shows heatmaps and log2 ratio distribution profiles of the changed protein ratios (-1.0 ≤ log2 (p45/p1 ratio) ≥ +1.0 and p value of < 0.05) detected by TMT sixplex and duplex analyses with the matching protein ratios (p value < 0.05) observed by SILAM-based quantitative approach. Of the 375 significant common proteins present in sixplex and SILAM, the SILAM quantification ratios of these proteins ranged from -3.07 to 3.71 while sixplex quantification ratio ranged from -2.65 to 2.59. For 225 significant proteins present in duplex and SILAM together, range of SILAM quantification ratio remained the same while TMT duplex exhibited the quantification ratio of -2.0 to 1.93.
Figure 5.
Scatter plots examining the comparison of fold change observed by the TMT-based quantitation and 15N metabolic labeling approach (SILAM) approach. The modest correlation of the log2-transformed p45/p1 protein ratios observed by the two varied quantification approach illustrate the reliability of detected changes in the overlapping proteins in sixplex vs SILAM and duplex vs SILAM, respectively.
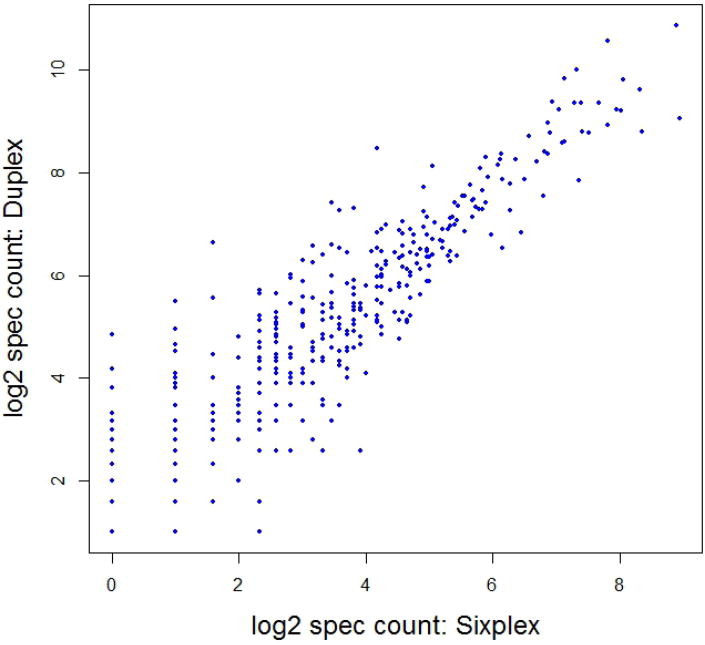
Multiply sequenced peptides show improved quantification during targeted data acquisition due to increasingly higher signal-to-noise values during subsequent sequencing event acquired near the apex of the chromatographic peak. A TMT based targeted studies have shown that multiple repeat counts, during which a peptide may be selected several times as a data-dependent mass and undergo fragmentation before it goes on the dynamic exclusion list, increased quantification precision.24 However, multiple repeat count settings are usually avoided in shotgun experiments because it negatively affects the protein coverage. In our hands, setting the repeat count to 4, to acquire multiple fragmentations of peptides, results in more than 55% drop in protein identifications with 65% fewer peptides identified compared to a repeat count of one. The correlation of fold change among commonly identified protein with repeat counts of 4 and 1 was 0.86 (Supplementary Figure 6). This indicates, for TMT quantitation using a shotgun approach, multiple fragmentation of peptides by using high repeat count settings may not be of much value in improving quantitative accuracy even after sacrificing the number of identified proteins. Nevertheless, many significantly altered proteins that are commonly identified in sixplex and duplex experiments have more than one spectral count, even with the default repeat count setting of 1 (Figure 6). More proteins were identified by higher spectral counts for the duplex experiment which could be due to the fact that three injections are performed for TMT duplex experiment compared to only single LC–MS/MS run of sixplex experiment. The minimum spectral count in duplex samples is two because three injections are combined and proteins whose p45/p1 ratios was observed in at least two of the three replicate MudPIT runs are included in the study. Moreover, of all the proteins that were quantified by sixplex, about 50% of the total quantified were obtained by four or less spectral counts, whereas, only 20% of the proteins have spectral count of four or less in the duplex experiment (Supplementary Figure 7a-d). The discussion of biological significance of the observed changes in rat neurodevelopment is beyond the scope of this project and the readers are referred to our other publications for the same.22, 23
Figure 6.
A base2 logarithmic representation of spectral counts observed for 357 proteins commonly detected in duplex and sixplex TMT experiments and demonstrate statistically significant regulation difference (-1.0 ≤ log2 (p45/p1 ratio) ≥ +1.0 and p value of < 0.05).
Discussion
Chemical peptide labeling with amine-reactive isobaric tags is gaining popularity in proteomics because it allows relative quantitation between multiple samples in the same experiment. Moreover, it is applicable to primary samples, e.g., human biofluids and disease tissues, circumventing the limitation associated with metabolic labeling studies which can only be performed in cell culture and model organisms where nutrients can be manipulated to provide stable isotopes for incorporation during growth. Also, unlike other multiplexing methods, there is no increase in sample complexity in the MS1 scan since peptides from all samples are efficaciously merged and appear as one peak. The coalescence of peptides in MS1 increases the total ion current for that peptide, simplifying the spectra, and requiring only one MS/MS spectrum per peptide for quantitation.25
Our goal in this study was to examine the performance of the sixplex and duplex TMT labeling approaches to assess the global protein changes in a typical quantitative proteomics experiment. We compared the strength of each TMT set to detect significant relative changes in protein expression at two different time points in rat brain development. Data coherence presented by both sixplex and duplex studies immensely increases the reliability of our observations. The number of proteins quantified as statistically significant were higher for the sixplex experiments. This could be because the design of the sixplex experiment was “triple duplex” and when a precursor ion is fragmented in sixplex, it corresponds to the same peptide species present in all labeled samples yielding quantitative information across samples within an experiment. However, when a TMT duplex is run multiple times, missing values are observed. In duplex, a precursor ion selected for fragmentation in one LC–MS/MS run may not be selected in subsequent runs, or spectra of suitable quality for identification and quantification are not obtained consistently, negatively affecting their quantitative potential. The sixplex multiplexing not only requires less MS analysis time when compared with multiple duplex runs, but it is also capable of minimizing errors associated with replicate experiments, thereby making quantification of more proteins possible. The ability to multiplex up to six different samples per experiment has resulted in TMT sixplex being employed in a variety of larger-scale proteomics studies. In one of the studies it was reported that the number of proteins identified were largest for iTRAQ fourplex, followed by TMT sixplex, and smallest with iTRAQ eightplex.18 This observation was mainly due to the manifestation of ions in MS/MS spectra as a consequence of loss of fragments of the labeled tag from the precursor ions. These fragment ions cannot be explained by current search engines and were observed to have a negative impact on peptide scores.18 A separate study compared two channel tags of iTRAQ eightplex (m/z 116 and 117) with two channel tags of TMT sixplex (m/z 126 and 127) by mixing 1:1 ratio of labeled peptides from two biological replicates of P. putida strain.26 They observed similar performance of both the isobaric labeling strategies in determining relative abundance ratios of the proteins.
It has been suggested that a significant number of MS/MS spectra remain unassigned in proteomic data sets due to the chimera spectra resulting from co-fragmentation of two or more different peptides.27 Isobaric tagging with standalone HCD method of data acquisition has been suggested to be a less accurate method of quantitation because the multiple ions are co-selected at the same time as a target peptide for fragmentation during LC–MS/MS. This results in an MS/MS spectrum containing reporter ions derived from mixed sources.28 Evidence exists in the literature claiming interference by the non-regulated background peptides that are co-isolated and co-fragmented in the same isolation window of the peptide of interest compromises the quantification results. The approaches that can be used to counteract ‘ratio compression’ effect from these phantom reporter ions have been discussed in more detail elsewhere.29-31 Nonetheless, our comparison of TMT labeling with metabolic labeling suggests that the proteins defined as enriched by the TMT quantitation method are undistorted, but the magnitude of change may be potentially suppressed. A reduction of precursor ion contamination may occur with more fractionation as occurs in MudPIT analyses.
Conclusion
In this study we examined the performance of sixplex and duplex TMT labeling approach to assess the global protein changes in a typical quantitative proteomics experiment. For this, we compared the strength of each TMT set to detect significant relative changes in protein expression at two different time points in rat brain development. Our study suggests that sixplex, instead of repetitive duplex experiments, should be used when performing isobaric quantitation strategy such as TMT. Even though the average of the three duplex runs resulted in 6% more identified proteins and 7% more quantified ones compared to sixplex experiment, sixplex was able to quantify more proteins that were statistically significant (p <0.05) than duplex. The restrictive quantitative efficiency of duplex is because multiple LC-MS runs are needed for analyzing replicate samples. This might compromise the number of statistically significant proteins because of run to run variations in the number of proteins that are identified. This is mainly attributed to the stochastic nature of data-dependent mass spectrometry where ions are selected for fragmentation on the basis of the intensity of the ion species and resolution from co-eluting precursor ions. Additionally, in sixplex the quantified proteins with fold change ≥ 2 were identified by two or more spectral counts thereby increasing confidence in their ratio measurements. Multiplexing with sixplex will enable analysis of several samples (analytical or biological replicates) simultaneously saving time on sample preparation and also the time demand on the high-end mass spectrometers that are high maintenance.
Supplementary Material
Acknowledgments
We thank Bryan Fonslow and Claire Delahunty for the helpful discussion and the critical reading of the manuscript. Financial support was provided by NIH grants P41 RR011823/GM103533, R01 MH067880-09, P01 AG031097-03, and R01 HL079442-07.
Footnotes
Notes: The authors declare no competing financial interest.
Supporting information: Supplementary figures and table. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR., 3rd Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Analytical Chemistry. 2004;76(17):4951–9. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 2.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 3.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci U S A. 1999;96(12):6591–6. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnology. 1999;17(10):994–9. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 5.Ji C, Guo N, Li L. Differential dimethyl labeling of N-termini of peptides after guanidination for proteome analysis. J Proteome Res. 2005;4(6):2099–108. doi: 10.1021/pr050215d. [DOI] [PubMed] [Google Scholar]
- 6.Heller M, Mattou H, Menzel C, Yao X. Trypsin catalyzed 16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J Am Soc Mass Spectrom. 2003;14(7):704–18. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 7.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3(12):1154–69. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Analytical Chemistry. 2003;75(8):1895–904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 9.Unwin RD. Quantification of proteins by iTRAQ. Methods Mol Biol. 2010;658:205–15. doi: 10.1007/978-1-60761-780-8_12. [DOI] [PubMed] [Google Scholar]
- 10.Dayon L, Hainard A, Licker V, Turck N, Kuhn K, Hochstrasser DF, Burkhard PR, Sanchez JC. Relative quantification of proteins in human cerebrospinal fluids by MS/MS using 6-plex isobaric tags. Anal Chem. 2008;80(8):2921–31. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 11.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nature Biotechnology. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 12.Fic E, Kedracka-Krok S, Jankowska U, Pirog A, Dziedzicka-Wasylewska M. Comparison of protein precipitation methods for various rat brain structures prior to proteomic analysis. Electrophoresis. 2010;31(21):3573–9. doi: 10.1002/elps.201000197. [DOI] [PubMed] [Google Scholar]
- 13.Unwin RD, Griffiths JR, Whetton AD. Simultaneous analysis of relative protein expression levels across multiple samples using iTRAQ isobaric tags with 2D nano LC-MS/MS. Nat Protoc. 2010;5(9):1574–82. doi: 10.1038/nprot.2010.123. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn K, Baumann C, Tommassen J, Prinz T. TMT labelling for the quantitative analysis of adaptive responses in the meningococcal proteome. Methods Mol Biol. 2012;799:127–41. doi: 10.1007/978-1-61779-346-2_8. [DOI] [PubMed] [Google Scholar]
- 15.Xu T, Venable JD, Park SK, Cociorva D, Lu B, Wohlschlegel J, Hewel J, Yates JR. 3rd, ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
- 16.Tabb DL, McDonald WH, Yates JR. 3rd, DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1(1):21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park SK, Venable JD, Xu T, Yates JR. 3rd, A quantitative analysis software tool for mass spectrometry-based proteomics. Nat Methods. 2008;5(4):319–22. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pichler P, Kocher T, Holzmann J, Mazanek M, Taus T, Ammerer G, Mechtler K. Peptide labeling with isobaric tags yields higher identification rates using iTRAQ 4-plex compared to TMT 6-plex and iTRAQ 8-plex on LTQ Orbitrap. Anal Chem. 2010;82(15):6549–58. doi: 10.1021/ac100890k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karp NA, Huber W, Sadowski PG, Charles PD, Hester SV, Lilley KS. Addressing accuracy and precision issues in iTRAQ quantitation. Mol Cell Proteomics. 2010;9(9):1885–97. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkhart JM, Vaudel M, Zahedi RP, Martens L, Sickmann A. iTRAQ protein quantification: a quality-controlled workflow. Proteomics. 2011;11(6):1125–34. doi: 10.1002/pmic.201000711. [DOI] [PubMed] [Google Scholar]
- 21.Collier TS, Randall SM, Sarkar P, Rao BM, Dean RA, Muddiman DC. Comparison of stable-isotope labeling with amino acids in cell culture and spectral counting for relative quantification of protein expression. Rapid Commun Mass Spectrom. 2011;25(17):2524–32. doi: 10.1002/rcm.5151. [DOI] [PubMed] [Google Scholar]
- 22.McClatchy DB, Liao L, Park SK, Venable JD, Yates JR. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Research. 2007;17(9):1378–88. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClatchy DB, Liao L, Lee JH, Park SK, Yates JR., 3rd Dynamics of subcellular proteomes during brain development. J Proteome Res. 2012;11(4):2467–79. doi: 10.1021/pr201176v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Savitski MM, Fischer F, Mathieson T, Sweetman G, Lang M, Bantscheff M. Targeted data acquisition for improved reproducibility and robustness of proteomic mass spectrometry assays. J Am Soc Mass Spectrom. 2010;21(10):1668–79. doi: 10.1016/j.jasms.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Unwin RD, Pierce A, Watson RB, Sternberg DW, Whetton AD. Quantitative proteomic analysis using isobaric protein tags enables rapid comparison of changes in transcript and protein levels in transformed cells. Mol Cell Proteomics. 2005;4(7):924–35. doi: 10.1074/mcp.M400193-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Adams RM, Chourey K, Hurst GB, Hettich RL, Pan C. Systematic comparison of label-free, metabolic labeling, and isobaric chemical labeling for quantitative proteomics on LTQ Orbitrap Velos. J Proteome Res. 2012;11(3):1582–90. doi: 10.1021/pr200748h. [DOI] [PubMed] [Google Scholar]
- 27.Ning K, Fermin D, Nesvizhskii AI. Computational analysis of unassigned high-quality MS/MS spectra in proteomic data sets. Proteomics. 2010;10(14):2712–8. doi: 10.1002/pmic.200900473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christoforou AL, Lilley KS. Isobaric tagging approaches in quantitative proteomics: the ups and downs. Anal Bioanal Chem. 2012 doi: 10.1007/s00216-012-6012-9. [DOI] [PubMed] [Google Scholar]
- 29.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Methods. 2011;8(11):937–40. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wenger CD, Lee MV, Hebert AS, McAlister GC, Phanstiel DH, Westphall MS, Coon JJ. Gas-phase purification enables accurate, multiplexed proteome quantification with isobaric tagging. Nat Methods. 2011;8(11):933–5. doi: 10.1038/nmeth.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savitski MM, Sweetman G, Askenazi M, Marto JA, Lang M, Zinn N, Bantscheff M. Delayed fragmentation and optimized isolation width settings for improvement of protein identification and accuracy of isobaric mass tag quantification on Orbitrap-type mass spectrometers. Anal Chem. 2011;83(23):8959–67. doi: 10.1021/ac201760x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.