Abstract
Aeromonas spp. (gram-negative, aquatic bacteria which include enteropathogenic strains) have two distinct flagellar systems, namely a polar flagellum for swimming in liquid and multiple lateral flagella for swarming over surfaces. Only ∼60% of mesophilic strains can produce lateral flagella. To evaluate flagellar contributions to Aeromonas intestinal colonization, we compared polar and lateral flagellar mutant strains of a diarrheal isolate of Aeromonas caviae for the ability to adhere to the intestinal cell lines Henle 407 and Caco-2, which have the characteristic features of human intestinal enterocytes. Strains lacking polar flagella were virtually nonadherent to these cell lines, while loss of the lateral flagellum decreased adherence by ∼60% in comparison to the wild-type level. Motility mutants (unable to swim or swarm in agar assays) had adhesion levels of ∼50% of wild-type values, irrespective of their flagellar expression. Flagellar mutant strains were also evaluated for the ability to form biofilms in a borosilicate glass tube model which was optimized for Aeromonas spp. (broth inoculum, with a 16- to 20-h incubation at 37°C). All flagellar mutants showed a decreased ability to form biofilms (at least 30% lower than the wild type). For the chemotactic motility mutant cheA, biofilm formation decreased >80% from the wild-type level. The complementation of flagellar phenotypes (polar flagellar mutants) restored biofilms to wild-type levels. We concluded that both flagellar types are enterocyte adhesins and need to be fully functional for optimal biofilm formation.
Aeromonas bacteria (aeromonads) comprise a complex genus of at least 14 recognized DNA hybridization groups (HG), or genomospecies (1). Representatives of these species are found in most aquatic environments and are common contaminants of many retail foods. Aeromonads colonize and form biofilms in water distribution and food-processing systems as well as on water-dwelling plants and animals (e.g., fish, leeches, and frogs). They are significant pathogens of reptiles, amphibians, and fish and are isolated as part of the fecal floras of a wide variety of other animals, including some used for human consumption, such as pigs, cows, sheep, and poultry. In humans, members of the genus cause water-associated wound infections and serious opportunistic infections (e.g., septicemia and meningitis) in immunocompromised individuals. Some strains, particularly of the species Aeromonas hydrophila (HG1), Aeromonas veronii bv. Sobria (HG8/10), and Aeromonas caviae (HG4), are also enteropathogens that cause diarrhea and dysenteric infections in children, the elderly and immunocompromised, and travelers in the summer (9, 11, 12). Little is yet known about how aeromonads adapt to colonize such a variety of inert and host cell surfaces.
A greater understanding of Aeromonas-host colonization mechanisms is needed, as at present it is not possible to identify strains in foods and water that are of public health significance. A type IV pilus (“bundle-forming pilus”) has been implicated as a critical human enterocyte adhesion (15). More recently, studies have shown that Aeromonas flagella facilitate adherence to (and possibly invasion of) the epithelial cell line HEp-2 as well as biofilm formation on plastic (microtiter plate assay), suggesting that these structures may also be virulence determinants for enteropathogenic strains (7, 8, 22). Aeromonas species, like the seafood pathogen Vibrio parahaemolyticus, possess two distinct flagellar systems (a polar flagellum [Fla] for swimming in liquid and a lateral flagellum [Laf], multiply expressed, for swarming over surfaces) (8, 19, 22). Members of our laboratory have previously shown that only approximately 60% of mesophilic Aeromonas strains possess lateral flagellar genes and are able to swarm on surfaces, thus increasing the interest in the possible role(s) of this flagellar type in Aeromonas-host interactions (16).
In this study, the role of flagella as adhesins for human enterocytes was investigated by using two intestinal cell lines (Henle 407 and Caco-2) and a selection of polar and lateral flagellar mutant strains of A. caviae strain Sch3 (a human diarrheal isolate). The Caco-2 cell line, in particular, is considered a good substitute for human intestinal epithelial cells. It is derived from a carcinoma of the colon, and in postconfluent culture, has the morphological and functional characteristics of normal small intestinal cells (20). As biofilms are a feature of persistent infections and characterize up to 30% of Aeromonas gastroenteric infections (6, 9, 23), we also used an in vitro model (the borosilicate glass tube model) to further investigate flagellar contributions to Aeromonas biofilm formation and to determine some of the variables that influence biofilm formation by this organism.
MATERIALS AND METHODS
Bacterial strains.
The genotypic and phenotypic characteristics of the wild-type and mutant strains used in this study are summarized in Table 1 and published more fully elsewhere (2, 8, 16, 22). In brief, the wild-type A. caviae strain Sch3 was originally isolated in the United Kingdom from the diarrheal feces of a child (27). A spontaneous nalidixic acid-resistant mutant of this strain (strain Sch3N) was used to construct insertion mutations in flagellar genes (Table 1). A kanamycin resistance cassette with an outward-reading promoter that ensured the transcription of the downstream genes was inserted into each gene. For the generation of tandem polar (AAR31 [flaA flaB]) or lateral (AAR6 [lafA1 lafA2]) flagellin knockout mutants, two insertions were required and a second chloramphenicol resistance cassette was used. Complemented polar flagellar mutants of strains AAR269, AAR27, AAR31, AAR150, AAR59, and AAR8 were prepared by using plasmid pARP1266 (flaA+ flaB+ flaG+ flaH+ flaJ+) and were constructed by using the broad-host-range mobilizable vector pBBRIMCS-3. The plasmid was introduced into the mutant strains by conjugation (22). In addition to the above fla and laf mutants, two mutants of the hook and basal body gene operon (flg locus) of the polar flagellum (AAR194 [flgH] and AAR460 [flgL]) and a chemotaxis mutant (AAR400 [cheA]), unable to swim or swarm in agar assays, were isolated by mini-Tn5::Cm transposon mutagenesis as described elsewhere (2, 10). For the mutant strain AAR400, the site of the transposon insertion was identified by direct genomic sequencing using 23-mer primers reading outwards from the O and I ends of the mini-transposon. Each primer was used separately in a 99-cycle sequencing reaction with genomic DNA from AAR400 and a Big Dye terminator kit (PE Applied Biosystems). The sequence obtained was analyzed as described in the above studies (2, 10).
TABLE 1.
Bacterial strains used for this studya
| Strain or mutant type | Genotype | Type of protein | Presence of phenotypeb,c
|
Reference | |||
|---|---|---|---|---|---|---|---|
| Fla | Laf | Swimming | Swarming | ||||
| Sch3 | Wild type | + | + | ++ | ++ | 27 | |
| Sch3N | Spontaneous Nalr | + | + | ++ | ++ | 22 | |
| Polar mutants | |||||||
| AAR269 | flaA::Kmr | Flagellin | + | + | + | ++ | 22 |
| AAR27 | flaB::Kmr | Flagellin | + | + | + | + | 22 |
| AAR31 | flaA::CmrflaB::Kmr | Flagellins | − | ± | − | − | 22 |
| AAR150 | flaG::Kmr | Flagellar protein | + | + | + | ++ | 22 |
| AAR59 | flaH::Kmr | Capping protein | − | ± | − | − | 22 |
| AAR8 | flaJ::Kmr | Chaperone | − | ± | − | − | 22 |
| AAR194 | flgH::mini-Tn5Cmr | L-ring | − | + | − | − | 2 |
| AAR460 | flgL::mini-Tn5Cmr | Hook protein | − | + | − | − | 2 |
| AAR400 | cheA::mini-Tn5Cmr | Chemotaxis protein | + | + | − | − | This study |
| Lateral mutants | |||||||
| AAR58 | lafA1::Kmr | Flagellin | + | + | + | + | 8 |
| AAR5 | lafA1::Kmr | Flagellin | + | + | + | + | 8 |
| AAR6 | lafA1::CmrlafA2::Kmr | Flagellins | + | − | + | − | 8 |
| AAR9 | lafB::Kmr | Capping protein | + | − | + | − | 8 |
| AAR20 | fliU::Kmr | N-lysine methylase | + | + | + | + | 8 |
Strains in bold form the “short panel” of key mutant strains tested in adhesion and biofilm assays.
Flagellar expression (all strains) was determined by transmission electron microscopy and Western blotting of sheared bacterial preparations. Although Laf was seen on the tandem polar mutant strain AAR31 and other polar mutant strains (contrary to the original report in which Western blots of whole-cell preparations were negative for Laf flagellin [8]), these strains are functionally Laf negative, suggesting overall that Laf expression is somehow impaired (16). Hence, they have been designated “±” here. (Note that there are other Laf+ strains [AAR194 and AAR460] that are also nonmotile in agar assays.) Swimming and swarming motilities were evaluated by agar assays, as summarized in Materials and Methods and described in full elsewhere (16). Symbols: ++, motility zone of ≥7 cm; +, motility zone of ≥2 cm but <7 cm; −, growth at the inoculum site but no movement. Strains were tested in duplicate in at least four separate experiments.
Complemented strains of the fla mutants, constructed by using plasmid pARP1266 (flA+ flaB+ flaG+ flaH+ flaJ+) and the broad-host-range mobilizable vector pBBRIMCS-3, restored flagellar expression and at least partially restored flagellar motilities (22).
Growth conditions.
The wild-type and mutant strains were routinely grown on tryptone soy agar (1.5% [wt/vol]) supplemented with 0.6% (wt/vol) yeast extract (TSAY; Oxoid, Basingstoke, United Kingdom) at 37°C for 16 to 18 h. Agar for the growth of the complemented mutant strains was supplemented with chloramphenicol at 25 μg/ml and tetracycline at 10 μg/ml. For adhesion assays, bacteria were taken directly from solid plates and placed into Eagle's minimal essential medium (MEM; Gibco, Invitrogen, Australia). For biofilm assays, colonies were inoculated into 10 ml of tryptone soy broth plus yeast extract (TSBY) and grown at 37°C for 16 to 18 h with shaking before further dilution in TSBY, unless otherwise stated. Short-term storage of isolates was done in minimal maintenance medium at room temperature. Long-term storage was done in glycerol-peptone (glycerol-1% bacteriological peptone L37 [1:4]) (Oxoid) at −70°C.
Motility assays (swimming and swarming).
Swimming motility was assessed by the measurement of motility zones in semisolid agar (0.3% agar in Luria-Bertani broth [Oxoid]) stabs inoculated into the center of the agar (30°C, 16 to 18 h). Growth from the edge of these plates provided the inoculum for the swarming assay. Swarm plates (0.5% Eiken agar [Eiken Chemical Co., Ltd., Tokyo, Japan] in Difco nutrient broth [Difco Laboratories, Detroit, Mich.]) were inoculated on the surface of the agar in the center, and swarming zones were measured after incubation at 30°C for 16 to 18 h (16).
Cell lines.
HEp-2 epithelial cells (American Type Culture Collection [ATCC] CCL23), Caco-2 cells (ATCC HTB37), and Henle 407 human intestinal epithelial cells (ATCC CCL6) were grown in MEM containing 5% (vol/vol) fetal calf serum. For standard adhesion assays, semiconfluent monolayers of these cell lines were grown on 12-mm-wide glass coverslips in 24-well tissue culture plates at 37°C with 5% CO2. Caco-2 cells were used at 12 to 14 days postseeding to allow cell differentiation and the expression of microvilli (20).
Adhesion assay.
Adherence to cell lines was determined as described elsewhere, with minor modifications (14). In brief, bacterial colonies from overnight TSAY plates were resuspended in MEM and adjusted (optical density at 570 nm [OD570]) to approximately 107 CFU/ml. A 1-ml aliquot of bacteria (i.e., ∼107 CFU) was inoculated onto coverslip cell monolayers which were then incubated for 90 min at 37°C with 5% CO2. Nonadherent bacteria were removed by washing (five times in phosphate-buffered saline). The monolayers were fixed with 1 ml of methanol-acetic acid (3:1) for 5 min, stained with May-Grünwald and Giemsa stains (BDH, Poole, United Kingdom), and mounted for the assessment of adherent bacteria by light microscopy. All strains were tested in triplicate in each assay.
Bacterial adherence was also assessed by quantitative bacterial counts, using a method adapted from Benítez and colleagues (5). For this assay, the cell lines were grown directly in the wells of 24-well tissue culture plates. After the standard 90-min adhesion step, the monolayers were washed twice with phosphate-buffered saline to remove nonadherent bacteria. The remaining cell-associated bacteria were then released from the cells by the addition of 1 ml of 0.1% (vol/vol) Triton X-100 solution (Sigma) (30 min on an orbital shaker). Serial dilutions (10−1 to 10−7) of the lysates were plated onto TSAY and the CFU were determined after overnight incubation. The percentage of bacteria recovered relative to the initial inoculum was determined for each strain.
Biofilm assay.
Biofilm formation in borosilicate glass tubes (10 by 75 mm) was assessed by the method of O'Toole and colleagues, with slight modifications (21, 26). In brief, the glass tubes were inoculated with 300 μl of a 1:100 dilution of bacteria from overnight (16 to 18 h) TSBY cultures (described above). These were then lightly covered with foil and incubated for up to 30 h at 37°C without shaking. For detection and quantification of biofilm formation, the tubes were rinsed thoroughly and vigorously with water and the remaining cells were stained with 0.5% crystal violet solution for 15 min at room temperature. The crystal violet-stained biofilm was then solubilized by the addition of 100% ethanol (600 μl for 10 min at room temperature), and the OD570 of 200 μl of the resultant suspension was measured in a microplate reader (Bio-Rad, Hercules, Calif.). Replicates of three to six tubes per organism were examined for each experiment, and an uninoculated broth control was included in each experiment. The growth rates of the flagellar mutant strains were assessed by measuring OD570 values and were shown not to differ significantly from that of the wild-type strain.
Statistical analysis.
Differences in adherence and biofilm formation between wild-type and mutant strains were analyzed by Student's t test with Microsoft Excel software.
RESULTS
Flagellar contributions to epithelial and intestinal cell adhesion.
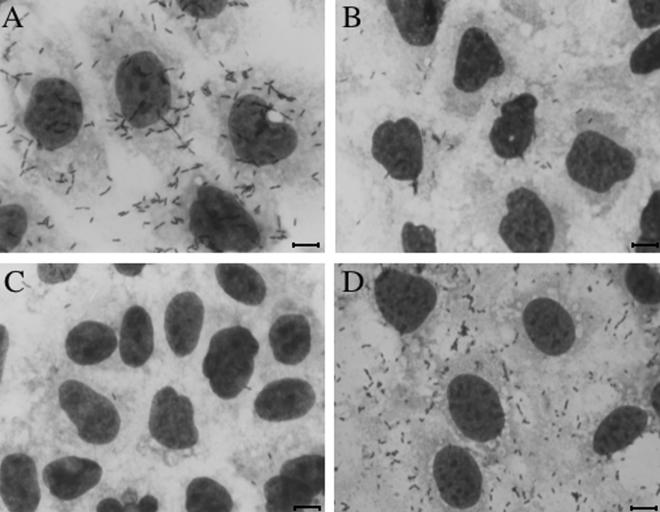
Our results confirmed previously published reports that the wild-type A. caviae strain Sch3 is highly adherent to the human laryngeal epithelial cell line HEp-2 (8, 22). Moreover, this strain was shown to be highly adhesive to both the intestinal cell lines that we studied (Henle 407 and Caco-2), with adhesion to the intestinal cell lines being significantly higher (P < 0.001) than adhesion to HEp-2 cells (Table 2 and Fig. 1). A comparison of the adherence ability of selected polar and lateral flagellar mutant strains showed that both polar and lateral flagellar types contribute to this cellular adhesion (Table 2). The same conclusion was reached irrespective of the cell line examined. The loss of the polar flagellum (mutant AAR31) virtually abolished adhesion, whereas the loss of the lateral flagellum (mutant AAR6) decreased adhesion by about 60%. Complementation of the flagellar defect (strain AAR31C) restored adhesive ability to >80% of wild-type adhesion levels (Fig. 1D). Motility is important in the adhesion process, as mutants expressing one or both flagella but with absent or altered swimming and swarming abilities (strains AAR194 and AAR400) showed reduced adhesion (∼50% of wild-type values), irrespective of their flagellar expression phenotypes. Unlike the diffuse adhesion pattern shown by the wild-type strain (Fig. 1A), adherence by these motility-impaired mutant strains was much more random over the monolayer (not shown).
TABLE 2.
Adhesion of A. caviae strain Sch3 wild-type and mutant strains to HEp-2 and intestinal cell linesa
| Strain | Mutation | Presence of polar flagellumb | Presence of lateral flagellab | Mean no. of bacteria per cell ± SD (% decrease from wild type)c
|
||
|---|---|---|---|---|---|---|
| HEp-2 | Henle 407 | Caco-2 | ||||
| Wild-type A. caviae Sch3 | + | + | 15.5 ± 6.2 | 19.5 ± 7.4 | 20.9 ± 5.1 | |
| AAR6 | lafA1::CmrlafA2::Kmr | + | − | 5.9 ± 3.2 (62) | 8.3 ± 2.1 (58) | 9.2 ± 3.6 (56) |
| AAR194d | flgH::mini-Tn5Cmr | − | + | 5.9 ± 3.2 (62) | 12.4 ± 3.6 (37) | 8.6 ± 3.9 (59) |
| AAR31 | flaA::CmrflaB::Kmr | − | ± | 0.5 ± 0.9 (97) | 0.2 ± 0.7 (99) | 2.9 ± 2.5 (88) |
| AAR31C | AAR31 plus pARP1266 (flA+flaB+flaG+flaH+flaJ+) | + | + | 14.8 ± 4.0 (4) | 16.9 ± 2.7 (13) | 17.5 ± 2.8 (17) |
| AAR400d | cheA::mini-Tn5Cmr | + | + | 8.8 ± 5.5 (43) | 9.9 ± 6.9 (49) | 8.9 ± 3.9 (57) |
Strains were tested on triplicate coverslips in 23, 16, and 10 separate experiments for HEp-2, Henle, and Caco-2 cells, respectively.
Flagellar expression was determined by transmission electron microscopy and Western blotting with whole-cell preparations (2, 8).
For the mutant strains AAR6, AAR194, AAR31, and AAR400, all adherence values were significantly reduced compared to the wild-type strain (P < 0.001). For two of the three cell lines (HEp-2 and Henle 407), adherence by the complemented mutant strain was not significantly different from adherence by the wild-type strain.
These mutants were unable to swim or swarm in agar motility assays.
FIG. 1.
Adherence of A. caviae strain Sch3 wild-type and flagellar mutant strains to Caco-2 cells (May-Grünwald- and Giemsa-stained coverslip culture cell monolayers). (A) Wild-type strain (Fla+ Laf+); (B) lateral flagellin double mutant strain AAR6 (Fla+ Laf−); (C) polar flagellin double mutant strain AAR31 (Fla− Laf±); (D) complemented mutant strain AAR31C (Fla+ Laf+). Bars, ∼10 μm.
Qualitative counts of bacterial adhesion (Triton X-100 assay) by selected mutant strains to HEp-2 cells confirmed the conclusions drawn from the light microscopic adhesion assays described above (data not shown). The Fla-negative mutants, AAR31 and AAR194, and the chemotactic motility mutant, AAR400, showed decreased adhesion levels (by >80%) compared to the wild-type strain. Adhesion by the complemented fla mutant, AAR31, was restored to >70% of the wild-type level.
Aeromonas biofilm formation and the roles of flagella.
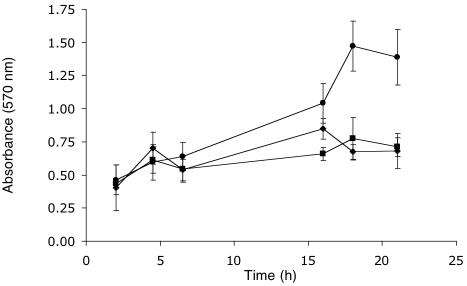
A. caviae strain Sch3 and the polar and lateral flagellar double mutant strains, called AAR31 (Fla− Laf±) and AAR6 (Fla− Laf+), respectively, were compared with respect to the ability to form biofilms in borosilicate glass tubes (n = 7 experiments). The microtiter plate biofilm assay model gave much less consistent results in our hands than the borosilicate glass tube model. In this latter system (using inocula from overnight broth cultures, with the medium unchanged), the time of peak biofilm formation by the wild-type strain varied between 16 and 30 h in different assays. Longer incubations resulted in a reduction in the biofilm reading. The highest reading occurred most commonly at about 16 to 20 h. The mean (± standard deviation [SD]) peak OD570 for the different experiments was 1.26 ± 0.47, a value comparable to peak biofilm readings obtained with Vibrio cholerae in published studies using the borosilicate assay (24, 25). Some variability was observed between assays, but biofilms formed by the flagellar mutant strains were significantly impaired (P ≤ 0.01) compared to wild-type biofilm levels in four of the seven time course experiments, while all experiments showed a trend toward this result. Figure 2 illustrates the data from one of these experiments. When biofilm formation by the double flagellar mutant strains was compared with the peak wild-type biofilm formation (normalized to 100%), biofilms were decreased by ∼60% for the polar mutant and ∼50% for the lateral flagellar mutant.
FIG. 2.
Biofilm development on borosilicate glass (37°C) by the A. caviae strain Sch3 wild type (•) and flagellar mutant strains AAR6 (Fla+ Laf−) (▪) and AAR31 (Fla− Laf±) (▴) over time. Values shown are the means of three replicate tubes ± SD from a representative experiment. The values at 18 h were significantly different from that of the wild-type strain (P < 0.01) for both mutant strains.
The above assays were performed at 37°C. At 30°C, the kinetics of biofilm formation were similar, but overall the maximum level of biofilm formation by the wild-type strain decreased by ∼30% when compared to the level formed at 37°C (n = 2 experiments). The preparation of inocula from 6- to 8-h cultures of bacteria on solid medium (conditions favoring maximum lateral flagellar expression [16]) did not result in more efficient biofilm formation by the wild-type strain than when inocula were taken from overnight broth cultures (as used in the above assays and for most assays below).
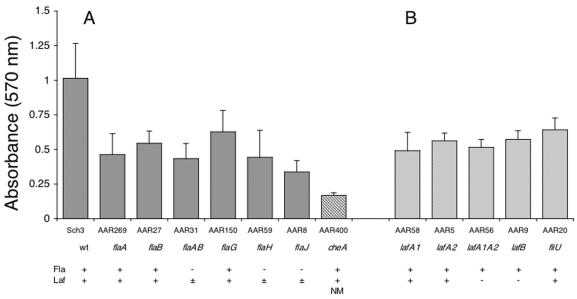
In other sets of experiments, biofilm formation by the extended range of polar and lateral flagellar mutant strains (Table 1) was compared with biofilm formation by the wild-type and double flagellar mutant strains described above at the fixed time point of 18 h (Fig. 3). Overall, the results showed that all mutants, irrespective of their phenotype, had decreased (by >30% of the wild-type level) abilities to form biofilms. For the polar mutant strains, there was a trend towards lower biofilm formation (mean of 58%) by those strains that were unable to produce the polar flagellum (AAR31, AAR59, and AAR8) compared to strains that were still able to express it (AAR269, AAR27, and AAR150) (mean of 45%), but overall the differences were not significant (P > 0.03) (Fig. 3A). Similarly for the lateral flagellar mutant strains, although all were significantly impaired in comparison to the wild-type strain, the fliU mutant (strain AAR20 [Fla+ Laf+]), which expressed functional lateral flagella, tended to form better biofilms than the lafA1 lafA2 double flagellin mutant AAR6 (Fla+ Laf−) (P < 0.05) (Fig. 3B).
FIG. 3.
Biofilm formation on borosilicate glass by polar (Fla) (A) and lateral (Laf) (B) flagellar mutant strains (18 h at 37°C). Each histogram represents the means ± SD of four experiments, with three to six replicate tubes per strain evaluated in each experiment. NM, nonmotile in swimming and swarming agar assays.
The motility function as well as chemotaxis appears to be important for biofilm formation (as was found for the cellular adhesion experiments [Table 2]), as the chemotactic mutant AAR400, which does not display directional swimming and is unable to form motility zones in either the swimming or swarming agar assay, was the most severely impaired mutant strain, with an 83% decrease in its ability to form biofilms. The flg mutant strains, AAR194 (flgH) and AAR460 (flgL) (both Fla− Laf+), were less extensively studied with the biofilm assay but showed decreases in biofilm formation (>40%) that were consistent with a loss of the polar flagellum and swimming motility (not illustrated).
Complementation of the fla mutant strains (AAR8, AAR59, and AAR31), which at least partially restored flagellar expression and motilities (22), restored biofilm formation in the borosilicate model. For strain AAR31, in two of three experiments performed with broth culture inocula, the complemented strain partially (60 to 70% of wild-type levels) restored the biofilm. When the assay was performed with inocula from solid medium, complete restoration was observed (n = 2 experiments), although as noted earlier, biofilm formation by solid medium inocula was less efficient (20 to 30% decrease) than biofilm formation by broth inocula. Introduction of the plasmid vector alone did not restore the phenotypes of the mutants.
DISCUSSION
This study has conclusively demonstrated that flagella (both polar and lateral) play critical roles in Aeromonas adherence to host cell surfaces. To our knowledge, it is the first to demonstrate that these flagella are adhesins for human intestinal cells. The evidence obtained also supports the conclusion that both flagellar types facilitate Aeromonas biofilm formation on surfaces. Thus, our findings are consistent with the hypothesis that the lateral flagellum is likely to be part of the as yet unknown combinations of virulence factors that contribute to persistent infections by virulent subsets of Aeromonas spp. in the gastrointestinal tract.
The results obtained with the fla double mutant strain AAR31 (flaA flaB) and the chemotactic motility mutant strain AAR400 (cheA) showed that the polar flagellar roles of swimming motility and initial adherence are likely to be vital for intestinal colonization. Lateral flagella are clearly also adhesins for intestinal cells, as shown by the markedly decreased cellular adherence of the laf double mutant stain AAR6 (lafA1 lafA2). In a previous investigation, members of our laboratory showed that Aeromonas lateral flagella appear to form bacterial linkages on cells and agar, as has been reported for Vibrio parahaemolyticus on agar (4, 16). This and swarming motility (discussed further below) are other mechanisms by which lateral flagella induced on surfaces or in viscous mucus could contribute to adhesion and more efficient colonization of the gastrointestinal tract by those strains which are able to produce them.
Knowledge of the factors influencing biofilm formation by Aeromonas spp. on surfaces is limited, although it is known that the development of the mature biofilm structure on inert surfaces is influenced by quorum-sensing signals (17). In this study, the influence of temperature and the kinetics of biofilm formation were investigated with a borosilicate glass model. Optimal biofilm formation occurred at 37°C, usually at about 18 h of incubation. Flagellar mutants were therefore evaluated under these conditions. The fact that biofilm formation was found to be more efficient at 37°C than at 30°C adds further weight to the possible in vivo pathogenic significance of the flagellar contributions identified in this study.
While the borosilicate glass tube model clearly demonstrated that swimming motility is essential for biofilm formation, results by this assay permit only a more cautious conclusion regarding the contribution of lateral flagella to biofilm formation. All flagellar mutations (fla and laf) resulted in a loss in the ability to form biofilms. In addition, strains that were unable to express lateral flagella were still able to form significant biofilms (∼50% of the wild-type level). Moreover, the limitations of the glass tube assay (experimental variation and screening at a single time point only) may also have masked effects. Our in vitro biofilm findings are at variance with recently published reports of Aeromonas biofilm formation in a plastic microtiter plate model which concluded that lateral flagella were essential for biofilm formation by Aeromonas species (7, 8). While the investigators who conducted those studies primarily examined an A. hydrophila strain (strain AH-3), they did report preliminary studies with A. caviae strain Sch3, the strain studied extensively in this investigation (8). In contrast to our findings, no differences were found in biofilm formation in the microtiter plate model between the wild-type A. caviae strain (strain Sch3N) and the mutants AAR58 (lafA1) and AAR5 (lafA2), while the Laf-negative mutants AAR6 (lafA1 lafA2) and AAR9 (lafB) showed 32 and 35% decreases in the capacity to form biofilms. The introduction of a plasmid containing the complete laf gene cluster from A. hydrophila strain AH-3 (pCOS-LAF) into these Laf-negative A. caviae mutants restored biofilm formation to wild-type levels. Similarly, the introduction of pCOS-LAF into other laf-negative Aeromonas species led to a twofold increase in the ability of these strains to form biofilms (7). These complementation studies add weight to the conclusion that lateral flagella do contribute to biofilm formation. Complementation of the fla mutants in our study also supported the conclusion that alterations in the flagellar function(s) overall were responsible for the impairment in biofilm formation seen with the mutant strains, although it is difficult to attribute a specific role to the lateral flagellum with the complemented strains we tested. Overall, our results do nevertheless indicate that fully functional lateral flagella seem to be required for optimal biofilm formation, and to this extent they are consistent with the conclusions that can be drawn from the published studies mentioned above.
The reasons for the differing findings of the two studies are unclear, but there are differences in the biofilm methodologies used. In the microtiter plate model, biofilms were quantitated after 48 h of incubation at 30°C, the initial bacterial inocula were from solid plates, and comparisons were made with a spontaneous nalidixic acid mutant of A. caviae strain Sch3 (strain Sch3N). (In our study, biofilm formation in glass tubes, which we found gave more reproducible results than the microtiter plate assay, was assessed after ∼18 h at 37°C, inocula were from broth cultures, and the wild-type strain A. caviae strain Sch3 was the control.) Interestingly, we observed more significant restoration of biofilm formation in complemented strains when they were grown on solid medium rather than in broth. Yet inocula from solid medium (6 to 8 h) showed less efficient biofilm formation in our initial optimization of the biofilm experiments. The A. caviae strains Sch3 and Sch3N behaved similarly in our adhesion and biofilm assays. There are no quantitative differences in the abilities of the two strains to adhere to HEp-2 cells. I. Gryllos, of the J. G. Shaw laboratory, extensively compared both strains in original cell adhesion studies, although only results with strain Sch3N were published (22). We also found no differences between the two strains in their abilities to swim and swarm in agar assays. Biofilm formation in the tube model was only compared on two occasions, but no significant difference in the levels of biofilms formed by the two strains was found. However, we observed (Table 1) and it was previously reported that mutations in flagellar genes can decrease motility functioning, even if flagella are expressed (22). Mutations that still allow flagellar expression appear to affect the delicate balance of the organelle and alter motility and adherence (22). The nonpolar antibiotic resistance cassette used to produce the mutant strains contains an outward-reading promoter that results in the constitutive expression of downstream genes. This could well upset the regulation in the flagellar hierarchy and hence alter the optimal functioning of the flagellum. Studies with such mutant strains therefore need careful interpretation.
It is clear also that significant biofilms can form in the absence of lateral flagella (as we found). Recently, a variant strain of A. caviae (not strain Sch3, which was studied in this investigation) which had increased type IV pilus expression and defective swimming and swarming motilities was reported to form rapid and strongly adherent biofilms on the surfaces of glass flasks (3). It was previously shown that at least one Aeromonas type IV pilus type (the bundle-forming pilus [Bfp]) is an important adhesin for enterocytes (15). A second type IV Aeromonas pilus, Tap, may also be able to substitute for flagellar roles in biofilm formation (13, 18). Biofilm formation is a multifactorial process involving both pili and flagella as well as other determinants. Therefore, mutation in any one factor, especially one involved in initial adherence, may slow down biofilm formation, but this lesion is probably able to be compensated for by one of the other components. Thus, the ability of Aeromonas spp. to adapt their biofilm formation mechanisms may account for some of the variability seen in our individual experiments and possibly for differing results after growth on solid or liquid medium.
The significance of Laf-mediated swarming motility for adhesion, colonization, and biofilm formation requires further investigation with A. caviae laf motility mutant strains lafT and lafU (when available). The lafT mutant of A. hydrophila strain AH-3 showed an ∼50% decrease in adhesion to HEp-2 cells which was recoverable by the restoration of motility by complementation, and biofilm results mirrored this result (17). These findings suggest that the motility function of Laf may be as significant as its adhesive and associated accessory roles in the colonization of surfaces.
The relationship between in vitro biofilm assays and biofilm formation on human tissues remains to be established. Further studies of the contribution of the lateral flagellum to the pathogenesis of Aeromonas gastrointestinal infections nevertheless seem warranted.
Acknowledgments
This work was supported by grants from the University of Tasmania Institutional Research Grants Scheme, the Royal Hobart Hospital Research Foundation, Tasmania, Australia, and the Wellcome Trust, United Kingdom.
We thank Ali Rabaan for allowing us to use his mutant strains in these studies, Bronwen Tassell and Lauri Salter for their assistance with the biofilm assays, and David Lees, TRSU, University of Tasmania, for his help in producing the figures.
Editor: V. J. DiRita
REFERENCES
- 1.Abbott, S. L., W. K. W. Cheung, and J. M. Janda. 2003. The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 41:2348-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altarriba, M., S. Merino, R. Gavín, R. Canals, A. Rabaan, J. G. Shaw, and J. M. Tomás. 2003. A polar flagella operon (flg) of Aeromonas hydrophila contains genes required for lateral flagella expression. Microb. Pathog. 34:249-259. [DOI] [PubMed] [Google Scholar]
- 3.Béchet, M., and R. Blondeau. 2003. Factors associated with the adherence and biofilm formation by Aeromonas caviae on glass surfaces. J. Appl. Microbiol. 94:1072-1078. [DOI] [PubMed] [Google Scholar]
- 4.Belas, M. R., and R. R. Colwell. 1982. Scanning electron microscope observation of the swarming phenomenon of Vibrio parahaemolyticus. J. Bacteriol. 150:956-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benítez, J. A., R. G. Spelbrink, A. Silva, T. E. Phillips, C. M. Stanley, M. Boesman-Kinkelstein, and R. A. Finkelstein. 1997. Adherence of Vibrio cholerae to cultured differentiated human intestinal cells: an in vitro colonization model. Infect. Immun. 65:3474-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 7.Gavín, R., S. Merino, M. Altarriba, R. Canals, J. G. Shaw, and J. M. Tomás. 2003. Lateral flagella are required for increased cell adherence, invasion and biofilm formation by Aeromonas spp. FEMS Microbiol. Lett. 224:77-83. [DOI] [PubMed] [Google Scholar]
- 8.Gavín, R., A. A. Rabaan, S. Merino, J. M. Tomás, I. Gryllos, and J. G. Shaw. 2002. Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Mol. Microbiol. 43:383-397. [DOI] [PubMed] [Google Scholar]
- 9.Gracey, M., V. Burke, and J. Robinson. 1982. Aeromonas-associated gastroenteritis. Lancet ii:1304-1306. [DOI] [PubMed] [Google Scholar]
- 10.Gryllos, I., J. G. Shaw, R. Gavín, S. Merino, and J. M. Tomás. 2001. Role of the flm locus in mesophilic Aeromonas species adherence. Infect. Immun. 69:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Janda, J. M., and S. L. Abbott. 1998. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin. Infect. Dis. 27:332-344. [DOI] [PubMed] [Google Scholar]
- 12.Kirov, S. M. 2001. Aeromonas and Plesiomonas species, p. 301-327. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. American Society for Microbiology Press, Washington, D.C.
- 13.Kirov, S. M., T. C. Barnett, C. Pepe, M. S. Strom, and M. J. Albert. 2000. Investigation of the role of type IV Aeromonas pilus (Tap) in the pathogenesis of Aeromonas gastrointestinal infection. Infect. Immun. 68:4040-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirov, S. M., L. J. Hayward, and M. A. Nerrie. 1995. Adhesion of Aeromonas sp. to cell lines used as models for intestinal adhesion. Epidemiol. Infect. 115:465-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirov, S. M., L. A. O'Donovan, and K. Sanderson. 1999. Functional characterization of type IV pili expressed on diarrhea-associated isolates of Aeromonas species. Infect. Immun. 67:5447-5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirov, S. M., B. C. Tassell, A. B. T. Semmler, L. A. O'Donovan, A. A. Rabaan, and J. G. Shaw. 2002. Lateral flagella and swarming motility in Aeromonas species. J. Bacteriol. 184:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. R. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 18.Masada, C. L., S. E. LaPatra, A. W. Morton, and M. S. Strom. 2002. An Aeromonas salmonicida type IV pilin is required for virulence in rainbow trout Oncorhynchus mykiss. Dis. Aquat. Organ. 51:13-25. [DOI] [PubMed] [Google Scholar]
- 19.McCarter, L. 1999. The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1:51-57. [PubMed] [Google Scholar]
- 20.Nishikawa, Y., A. Hase, J. Ogawasara, S. M. Scotland, H. R. Smith, and T. Kimura. 1994. Adhesion to and invasion of human colon carcinoma Caco-2 cells by Aeromonas strains. J. Med. Microbiol. 40:55-61. [DOI] [PubMed] [Google Scholar]
- 21.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 22.Rabaan, A. A., I. Gryllos, J. M. Tomás, and J. G. Shaw. 2001. Motility and the polar flagellum are required for Aeromonas caviae adherence to HEp-2 cells. Infect. Immun. 69:4257-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rautelin, H., M. L. Hänninen, A. Sivonen, U. Turunen, and V. Valtonen. 1995. Chronic diarrhea due to a single strain of Aeromonas caviae. Eur. J. Clin. Microbiol. Infect. Dis. 14:51-53. [DOI] [PubMed] [Google Scholar]
- 24.Watnick, P. I., K. J. Fullner, and R. Kolter. 1999. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J. Bacteriol. 181:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watnick, P. I., and R. Kolter. 1999. Steps in the development of Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watnick, P. I., C. M. Lauriano, K. E. Klose, L. Croal, and R. Kolter. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilcox, M. H., A. M. Cook, A. Eley, and R. C. Spencer. 1992. Aeromonas spp. as a potential cause of diarrhoea in children. J. Clin. Pathol. 45:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]