Abstract
Mice deficient in interleukin-2 are well suited for use as an animal model for inflammatory bowel disease. Raised under specific-pathogen-free conditions, interleukin-2-deficient mice develop an inflammatory bowel disease resembling ulcerative colitis in humans. The finding that colitis was attenuated when the mice were kept under germfree conditions implies that the resident intestinal flora is involved in the pathogenesis of colitis. The present study addresses the composition of the mucosa-associated bacterial flora in colon samples from interleukin-2-deficient mice that developed colitis. This was investigated by comparative 16S ribosomal DNA (rDNA) sequence analysis and fluorescence in situ hybridization using rRNA-targeted fluorescent probes to quantify the bacterial populations of the mucosa-associated flora. The investigations revealed distinct differences in the bacterial composition of the mucosa-associated flora between interleukin-2-deficient mice and healthy controls. Fluorescence in situ hybridization identified up to 10% of the mucosa-associated flora in interleukin-2-deficient mice as Escherichia coli, whereas no E. coli was detected in the mucosa from healthy wild-type mice. This finding was consistent with the results from comparative 16S rDNA analysis. About one-third of the clones analyzed from 16S rDNA libraries of interleukin-2-deficient mice represented Enterobacteriaceae, whereas none of the clones analyzed from the healthy controls harbored 16S rDNA from Enterobacteriaceae. The abundance of E. coli in the colonic mucosa of interleukin-2-deficient mice strongly suggests a participation in the pathogenesis of colitis in the interleukin-2-deficient mouse model for inflammatory bowel disease.
Ulcerative colitis and Crohn's disease are chronic diseases of the intestinal tract which are characterized by a strong activation of the intestinal mucosa-associated immune system due to a complex interaction of genetic, immunologic, and environmental factors. Despite numerous investigations over the last half-century, the etiology of inflammatory bowel diseases (IBD) remains unknown (32). Today several animal models for IBD are established and allow the investigation of factors which are involved in the pathogenesis of the disease (6). Interleukin-2 (IL-2)-deficient mice represent an animal model in which mice develop a colitis resembling ulcerative colitis in humans (4). Colitis developed spontaneously when the mice were kept under specific-pathogen-free (SPF) housing conditions but was greatly attenuated when they were raised under germfree conditions (29). This finding is consistent in various rodent models of IBD and demonstrates the pivotal role that the endogenous intestinal bacterial flora plays in the initiation and perpetuation of the chronic inflammation (23, 27, 36) Results from studies of T-cell-receptor-deficient mice and of rats with induced experimental colitis indicate that the species composition of the microbial flora, rather than the intestinal flora per se, seems to be of particular importance for the initiation of the disease (12, 15, 24, 28). Consequently, knowledge about the microbial composition of the intestinal microbiota is crucial for future investigations of the potential role that enteric bacteria play in the etiology and pathogenesis of IBD.
The gastrointestinal tract of rodents harbors a complex microbial community comprising a multitude of bacterial species (31). As with other mammals, the numerically predominant microbes are strictly anaerobic bacteria showing fastidious growth properties. Due to the limitations of culture-based techniques, most members of the intestinal microbiota are still unknown (5). Today molecular techniques such as PCR amplification, cloning and sequencing of 16S rRNA genes, and fluorescence in situ hybridization (FISH) provide suitable tools for the culture-independent detection and identification of bacteria in complex microbial communities (3, 20). Therefore, we combined molecular 16S rRNA-based identification techniques and the IL-2-deficient mouse model in order to analyze the bacterial composition of the mucosa-associated flora in mice developing colitis. The goal of this study was to analyze and compare the mucosa-associated flora of IL-2-deficient mice and wild-type mice (used as healthy controls) in order to obtain evidence for an association of distinct bacterial populations of the indigenous microbiota with colitis in the IL-2-deficient mouse model.
MATERIALS AND METHODS
Mice.
Heterozygous IL-2+/− mice from a C57BL/6 H129/Ola background were crossed to obtain homozygous IL-2-deficient (IL-2−/−) and wild-type (IL-2+/+) mice (30). Mice were bred under SPF conditions in a barrier-sustained facility. Offspring were screened for IL-2 mutation by PCR (28). Three IL-2−/− mice and three IL-2+/+ mice, all about 22 weeks old, were investigated in this study. Physical properties of the IL-2-deficient mice—body weight, posture, and consistency of feces—exhibited obvious impairment.
Collection of colonic mucosa.
The animals were killed by CO2 asphyxiation, and their colons were removed aseptically. The colons were flushed thoroughly with sterile phosphate-buffered saline (PBS; pH 7.4). Prior to snap-freezing and storage in liquid nitrogen at −80°C, the colon samples were sectioned into proximal and distal parts of the colon.
Oligonucleotide primers and probes.
Oligonucleotide primers and probes were obtained from Interactiva Biotechnology GmbH (Ulm, Germany). Oligonucleotide probes for in situ hybridization experiments were labeled with either Cy3 or fluorescein. Table 1 displays the specificities and hybridization conditions of the fluorescent probes used in this study.
TABLE 1.
Oligonucleotide probes used for quantitative FISH analysis of mucosa-associated flora
| Probe | Sequence (5′-3′) | Specificity | % Formamidea | Reference or source |
|---|---|---|---|---|
| EUB338 | GCTGCCTCCCGTAGGAGT | Bacteria | 0 | 2 |
| NON338 | ACTCCTACGGGAGGCAGC | Negative control | 0 | 2 |
| BAC303 | CCAATGTGGGGGACCTT | Bacteroides and Prevotella | 15 | 21 |
| EREC482 | GCTTCTTAGTCA[A/G]GTACCG | C. coccoides-E. rectale group | 20 | 14 |
| SRB385 | CGGCGTCGCTGCGTCAGG | SRBb | 45 | 1 |
| VEIL501 | CGTGGCTTTCTATTCCGG | Veillonella | 40 | This study |
| ASF500 | CAATGTGGCCGGCCAACC | Bacterium ASF500c | 0 | This study |
| ASF519 | TGGAACCCCTGTTTTATGCG | Bacteroides sp. ASF519c | 0 | This study |
| ECO1531 | CACCGTAGTGCCTCGTCA | E. coli | 35 | Modified from reference 25 |
Respective concentration of formamide in the hybridization buffer.
SRB, sulfate-reducing bacteria of the delta subclass of Proteobacteria.
Part of the “altered Schaedler flora” (ASF).
Preparation of DNA.
Bulk DNA was extracted from colon samples by using either an enzymatic lysis method (DNeasy Tissue kit; QIAGEN, Hilden, Germany) according to the manufacturer's recommendations or a combination of mechanical disruption and enzymatic lysis using an MM2 mixer mill (Retsch, Haan, Germany). In brief, 25 mg of tissue was cut into small pieces and incubated overnight at 55°C in 500 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM NaCl, 10 mM EDTA, 1.0% [wt/vol] sodium dodecyl sulfate, and 0.5 μg of proteinase K/μl). To 500 μl of the lysis reaction mixture, 500 μl of zirconium beads (diameter, 0.1 mm) and 500 μl of phenol (equilibrated with 0.5 M Tris-HCl [pH 8.0]) were added and mixed for 4 min at maximum speed in an MM2 mixer mill. After phase separation (10 min, 15,000 × g), the aqueous phase was extracted twice by adding 1 volume of phenol-chloroform (1:1, vol/vol) and 1 volume of chloroform-isoamyl alcohol (24:1, vol/vol). DNA was precipitated by adding 2 volumes of ethanol (96%) and 1/10 volume of sodium acetate (3 M; pH 4.8). After the precipitated DNA was washed twice with ethanol (75 and 100%, respectively), the pellet was dried under a vacuum and dissolved in 50 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA).
Amplification of 16S rRNA genes.
Two sets of broad-range primers were used for amplification of 16S rRNA genes. Primer pairs 27F (5′-AGA GTT TGA TCM TGG CTC AG-3′)-1492R (5′-TAC GGY TAC CTT GTT ACG ACT T-3′) and 63F (5′-GAG GCC TAA CAC ATG CAA GTC-3′)-1387R (5′-GGG CGG WGT GTA CAA GGC-3′) were used to amplify almost-complete 16S rRNA genes from the mixed template preparations (18, 22). Amplification was performed on a GeneAmp PCR system 2400 thermal cycler (Perkin-Elmer [PE] Applied Biosystems, Foster City, Calif.). In brief, serial dilutions of the extracted DNA were used in 50-μl PCR mixtures containing 1× GeneAmp PCR buffer with 1.5 mM MgCl2 (PE Applied Biosystems), 200 μM each deoxynucleoside triphosphate, 1 μM each primer, and 1.25 U of AmpliTaq Gold DNA polymerase (PE Applied Biosystems). To remove contaminating DNA, the master mixes were filtered through Microcon YM-100 filters (Millipore, Bedford, Mass.) before addition of the template DNA. PCR cycling conditions were as follows: an initial denaturation step of 9 min at 95°C; 30 cycles of 60 s of denaturation at 95°C, 60 s of annealing at 55°C, and 2 min of extension at 72°C; and a single final extension step of 10 min at 72°C. Amplified 16S rRNA gene fragments were analyzed by agarose gel electrophoresis (1.5%) and purified by using the QIAquick PCR Purification kit (QIAGEN) according to the manufacturer's specifications.
16S rDNA analysis.
Representative 16S ribosomal DNA (rDNA) libraries were established on the basis of the 16S rDNA fragments amplified from nucleic acid extracts of the colon samples of three healthy control mice with an IL-2+/+ genotype and three mice with an IL-2−/− genotype that developed colitis. Cloning of the PCR fragments in Escherichia coli was performed by using the TOPO TA Cloning kit (Invitrogen Corporation, Carlsbad, Calif.) as recommended by the manufacturer.
The 16S rDNA inserts of recombinant clones were amplified by using the forward primer M13f24 (5′-CGC CAG GGT TTT CCC AGT CAC GAC-3′) and the reverse primer M13r22 (5′-AGC GGA TAA CAA TTT CAC ACA GGA-3′). Briefly, a small amount of colony material from randomly chosen recombinant clones was picked with a sterile toothpick and directly added to 50-μl PCR mixtures of 1× GeneAmp PCR buffer with 1.5 mM MgCl2 (PE Applied Biosystems), 200 μM each deoxynucleoside triphosphate, 1.25 U of AmpliTaq Gold DNA polymerase (PE Applied Biosystems), and 1 μM primers. PCR cycling conditions consisted of an initial denaturation step (9 min at 95°C); 30 cycles of denaturation (60 s at 95°C), annealing (60 s at 55°C), and extension (2 min at 72°C); and a single final extension step (10 min at 72°C). The resulting PCR fragments were analyzed for correct length on agarose gels (1.5%) and were purified by using the QIAquick PCR purification kit (QIAGEN) according to the manufacturer's recommendations. Amplicons were sequenced by using the ABI PRISM BigDye Ready Reaction Terminator cycle sequencing kit (PE Applied Biosystems) and the broad-range primer 63F as a sequencing primer on a GeneAmp PCR system 2400 thermal cycler (PE Applied Biosystems). Sequence reaction products were purified by using AutoSeq G50 columns (Amersham Pharmacia Biotech, Braunschweig, Germany) and analyzed on an ABI PRISM system 377 automated sequencer (PE Biosystems). To determine the approximate phylogenetic affiliations, the partial 16S rDNA sequences were compared to the EMBL and GenBank databases as described previously (34). The 16S rDNA sequences were added to the rDNA sequence database of the Technical University of Munich by using the ARB program package (Lehrstuhl für Mikrobiologie, Technische Universität München [http://www.mikro.biologie.tu-muenchen.de]). The resulting alignments were checked and corrected manually by considering the secondary structure of the rRNA molecule. The partial sequences were inserted into an existing tree by parsimony criteria, without allowing changes in the overall tree topology.
FISH.
Each colon sample was divided and fixed by adding PBS-ethanol (1:1 [vol/vol]; recommended for gram-positive bacteria) or paraformaldehyde (4% [wt/vol] in PBS; recommended for gram-negative bacteria). After overnight incubation at 4°C, the paraformaldehyde-fixed specimens were washed twice with sterile PBS (pH 7.4) and finally stored in PBS-ethanol (1:1 [vol/vol]). The fixed samples were disintegrated by using a tissue grinder (Dstroy-SR-16; Biozym, Hessisch Oldendorf, Germany) and stored in PBS-ethanol (1:1 [vol/vol]) at −20°C.
Fixed samples were spotted onto the wells of glass slides, dried at 37°C, and dehydrated in 50, 80, and 96% (vol/vol) ethanol (3 min each). In situ hybridization was performed at a constant temperature of 46°C in an isotonically equilibrated humidity chamber as described previously (35). Stringent hybridization conditions for the specific oligonucleotide probes listed in Table 1 were adjusted by different formamide concentrations in the hybridization buffer. After in situ hybridization, samples were counterstained with DAPI (4′,6-diamidine-2′-phenylindole). DAPI solution (100 ng ml−1) was added to the wells, and slides were incubated for 10 min at room temperature in the dark. After a rinse with distilled water, the slides were dried in the dark.
Slides were examined by using an Axiovert S100 epifluorescence microscope equipped with a 63× Plan-Apochromat Ph3 1.40 oil objective (both from Zeiss, Jena, Germany). Probe EUB338 was used as a universal probe to detect all members of the domain Bacteria and to determine the total numbers of bacteria accessible by FISH. Probe NON338 (a complement of universal probe EUB338) was used as a negative control and was included for every sample. First, the fraction of bacterial cells accessible by FISH was determined for all samples as the ratio of EUB338-positive bacterial cells to the total cell counts determined by DAPI staining. Labeling of the specific probes with one fluorochrome (Cy3; red emission) and the universal bacterial probe EUB338 with a second fluorochrome (fluorescein; green emission) allowed the probes to be applied simultaneously. Microscopic counts were determined in duplicate by performing two independent hybridizations for each sample investigated. To quantify probe-positive cells, 10 optical fields containing 50 to 100 bacterial cells per field were counted for each probe applied in each sample. The fraction of probe-positive cells was calculated as the ratio of the number of bacterial cells positive for the specific probe to the total counts of cells determined with the universal probe EUB338 in the same optical field. Upon hybridization, monochrome images were captured for each probe by using a Spot2 cooled digital charge-coupled device camera (Diagnostic Instruments Inc., Sterling Heights, Mich.) and were processed by using the MetaView software package (Universal Imaging Corporation, West Chester, Pa.).
Nucleotide sequence accession numbers.
Representative sequences determined in this study have been deposited in the EMBL nucleotide sequence database under accession numbers AJ418942 to AJ419087.
RESULTS
Influence of nucleic acid isolation methods and different primer sets on the composition of 16S rDNA clone libraries.
In preliminary experiments we investigated the effects of the nucleic acid isolation method and of different primer pairs used for amplification of 16S rRNA genes on the composition of the resulting 16S rDNA clone libraries. Two different strategies, an enzymatic and a combined mechanic-enzymatic procedure, were tested. Analysis of clone libraries from different DNA extracts of identical colon samples by sequencing randomly chosen recombinant clones revealed only minor differences. The overall composition of the clone libraries was not affected by the different isolation protocols (data not shown). In further experiments we used the combination of mechanical and enzymatic treatment for isolation of nucleic acids.
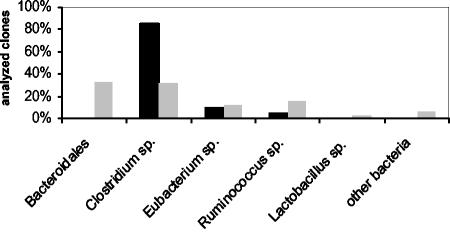
In contrast to this observation, the use of different pairs of broad-range primers for amplification of 16S rRNA genes from a colon sample of a wild-type control mouse resulted in 16S rDNA clone libraries showing differences in bacterial composition (Fig. 1). The library established from an amplicon of primers 27F and 1492R harbored exclusively sequences from members of Clostridiales. Most clones analyzed represented Clostridium spp., accompanied by a few sequences from Eubacterium spp. and Ruminococcus spp. Use of primers 63F and 1387R resulted in a 16S rDNA clone library representing a much broader bacterial diversity. In addition to Clostridiales, the library included sequences from Bacteroidales, Lactobacillaceae, and other bacterial lineages. Consequently, primers 63F and 1387R were used for all subsequent studies.
FIG. 1.
Effects of different primers on the composition of the resulting 16S rDNA clone libraries. The broad-range primer pairs 27F-1492R (solid bars) and 63F-1387R (shaded bars) were used to generate two different 16S rDNA clone libraries from the same colon sample of a wild-type control mouse. The resulting 16S rDNA clone libraries were analyzed by comparative sequence analysis of about 60 randomly chosen recombinant clones. Each bar indicates the percentage of clones representing a specific bacterial group in relation to the total number of clones analyzed.
Comparison of proximal and distal sections of mouse colon.
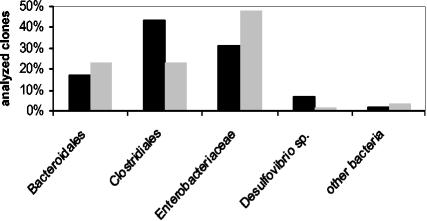
To investigate the bacterial composition of proximal and distal sections of the mouse colon, we established two different 16S rDNA clone libraries, one from a proximal and one from a distal colon sample of an IL-2-deficient mouse. The analysis revealed similar bacterial compositions in proximal and distal parts of the colon. Only slight differences were observed in the numbers of sequences representing the different bacterial subgroups harbored by the clone libraries (Fig. 2). Both libraries contained predominantly sequences from Bacteroidales, Clostridiales, and Enterobacteriaceae. Sequences from Desulfovibrionaceae and other bacterial groups were also found in both libraries.
FIG. 2.
Bacterial composition in 16S rDNA clone libraries from proximal (solid bars) and distal (shaded bars) parts of the colon from an IL-2-deficient mouse. Bars indicate the percentages of sequences from Bacteroidales, Clostridiales, Enterobacteriaceae, Desulfovibrio, and other bacteria in relation to total clones analyzed. The analysis was performed for 60 recombinant clones from each of the two clone libraries.
Comparative 16S rDNA analysis of the mucosa-associated bacterial flora from IL-2-deficient and healthy wild-type control mice.
To study and compare the composition of the mucosa-associated bacterial flora of IL-2-deficient mice and healthy wild-type controls, we established 16S rDNA clone libraries from colon samples of three IL-2-deficient mice and three wild-type control mice. Sequence analysis of clones from the libraries revealed a broad diversity of 16S rDNA sequences from gram-positive and gram-negative bacterial lineages (Table 2). Overall, the majority of sequences represented in the clone libraries from the mice investigated derived from members of Bacteroidales, Clostridiales, and Enterobacteriaceae. The libraries harbored numerous well-known members of the intestinal flora of mice but also sequences representing putative new, uncultured bacteria. A considerable fraction of the sequences obtained by this study showed the closest relationship to recently submitted “uncultured mouse clone” sequences from a 16S rDNA analysis of mouse gastrointestinal microbiota (31). Several sequences revealed homology to 16S rDNA sequences from strains ASF356, ASF457, ASF500, and ASF519. These strains belong to the “altered Schaedler flora” (ASF) and are well-known members of mouse intestinal microflora (11). Only a few sequences represented Desulfovibrio spp. and relatives such as Bilophila wadsworthia or Lawsonia intracellularis; they are therefore considered to represent sulfate-reducing bacteria (SRB) of the delta subclass of Proteobacteria (10, 13, 16). Sequences from Lactobacillus spp., Enterococcus spp., or other known members of mouse intestinal microflora were sporadically found within the clone libraries analyzed (Table 2).
TABLE 2.
Bacterial groups identified by comparative 16S rDNA analysis of mucosa-associated flora of wild-type mice and IL-2-deficient mice
| Bacterial group | No. of clones from the 16S rDNA clone library of the indicated mousea
|
|||||
|---|---|---|---|---|---|---|
| Wild type
|
IL-2 deficient
|
|||||
| 5096 | 0065 | 0076 | 5099 | 0067 | 0073 | |
| Bacteroidaceae | 17 | 35 | 37 | 25 | 16 | 10 |
| Clostridiaceae | 31 | 16 | 12 | 1 | 23 | 25 |
| Enterobacteriaceae | 25 | 19 | 18 | |||
| Desulfovibrionaceae | 1 | 1 | 4 | |||
| Acinetobacter sp. | 1 | |||||
| Denitrobacterium detoxificans | 3 | 1 | 1 | |||
| Enterococcus faecium | 1 | |||||
| Lactobacillus sp. | 1 | 1 | ||||
| Flexistipes (bacterium ASF457) | 2 | 1 | ||||
| Total | 52 | 56 | 51 | 52 | 59 | 58 |
Mice are identified by number.
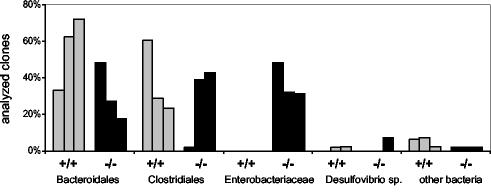
Comparison of the libraries from IL-2-deficient mice and wild-type control mice showed that the most striking difference was the lack of sequences from Enterobacteriaceae in all libraries from wild-type control mice. As indicated by Fig. 3, libraries from wild-type control mice were dominated by sequences from members of Bacteroidales and Clostridiales and harbored no sequences from members of Enterobacteriaceae. Conversely, libraries from IL-2-deficient mice suffering from colitis harbored up to 50% sequences from Enterobacteriaceae that were almost identical to the 16S rDNA from E. coli. This result points to a preferential occurrence of E. coli in the colonic mucosae of the IL-2-deficient mice. To verify this observation, we quantified the respective bacterial populations in colon samples by FISH using 16S rRNA-targeted oligonucleotide probes.
FIG. 3.
Bacterial composition of the mucosa-associated flora determined by comparative 16S rDNA analysis of wild-type control mice and IL-2-deficient mice. 16S rDNA clone libraries were established from three wild-type control mice (+/+) (shaded bars) and three IL-2-deficient mice (−/−) (solid bars). About 60 randomly chosen clones of each library were sequenced and compared to databases. The amount of each bacterial subgroup is expressed as a percentage of the total number of analyzed clones represented in the 16S rDNA clone library for each mouse.
Quantification of bacterial populations by FISH.
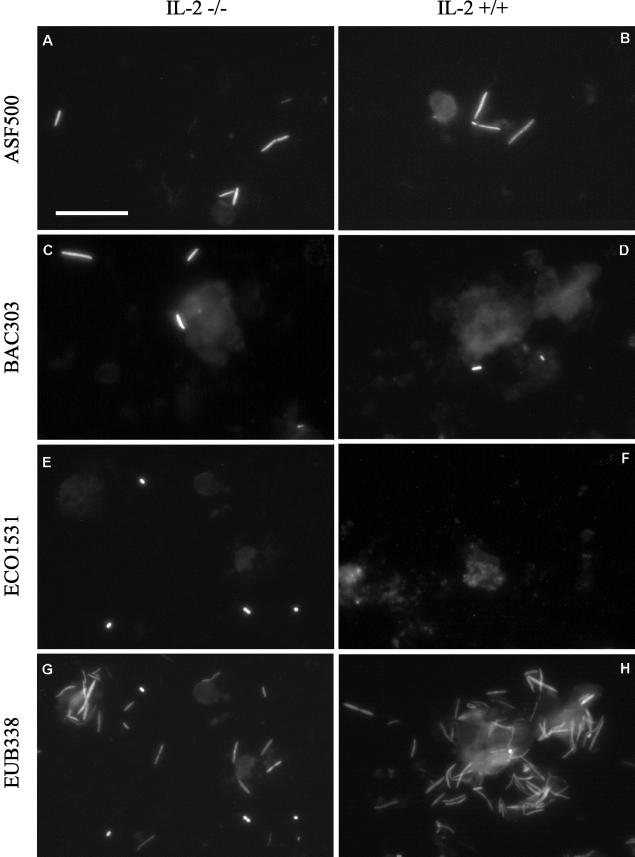
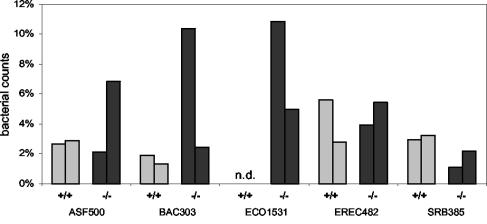
FISH was performed by using the oligonucleotide probes and hybridization conditions given in Table 1. In a first step, we assessed the proportion of bacteria accessible by FISH. For that purpose, we determined for each colon sample the counts obtained by FISH using the universal probe EUB338 and compared them with the values obtained after counterstaining with DAPI. Use of the universal probe EUB338 revealed that about 95% of the DAPI-stained bacteria in the samples investigated were accessible by FISH. In general, use of the universal probe EUB338 showed a great domination of fusiform bacteria or tapered rods presumably belonging to the extremely oxygen sensitive (EOS) bacteria. In a second step, we used group-specific probes to quantify defined bacterial populations represented by the 16S rDNA clone libraries. Figure 4 shows examples of the use of the specific oligonucleotide probes with colon samples from a wild-type mouse and an IL-2-deficient mouse. The relative levels of certain bacterial populations in the colon samples from wild-type and IL-2-deficient mice determined by FISH are summarized in Fig. 5. An estimate of the field-to-field and sample-to-sample variation is provided by Table 3.
FIG.4.
Whole-cell in situ hybridization of mucosa-associated microflora from IL-2-deficient mice (−/−) (left) and wild-type control mice (+/+) (right) by use of the fluorescent rRNA-targeted oligonucleotide probes shown. Bar, 20 μm (applies to all photomicrographs). (A and B) Probe ASF500 identified tapered rods as part of the mucosa-associated microflora in both groups of mice. (C and D) Probe BAC303 facilitated the specific detection of members of Bacteroides and Prevotella in all samples analyzed. (E and F) Probe ECO1531 identified E. coli cells in samples from IL-2-deficient mice, whereas no signals were observed in samples from wild-type mice. (G and H) The Bacteria-specific probe EUB338 was used as a positive control to prove that bacteria were accessible for the fluorescent rRNA-targeted oligonucleotide probes used.
FIG. 5.
Quantification of bacterial populations in colon samples from two healthy wild-type control mice (+/+) (shaded bars) and two IL-2-deficient mice (−/−) (solid bars) by whole-cell hybridization using a set of fluorescent rRNA-targeted oligonucleotide probes. The specificities of the oligonucleotide probes used are shown in Table 1. Each bar represents an individual animal and reflects the mean percentage of specific bacterial populations relative to the total bacterial counts from two independent hybridizations. n.d., no bacteria detected.
TABLE 3.
Quantification of bacteria in wild-type and IL-2-deficient mice by FISH
| Probea | No. of bacteria in the indicated sample from the indicated mouseb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild type
|
IL-2 deficient
|
|||||||
| 0065
|
0076
|
0073
|
5099
|
|||||
| Sample 1 | Sample 2 | Sample 1 | Sample 2 | Sample 1 | Sample 2 | Sample 1 | Sample 2 | |
| ASF500 | 1.5 ± 0.7 | 1.4 ± 0.8 | 1.5 ± 0.5 | 1.6 ± 0.7 | 4.4 ± 1.4 | 5.0 ± 1.6 | 1.3 ± 0.5 | 1.5 ± 0.7 |
| BAC303 | 1.1 ± 0.3 | 1.0 ± 0.0 | 0.7 ± 0.5 | 0.6 ± 0.7 | 1.6 ± 0.7 | 1.7 ± 0.7 | 9.2 ± 2.4 | 9.3 ± 2.4 |
| ECO1531 | ND | ND | ND | ND | 3.2 ± 1.6 | 3.0 ± 1.1 | 10.3 ± 3.8 | 9.4 ± 2.6 |
| EREC482 | 2.9 ± 1.0 | 3.0 ± 0.9 | 1.7 ± 0.8 | 1.5 ± 0.7 | 3.4 ± 1.0 | 3.9 ± 1.7 | 2.0 ± 0.7 | 2.0 ± 0.7 |
| SRB385 | 1.6 ± 0.7 | 1.5 ± 0.7 | 1.3 ± 0.5 | 1.4 ± 0.5 | 1.3 ± 0.5 | 1.3 ± 0.5 | 1.2 ± 0.4 | 0.7 ± 0.7 |
Specificities of probes are given in Table 1.
Mice are identified by number. Values are average counts ± standard deviations from 10 optical fields containing 50 to 100 bacterial cells per field. ND, no bacteria detected.
According to the results of the comparative 16S rDNA analysis, the most obvious difference between wild-type control mice and IL-2-deficient mice was observed by use of probe ECO1531. In IL-2-deficient mice, probe ECO1531 recognized up to 10% of total bacterial counts, whereas no bacteria were detected in samples from wild-type controls. Probe SRB385 identified about 2% of the total bacterial counts in both groups of mice as SRB of the delta subclass of Proteobacteria. About 4% of total bacteria in each group of mice were identified as members of the Clostridium coccoides-Eubacterium rectale subgroup of Clostridiaceae by probe EREC482. Populations of Bacteroides spp. and Prevotella spp. recognized by probe BAC303 were detected in different amounts. Wild-type control mice harbored about 2% Bacteroides and Prevotella spp., whereas one sample from an IL-2-deficient mouse revealed higher numbers, up to 10%. About 3% of total bacteria in both groups of mice hybridized with probe ASF500, whereas ASF519 and VEIL501 did not recognize any bacteria in the colon samples from the mice investigated in this study.
In conclusion, E. coli was detected exclusively in samples from IL-2-deficient mice and not in samples from healthy wild-type control mice. All other probes used in this study identified populations of the respective target bacteria in both groups of mice. The differences in the presence of E. coli found by the quantitative FISH analysis strongly underline the findings from the comparative 16S rDNA analysis and support the differential occurrence of E. coli in the colonic mucosae of IL-2-deficient mice and wild-type control mice.
DISCUSSION
Various studies with animal models indicate that the enteric microflora represents an essential cofactor involved in the pathogenesis of IBD (12, 15, 24, 27, 36). However, it is a matter of controversy whether certain bacteria play a key role and how they contribute to the pathogenesis of IBD. In this study, to our knowledge the first one combining a culture-independent 16S rRNA-based approach and IL-2-deficient mice as an animal model for IBD, we analyzed and compared the mucosa-associated microflora in colon samples from IL-2-deficient mice suffering from colitis and healthy wild-type control mice.
Although today the 16S rRNA approach represents the ultimate technique for a culture-independent analysis of complex microbial communities, even this method is subject to bias (41). Therefore, we performed preliminary experiments to study the effects of different methods for isolation of nucleic acids or different primers used for the amplification of 16S rRNA genes. The particular method used for the isolation of DNA had no major effect on the composition of the resulting 16S rDNA libraries. In contrast, the results from testing different broad-range primers supported the observation that primers 27F and 1492R fail to work with difficult samples (22). Furthermore, we investigated proximal and distal parts of a colon sample from an IL-2-deficient mouse in order to exclude differences in bacterial composition due to differences in the anatomic provenance of the samples. Analysis of the respective 16S rDNA libraries revealed only slight differences, indicating that the proximal and distal parts of the colonic mucosa harbor a similarly structured microbiota.
Analysis of the 16S rDNA libraries established from IL-2-deficient mice and healthy wild-type controls revealed a diverse microbiota representing a wide variety of bacterial lineages. As expected for gastrointestinal samples, most sequences represented members of Bacteroidales, Clostridiales, or Enterobacteriaceae. The presence of numerous 16S rDNA sequences showing only weak homologies to database sequences from known bacterial species supports the hypothesis that most bacteria that inhabit the gastrointestinal tracts of mammals or other anaerobic ecosystems represent hitherto unknown bacterial species (17, 26, 38). This observation was further supported by the occurrence of numerous sequences from members of Bacteroidales or Clostridiales which showed by far the highest homology to recently submitted 16S rDNA sequences retrieved from mouse gastrointestinal microbiota (31). Together with the identification of sequences from members of the ASF, these findings give further support to the plausibility of the results of the 16S rDNA analysis and demonstrate that the 16S rDNA libraries established in this study covered a broad spectrum of the mouse intestinal microbiota.
The most striking difference in the composition of the 16S rDNA clone libraries from wild-type and IL-2-deficient mice was the lack of sequences from Enterobacteriaceae in libraries from wild-type control mice. In contrast, the libraries from IL-2-deficient mice were dominated by 16S rDNA sequences from Enterobacteriaceae resembling E. coli. This was a surprising observation, because E. coli is a known member of the luminal flora of mice (40). To confirm the results obtained by comparative 16S rDNA analysis, we performed a FISH analysis to quantify the respective bacterial populations in the colon samples. The quantitative results from FISH clearly supported our data from the 16S rDNA analysis. The colonic biopsy specimens of IL-2-deficient mice harbored up to 10% E. coli, but not a single E. coli cell was detectable in the samples from wild-type control mice. Probe ECO1531 shows no exclusive specificity for E. coli. In fact, closely related members of Salmonella and Shigella spp., as well as Citrobacter freundii and Klebsiella pneumoniae, also harbor the target sequence. But the knowledge that the mice in this study were SPF renders an occurrence of these pathogenic bacteria in the samples from these mice rather improbable. Moreover, the absence of these bacteria is supported by the results of the 16S rDNA analysis. The libraries harbored exclusively sequences from Enterobacteriaceae which could be assigned to E. coli or Shigella spp. This observation legitimates the assumption that the bacteria detected in the colon samples of the mice investigated represent E. coli.
These results suggest that E. coli, although present in high numbers in the feces of mice, is absent or below the detection limit in the colonic mucosae of wild-type mice, an observation which is concordant with reports from the investigation of thin sections from large intestines of healthy conventional mice by in situ hybridization (25). The single E. coli cells identified in the thin frozen sections were seen embedded in the mucosal material overlying the epithelial cells of the large intestine, and no direct attachment to the epithelium was observed. This leads to the assumption that the absence of E. coli in the samples from healthy controls in this study was due to the pretreatment of the colon samples. After removal of the intestines, the colon was flushed thoroughly with PBS. The presence of numerous E. coli organisms in samples from IL-2-deficient mice would therefore point to a tighter attachment or invasion of the mucosa. The question arises whether the different results for IL-2-deficient and wild-type mice could be due to differences in the relative levels of E. coli in the feces of wild-type versus IL-2-deficient mice. Culture-based enumeration of E. coli in fecal samples of IL-2-deficient and wild-type mice revealed an insignificantly higher abundance of E. coli in colonic feces of IL-2-deficient mice than in those of wild-type controls (42). This result does not suggest that different relative levels of E. coli in the feces of IL-2-deficient mice and wild-type controls represent the cause for the different occurrence of E. coli in the colonic mucosae.
In conclusion, our results from 16S rDNA analysis and FISH demonstrate the high incidence of E. coli in the colonic mucosae of IL-2-deficient mice. The numbers of E. coli bacteria in the colonic mucosae of healthy wild-type mice were not only significantly lower; E. coli was almost absent. The abundance of E. coli in the mucosa-associated flora of IL-2-deficient mice, together with the observed lack of E. coli in the mucosa-associated flora of wild-type mice, implies a participation of E. coli in the development of colitis in this animal model.
The hypothesis that E. coli contributes to inflammatory disease is further emphasized by several studies. Strong evidence for a pivotal role of E. coli in the pathogenesis of colitis in IL-2-deficient mice also appears from experiments demonstrating that gnotobiotic IL-2-deficient mice monoassociated with E. coli develop severe colitis (42). Onderdonk et al. (23) analyzed the cecal flora of HLA-B27 transgenic rats with IBD and reported a rise in the numbers of E. coli and Enterococcus spp. corresponding to the presence and severity of IBD in these rats. The investigation of sera from IBD patients and matched control subjects revealed that the majority of patients with IBD had agglutinating antibodies to a higher number of E. coli O antigens, and in higher titers, than the control group (39). In addition, it was shown that E. coli and some Bacteroides strains express proteins that cross-react with pANCA autoantibodies (8). These antibodies are found in most cases of ulcerative colitis and hence reflect an immune response associated with the disease. The interpretation of immunocytochemical data gives further support to the findings described above (19). Beyond data from immunology, evidence for a pathogenic role of E. coli in IBD is also based on results from studies that demonstrate the colonization of early and chronic ileal lesions of Crohn's disease by E. coli strains. These strains were found to be devoid of virulence genes so far described for pathogenic strains involved in acute gastrointestinal diseases (37). Furthermore, the presence of adherent E. coli strains in ileal mucosae of patients with Crohn's disease was demonstrated, and an E. coli strain isolated from the ileal mucosa of a patient with Crohn's disease was proved to possess invasive ability (7, 9). These observations point to the existence of a new potentially pathogenic group of E. coli, for which the designation “adherent-invasive E. coli” has been proposed.
Despite these observations, the primary pathogenic role of E. coli in IBD is the subject of controversy due to studies reporting conflicting data. The presence of E. coli antigens in ulcers suggests a secondary infection in these lesions and does not support a primary pathogenic role (33). Furthermore, it has been shown that E. coli strains which possess adherence factors reside in the large intestine and adhere to the rectal mucosa, irrespective of the presence of colitis (43).
In addition, while monocolonization of IL-2-deficient mice with E. coli strain mpk induced colitis, monocolonization with E. coli Nissle did not (42), suggesting that specific features of a given strain determine its potential to induce colitis. Therefore, we can conclude that 16S rRNA-based approaches might be useful for addressing the composition of the microflora in IBD. However, to understand the role of a given bacterial species in the pathogenesis of IBD, additional molecular approaches including pathotyping are required.
Editor: B. B. Finlay
REFERENCES
- 1.Amann, R., W. Ludwig, and K. H. Schleifer. 1992. Identification and in situ detection of individual bacterial cells. FEMS Microbiol. Lett. 100:45-50. [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Autenrieth, I. B., N. Bucheler, E. Bohn, G. Heinze, and I. Horak. 1997. Cytokine mRNA expression in intestinal tissue of interleukin-2 deficient mice with bowel inflammation. Gut 41:793-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, R. D. 1996. The indigenous gastrointestinal microflora. Trends Microbiol. 4:430-435. [DOI] [PubMed] [Google Scholar]
- 6.Blumberg, R. S., L. J. Saubermann, and W. Strober. 1999. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr. Opin. Immunol. 11:648-656. [DOI] [PubMed] [Google Scholar]
- 7.Boudeau, J., A. L. Glasser, E. Masseret, B. Joly, and A. Darfeuille-Michaud. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cohavy, O., D. Bruckner, L. K. Gordon, R. Misra, B. Wei, M. E. Eggena, S. R. Targan, and J. Braun. 2000. Colonic bacteria express an ulcerative colitis pANCA-related protein epitope. Infect. Immun. 68:1542-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darfeuille-Michaud, A., C. Neut, N. Barnich, E. Lederman, P. Di Martino, P. Desreumaux, L. Gambiez, B. Joly, A. Cortot, and J. F. Colombel. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405-1413. [DOI] [PubMed] [Google Scholar]
- 10.Deplancke, B., K. R. Hristova, H. A. Oakley, V. J. McCracken, R. Aminov, R. I. Mackie, and H. R. Gaskins. 2000. Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 66:2166-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewhirst, F. E., C. C. Chien, B. J. Paster, R. L. Ericson, R. P. Orcutt, D. B. Schauer, and J. G. Fox. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dianda, L., A. M. Hanby, N. A. Wright, A. Sebesteny, A. C. Hayday, and M. J. Owen. 1997. T cell receptor-alpha beta-deficient mice fail to develop colitis in the absence of a microbial environment. Am. J. Pathol. 150:91-97. [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, J. G., F. E. Dewhirst, G. J. Fraser, B. J. Paster, B. Shames, and J. C. Murphy. 1994. Intracellular Campylobacter-like organism from ferrets and hamsters with proliferative bowel disease is a Desulfovibrio sp. J. Clin. Microbiol. 32:1229-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Lafuente, A., M. Antolín, F. Guarner, E. Crespo, A. Salas, P. Forcada, M. Laguarda, J. Gavaldá, J. A. Baena, J. Vilaseca, and J. R. Malagelada. 1997. Incrimination of anaerobic bacteria in the induction of experimental colitis. Am. J. Physiol. 272:G10-G15. [DOI] [PubMed] [Google Scholar]
- 16.Gebhart, C. J., S. M. Barns, S. McOrist, G. F. Lin, and G. H. Lawson. 1993. Ileal symbiont intracellularis, an obligate intracellular bacterium of porcine intestines showing a relationship to Desulfovibrio species. Int. J. Syst. Bacteriol. 43:533-538. [DOI] [PubMed] [Google Scholar]
- 17.Godon, J. J., E. Zumstein, P. Dabert, F. Habouzit, and R. Moletta. 1997. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63:2802-2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acids techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 19.Liu, Y., H. J. van Kruiningen, A. B. West, R. W. Cartun, A. Cortot, and J. F. Colombel. 1995. Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn's disease. Gastroenterology 108:1396-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K. H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 21.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 22.Marchesi, J. R., T. Sato, A. J. Weightman, T. A. Martin, J. C. Fry, S. J. Hiom, D. Dymock, and W. G. Wade. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 64:795-799. (Erratum, 64:2333.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onderdonk, A. B., J. A. Richardson, R. E. Hammer, and J. D. Taurog. 1998. Correlation of cecal microflora of HLA-B27 transgenic rats with inflammatory bowel disease. Infect. Immun. 66:6022-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitcher, M. C., E. R. Beatty, and J. H. Cummings. 2000. The contribution of sulphate reducing bacteria and 5-aminosalicylic acid to faecal sulphide in patients with ulcerative colitis. Gut 46:64-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulsen, L. K., F. Lan, C. S. Kristensen, P. Hobolth, S. Molin, and K. A. Krogfelt. 1994. Spatial distribution of Escherichia coli in the mouse large intestine inferred from rRNA in situ hybridization. Infect. Immun. 62:5191-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rath, H. C., H. H. Herfarth, J. S. Ikeda, W. B. Grenther, T. E. J. Hamm, E. Balish, J. D. Taurog, R. E. Hammer, K. H. Wilson, and R. B. Sartor. 1996. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human β2 microglobulin transgenic rats. J. Clin. Investig. 98:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rath, H. C., K. H. Wilson, and R. B. Sartor. 1999. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infect. Immun. 67:2969-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadlack, B., H. Merz, H. Schorle, A. Schimpl, A. C. Feller, and I. Horak. 1993. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell 75:253-261. [DOI] [PubMed] [Google Scholar]
- 30.Sadlack, B., R. Kuhn, H. Schorle, K. Rajewski, W. Muller, and I. Horak. 1994. Development and proliferation of lymphocytes in mice deficient for both interleukins-2 and 4. Eur. J. Immunol. 24:281-284. [DOI] [PubMed] [Google Scholar]
- 31.Salzman, N. H., H. de Jong, Y. Paterson, H. J. M. Harmsen, G. W. Welling, and N. A. Bos. 2002. Analysis of 16S rDNA libraries of mouse gastrointestinal microbiota reveals a large new taxon of mouse intestinal bacteria. Microbiology 148:3651-3660. [DOI] [PubMed] [Google Scholar]
- 32.Sartor, R. B. 1995. Current concepts of the etiology and pathogenesis of ulcerative colitis and Crohn's disease. Gastroenterol. Clin. N. Am. 24:475-507. [PubMed] [Google Scholar]
- 33.Schultsz, C., M. Moussa, R. van Ketel, G. N. Tytgat, and J. Dankert. 1997. Frequency of pathogenic and enteroadherent Escherichia coli in patients with inflammatory bowel disease and controls. J. Clin. Pathol. 50:573-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuppler, M., F. Mertens, G. Schön, and U. B. Göbel. 1995. Molecular characterization of nocardioform actinomycetes in activated sludge by 16S rRNA analysis. Microbiology 141:513-521. [DOI] [PubMed] [Google Scholar]
- 35.Schuppler, M., M. Wagner, G. Schön, and U. B. Göbel. 1998. In situ identification of nocardioform actinomycetes in activated sludge using fluorescent rRNA-targeted oligonucleotide probes. Microbiology 144:249-259. [DOI] [PubMed] [Google Scholar]
- 36.Sellon, R. K., S. Tonkonogy, M. Schultz, L. A. Dieleman, W. Grenther, E. Balish, D. M. Rennick, and R. B. Sartor. 1998. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect. Immun. 66:5224-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen, W., H. Steinruck, and A. Ljungh. 1995. Expression of binding of plasminogen, thrombospondin, vitronectin, and fibrinogen, and adhesive properties by Escherichia coli strains isolated from patients with colonic diseases. Gut 36:401-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tabaqchali, S., D. P. O'Donoghue, and K. A. Bettelheim. 1978. Escherichia coli antibodies in patients with inflammatory bowel disease. Gut 19:108-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tannock, G. W. 1979. Coliforms and enterococci isolated from the intestinal tract of conventional mice. Microb. Ecol. 5:27-34. [DOI] [PubMed] [Google Scholar]
- 41.von Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 42.Waidmann, M., O. Bechtold, J. Frick, H. Lehr, S. Schubert, U. Dobrindt, J. Loeffler, E. Bohn, and I. B. Autenrieth. 2003. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology 125:162-178. [DOI] [PubMed] [Google Scholar]
- 43.Walmsley, R. S., A. Anthony, R. Sim, R. E. Pounder, and A. J. Wakefield. 1998. Absence of Escherichia coli, Listeria monocytogenes, and Klebsiella pneumoniae antigens within inflammatory bowel disease tissues. J. Clin. Pathol. 51:657-661. [DOI] [PMC free article] [PubMed] [Google Scholar]