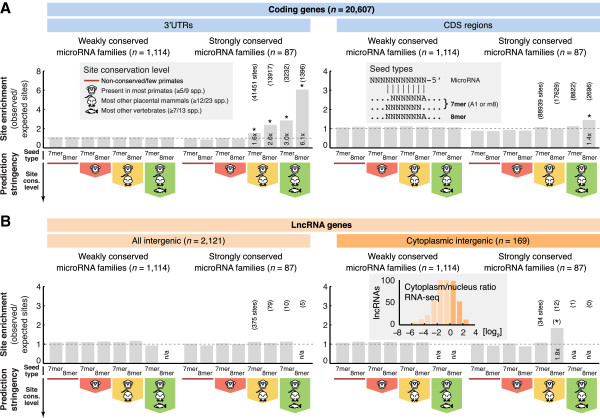
Figure 2.
Ratios between observed and expected microRNA target site frequencies in coding genes and long non-coding RNAs (lncRNAs). (A) Our methodology was first established on coding genes. The 3’ untranslated regions (UTRs) and coding sequences (CDS) were analyzed separately. We compared observed numbers of seed matches (in parentheses) to randomly expected numbers based on sets of synthetic seeds that preserved the dinucleotide frequencies of the actual seeds. Different prediction stringencies (site conservation level and seed quality) were applied, further explained within gray boxes. The analysis focused on highly conserved microRNA families (n = 87), but non-conserved families were included as a control. Bars show mean observed-to-expected ratios from 20 repeated trials. (B) Similar analysis based on intergenic lncRNAs and cytoplasmic intergenic lncRNAs. Placental mammal conserved 8-mer sites were present above expectation in a small subset of cytoplasmic intergenic lncRNAs (12 sites for 11 microRNA families, in 8 lncRNA genes). Subcellular localization was determined based on RNA-seq libraries from seven fractionated cell lines. *, empirical P <0.05 for ratio being greater than 1; (*), P = 0.10; n/a, observed counts to low.