Abstract
C1 inhibitor (C1INH) prevents endotoxin shock in mice via a direct interaction with lipopolysaccharide (LPS). This interaction requires the heavily glycosylated amino-terminal domain of C1INH. C1INH in which N-linked carbohydrate was removed by using N-glycosidase F was markedly less effective in protecting mice from LPS-induced lethal septic shock. N-deglycosylated C1INH also failed to suppress fluorescein isothiocyanate (FITC)-LPS binding to and LPS-induced tumor necrosis factor alpha mRNA expression by the murine macrophage-like cell line, RAW 264.7, and cells in human whole blood. In an enzyme linked immunosorbent assay, the N-deglycosylated C1INH bound to LPS very poorly. In addition, C1INH was shown to bind to diphosphoryl lipid A (dLPA) but only weakly to monophosphoryl lipid A (mLPA). As with intact LPS, binding of N-deglycosylated C1INH to dLPA and mLPA was diminished in comparison with the native protein. Removal of O-linked carbohydrate had no effect on any of these activities. Neither detoxified LPS, dLPA, nor mLPA had any effect on the rate or extent of C1INH complex formation with C1s or on cleavage of the reactive center loop by trypsin. These data demonstrate that N-linked glycosylation of C1INH is essential to mediate its interaction with the LPA moiety of LPS and to protect mice from endotoxin shock.
Septic shock caused by gram-negative bacteria is due primarily to endotoxin lipopolysaccharide (LPS), which is a complex glycolipid found in the outer membrane of all gram-negative bacteria (6). Treatment of gram-negative bacterial infections would be greatly aided by substances that can effectively block production of inflammatory mediators from LPS-induced mononuclear phagocytes. Administration of LPS to humans or experimental animals results in many of the physiological changes observed during gram-negative bacterial infection, including fever, hypotension, hypoglycemia, disseminated intravascular coagulation, and shock (5). LPS is composed of two chemically dissimilar structural regions: the hydrophilic repeating polysaccharide of the core and O-antigen structures and a hydrophobic domain known as lipid A (LPA) (40). LPA is the toxic principle of gram-negative bacterial LPS and has full endotoxin activity (15, 38). Virtually all LPS-induced biological responses are LPA dependent (33). Therefore, recognition of LPA by cells must be the initial step in LPS-induced cellular responses. The general chemical structure of LPA from diverse gram-negative bacteria is highly conserved (40). LPA has the biological function to induce nuclear factor-κB activation in monocytes (24) and the production of proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α) and interleukin-1 (IL-1) from macrophages (2, 16). The acute-phase reactant LPS binding-protein (LBP) binds to the LPA moiety with high affinity and facilitates the transfer of LPS to CD14 (38, 40, 44). LPA contains the binding domain recognized by LPS-binding proteins, but the polysaccharide O chain and oligosaccharide core structure of endotoxin LPS may interfere with the interaction of these proteins with LPA (1, 8).
Both the complement system and the contact system are implicated in the pathophysiology of sepsis. C1 inhibitor (C1INH), a plasma glycoprotein that belongs to the superfamily of serine proteinase inhibitors, regulates these two systems. Levels of proteolytically inactivated C1INH are increased in fatal septic shock, which suggests an increased turnover and a relative secondary deficiency of biologically active C1INH (27). C1INH can be inactivated by limited proteolytic cleavage by elastase released from activated neutrophils (7, 9). The inactivation of C1INH may occur locally in inflamed tissue and thereby contribute to increased local complement activation (7). Both active C1INH and reactive center cleaved, inactive C1INH (iC1INH) protected mice from lethal gram-negative endotoxin shock (22). C1INH blocked the binding of Salmonella enterica serovar Typhimurium LPS to the murine macrophage cell line, RAW 264.7, and to human blood cells. It directly interacted with LPS and suppressed LPS-induced TNF-α mRNA expression. Deletion of the amino-terminal 97 amino acid residues abolished the ability of C1INH to bind to LPS. Therefore, C1INH, in addition to its function as a serine protease inhibitor, has a novel anti-inflammatory function mediated via the heavily glycosylated amino-terminal nonserpin domain (22).
We provide here evidence that N deglycosylation significantly reduced C1INH-mediated protection of mice from lethal LPS-induced shock. The data also demonstrate that N-deglycosylated C1INH did not bind to LPS, could not inhibit the binding of LPS to RAW 264.7 cells or to human blood cells, and could not prevent the activation of these cells by LPS. Furthermore, additional experiments demonstrate that C1INH binding to LPS was mediated via LPA and that this binding also was reduced by removal of N-linked carbohydrate from C1INH.
MATERIALS AND METHODS
Reagents.
C1INH and C1s were from Advanced Research Technologies (San Diego, Calif.). Moloney murine leukemia virus (M-MuLV) reverse transcriptase (RT), N-glycosidase F (from Flavobacterium meningosepticum), and neuraminidase (from Arthrobacter ureafaciens) were purchased from New England BioLabs (Beverly, Mass.). O-Glycosidase (from Diplococcus pneumoniae) was from Roche Diagnostics GmbH (Mannheim, Germany). ο-Phenylenediamine dihydrochloride substrates, LPS, and fluorescein isothiocyanate (FITC)-conjugated LPS from Salmonella enterica serovar Typhimurium were obtained from Sigma Chemical Co. (St. Louis, Mo.), as were serovar Typhimurium LPS-derived monophosphoryl lipid A (mLPA), diphosphoryl lipid A (dLPA), and detoxified lipopolysaccharide (dLPS). Rabbit anti-human C1INH antibody was purchased from Dako (Glostrup, Denmark). Goat anti-rabbit immunoglobulin G (H+L) conjugated with horseradish peroxidase was purchased from Pierce Biotechnology (Rockford, Ill.).
Enzymatic deglycosylation of C1INH.
Deglycosylation was performed as described previously (12). N-linked carbohydrate was removed from plasma-derived C1INH by treatment with 50 U of N-glycosidase F/ml for 2 h at 37°C. O-linked carbohydrate was removed by incubation with 0.1 U of O-glycosidase and 0.1 U of neuraminidase/ml overnight at 37°C. To remove both N- and O-linked carbohydrates, C1INH (10 μg/ml) was treated with 50 U of N-glycosidase F, 0.1 U of O-glycosidase, and 0.1 U of neuraminidase/ml overnight at 37°C. Each of these reactions was performed in sodium phosphate buffer (50 mM, pH 7.5).
Mouse endotoxin shock model.
C57BL/6J mice (male, 6 to 8 weeks, 18 to 22 g) (Charles River Laboratories, Wilmington, Mass.) were injected intraperitoneally (i.p.) with a lethal dose (20 mg/kg) of serovar Typhimurium LPS mixed with either normal intact, N-deglycosylated, O-deglycosylated, or both N- and O-deglycosylated C1INH (200 μg/mouse). Control mice were injected (i.p.) with LPS (20 mg/kg) alone, with a mixture of LPS (20 mg/kg) and the buffer used for deglycosylation, with the deglycosylation buffer alone, with a mixture of LPS and the glycosidase enzymes, or with the glycosidases alone. Mice were monitored for 5 days. Kaplan-Meier survival analysis was performed by using GraphPad Prism 4.00 (GraphPad Software, Inc., San Diego, Calif.). Survival curves of the different groups were compared by using the log-rank test (GraphPad Prism 4.00). All experiments were performed in compliance with relevant laws and institutional guidelines and were approved by the CBR Institute for Biomedical Research Animal Care and Use Committee.
Cell culture.
The murine RAW 264.7 macrophage cell line (American Type Culture Collection [ATCC], Manassas, Va.) was cultured in Dulbecco minimal essential medium (DMEM; ATCC) supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, Calif.) at 37°C in 5% CO2. Confluent macrophages were detached by washing with phosphate-buffered saline (PBS; pH 7.4).
FACS.
The murine RAW 264.7 macrophages were incubated with FITC-conjugated LPS (175 ng/ml) in the absence or presence of the various deglycosylated C1INH (150 μg/ml) preparations in DMEM containing 10% FBS for 15 min at 37°C. In other experiments, human peripheral venous blood from a normal volunteer was collected into a tube containing EDTA (1 mg/ml of whole blood). Aliquots of the whole blood were treated with LPS at a final concentration of 175 ng/ml in the absence or presence of added N-deglycosylated C1INH (150 μg/ml), O-deglycosylated C1INH (150 μg/ml), or both N- and O-deglycosylated C1INH (150 μg/ml) for 15 min at 37°C. Cells were fixed with fluorescence-activated cell sorter (FACS) solution after being washed with PBS three times. The binding of FITC-conjugated LPS was analyzed on a FACSCalibur apparatus (Becton Dickinson, San Jose, Calif.) by using CellQuest software.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
C1INH was incubated with C1s (ratio, 1:1) for 20 min at 37°C after the addition of mLPA, dLPA, or dLPS from serovar Typhimurium LPS. Reactions were subjected to electrophoresis with a 10% SDS-Tris-glycine polyacrylamide gel (Invitrogen). The gels were stained with Coomassie blue and then dried for 1 h at 80°C.
RT-PCR.
Total RNA was isolated from the RAW 264.7 macrophages induced with LPS (175 ng/ml) in the presence or absence of N-deglycosylated C1INH, O-deglycosylated C1INH, or both N- and O-deglycosylated C1INH and then reverse transcribed by using M-MuLV RT with oligo(dT)20 primers (Invitrogen) for 1 h at 37°C. PCR primers were designed to generate mouse TNF-α and β-actin fragments, each with lengths of 200 bp (TNF-α, sense [5′-ATGAGCACAGAAAGCATGATCC-3′] and antisense [5′-GAGGCCATTTGGGAACTTCTC-3′]; β-actin, sense [5′-TGGATGACGATATCGCTGC-3′] and antisense [5′-AGGGTCAGGATACCTCTCTT-3′]). PCR products were analyzed on 1.2% (wt/vol) agarose gels containing 0.5 μg of ethidium bromide/ml and visualized under UV light. Band density was analyzed and quantified by using ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). In addition, human peripheral venous blood was collected from a normal volunteer and anticoagulated with EDTA (1 mg/ml of whole blood). Aliquots of the whole blood were treated with LPS at a final concentration of 175 ng/ml with or without added C1INH (5 to 150 μg/ml) for 15 min at 37°C. In other experiments, total RNA was isolated from the blood leukocytes and was reverse transcribed by using M-MuLV RT with oligo(dT)20 primers. PCR primers were designed for human TNF-α (sense [5′-ATGAGCACTGAAAGCATGATCCGGGACGTG-3′] and antisense [5′-AGGTCCCTGGGGAACTCTTCCCTCTG-3′]) and human β-actin (sense [5′-ATGGATGATGATATCGCCGCGCTCGTCGTC-3′] and antisense [5′-AGGGTGAGGATGCCTCTCTTGCTCTG-3′]).
Enzyme-linked immunosorbent assay (ELISA).
Plates (polyvinyl chloride, 96 U-bottom wells; Becton Dickinson, Franklin Lakes, N.J.) were coated with mLPA, dLPA, dLPS, and LPS at room temperature overnight. C1INH (150 μg/ml) was incubated with mLPA-, dLPA-, dLPS-, and LPS-coated plates for 1 h at room temperature, respectively. In other experiments, deglycosylated C1INH (150 μg/ml) was treated for 2 h at 37°C or overnight at 37°C and was stopped by chilling, followed by incubation in LPS (175 ng/ml)-coated plates for 1 h at room temperature. Rabbit anti-human C1INH antibody (1:1,000) was incubated for 1 h at room temperature. After being washed, plates were incubated with ImmunoPure goat anti-rabbit immunoglobulin G (H+L) conjugated with horseradish peroxidase (1:100,000). Color reactions were developed for 3 min at room temperature, and reactions were terminated with 3 N HCl. The absorbance of each well was measured at 490 nm by using Revelation Microsoft in an MRX Microplate reader (Dynex Technologies, Chantilly, Va.). Standard samples were detected with the different concentrations of human C1INH binding to rabbit anti-human C1INH antibody.
RESULTS
Analysis of deglycosylated C1INH.
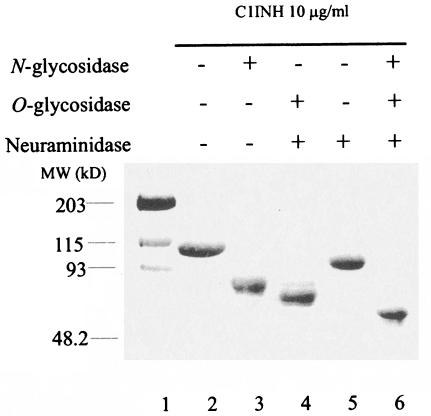
Treatment of plasma-derived intact C1INH with neuraminidase reduced the molecular mass, as judged by SDS-PAGE, from 106 to 96 kDa (Fig. 1). Removal of N- and O-linked carbohydrate resulted in decreases to 89 and 83 kDa, respectively, while the combination of N- and O-glycosidase treatment reduced the molecular mass to 75 kDa. These apparent size differentials are consistent with previously published data (30).
FIG. 1.
SDS-PAGE analysis of deglycosylated C1INH. C1INH (10 μg) was incubated with N-glycosidase F, O-glycosidase, and neuraminidase as described in Materials and Methods. The various forms of C1INH were analyzed by SDS-PAGE and stained with Coomassie brilliant blue.
Removal of N-linked carbohydrate abrogates the ability of C1INH to protect mice from lethal endotoxin shock.
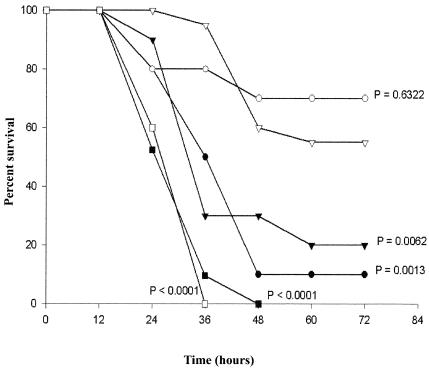
C1INH prevented the adverse biologic effects of LPS via a mechanism unrelated to protease inhibition and this protection appeared to be secondary to a direct interaction with endotoxin (22). Deletion of the amino-terminal 97 amino acid residues abolished the ability of C1INH to bind to LPS in vitro. However, the available quantities of this recombinant truncated C1INH are insufficient for testing in animals. We previously demonstrated, by using the lowest dose of LPS (20 mg/kg), that resulted in 100% mortality of C57BL/6J mice within 48 h that a single dose of C1INH (200 μg) improved survival to 45 and 50% when administered via the i.p. and intravenous routes, respectively. A mixture of LPS (20 mg/kg) and C1INH (200 μg/per mouse) given i.p. increased survival to 65% (22). In order to evaluate the role of carbohydrate in C1INH-mediated protection from endotoxin shock, mice were injected i.p. with LPS mixed with either native C1INH, N-deglycosylated C1INH, O-deglycosylated C1INH, or both N- and O-deglycosylated C1INH (200 μg/mouse). Treatment with native C1INH and O-deglycosylated C1INH resulted in 55 and 70% survival, respectively (Fig. 2). These survival curves were not significantly different (P = 0.6322). N-deglycosylated C1INH and both N- and O-deglycosylated C1INH, on the other hand, resulted in survival rates of only 20 and 10%, respectively, at 72 h. The survival of each of these groups was significantly less than that of mice treated with native C1INH (P = 0.0062 and 0.0013, respectively). However, although only 2 of 10 and 1 of 10 mice survived in the groups treated with N-deglycosylated C1INH and with N- and O-deglycosylated C1INH, respectively, their survival rates were significantly different from mice treated with LPS alone (no mice survived of 21 given LPS alone; P = 0.0087 and 0.0151). Therefore, N-deglycosylated C1INH and N- and O-deglycosylated C1INH may provide some protection from endotoxin shock, but the level of protection is much less than that with either native or O-deglycosylated C1INH. Control mice treated with either N- or O-glycosidase alone all survived, whereas none of five mice treated with N-glycosidase plus LPS and one of five mice treated with both N- and O-glycosidase plus LPS survived (data not shown). These data indicate that N-linked glycosylation of C1INH is essential to protect mice from lethal LPS-induced endotoxin shock.
FIG. 2.
Effect of deglycosylated C1INH on survival of mice in gram-negative endotoxin LPS-induced lethal endotoxemia. C57BL/6J mice were injected i.p. with a mixture of LPS (20 mg/kg) with native, plasma-derived C1INH (▵, n = 20), with N-deglycosylated C1INH (▴, n = 10), with O-deglycosylated C1INH (○, n = 10), or with both N- and O-deglycosylated C1INH (•, n = 10) (200 μg/per mouse). Control mice were injected (i.p.) with LPS (20 mg/kg) alone (▪, n = 21) or a mixture of LPS (20 mg/kg) with the glycosidase buffer (□, n = 5). The indicated P values are for each group in comparison with the group treated with native, plasma-derived C1INH.
C1INH lacking N glycosylation failed to suppress LPS binding to the murine macrophage cell line, RAW 264.7, and to human blood cells.
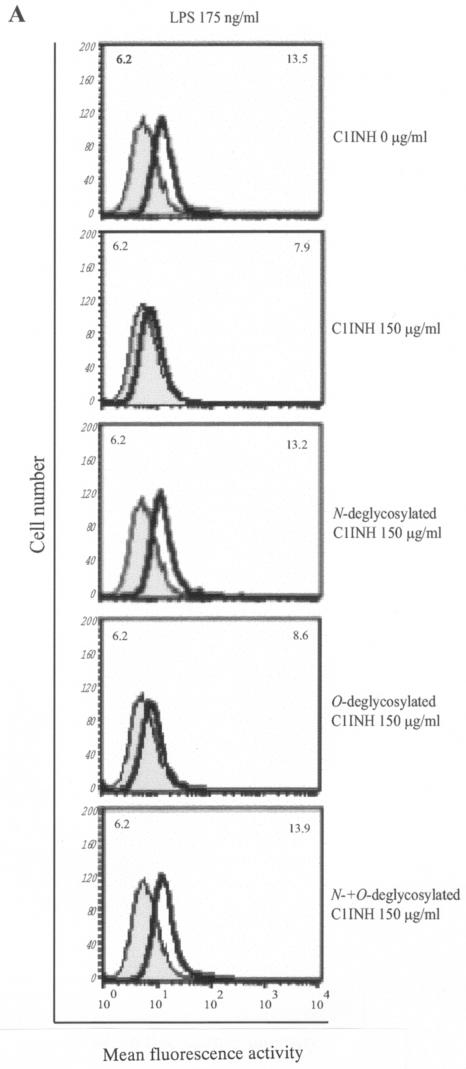
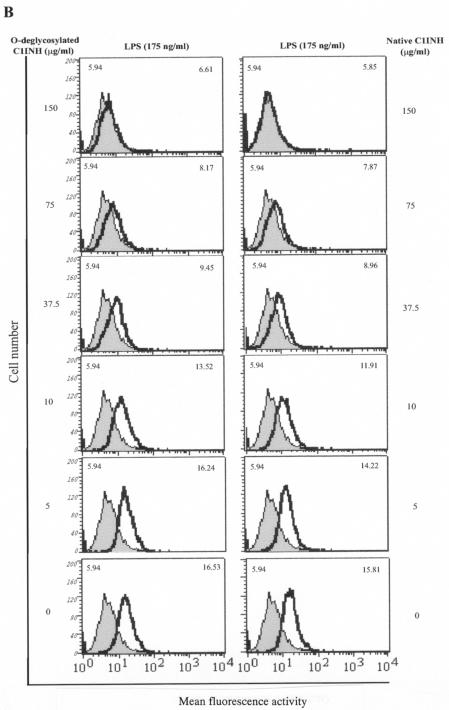
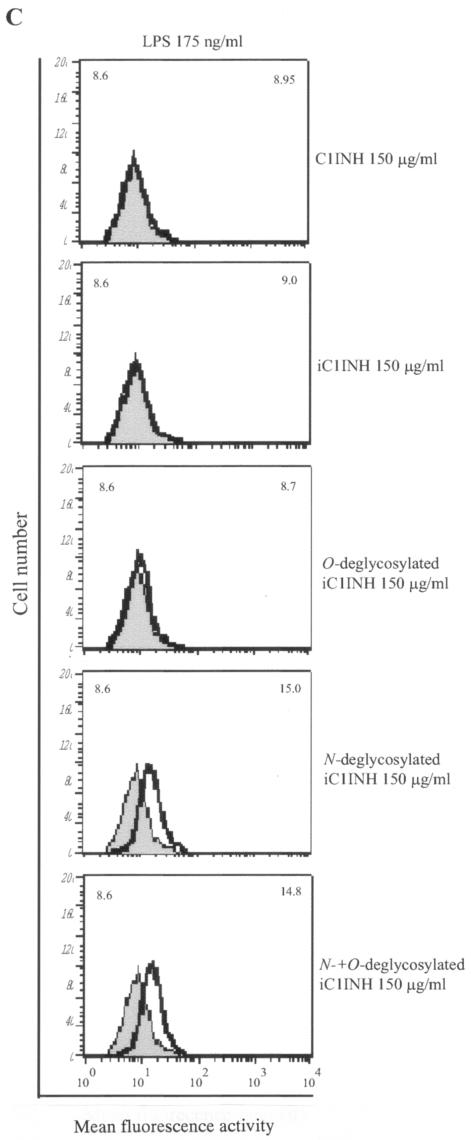
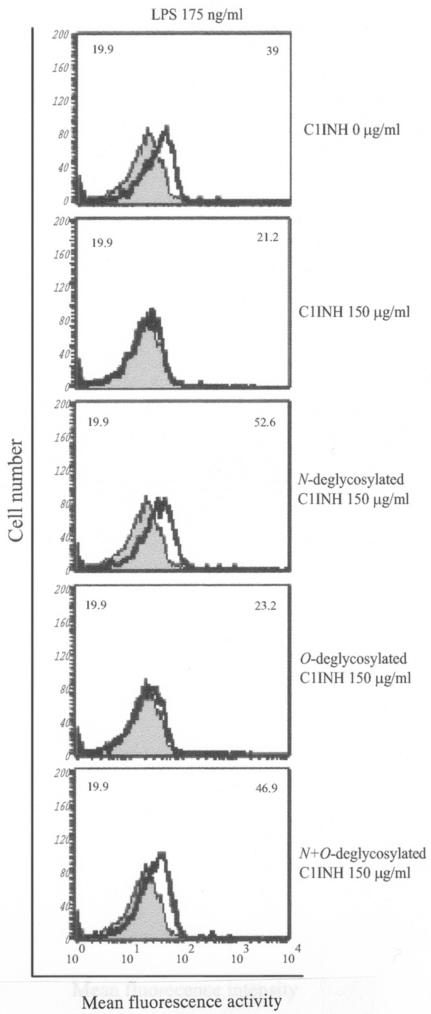
The finding that C1INH has the ability to block the binding of serovar Typhimurium LPS to macrophages prompted us to further investigate whether N- or O-linked glycosylation participates in this suppression of LPS binding. We tested by FACS the ability of deglycosylated C1INH to inhibit LPS binding to RAW 264.7 macrophages in the presence of 10% FBS. N-deglycosylated C1INH (150 μg/ml) lost the ability to suppress FITC-LPS binding to RAW 264.7 cells (Fig. 3A). However, removal of O-linked carbohydrate had no effect on the inhibition of LPS binding (Fig. 3B). As described previously, C1INH in which the reactive center loop has been cleaved with trypsin (iC1INH) also inhibits the binding of LPS to RAW 264.7 cells. As with intact C1INH, removal of N-linked carbohydrate eliminates this activity, whereas O-deglycosylation has no effect (Fig. 3C). Neither the N-glycosidase nor the O-glycosidase interfered with FITC-LPS binding to RAW 264.7 cells (data not shown). In addition, whole human blood was incubated with LPS (175 ng/ml) in the presence of C1INH (150 μg/ml) and the three different forms of deglycosylated C1INH. N deglycosylation eliminated the ability of C1INH to block the binding of LPS to human blood cells, whereas C1INH lacking O-linked glycosylation retained the ability to suppress LPS binding (Fig. 4).
FIG. 3.
Effect of deglycosylated C1INH on the binding of FITC-conjugated LPS to the murine macrophage cell line, RAW 264.7. LPS binding, thick line; control, shaded field; mean fluorescence intensities for the control and treated cells are indicated in the upper left and upper right corners, respectively, in each panel. (A) RAW 264.7 macrophages were incubated with FITC-LPS (175 ng/ml) in the absence or presence of C1INH, N-deglycosylated C1INH, O-deglycosylated C1INH, or both N- and O-deglycosylated C1INH (each at 150 μg/ml) in DMEM containing 10% FBS for 15 min at 37°C. (B) RAW 264.7 macrophages were incubated with FITC-LPS (175 ng/ml) in the presence of different doses of O-deglycosylated C1INH or untreated native C1INH (150, 75, 37.5, 10, or 5 μg/ml) in DMEM containing 10% FBS for 15 min at 37°C. (C) RAW 264.7 macrophages were incubated with FITC-LPS (175 ng/ml) in the presence of iC1INH (150 μg/ml) lacking N-, O-, or both N- and O-linked glycosylation. Cells were fixed with FACS solution after being washed with PBS three times and were analyzed by FACS.
FIG. 4.
Effect of deglycosylated C1INH on the binding of FITC-conjugated LPS to human blood cells. LPS binding, thick line; control, shaded field. Mean fluorescence intensities for the control and treated cells are indicated in the upper left and upper right corners, respectively, in each panel. The human blood cells were incubated for 15 min at 37°C with FITC-LPS (175 ng/ml) in the presence of C1INH (150 μg/ml) lacking N-, O-, or both N- and O-linked glycosylation. Cells were fixed with FACS solution after being washed with PBS three times and were analyzed by FACS.
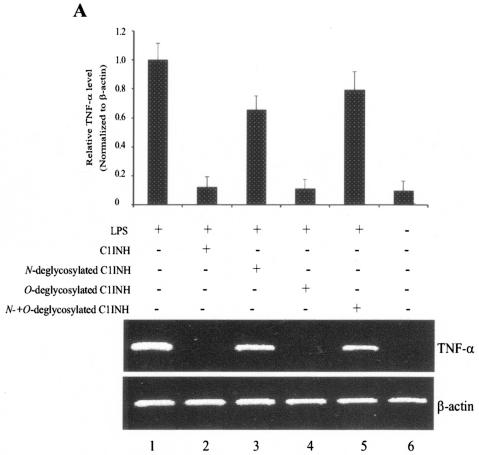
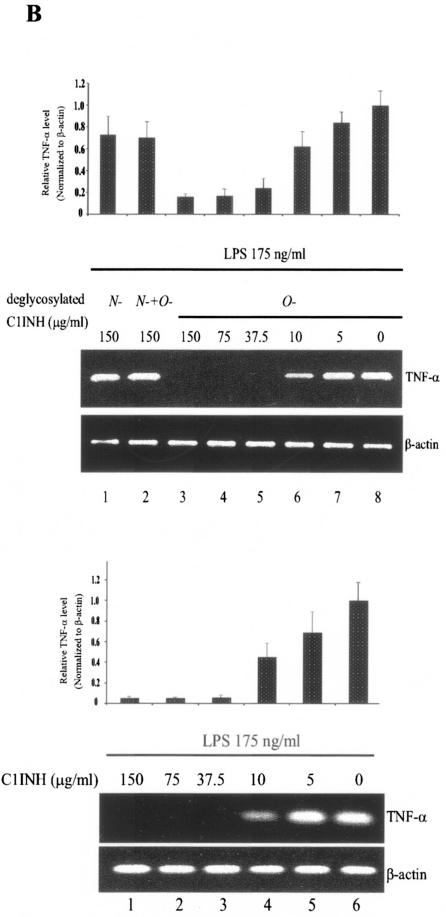
In addition, TNF-α mRNA expression induced by LPS was detected in RAW 264.7 cells by RT-PCR. LPS-mediated upregulation of TNF-α mRNA was completely suppressed after treatment with native plasma-derived C1INH (150 μg/ml) or with O-deglycosylated C1INH (150 μg/ml) (Fig. 5A). Similarly, C1INH with O-linked carbohydrate removed (150 μg/ml) also suppressed LPS-induced TNF-α mRNA expression by cells in whole human blood (Fig. 5B). However, removal of N-linked carbohydrate completely destroyed the ability of C1INH to inhibit LPS-mediated upregulation of TNF-α by both RAW 264.7 cells and by human peripheral blood cells (Fig. 5A and B).
FIG. 5.
Effect of deglycosylated C1INH on LPS-induced TNF-α mRNA expression in the murine macrophage cell line, RAW 264.7 and human blood cells. (A) total RNA from RAW 264.7 macrophages was isolated after treatment with LPS (175 ng/ml) in the presence of C1INH or with N-, O-, or both N- and O-deglycosylated C1INH (all at 150 μg/ml) for 30 min at 37°C. RT-PCR was performed with mouse TNF-α cDNA and β-actin cDNA primers. The band intensity was quantitated by using ImageQuant software (Molecular Dynamics), was normalized to the β actin level, and is expressed relative to the amount of PCR product present in the samples treated with LPS alone. (B) Total RNA from human blood cells was isolated after treatment of whole human blood with LPS (175 ng/ml) in the presence of C1INH (5, 10, 37.5, 75, or 150 μg/ml) or with N-deglycosylated C1INH (150 μg/ml), O-deglycosylated C1INH (5, 10, 37.5, 75, or 150 μg/ml), or N- and O-deglycosylated C1INH (150 μg/ml) (upper panel) for 30 min at 37°C. RT-PCR was performed with human TNF-α cDNA and β-actin cDNA primers. PCR products were quantitated as in panel A.
N-linked glycosylation of C1INH is crucial for its interaction with LPS.
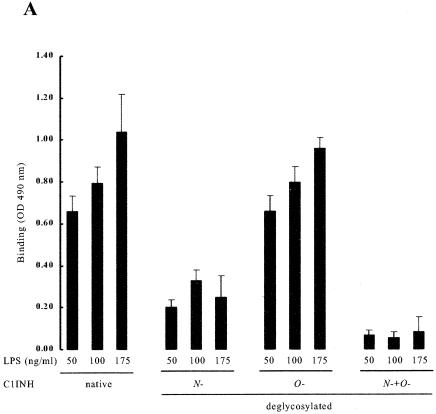
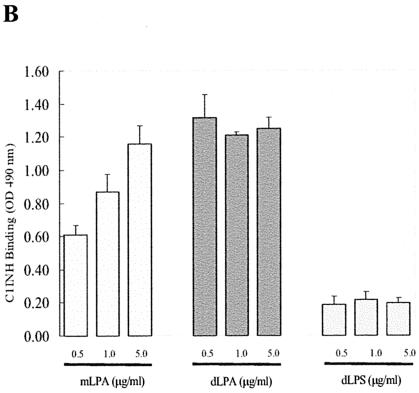
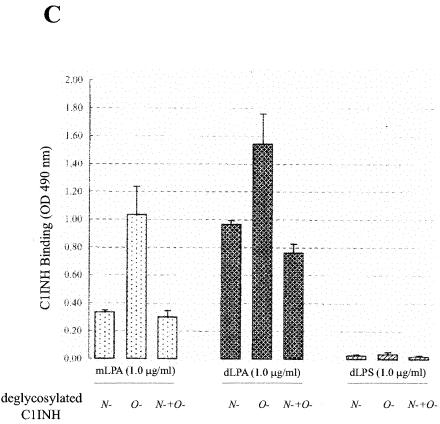
LPS is composed of two structural regions: the hydrophilic repeating polysaccharide of the core and O-antigen structures and the hydrophobic LPA domain (40). Virtually all of the biological responses induced by LPS are dependent upon LPA (15, 33, 38). As expected from the experiments described above, deletion of N-linked carbohydrate or of N- and O-linked carbohydrate from C1INH completely eliminated its ability to bind to LPS, but the binding of O-deglycosylated C1INH (150 μg/ml) was unaltered in comparison with native C1INH (Fig. 6A). To further investigate the mechanism of interaction of C1INH with endotoxin, intact C1INH and the different forms of deglycosylated C1INH were analyzed for binding to immobilized mLPA, dLPA, or dLPS by using an ELISA. Analysis of C1INH binding to mLPA (0.5, 1.0, and 5.0 ng/ml), dLPA (0.5, 1.0, and 5.0 ng/ml), and dLPS (0.5, 1.0, and 5.0 ng/ml) showed binding to both dLPA and mLPA but very little binding to dLPS (Fig. 6B). Similarly, O-deglycosylated C1INH bound to both mLPA and dLPA (Fig. 6C). N-deglycosylated (and N/O-deglycosylated) C1INH showed very little binding to mLPA. Both, however, did bind to dLPA, although not as well as did native or O-deglycosylated C1INH. These data suggested that N-linked glycosylation is essential to the interaction of C1INH with LPS. As with most other proteins that bind to LPS, LPA likely represents the primary structure in the LPS molecule that is recognized by C1INH.
FIG. 6.
Analysis of the binding of deglycosylated C1INH to LPS and LPA. The interaction of C1INH or deglycosylated C1INH with immobilized LPS, mLPA, dLPA, and dLPS was analyzed by ELISA. (A) Binding of deglycosylated C1INH (150 μg/ml) to LPS (50, 100, or 175 ng/ml) in comparison with native C1INH (150 μg/ml); (B) binding of C1INH (150 μg/ml) to mLPA (0.5, 1.0, or 5.0 μg/ml), dLPA (0.5, 1.0, or 5.0 μg/ml), and dLPS (0.5, 1.0, and 5.0 μg/ml); (C) binding of deglycosylated C1INH (150 μg/ml) to mLPA (1.0 μg/ml), dLPA (1.0 μg/ml), or dLPS (1.0 μg/ml).
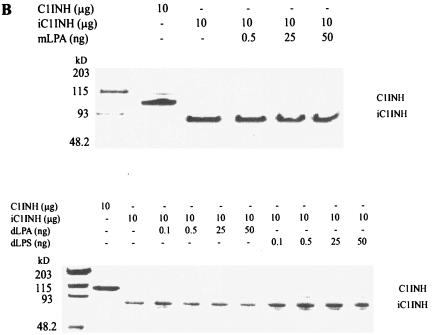
Neither LPA nor dLPS alter the ability of C1INH to complex with C1s.
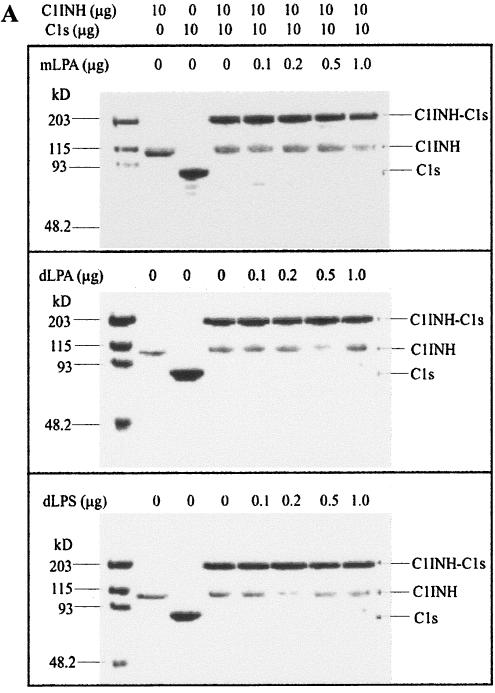
To examine whether mLPA, dLPA, and dLPS had an effect on the formation of C1INH-C1s complexes, native, active C1INH (10 μg) was incubated with mLPA, dLPA, and dLPS, respectively, followed by the addition of C1s (10 μg). The C1INH treated with mLPA, dLPA, and dLPS retained the ability to form C1INH-C1s complexes (Fig. 7A). In addition, mLPA, dLPA, and dLPS did not interfere with the susceptibility of the reactive center loop to cleavage with trypsin (Fig. 7B).
FIG. 7.
Effects of mLPA, dLPA, and dLPS on the formation of C1INH-C1s complexes and C1INH cleavage by trypsin. (A) mLPA (0.1, 0.2, 0.5, or 1.0 μg), dLPA (0.1, 0.2, 0.5, or 1.0 μg), and dLPS (0.1, 0.2, 0.5, or 1.0 μg) have no effect on the rate or extent of C1INH-C1s (10 μg: 10 μg) complex formation, as assessed by SDS-PAGE stained with Coomassie brilliant blue. (B) mLPA (0.5, 25, or 50 ng), dLPA (0.1, 0.5, 25, or 50 ng), and dLPS (0.1, 0.5, 25, or 50 ng) have no effect on the formation of cleaved C1INH (10 μg) by trypsin, as assessed by SDS-PAGE stained with Coomassie brilliant blue.
DISCUSSION
The data presented here demonstrate that N-linked glycosylation of C1INH is required for the interaction of C1INH with endotoxin. Removal of N-linked carbohydrate, but not the removal of O-linked carbohydrate, resulted in a significant reduction in the ability of C1INH to protect mice from LPS-mediated endotoxin shock (Fig. 2). Consistent with this observation, N-linked carbohydrate also was required for inhibition of LPS binding to RAW 264.7 cells (Fig. 3) and to human blood cells (Fig. 4) and for the induction of TNF-α synthesis by these cells (Fig. 5). Lastly, in an ELISA, the direct binding of C1INH to LPS was dependent on N glycosylation (Fig. 6A). Previous data demonstrated that iC1INH, which has no protease inhibitory activity, retained the ability to bind to LPS but that binding required the presence of the amino-terminal nonserpin domain (22). A recombinant C1INH variant with deletion of the amino-terminal 97 amino acids retained the ability to inactivate proteases but did not bind to LPS (11, 22). This domain consists of 120 amino acids and is extremely heavily glycosylated with three N-linked and at least seven O-linked carbohydrate groups (4). Three additional N-linked groups are within the serpin domain. Deglycosylation of C1INH with N-glycanase, O-glycanase, or both has no significant effect on protease inhibitory function (30). The combination of the current and previous findings, therefore, indicate that the three N-linked carbohydrates within the amino-terminal domain at Asn residues 3, 47, and 59 and not those within the serpin domain are required for the reactivity of C1INH with endotoxin.
LPS interacts with a number of glycoproteins, including the three components of the cell membrane receptor for LPS, CD14, MD-2 and Toll-like receptor 4 (TLR4) (12, 19, 36, 37, 42, 43). LPS is transported by the plasma protein, LPS-binding protein (LBP), and is transferred to the LPS receptor on the cell surface. Engagement of this receptor ultimately results in triggering of innate immune responses, in particular, increased expression of TNF-α and a variety of other cytokines. MD-2 has two N glycosylation sites at Asn26 and Asn114, whereas TLR4 has nine N-linked sites in its amino-terminal extracellular domain (12). Binding of LPS to the LPS receptor requires N-linked glycosylation of both MD-2 and TLR4 (12) but not of CD14 (36). It is, however, not known whether the glycosylation of TLR4 is required to directly mediate the binding of LPS or if it is necessary for stabilization of the complex receptor (12). In the case of MD-2, however, the binding site for LPS has been shown to consist of a positively charged domain that is similar to the binding domains described in a number of other LPS-binding proteins (23, 29). Therefore, with MD-2, as might be expected, the role of glycosylation must be to maintain the conformation of the protein rather than to participate directly in binding. Although a number of residues within CD14 have been shown to be required for binding of LPS, the precise biochemical basis of this binding also remains to be defined (19, 36, 37, 42, 43).
LPS is released from the outer membrane of gram-negative bacteria and is largely responsible for the symptoms of septic shock. The active component of endotoxin, LPA, is a phosphoglycolipid with an acylated and phosphorylated dihexosamine headgroup. The polysaccharide component contains antigenic determinants but does not contribute to endotoxin activity (32, 33). The removal of an acid labile phosphate group and normal fatty acid groups from dLPA diminishes endotoxic activity. Although mLPA lacks many of the endotoxic properties of LPS (2), in vitro it induces the production of proinflammatory cytokines from macrophages (16) and gamma interferon and IL-2 from lymphocytes (10). However, mLPA is markedly less active than is LPS in the induction of these cytokines. In addition, mLPA and LPS may differentially regulate the production of some cytokines. For example, mLPA induced IL-10 to a greater extent than did LPS (34). In addition, mLPA activates human dendritic cells and T cells and enhances the generation of both Th1- and Th2-specific immune responses in mice (13, 18). Previously, we demonstrated that C1INH inhibits the interaction of LPS with LBP and, ultimately, with macrophages (22). The data presented here show that C1INH interacts with the LPA moiety within LPS to prevent this interaction. C1INH at a fixed concentration (150 μg/ml) bound to immobilized dLPA but did not bind as efficiently to mLPA, and bound extremely poorly to dLPS (Fig. 6B). As with binding to LPS, the binding of C1INH to LPA also was reduced by N deglycosylation but not by removal of O-linked carbohydrate (Fig. 6C). However, the reduction in binding of N-deglycosylated versus native C1INH to dLPA (Fig. 6C) was not as striking as the reduction in binding of N-deglycosylated C1INH to LPS (Fig. 6A). This could be a result of variable and incomplete deglycosylation in the two experiments. However, a more likely explanation is that, analogous to the apparent situation with MD-2, the binding site on the C1INH amino-terminal domain does not reside on the N-linked carbohydrate itself, but is on the peptide backbone. Removal of carbohydrate may result in alteration of the conformation of this domain and partially mask the binding site. The smaller dLPA molecule may be able to more readily gain access to the binding site, whereas the larger LPS molecule would be unable to interact with the site. Binding of LPS (or dLPA) to many LPS-binding proteins is via interaction of the phosphate groups on the LPA with specific positively charged residues within the LPS-binding protein (1, 3, 14, 17, 20, 21, 25, 26, 28, 31, 35, 39, 41). C1INH has one Arg (at position 18) and 3 Lys residues (at positions 22, 30, and 55) within the amino-terminal domain. Mutagenesis studies are in progress to test this hypothesis and to define which of the three carbohydrate groups within the amino-terminal domain are required for binding to endotoxin.
In summary, the binding of C1INH to LPS prevents the interaction of LPS with LBP which in turn prevents the delivery of LPS to cells that express the LPS receptor complex. The characteristics of this binding are that it is mediated by the LPA moiety of LPS, it does not require an intact C1INH reactive center loop (and therefore is not dependent on protease inhibitory activity), and it requires the C1INH amino-terminal nonserpin domain and intact N-linked carbohydrate.
Acknowledgments
This study was supported by Public Health Service grants HD22082 and HD33727 from the National Institute of Child Health and Human Development.
Editor: A. D. O'Brien
REFERENCES
- 1.Appelmelk, B. J., Y.-Q. An, M. Geerts, B. G. Thijs, H. A. De Boer, D. M. MacLaren, J. de Graaff, and J. H. Juijens. 1994. Lactoferrin is a lipid A-binding proten. Infect. Immun. 62:2628-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Astiz, M. E., R. C. Rackow, J. G. Still, S. T. Howell, A. Cato, K. B. Von Eschen, J. T. Ulrich, J. A. Rudbach, G. McMahon, R. Vargas, and W. Stern. 1995. Pretreatment of normal humans with monophosphoryl lipid A induces tolerance to endotoxin: a prospective, double-blind, randomized, controlled trial. Crit. Care Med. 23:9-17. [DOI] [PubMed] [Google Scholar]
- 3.Battafarano, R. J., P. S. Dahlberg, C. A. Ratz, J. W. Johnston, B. H. Gray, J. R. Haseman, K. H. Mayo, and D. L. Dunn. 1995. Peptide derivatives of three distinct lipopolysaccharide binding proteins inhibit lipopolysaccharide-induced tumor necrosis factor-alpha secretion in vitro. Surgery 118:318-324. [DOI] [PubMed] [Google Scholar]
- 4.Bock, S. C., K. Skriver, E. Nielsen, H. C. Thogersen, B. Wiman, V. H. Donaldson, R. L. Eddy, J. Marrinan, E. Radziejewska, R. Huber, T. B. Shows, and S. Magnussen. 1986. Human C1 inhibitor: primary structure, cDNA cloning, and chromosomal localization. Biochemistry 25:4292-4301. [DOI] [PubMed] [Google Scholar]
- 5.Bone, R. C. 1993. gram-negative sepsis: a dilemma of modern medicine. Clin. Microbiol. Rev. 6:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bone, R. C. 1991. The pathogenesis of sepsis. Ann. Intern. Med. 115:457-469. [DOI] [PubMed] [Google Scholar]
- 7.Brower, M. S., and P. C. Harpel. 1982. Proteolytic cleavage and inactivation of α2 plasmin inhibitor and C1 inactivator by human polymorphonuclear leukocyte elastase. J. Biol. Chem. 257:9849-9854. [PubMed] [Google Scholar]
- 8.Caccavo, D., A. Afeltra, S. Pece, G. Giuliani, M. Freudenberg, C. Galanos, and E. Jirillo. 1999. Lactoferrin-lipid A-lipopolysaccharide interaction: inhibition by anti-human lactoferrin monoclonal antibody AGM 10.14. Infect. Immun. 67:4668-4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caliezi, C., W. A. Wuillemin, S. Zeerleder, M. Redondo, B. Eisele, and C. E. Hack. 2001. C1-esterase inhibitor: an anti-inflammatory agent and its potential use in the treatment of diseases other than hereditary angioedema. Pharmacol. Rev. 52:91-112. [PubMed] [Google Scholar]
- 10.Carozzi, S., M. Salit, A. Cantaluppi, M. G. Nasini, S. Barocci, S. Cantarella, and S. Lamperi. 1989. Effect of monophosphoryl lipid A on the in vitro function of peritoneal leukocytes from uremic patients on continuous ambulatory peritoneal dialysis. J. Clin. Microbiol. 27:1748-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coutinho, M., K. S. Aulak, and A. E. Davis III. 1994. Functional analysis of the serpin domain of C1 inhibitor. J. Immunol. 153:3648-3654. [PubMed] [Google Scholar]
- 12.da Silva Correia, J., and R. J. Ulevitch. 2002. MD-2 and TLR4 N-linked glycoproteins are important for a functional lipopolysaccharide receptor. J. Biol. Chem. 277:1845-1854. [DOI] [PubMed] [Google Scholar]
- 13.De Becker, G., V. Moulin, B. Pajak, C. Bruck, M. Francotte, C. Thiriart, J. Urbain, and M. Moser. 2000. The adjuvant monophosphoryl lipid A increases the function of antigen-presenting cells. Int. Immunol. 12:807-815. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson, A. D., W. Welte, E. Hofmann, B. Lindner, O. Holst, J. W. Coulton, and K. Diederichs. 2000. A conserved structural motif for lipopolysaccharide recognition by procaryotic and eucaryotic proteins. Structure 8:585-592. [DOI] [PubMed] [Google Scholar]
- 15.Grabarek, J., G. Her, V. N. Reinhold, and J. Hawiger. 1990. Endotoxic lipid A interaction with human platelets: structure-function analysis of lipid A homologs obtained from Salmonella Minnesota Re595 lipopolysaccharide. J. Biol. Chem. 265:8117-8121. [PubMed] [Google Scholar]
- 16.Henricson, B. E., C. L. Mnathey, P. Y. Perera, T. A. Hamilton, and S. N. Vogel. 1993. Dissociation of lipopolysaccharide (LPS)-inducible gene expression in murine macrophages pretreated with smooth LPS versus monophosphoryl lipid A. Infect. Immun. 61:2325-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoess, A., S. Watson, G. R. Siber, and R. Liddington. 1993. Crystal structure of an endotoxin-neutralizing protein from the horseshoe crab, Limulus anti-LPS factor, at 1.5 Å resolution. EMBO J. 12:3351-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ismaili, J., J. Rennesson, E. Aksoy, J. Vekemans, B. Vincart, Z. Amraoui, F. Van Laethem, M. Goldman, and P. M. Dubois. 2002. Monophosphoryl lipid A activates both human dendritic cells and T cells. J. Immunol. 168:926-932. [DOI] [PubMed] [Google Scholar]
- 19.Juan, T. S., E. Hailman, M. J. Kelley, L. A. Busse, E. Davy, C. J. Empig, L. O. Narhi, S. D. Wright, and H. S. Lichenstein. 1995. Identification of a lipopolysaccharide binding domain in CD14 between amino acids 57 and 64. J. Biol. Chem. 270:5219-5224. [DOI] [PubMed] [Google Scholar]
- 20.Lamping, N., A. Hoess, B. Yu, T. C. Park, C.-J. Kirschning, C. Pfeil, D. Reuter, S. D. Wright, F. Herrman, and R. R. Schumann. 1996. Effects of site-directed mutagenesis of basic residues (Arg 94, Lys 95, Lys 99) of LPS-binding protein on binding and transfer of LPS and subsequent immune cell activation. J. Immunol. 157:4648-4656. [PubMed] [Google Scholar]
- 21.Little, R. G., D. N. Kelner, E. Lim, D. J. Burke, and P. J. Conlon. 1994. Functional domains of recombinant bactericidal/permeability increasing protein (rBPI23). J. Biol. Chem. 269:1865-1872. [PubMed] [Google Scholar]
- 22.Liu, D., S. Cai, X. Gu, J. Scafidi, X. Wu, and A. E. Davis III. 2003. C1 inhibitor prevents endotoxin shock via a direct interaction with lipopolysaccharide. J. Immunol. 171:2594-2601. [DOI] [PubMed] [Google Scholar]
- 23.Mancek, M., P. Pristovsek, and R. Jerala. 2002. Identification of LPS-binding peptide fragment of MD-2, a Toll-receptor accessory protein. Biochem. Biophys. Res. Commun. 292:880-885. [DOI] [PubMed] [Google Scholar]
- 24.Mansell, A., A. Reinicke, D. Marharet Worral, and L. A. J. O'Neill. 2001. The serine protease inhibitor antithrombin III LPS-mediated NF-κB activation by TLR-4. FEBS Lett. 508:313-317. [DOI] [PubMed] [Google Scholar]
- 25.Nagaoka, I., S. Hirota, F. Niyonsaba, M. Hirata, Y. Adachi, H. Tamura, and D. Heumann. 2001. Cathelicidin family of antibacterial peptides CAP18 and CAP11 inhibit the expression of TNF-α by blocking the binding of LPS to CD14+ cells. J. Immunol. 167:3329-3338. [DOI] [PubMed] [Google Scholar]
- 26.Nagaoka, I., S. Hirota, F. Niyonsaba, M. Hirata, Y. Adachi, H. Tamura, S. Tanaka, and D. Heumann. 2002. Augmentation of the lipopolysaccharide-neutralizing activities of human cathelicidin CAP18/LL-37-derived antimicrobial peptides by replacement with hydrophobic and cationic amino acid residues. Clin. Diagn. Lab. Immunol. 9:972-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuijens, J. H., A. J. M. Eerenberg-Belmer, C. C. M. Huijbregts, W. O. Schreuder, R. J. F. Felt-Bersma, J. J. Abbink, L. G. Thijs, and C. E. Hack. 1989. Proteolytic inactivation of plasma C1 inhibitor in sepsis. J. Clin. Investig. 84:443-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Odell, E. W., R. Sarra, M. Foxworthy, D. S. Chapple, and R. W. Evans. 1996. Antibacterial activity of peptides homologous to a loop region in human lactoferrin. FEBS Lett. 382:175-178. [DOI] [PubMed] [Google Scholar]
- 29.Re, F., and J. L. Strominger. 2003. Separate functional domains of human MD-2 mediate Toll-like receptor 4-binding and lipopolysaccharide responsiveness. J. Immunol. 171:5272-5276. [DOI] [PubMed] [Google Scholar]
- 30.Reboul, A., M. Prandini, and M. Colomb. 1987. Proteolysis and deglycosylation of human C1 inhibitor: effect on functional properties. Biochem. J. 24:117-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reyes, O., M. G. Vallespi, H. E. Garay, L. J. Cruz, L. J. Gonzalez, G. Chinea, W. Buurman, and M. J. Arana. 2002. Identification of single amino acid residues essential for the binding of lipopolysaccharide (LPS) to LPS binding protein (LBP) residues 86-89 using an Ala-scanning library. J. Peptide Sci. 8:144-150. [DOI] [PubMed] [Google Scholar]
- 32.Ribi, E. 1986. Structure-function relationship of bacterial adjuvants, p. 35. In R. M. Nervig, P. M. Gough, M. L. Kaeverle, and C. A. Whetstone (ed.), Advances in carries and adjuvants for veterinary biologicals. Iowa State University Press, Ames.
- 33.Rietschel, E. T., T. Kirikae, F. U. Schade, U. Mamat, G. Schmidt, H. Loppnow, A. J. Ulmer, U. Zahringer, U. Seydel, F. di Padova, M. Schreier, and H. Brade. 1994. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 8:217-225. [DOI] [PubMed] [Google Scholar]
- 34.Salkowski, C. A., G. R. Detore, and S. N. Vogel. 1997. Lipopolysaccharide and monophosphoryl lipid A differentially regulate interleukin-12, gamma interferon, and interleukin-10 mRNA production in murine macrophages. Infect. Immun. 65:3239-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scott, M. G., A. C. E. Vreugdenhil, W. A. Buurman, R. E. W. Hancock, and M. R. Gold. 2000. Cationic antimicrobial peptides block the binding of lipopolysaccharide (LPS) to LPS binding protein. J. Immunol. 164:549-553. [DOI] [PubMed] [Google Scholar]
- 36.Stelter, F., M. Bernheiden, R. Menzel, R. S. Jack, S. Witt, X. Fan, M. Pfister, and C. Schutt. 1997. Mutation of amino acids 39-44 of human CD14 abrogates binding of lipopolysaccharide and Escherichia coli. Eur. J. Biochem. 243:100-109. [DOI] [PubMed] [Google Scholar]
- 37.Stelter, F., H. Loppnow, R. Menzel, U. Grunwald, M. Bernheiden, R. S. Jack, A. J. Ulmer, and C. Schutt. 1999. Differential impact of substitution of amino acids 9-13 and 91-101 of human CD14 on soluble CD14-dependent activation of cells by lipopolysaccharide. J. Immunol. 163:6035-6044. [PubMed] [Google Scholar]
- 38.Takada, H., and S. Kotani. 1989. Structural requirements of lipid A for endotoxicity and other biological activities. CRC Crit. Rev. Microbiol. 16:423-477. [DOI] [PubMed] [Google Scholar]
- 39.Taylor, A. H., G. Heavner, M. Nedelman, D. Sherris, E. Brunt, D. Knight, and J. Ghrayeb. 1995. Lipopolysaccharide neutralizing peptides reveal a lipid A binding site of LPS binding protein. J. Biol. Chem. 270:17934-17938. [DOI] [PubMed] [Google Scholar]
- 40.Ulevitch, R. J., and P. S. Tobias. 1995. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu. Rev. Immunol. 13:437-457. [DOI] [PubMed] [Google Scholar]
- 41.van Berkel, P. H., M. E. Geerts, H. A. van Veen, M. Mericskay, H. A. de Boer, and J. H. Nuijens. 1997. N-terminal stretch Arg2, Arg3, Arg4 and Arg5 of human lactoferrin is essential for binding to heparin, bacterial lipopolysaccharide, human lysozyme and DNA. Biochem. J. 328:145-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Viriyakosol, S., and T. N. Kirkland. 1995. A region of human CD14 required for lipopolysaccharide binding. J. Biol. Chem. 270:361-368. [DOI] [PubMed] [Google Scholar]
- 43.Viriyakosol, S., J. C. Mathison, P. S. Tobias, and T. N. Kirkland. 2000. Structure-function analysis of CD14 as a soluble receptor for lipopolysaccharide. J. Biol. Chem. 275:3144-3149. [DOI] [PubMed] [Google Scholar]
- 44.Wilde, C. G., J. J. Seilhamer, M. McGrogan, N. Ashton, J. L. Snable, J. C. Lane, S. R. Leong, M. B. Thornton, K. L. Miller, R. W. Scott, and M. N. Marra. 1994. Bactericidal/permeability-increasing protein and lipopolysaccharide (LPS)-binding protein. LPS binding properties and effects on LPS-mediated cell activation. J. Biol. Chem. 269:17411-17416. [PubMed] [Google Scholar]