Abstract
Background
Stable expression of transgenes is an important technique to analyze gene function. Various drug resistance genes, such as neo, pac, hph, zeo, bsd, and hisD, have been equally used as selection markers to isolate a transfectant without considering their dose-dependent characters.
Results
We quantitatively measured the variation of transgene expression levels in mouse embryonic stem (mES) cells, using a series of bi-cistronic expression vectors that contain Egfp expression cassette linked to each drug resistant gene via IRES with titration of the selective drugs, and found that the transgene expression levels achieved in each system with this vector design are in order, in which pac and zeo show sharp selection of transfectants with homogenously high expression levels. We also showed the importance of the choice of the drug selection system in gene-trap or gene targeting according to this order.
Conclusions
The results of the present study clearly demonstrated that an appropriate choice of the drug resistance gene(s) is critical for a proper design of the experimental strategy.
Keywords: Transgene, Expression, Marker, Gene targeting, Vector
Background
The introduction of exogenous transgene cassettes into culture cells to direct their expressions is an important strategy in molecular biology to analyze the functions of the genes. However, a simple introduction of the DNA fragment into cells by either electroporation or lipofection results in its stable integration into the genome of the host cells only at a low frequency. Therefore, it is always required to select the cells carrying the integrated copies of the transgenes by using dominant selection markers. The combinations of the antibiotics that kill the mammalian cells and the genes that establish the resistance against them have been preferentially applied for this purpose: such as neomycin phosphotransferase II from transposon Tn5 (designated as neo in this paper) against the neomycin derivative G418, puromycin N-acetyltransferase from Streptomyces alboniger (pac) against puromycin, hygromycin B phosphotransferase from Escherichia coli (hph) aginst hygromycin B, Streptoalloteichus hindustanus ble (Sh ble: designated as zeo in this paper) against the bleomycin derivative zeocin, blasticidin S deaminase from Aspergillus terreus (bsd) against blasticidin S, and histidinol dehydrogenase from Salmonella typhimurium (hisD) against histidinol [1-6]. These drugs and the resistance genes have equally been regarded as dominant selection markers that reflect the introduction of the transgenes into mammalian cells. Transfection of drug resistance genes together with transgenes, each in separate expression cassette, to obtain stable transfectants has been a commonly used method. However, in this strategy, the drug resistance does not always appropriately reflect the expression level of the transgene because generally the stable expression levels of exogenous expression cassettes are highly sensitive to thier sites of integration, as a result of the local chromatin environment when the transgenes are randomly integrated into the host genome [7], which affect the expression levels of the drug resistance gene cassette and the transgene cassette separately.
The bi-cistronic expression of the transgene and the drug resistance gene using an internal ribosome entry site (IRES) is able to confer a tight correlation between the transgene expression and the drug resistance because the IRES-mediated cap-independent translation ensures parallel expressions of the transgene and the drug resistance gene [8]. This vector design is particularly important to drive transgene expressions in mouse embryonic stem (mES) cells since the silencing effect to the stably integrated transgene cassette is problematic in these cells [9]. In this vector design, the expression levels of the transgenes depend on the threshold expression levels of the drug resistance genes that confer the survival of the transfectants in the presence of the drugs. The expression levels of some transgenes can also possess unique thresholds based on their effect on cell viability. The combination of the effect of the transgene with the range of selection generated by the antibiotic resistance marker can produce a narrow expression range that can mimic the physiological function range of expression. Therefore, a proper choice of drug resistance genes is important to achieve the optimal range of transgene expression levels.
Here we demonstrated the parallel comparison of the kinetics among each drug selection system determined by the expression levels from the IRES-based expression vectors. We found obvious differences in their kinetics and the impact on various experimental situations.
Results
Kinetics of the drug-selection systems in the IRES-based expression vectors
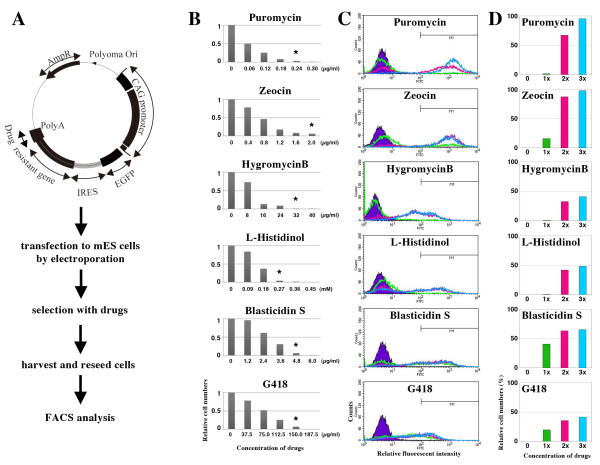
To evaluate the kinetics of the bi-cistronic expression vectors carrying various drug resistance genes in mES cells, we constructed a bi-cistronic expression vector system using enhanced green fluorescent protein (Egfp) as an indicator of the expression level and drug resistance genes under the control of the IRES from encephalomyocarditis virus (EMCV) driven by the CAG expression unit [10] (Figure 1A). To apply comparable selection pressures in different drug-selection systems, we first determined the minimal doses of each drugs to kill mES cells by serial titrations (Figure 1B). We determined the minimal doses as killing more than 93% of mES cells at low cell density (1×103 cells per 90 mm dish) within 6 days, indicated with asterisks (Figure 1B). Each vector, that is, pCAG-Egfp-IRES-neo, -hph, -zeo, -hisD, -bsd, and -pac, was transfected independently into mES cells via random integration into the genome by electroporation, and the transfected cells were seeded at high density (1×106 cells per 90 mm dish) and cultured in the presence of each drug at the concentrations of the minimal doses, twice higher or three times higher, from the next day. After the selection for 7 days, cells were harvested and seeded on new plates followed by the culture with the same concentrations of the drugs. Then the expression levels of the transgenes in the proliferated cells were estimated by the intensities of the fluorescence measured by flow-cytometry (Figure 1C) and the Egfp-positive proportions were scored (Figure 1D). Selection by neo showed broad and lower levels of fluorescence even at the highest dose (3 times higher than the minimal dose), in which less than 50% of the selected cells were diagnosed as Egfp-positive and strong positive cells expressing Egfp more than the relative intensity value of 103 were merely observed. This situation may refer to the original distribution of the Egfp expression under the CAG unit at different integration sites in the genome. Similar tendency was observed for the selection with hph, hisD and bsd although the hisD selection gave higher levels of Egfp expressions and the bsd selection conferred higher proportion of the Egfp-positive cells than the neo selection. In contrast, selection with pac and zeo gave high proportion of Egfp-positive cells with high levels of fluorescent signals, indicating the enrichment of the transfectants expressing high levels of the transgene. This is not due to the application of the high doses of drugs because once the selection systems started to work at the minimal dose at high density culture condition, which was twice of the minimal doses we determined in the pilot experiments at low density culture condition, both pac and zeo selection systems gave the sharp enrichment of high expressants. Therefore, we suppose that this observation reflects the different threshold expression levels of the drug resistant genes to confer the drug-resistant phenotypes in mES cells. Pac and zeo require higher levels of expression to support the proliferation in the presence of the lethal amount of drugs than others, which allowed us to obtain the transfectants with homogenously high levels of transgene expression. These evidences accorded to our experiences that zeo and pac efficiently worked to select mES cell lines expressing fluorescent markers ubiquitously and strongly in chimeric embryos [11], and that neo gave higher numbers of LIF-independent colonies than pac when applied to select drive the expression of Tbx3 transgene, of which the expression at high level is toxic in mES cells [12].
Figure 1.
Expression levels of the transgenes from the stably integrated bi-cistronic transgene cassettes with various drug selection systems. (A) Design of the bi-cistronic expression vectors containing the drug resistance genes. The drug resistance genes for G418 (neo), puromycin (pac), hygromycin B (hph), zeocin (zeo), blasticidin S (bsd), and histidinol (hisD) were fused to EMCV-IRES and placed downstream of Egfp under the control of the CAG expression unit [10]. (B) Determination of the minimal concentrations of the drugs. The D3 mES were seeded at low cell density (1000 cells per 90 mm dish) with various concentrations of drugs in triplicate, and the cell numbers were counted. The relative number of the cells to that obtained without the drug, which was set at 1.0, are shown. Asterisks show the doses of 1× of the selection in FACS analyses, by which more than 93% of the wild-type mES cells were killed. (C) The FACS analyses for the EGFP expression levels. In these FACS analyses, the EGFP expression level (FL1) under the selection with the minimal concentration (1×, green line), twice higher than the minimal concentration (2×, red line) and three times higher than the minimal concentration (3×, blue line) were analyzed by flow cytometry using FACSCalibur (BD). Blue filled peaks indicate the signals from wild-type mES cells. (D) Quantification of the relative cell numbers expressing EGFP. The proportions within M1 indicated in C are shown.
Generation of new fusion genes of fluorescent markers and drug-resistant genes
If the modulation of the transgene expression levels by different drug concentration with the bi-cistronic expression vector enables us the precise control of transgene expression, the monitoring of the expression levels of the drug resistance genes in living cells will be ideal. The functional fusion genes of Egfp and neo, and Egfp and hph have already been reported [13]. Here we made two novel chimeric selection markers composed of drug resistance genes and fluorescent markers and tested their functions. Using the same system as described above, we confirmed that both hisDsRed (hisD + DsRedT4) and pacEgfp (pac + Egfp) are bi-functional as drug resistance markers comparable to the wild-type genes and as fluorescent markers (Figure 2A, E): The fusion genes worked as drug-resistant genes as efficiently as the wild-type genes since they gave comparable numbers of the drug-resistant colonies (Figure 2B, F); They also worked as the fluorescent markers that were expressed at the levels correlating to the transgenes placed at the upstream (Figure 2C, D, G, H). Therefore, these fusion genes will be useful for monitoring the gene expression levels in the stable transfectants.
Figure 2.
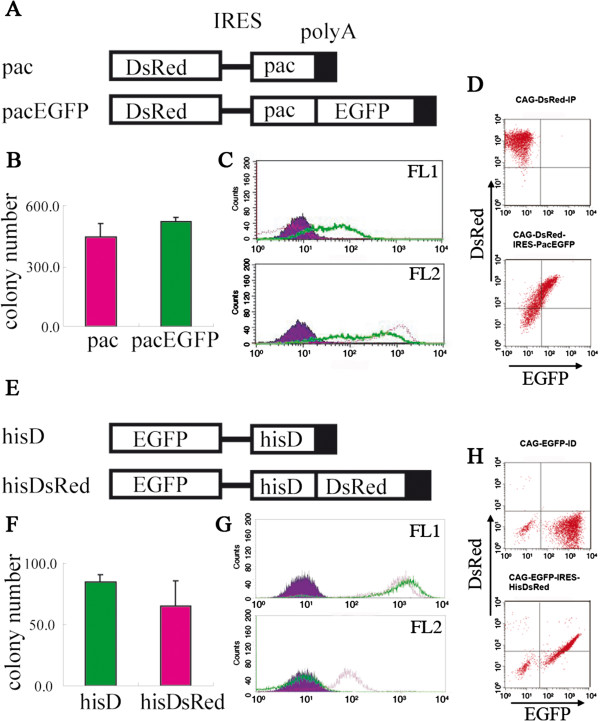
Bi-functional chimeric proteins of drug resistance genes and fluorescent markers. (A, E) Design of the vectors to test the functions of the fusion proteins driven by the CAG promoter. All constructs were electroporated into D3 mES cells, and the transfectants were selected with the standard concentrations of puromycin (A) or histidinol (D) as in Figure 1. (B, F) The numbers of the primary colonies from pac (red) and pacEGFP (green) after puromycin selection (B), and hisD (green) and hisDsRed (red) after histidinol selection. Comparable numbers of drug resistant colonies were obtained in both cases by the fusion constructs. (C) FACS analyses of the transfectants with pac vectors. EGFP fluolescence could be detected in the transfectant containing pacEGFP (green line) but not pac (red line) (upper panel: FL1). Levels of DsRed fluorescence in ES cells with pacEGFP (green line) were comparable to that with pac (red line) (lower panel: FL2). (D) Correlation between the expression levels of DsRed and Egfp from CAG-DsRed-IRES-pac-pA (upper) and CAG-DsRed-IRES-pacEGFP-pA (lower). (G) FACS analyses of the transfectants with hisD vectors. Levels of EGFP fluorescence in mES cells with hisDsRed (red line) were comparable to that with hisD (green line) (upper panel: FL1). DsRed fluolescence could be detected in the transfectant containing hisDsRed (red line) but not hisD (green line) (lower panel: FL2). (H) Correlation between the expression levels of DsRed and Egfp from CAG-EGFP-IRES-hisD-pA (upper) and CAG-EGFP-IRES-hisDsRed-pA (lower).
Application of the drug-resistant systems to gene targeting
The IRES-mediated drug resistance gene cassettes are also useful for the gene targeting since this strategy is able to enrich the homologous recombinants [14]. When the promoter-less knockout vectors carrying the IRES-mediated drug resistance gene cassettes are integrated into the genome via random insertion, only the clones in which the IRES-mediated drug resistance gene cassettes are accidentally inserted into the genes and driven by the upstream promoter will survive, whereas all homologous recombinants are alive if the transcription levels of the IRES-mediated drug resistance gene cassettes reach the threshold levels to confer the drug resistance. To evaluate the expression levels achieved by these selection systems in comparison to the endogenous genes, we next tested their functions by the gene-trap method [15]. We placed the drug resistance genes fused to IRES (Figure 3A) downstream of the splice acceptor and introduced them into the host cells. With the pac selection system, we obtained very few drug-resistant colonies—less than 10% of the number obtained with the neo selection system (Figure 3B)—indicating that the threshold expression level of the pac selection system could be achieved only with strong endogenous promoters. Previous applications of the neo selection system to the gene-trap strategy in mES cells revealed that ~15,000 of ~20,000 genes expressed in mES cells could be trapped [16], indicating that this selection system can work under the control of endogenous promoters with weak transcriptional activities. We succeeded to obtain homologous recombinants for Nrp1, Nrp2, Sema4d, Cry1, Cry2 and Rad18 using the promoter-less targeting vector with neo at high efficiency (>10% of G418 resistant clones), confirming the high sensitivity of the neo selection system because the expression levels of these neuronal and housekeeping genes are moderate or low in ES cells (Table 1). In contrast, the promoter-less targeting vector with pac looks only applicable for the genes expressed at high levels, such as Zfp42 and Oct3/4, although the knockout efficiency is high (>50% of puromycin resistant clones). In the case of Estrogen-related receptor β (Esrrb), we got the homologous recombinants using the promoter-less targeting vector with neo but not with pac, indicating the difference of their threshold levels as well as the importance of their choice ( [17] and Sugimoto, Adachi and Niwa, unpublished). Other markers are also applicable for the promoter-less targeting vectors like in the cases of Zfp42, Oct3/4 and Sox2, which are all expressed at high levels (Table 1).
Figure 3.
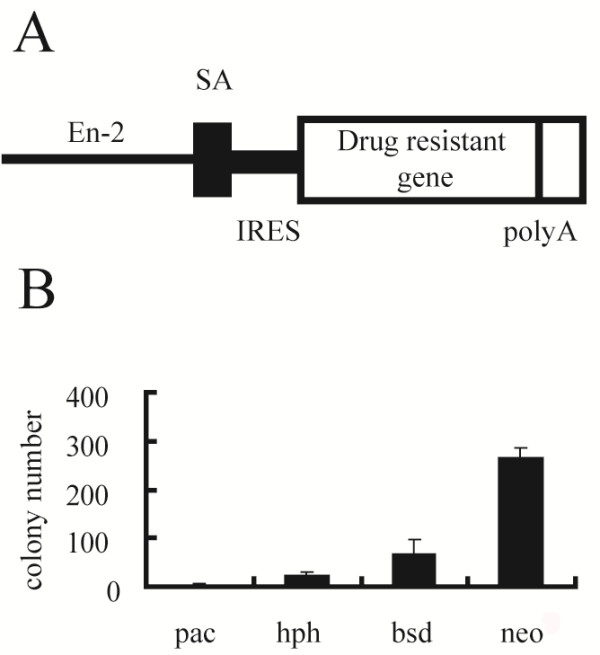
Comparison of the efficiency of gene trap with different drug selection systems. (A) Design of the gene trap vectors containing the different drug resistance genes. The drug resistance genes were fused to EMCV-IRES and placed downstream of the splice acceptor (SA) of Engrailed (En)-2 gene [15]. (B) The numbers of drug-resistant colonies in gene-trap screening were counted in each drug selection. Columns and bars represent average colony numbers and standard error means (s.e.m.) among the triplicate per 2 × 107 mES cells transfected with 100 μg of each promoter trap vector, respectively.
Table 1.
The expression levels of the genes targeted by the promoter-less vectors in ES cells
| Symbol | Average | Applied selection | Ref |
|---|---|---|---|
| |
log intensity* |
markers |
|
|
Zfp42 |
4.8854 |
pac, hph, bsd |
Toyooka et al., 2008 [[18]] |
| |
|
|
Masui et al., 2008 [[11]] |
|
Socs3 |
4.4746 |
pac |
(this paper) |
|
Pou5f1 |
4.4563 |
pac, neo, zeo |
Nichols et al., 1998 [[19]] |
| |
|
hph, bsd |
Toyooka et al., 2008 [[18]] |
|
Klf4 |
4.2232 |
pac |
(unpublished) |
|
Sox2 |
4.2167 |
hph, hisD |
Masui et al., 2007 [[20]] |
|
Sema4d |
3.8042 |
neo |
Taniguchi et al., 2009 [[21]] |
|
Nrp2 |
3.6699 |
neo |
Takashima et al., 2002 [[22]] |
|
Cry1 |
3.5175 |
neo |
Vitaterna et al., 1999 [[23]] |
|
Rad18 |
3.4387 |
neo |
Tateishi et al., 2003 [[24]] |
|
Cry2 |
3.2127 |
neo |
Vitaterna et al., 1999 [[23]] |
|
Esrrb |
2.0747 |
neo |
Martello et al, 2012 [[17]] |
| Nrp1 | 1.6905 | neo | Takashima et al., 2002 [[22]] |
*Microarray data of 50 ES cell lines (Nishiyama et al., [25]). Hprt: 4.9305, Gapdh: 5.2380, Actb: 5.2452.
Efficient selection of the gene conversion event by pac
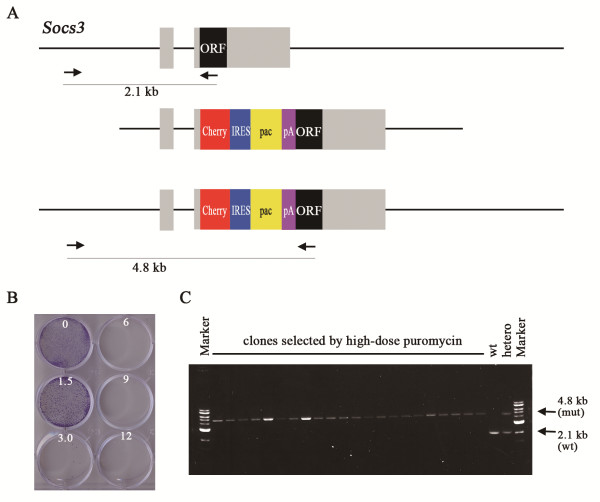
Sensitive dosage effect of the puromycin selection system could be advantageous for the tight selection of the expression level of pac. To select the homozygous mutant mES cells spontaneously appearing in the heterozygous pools via gene conversion, the efficient selection of the cells expressing two copies of the selection marker genes from those expressing one copy is required. The neo system was reported to be applicable for such selection, but the efficiency is not so high in general as originally reported [26]. We previously applied the puromycin selection system in the same way to get the homozygous mES cells for the Zfp42 knockout allele, which resulted in the 100% efficiency of the selection (4 in 4 clones analyzed) [11]. Here we tested the general applicability of this strategy in heterozygous mES cells carrying the promoter-less pac cassette. In the case of the heterozygous mES cells for the Socs3 knockout allele, the titration of puromycin concentration determined the maximal dose allowing the survival of the majority of the heterozygotes at 4 μg/ml, and the high-dose (6 μg/ml) puromycin selection resulted in the 100% efficiency of the homozygous mutant selection (46 in 46 clones analyzed; Figure 4). Similarly, in the case of the heterozygous mES cells for the floxed Klf4 allele, the high-dose (6 μg/ml) puromycin selection resulted in 60% efficiency of the homozygous mutant selection (12 in 20 clones analyzed; data not shown). These data indicated that the tight dosage-dependency of the puromycin selection system is suitable for this strategy.
Figure 4.
Generation of mES cells homozygous for the Socs3 knock-out allele by high-dose puromycin selection. (A) Strategy for the generation of Socs3-KO ES cells. The schematic maps of the Socs3 allele (top), the KO vector carrying the Cherry-IRES-pac-pA cassette (middle), and the KO allele generated by homologous recombination (bottom). The PCR with the primers set at the 5′ external genome and the open reading frame (ORF) in the second exon provides the polymorphism between the wild-type and mutant alleles, 2.1 kb and 4.8 kb, respectively. (B) Sensitivity of the heterozygous clone to various concentration of puromycin. Most of the cells were killed by 3.0 μg/ml of puromycin for 5 days. (C) PCR analysis of the genotypes of the clones selected by 6 μg/ml of puromycin from the heterozygotes. All clones possess the mutant allele only, indicating that they are homozygotes for the Socs3-KO allele.
Discussion
The drug resistance genes described here can be used simultaneously for multiple transgene expression and gene targeting in mES cells. There is no interference between any of these positive selection markers because we succeeded to obtain mES cells carrying multiple drug-resistance genes in various combinations [20,27]. Here we described the different kinetics of the drug selection systems pac, zeo, hph, hisD, bsd and neo. Pac and zeo require high levels of expression to confer the proper drug resistance to mES cells, whereas neo establishes the resistance to G418 with minimal expression level in mES cells.
In the conventional transgene expression strategy, the drug resistant genes were driven under the control of strong viral enhancer/promoters derived from simian virus 40 and human cytomegalovirus. However, separation of the expression units of the transgene and the drug-resistant genes failed to ensure the transgene expression in the drug-resistant cells and it was required to screen a large number of drug-resistant clones to obtain the stable transfectants with ideal expression levels. The IRES-mediated bi-cistronic expression vector directs the expression of the transgene and the drug-resistant gene from the same promoter, in which the drug selection always confirms the expression of the transgene. If we apply the rule we identified in this report, it is possible to design an expression vector with an ideal expression level of the transgene by a proper choice of the drug selection system. To obtain the homogenous expression of the fluorescent markers in mES cells, pac and zeo are recommended as we succeeded previously [11,20]. However, when a moderate or low level of expression is ideal like in the cases of tetracycline-dependent transcriptional activator tTA/rtTA, of which the expression at a high level is toxic for mES cells (Niwa, unpublished), and Tbx3 [12], neo is the first choice.
We recently reported the function of Esrrb as a target of glycogen synthase kinase-3 (Gsk3)-Tcf3 pathway [17]. In this report we first applied the bi-cistronic expression vector with CAG and IRES-hph to direct the expression of Esrrb in mES cells, resulting in 6–8 fold higher expression of Esrrb transgene than the endogenous levels. Since this situation creates the possibility of neomorphic effects, we switched to the expression vector with IRES-neo and succeeded to confirm that the constitutive expression of the exogenous Esrrb at endogenous levels is sufficient to sustain self-renewal of mES cells in Gsk3 inhibitor-independent manner. This is a good example demonstrating the importance of the choice of a proper drug selection system to obtain appropriate levels of transgene expression. Well-designed strategy for transgene expression will provide clear results in cell biological analyses.
Conclusions
The expression levels of the transgenes using the bi-cistronic expression vectors depend on the drug selection systems. Appropriate choices of the systems will give clean results. This is also applicable to the gene targeting with bi-cistronic durg-resistant genes. The principle shown here in mES cells should be applicable to mouse induced pluripotent stem (iPS) cells directly and most likely to human ES cells after modification.
Methods
Plasmid constructions
Initial Methionine of all drug resistance genes in Egfp expression vectors were fused in frame to ATG sequence of the NcoI site in 3′ terminus of EMCV-IRES sequence derived from pCITE-1 (Novagen). pCAG-IP and pCAG-IZ plasmid was constructed for puromycin and zeocin selection as described [9,28]. pCAG-IB was constructed by replacing the NcoI-XbaI fragment in pCAG-IZ with the BsaI-SpeI fragment containing the bsd gene derived from pUC-SV-BSD (Funakoshi). hisD ORF was amplified from pAGHisD plasmid (a gift from S. Takeda, Kyoto university) by PCR method with sense primer fused to BspHI recognition site and antisense primer fused to XbaI site and exchanged NcoI and XbaI fragment of pGTIRESβgeopA[14], resulting pGTIRESHisDpA. pCAG-ID was made by replacement of KpnI-BamHI fragment between the pGTIRESHisD and pCAG-IP. PvuI-MscI fragment of pBR322 was ligated to blunted BamHI and PvuI digested pCAG-IP, resulting pBRCAG-IP. pGTIRESHygropA was made by exchanging BspHI-BglII hygro-PGKpA fragment from pSP72-tkphygropA with NcoI-BamHI fragment of pGTIRESβgeopA in which BamHI and BstXI sites within PGKpA are disrupted. pCAG-IH was constructed by exchanging BamHI-KpnI fragment between pBRCAG-IP and pGTIRESHygropA. The puromycin resistance gene of pBRCAG-IP was also exchanged with PCR fragment amplified from pMC1-neo-pA (Stratagene) with BspHI site attached sense primer and both BamHI site and SV40 polyA attached antisense primer, resulting pCAG-IN. pPyCAGIHisDsRedT4 was made by fusing of DsRedT4[29] open reading frame (ORF) to C-terminus of hisD gene linked with 5′-gagcaagcaagatcgaccaccatg-3′ sequence. The Egfp fragment with XhoI and NotI site obtained from pEGFP-N1 (Clonetech) by PCR was inserted between XhoI and NotI site upstream of IRES in each expression vector backbone, pCAG-IP, -IZ, -ID, -IH, -IB, and –IN, examined for puromycin, zeocin, histidinol, Hygromycin B, blasticisin S, and G418 selection, respectively. pCAG-IpacEGFP was constructed by replacement pac fragment of pCAG-IP with pacEGFP fusion fragment ligated between SalI/NotI digested Egfp fragment from pEGFP-N1 and pac fragment amplified by PCR (sense: 5′-CCTCATGACCGAGTACAAGCCCA-‘3 antisense: 5′-CGGATCCGGCACCGGGCTTGCGGGTCAT-3′) that was digested with BspHI/BamHI, linked between partially filled BamHI and SalI site. hisDsRedT4 ORF was inserted between XhoI and NotI site of pCAG-IP and –IpacEGFP. Full sequence information’s of all expression vectors are available on our web site (http://www.cdb.riken.jp/pcs/protocol/vector/vector_top.html).
Cell culture and electroporation
D3, E14tg2a and EB3 ES cells were maintained in the absence of feeder cells in Glasgow minimal essential medium (GMEM) supplemented with 10% fetal calf serum, 10-4 M 2-mercaptoethanol, and 1000 unit/ml of LIF at normal condition 37°C, 5% CO2 [30]. 2 × 107 ES cells were electroporated with 30 μg of linearized plasmid DNA at 800 V and 3 μF in a 0.4 cm cuvette using a Gene-Pulser (Bio-Rad) and then cultured in the presence of the drugs for selection, Puromycin (Nacalai tesque) Zeocin (Invivogen), Hygromycin B (HygroGold, Invivogen), L-Histidinol (Sigma), Blasticidin S (Invivogen) and G418 (Nacalai tesque), at indicated concentrations. Colonies were identified by Leishman (SIGMA) staining, and counted.
Flow cytometric analysis
Transfectants grown in the presence of each drug concentrations were harvested and 10.000 data points were collected for each sample in flow cytometry, using FACSCALIBUR (Becton Dickinson). Data were analysed using CellQuest Pro Software ver.5.2 (Becton Dickinson).
Gene targeting of Socs3
Genomic DNA sequences were amplified using the primers 5′- ataaatCGatGGCGGCTCTAACTCTGACTCTACACTC-3′ and 5′- ttaagctTGGCGCACGGAGCCAGCGTGGATCTG-3′ (for the left arm of the KO construct); and 5′- CCGGGATcCGGTAGCGGCCGCTGTGCGGAG-3′ and 5′- CAGAGCTCgtcgaCTCCTGTCTGTACAGAAGGAAAGAGAGAG-3′ (for the right arm of the KO construct). Amplified PCR products were cloned into pBR-blue vecotor. The 1.0 kb ClaI (in primer)–NotI (in genomic DNA) fragment from the left arm and the 3.0 kb NotI (in genomic DNA)–SacI (in primer) fragment were cloned into pBR-MC1DTApA. The NotI site was used to clone the marker gene cassette dCherry-IRES-pac-pA. 1 × 107 EB3 ES cells were electroporated with 100 μg of plasmid DNA linealized by XhoI. From the next day, these transfectants were selected with 1.5 μg/ml of puromycin for 8 days. 16 puromycin-resistant colonies were picked, expanded and analyzed their genotype by PCR using the primers 5′- CAGTCCTCCTAGTCGACATTCCTTCTC-3′ 5′- ttaagctTGGCGCACGGAGCCAGCGTGGATCTG-3′ with KOD-Fx (Toyobo) that amplify 2.1 kb fragment from the wild-type allele and 4.8 kb fragment from the targeted allele. One of three homologous recombinants (sKO2) was examined for their ability to survive higher concentration of puromycin, and 1 × 106 sKO2 ES cells were selected with 6 μg/ml of puromycin for 4 days followed by the culture with 1.5 μg/ml of puromycin for 6 days. About 100 colonies were formed and 46 clones were picked, expanded and analyzed their genotype by PCR using the primers shown above.
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
YN and HN conceived the project and designed the experiments along with all authors performed the experiments. YN and HN wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Yuhki Nakatake, Email: nakatakey@a2.keio.jp.
Setsuko Fujii, Email: s-fujii@cdb.riken.jp.
Shinji Masui, Email: smasui@cira.kyoto-u.ac.jp.
Toshimi Sugimoto, Email: sutoshimi@cdb.riken.jp.
Satomi Torikai-Nishikawa, Email: torikai@cdb.riken.jp.
Kenjiro Adachi, Email: adachi710@gmail.com.
Hitoshi Niwa, Email: niwa@cdb.riken.jp.
Acknowledgement
We thank Dr Futatsugi in our laboratory for editing English. This research was supported by a RIKEN grant.
References
- Hartman SC, Mulligan RC. Two dominant-acting selectable markers for gene transfer studies in mammalian cells. Proc Natl Acad Sci USA. 1988;85(21):8047–8051. doi: 10.1073/pnas.85.21.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, Kamakura T, Tao QZ, Kaneko I, Yamaguchi I. Cloning of the blasticidin S deaminase gene (BSD) from Aspergillus terreus and its use as a selectable marker for Schizosaccharomyces pombe and Pyricularia oryzae. Mol Gen Genet. 1994;242(2):121–129. doi: 10.1007/BF00391004. [DOI] [PubMed] [Google Scholar]
- Mulsant P, Gatignol A, Dalens M, Tiraby G. Phleomycin resistance as a dominant selectable marker in CHO cells. Somat Cell Mol Genet. 1988;14(3):243–252. doi: 10.1007/BF01534585. [DOI] [PubMed] [Google Scholar]
- Sugden B, Marsh K, Yates J. A vector that replicates as a plasmid and can be efficiently selected in B-lymphoblasts transformed by Epstein-Barr virus. Mol Cell Biol. 1985;5(2):410–413. doi: 10.1128/mcb.5.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vara JA, Portela A, Ortin J, Jimenez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986;14(11):4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenofsky RL, Fine M, Pellow JW. A mutant neomycin phosphotransferase II gene reduces the resistance of transformants to antibiotic selection pressure. Proc Natl Acad Sci USA. 1990;87(9):3435–3439. doi: 10.1073/pnas.87.9.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitman M, Lee E, Westphal H, Felsenfeld G. Site-independent expression of the chicken beta A-globin gene in transgenic mice. Nature. 1990;348(6303):749–752. doi: 10.1038/348749a0. [DOI] [PubMed] [Google Scholar]
- Jang SK, Davies MV, Kaufman RJ, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63(4):1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12(13):2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-D. [DOI] [PubMed] [Google Scholar]
- Masui S, Ohtsuka S, Yagi R, Takahashi K, Ko MS, Niwa H. Rex1/Zfp42 is dispensable for pluripotency in mouse ES cells. BMC Dev Biol. 2008;8:45. doi: 10.1186/1471-213X-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460(7251):118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Tsakiridis A, Tzouanacou E, Larralde O, Watts TM, Wilson V, Forrester L, Brickman JM. A novel fusion reporter system for use in gene trap mutagenesis. Genesis. 2007;45(6):353–360. doi: 10.1002/dvg.20301. [DOI] [PubMed] [Google Scholar]
- Mountford P, Zevnik B, Duwel A, Nichols J, Li M, Dani C, Robertson M, Chambers I, Smith A. Dicistronic targeting constructs: reporters and modifiers of mammalian gene expression. Proc Natl Acad Sci USA. 1994;91(10):4303–4307. doi: 10.1073/pnas.91.10.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244(4903):463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- Skarnes WC, von Melchner H, Wurst W, Hicks G, Nord AS, Cox T, Young SG, Ruiz P, Soriano P, Tessier-Lavigne M. et al. A public gene trap resource for mouse functional genomics. Nat Genet. 2004;36(6):543–544. doi: 10.1038/ng0604-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Sugimoto T, Diamanti E, Joshi A, Hannah R, Ohtsuka S, Gottgens B, Niwa H, Smith A. Esrrb is a pivotal target of the gsk3/tcf3 axis regulating embryonic stem cell self-renewal. Cell Stem Cell. 2012;11(4):491–504. doi: 10.1016/j.stem.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135(5):909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95(3):379–391. doi: 10.1016/S0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA. et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9(6):625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Amazaki M, Furuyama T, Yamaguchi W, Takahara M, Saino O, Wada T, Niwa H, Tashiro F, Miyazaki J. et al. Sema4D deficiency results in an increase in the number of oligodendrocytes in healthy and injured mouse brains. J Neurosci Res. 2009;87(13):2833–2841. doi: 10.1002/jnr.22124. [DOI] [PubMed] [Google Scholar]
- Takashima S, Kitakaze M, Asakura M, Asanuma H, Sanada S, Tashiro F, Niwa H, Miyazaki Ji J, Hirota S, Kitamura Y. et al. Targeting of both mouse neuropilin-1 and neuropilin-2 genes severely impairs developmental yolk sac and embryonic angiogenesis. Proc Natl Acad Sci USA. 2002;99(6):3657–3662. doi: 10.1073/pnas.022017899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J. et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96(21):12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi S, Niwa H, Miyazaki J, Fujimoto S, Inoue H, Yamaizumi M. Enhanced genomic instability and defective postreplication repair in RAD18 knockout mouse embryonic stem cells. Mol Cell Biol. 2003;23(2):474–481. doi: 10.1128/MCB.23.2.474-481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Xin L, Sharov AA, Thomas M, Mowrer G, Meyers E, Piao Y, Metha S, Yee S, Nakatake Y. et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem Cell. 2009;5(4):420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen RM, Conner DA, Chao S, Geisterfer-Lowrance AA, Seidman JG. Production of homozygous mutant ES cells with a single targeting construct. Mol Cell Biol. 1992;12(5):2391–2395. doi: 10.1128/mcb.12.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol Cell Biol. 2002;22(5):1526–1536. doi: 10.1128/MCB.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20(1):83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- Nichols J, Evans EP, Smith AG. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development. 1990;110(4):1341–1348. doi: 10.1242/dev.110.4.1341. [DOI] [PubMed] [Google Scholar]