Abstract
We found that oral immunization with flagellum-defective mutant strains of Salmonella enterica serovar Typhimurium with the ClpXP-deficient background protected mice against oral challenge with the virulent strain. These data indicate that Salmonella flagellin is not a dominant protective antigen in oral immunization with attenuated live vaccine strains.
Growing Escherichia coli cells are capable of protein degradation, since a variety of “abnormal” and “misfolded” proteins can be rapidly and specifically degraded during proliferation. Most intracellular proteolysis is initiated by energy-dependent proteases, mainly Lon and Clp proteases. The serine protease ClpP is normally associated with ClpX, ClpA, or both, which act as molecular chaperones (2, 12). In Salmonella enterica serovar Typhimurium, ClpXP protease is also involved in the stress response and degradation of misfolded proteins (15). It was previously reported that the ClpXP protease-depleted mutant of serovar Typhimurium loses virulence and persistently resides in BALB/c mice for long periods after either intraperitoneal (18) or oral (10) infection without causing an overwhelming systemic infection. In a previous study, the mice developed strong protective immunity after a single oral administration of ClpXP-deficient serovar Typhimurium. Consequently, at week 4 after immunization, the immunized mice were completely protected against oral challenge with serovar Typhimurium (10). We have observed that a certain amount of serovar Typhimurium lipopolysaccharide-specific antibodies are present in ClpXP-deficient-serovar Typhimurium-immunized mice and that these mice have the ability to resist systemic infections with the virulent strain of serovar Typhimurium for more than a year after a single oral immunization (data not shown). On the other hand, Tomoyasu et al. found that the ClpXP protease of serovar Typhimurium affects flagellar formation and that bacterial cells with the clpP gene deleted show a “hyperflagellate” phenotype in vitro (16). ClpXP-deficient serovar Typhimurium overproduces the flagellar protein and shows a fourfold increase in the rate of transcription of the fliC gene encoding the flagellar filament protein (16), since the ClpXP protease negatively regulates transcription of the flagellar regulon by controlling the turnover of the FlhD2FlhC2 master regulators (17). Under these circumstances, we hypothesized that ClpXP-deficient serovar Typhimurium may overproduce the flagellar protein in mice, with the result that the produced flagellar protein may work as a dominant protective antigen. In order to verify this hypothesis, we evaluated the flagellum-defective mutant strains with the ClpXP-deficient background in terms of their efficacy as live oral vaccine strains for use against Salmonella infection. The flagellar operons are divided into three classes with respect to their transcription hierarchy (6). Class 3 contains five operons, including a filament formation operon. In addition, most Salmonella serovars have two genes for a major component protein of the filament at different locations on the chromosome that code for the antigenically distinct flagellar types, H1 (phase 1 [FliC]) and H2 (phase 2 [FljB]) (6, 9). The expression of the class 3 operons requires FliA (the class 3 operon-specific sigma factor). The fliA gene is included in class 2, and it has been found to positively regulate expression by the activator proteins, FlhD and FlhC, which are encoded by the flhD class 1 operon lying at the top of the transcription hierarchy (7, 8). Each class-specific flagellum-defective mutant strain of serovar Typhimurium was previously constructed with or without the ClpXP-deficient background (10). Table 1 shows the serovar Typhimurium strains used in this study. CS2007 is the ClpXP-deficient mutant strain of serovar Typhimurium. CS2056, CS2062, and CS2086 are the fliC- and fljB-, fliA-, and flhD-defective strains derived from CS2007, respectively. The virulence levels in splenic infection were the same among the flagellum-defective mutant strains with the ClpXP-deficient background (CS2056, CS2062, and CS2086), and it appeared that there were also no discrepancies in the numbers of splenic CFU among CS2007-, CS2056-, CS2062-, and CS2086-inoculated mice. Therefore, it was concluded that when mice are orally inoculated, the flagellar structures do not affect the virulence of CS2007, and splenic infection is thus enabled (10).
TABLE 1.
S. enterica serovar Typhimurium SR-11 strains
| Strain | Relevant characteristic(s)a | Reference |
|---|---|---|
| χ3306 | Virulent strain, Nalr | 3 |
| χ3456 | Virulent strain, Tetr | 3 |
| CS2007 | clpP::Chlr in χ3306 | 18 |
| CS2056 | fliC::MudI(Amprlac), fljB::Tn10 in CS2007 | 10 |
| CS2062 | fliA::MudI(Amprlac) in CS2007 | 10 |
| CS2086 | flhD::MudI(Amprlac) in CS2007 | 10 |
Nalr, nalidixic acid resistance; Tetr, tetracycline resistance; Chlr, chloramphenicol resistance; Ampr, ampicillin resistance.
Oral immunization with the ClpXP- and flagellum-defective mutants protects mice against oral challenge with the virulent strain.
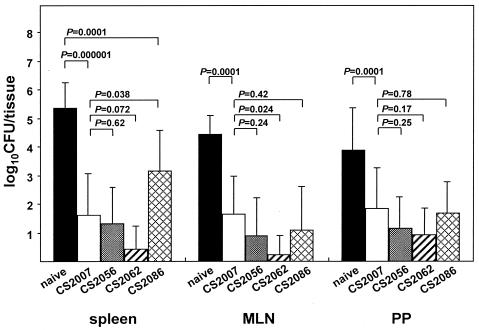
In the present study, 7-week-old female BALB/c mice (Charles River Japan, Yokohama, Japan) were orally immunized with 5 × 108 CFU of salmonellae. Four weeks later, immunized and naïve (unimmunized) mice were orally infected with 5 × 108 CFU of χ3456 (the virulent strain). The levels of recovery (numbers of CFU) of infecting salmonellae colonizing the spleens, mesenteric lymph nodes (MLN), and Peyer's patches (PP) were determined 5 days after the infection. In the same tissue sample, mixed salmonellae were distinguished as belonging to the avirulent vaccine strain (CS2007, CS2056, CS2062, or CS2086) or the infecting virulent strain (χ3456) on Luria-Bertani agar plates (Difco Laboratories, Detroit, Mich.) containing 25 μg of nalidixic acid (Sigma, St. Louis, Mo.) per ml or 15 μg of tetracycline (Sigma) per ml. As shown in Fig. 1, a small number of CFU of a virulent strain of salmonellae (χ3456) was detected in each tissue sample from immunized mice, although a large number of CFU of salmonellae was detected in each tissue sample from naïve mice. From the spleens, the levels of recovery of bacterial cells were 5.35 ± 0.89, 1.61 ± 1.44, 1.30 ± 1.27, 0.44 ± 0.77, and 3.14 ± 1.43 (log10 numbers of CFU ± standard deviations for naïve and CS2007-, CS2056-, CS2062-, and CS2086-immunized mice, respectively). The number of splenic CFU in the CS2086-immunized mice was significantly higher than that in the CS2007-immunized mice (P = 0.038). However, this number was still significantly lower than that in naïve mice (P = 0.0001). In the other tissue samples, there was a significant difference in numbers of bacterial CFU between the naïve and immunized mice, and there was no significant difference in numbers of bacterial CFU among the CS2007-, CS2056-, CS2062-, and CS2086-immunized mice, except for that in MLN of the CS2062-immunized mice (P = 0.024). The data clearly showed that a single oral immunization with ClpXP- or ClpXP-, FliC-, FljB-deficient serovar Typhimurium protected mice against oral challenge with the virulent strain. By contrast, we detected very few salmonellae of each avirulent vaccine strain in the PP and a small number of salmonellae in the spleens (less than 102 CFU/tissue) and MLN (less than 101 CFU/tissue) of the CS2007-, CS2056-, CS2062-, and CS2086-immunized mice 5 days after the challenge (data not shown). It seems that salmonellae of every vaccine strain that resided in mice were eliminated by the immunity induced after the challenge.
FIG. 1.
Protection against the virulent serovar Typhimurium strain. Seven-week-old female BALB/c mice were orally immunized with 5 × 108 CFU of CS2007, CS2056, CS2062, or CS2086. Four weeks later, the mice were challenged orally with 5 × 108 CFU of the virulent strain χ3456. After an additional 5 days, the levels of recovery of two strains (CS2007, CS2056, CS2062, or CS2086 and χ3456) were distinguishably measured in the spleens, MLN, or PP of challenged immune mice. Naïve (unimmunized) mice were also infected with χ3456 as a control. Shown are the combined results from two experiments, with five mice from each group. Significant differences between the means ± standard deviations were examined using a two-tailed Student t test (n = 10). A P value of <0.05 was regarded as statistically significant.
It was previously reported that Salmonella flagellin is one of the most relevant antigens for the generation of protective immunity in mice (4, 11). Therefore, Salmonella flagellin has been used as a carrier for heterologous peptide epitopes, which are exposed at the surface of the flagellar filament in live attenuated vaccine strains (14). In contrast, it was shown that Salmonella flagellin does not represent an efficient peptide carrier for the activation of antibody responses in mice orally immunized with live, attenuated Salmonella strains (1). Moreover, Salmonella flagellin-specific serum antibody (immunoglobulin G) responses elicited in mice following intranasal or oral inoculation with attenuated Salmonella strains were marginal (13). We also detected low levels of serum flagellin-specific immunoglobulin G responses in mice orally immunized with CS2007 (data not shown). Most recently, Harada et al. demonstrated that oral immunization with flagellin purified from serovar Typhimurium LT2 along with cholera toxin induces flagellin-specific humoral and cell-mediated immunities in mice; however, this vaccine confers at most 50 to 60% survival rates in the case of oral challenge with 106 CFU of virulent serovar Typhimurium (5). In this study, the CS2056, CS2062, and CS2086 strains were genetically deficient in flagellin (in fact, we detected no flagellar filaments on the bacterial surface of CS2056, CS2062, or CS2086 by using transmission electron microscopy [data not shown]); however, oral immunization with each vaccine strain was able to protect mice against oral challenge with 5 × 108 CFU (more than 1,500 50% lethal doses) of virulent serovar Typhimurium. Actually, immune, challenged mice did not become sick and all of these mice survived after the challenge. Based on these data, we came to the conclusion that the Salmonella flagellar filament proteins (FliC and FljB) are not dominant protective antigens in oral immunization with attenuated live vaccine strains.
Acknowledgments
We thank A. Takaya, T. Tomoyasu, and T. Yamamoto at Chiba University for providing mutant strains of serovar Typhimurium and for their helpful advice, and we thank Y. Kikuchi at Kitasato University for engaging in fruitful discussions.
This work was supported by a Grant-in-Aid for Scientific Research C (15590398) and in part by a grant from the 21st-Century COE Program from the Ministry of Cultures, Sciences, and Technology of the Japanese Government.
Editor: B. B. Finlay
REFERENCES
- 1.De Almeida, M. E., S. M. Newton, and L. C. Ferreira. 1999. Antibody responses against flagellin in mice orally immunized with attenuated Salmonella vaccine strains. Arch. Microbiol. 172:102-108. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman, S. 1996. Proteases and their targets in Escherichia coli. Annu. Rev. Genet. 30:465-506. [DOI] [PubMed] [Google Scholar]
- 3.Gulig, P. A., and R. Curtiss III. 1987. Plasmid-associated virulence of Salmonella typhimurium. Infect. Immun. 55:2891-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hackett, J., S. Attridge, and D. Rowley. 1988. Oral immunization with live, avirulent fla+ strains of Salmonella protects mice against subsequent oral challenge with Salmonella typhimurium. J. Infect. Dis. 157:78-84. [DOI] [PubMed] [Google Scholar]
- 5.Harada, H., F. Nishikawa, N. Higashi, and E. Kita. 2002. Development of a mucosal complex vaccine against oral Salmonella infection in mice. Microbiol. Immunol. 46:891-905. [DOI] [PubMed] [Google Scholar]
- 6.Kutsukake, K., Y. Ohya, and T. Iino. 1990. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J. Bacteriol. 172:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kutsukake, K., Y. Ohya, S. Yamaguchi, and T. Iino. 1988. Operon structure of flagellar genes in Salmonella typhimurium. Mol. Gen. Genet. 214:11-15. [DOI] [PubMed] [Google Scholar]
- 8.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 10.Matsui, H., M. Suzuki, Y. Isshiki, C. Kodama, M. Eguchi, Y. Kikuchi, K. Motokawa, A. Takaya, T. Tomoyasu, and T. Yamamoto. 2003. Oral immunization with ATP-dependent protease-deficient mutants protects mice against subsequent oral challenge with virulent Salmonella enterica serovar Typhimurium. Infect. Immun. 71:30-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McSorley, S. J., B. T. Cookson, and M. K. Jenkins. 2000. Characterization of CD4+ T cell responses during natural infection with Salmonella typhimurium. J. Immunol. 164:986-993. [DOI] [PubMed] [Google Scholar]
- 12.Miller, C. G. 1996. Protein degradation and proteolytic modification, p. 938-954. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 13.Sbrogio-Almeida, M. E., and L. C. Ferreira. 2001. Flagellin expressed by live Salmonella vaccine strains induces distinct antibody responses following delivery via systemic or mucosal immunization routes. FEMS Immunol. Med. Microbiol. 30:203-208. [DOI] [PubMed] [Google Scholar]
- 14.Stocker, B. A., and S. M. Newton. 1994. Immune responses to epitopes inserted in Salmonella flagellin. Int. Rev. Immunol. 11:167-178. [DOI] [PubMed] [Google Scholar]
- 15.Thomsen, L. E., J. E. Olsen, J. W. Foster, and H. Ingmer. 2002. ClpP is involved in the stress response and degradation of misfolded proteins in Salmonella enterica serovar Typhimurium. Microbiology 148:2727-2733. [DOI] [PubMed] [Google Scholar]
- 16.Tomoyasu, T., T. Ohkishi, Y. Ukyo, A. Tokumitsu, A. Takaya, M. Suzuki, K. Sekiya, H. Matsui, K. Kutsukake, and T. Yamamoto. 2002. The ClpXP ATP-dependent protease regulates flagellum synthesis in Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:645-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomoyasu, T., A. Takaya, E. Isogai, and T. Yamamoto. 2003. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 48:443-452. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto, T., H. Sashinami, A. Takaya, T. Tomoyasu, H. Matsui, Y. Kikuchi, T. Hanawa, S. Kamiya, and A. Nakane. 2001. Disruption of the genes for ClpXP protease in Salmonella enterica serovar Typhimurium results in persistent infection in mice, and development of persistence requires endogenous gamma interferon and tumor necrosis factor alpha. Infect. Immun. 69:3164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]