Abstract
The infected cell protein 0 (ICP0) of herpes simplex virus 1, a promiscuous transactivator shown to enhance the expression of genes introduced into cells by infection or transfection, interacts with numerous cellular proteins and has been linked to the disruption of ND10 and degradation of several proteins. ICP0 contains a RING finger domain characteristic of a class of E3 ubiquitin ligases. We report that: (i) in infected cells, ICP0 interacts dynamically with proteasomes and is bound to proteasomes in the presence of the proteasome inhibitor MG132. Also in infected cells, cdc34, a polyubiquitinated E2 ubiquitin-conjugating enzyme, exhibits increased ICP0-dependent dynamic interaction with proteasomes. (ii) In an in vitro substrate-independent ubiquitination system, the RING finger domain encoded by exon 2 of ICP0 binds cdc34, whereas the carboxyl-terminal domain of ICP0 functions as an E3 ligase independent of the RING finger domain. The results indicate that ICP0 can act as a unimolecular E3 ubiquitin ligase and that it promotes ubiquitin-protein ligation and binds the E2 cdc34. It differs from other unimolecular E3 ligases in that the domain containing the RING finger binds E2, whereas the ligase activity maps to a different domain of the protein. The results also suggest that ICP0 shuttles between nucleus and cytoplasm as a function of its dynamic interactions with proteasomes.
Infected cell protein 0 (ICP0) of herpes simplex virus 1 (HSV-1) acts as a promiscuous transactivator of viral and cellular genes (reviewed in ref. 1). ICP0 is critical for viral replication in cells infected at low multiplicity but is not essential in cells infected at high multiplicity (2, 3). In euploid human embryonic lung (HEL) fibroblasts, ICP0 is transported into the cytoplasm between 5 and 7 h after infection. The protein localizes with the promyelocytic leukemia protein, a component of a nuclear structure known as ND10 (4), and causes its disruption (5–7). It also interacts with several proteins such as the BMAL1 transactivator (8), the translation elongation factor 1δ (9), cyclin D3 (10), and a ubiquitin-specific protease USP7 (11–13).
Several lines of investigation have led to the suggestion that ICP0 also interacts with the ubiquitin-proteasomal degradation pathway. The evidence includes the association with USP7 and the functional association with the degradation of sumoylated promyelocytic leukemia protein and other as-yet-unidentified sumoylated proteins (14), the regulatory and catalytic subunits of DNA-dependent protein kinase (15, 16), centromeric proteins C and A (17, 18), and Sp100 (19, 20). In addition, this laboratory demonstrated that ICP0 is dynamically associated with proteasomes in untreated cells but remains bound to proteasomes in cells treated with proteasomal inhibitor MG132. Last, the 775-aa ICP0 is translated from a spliced mRNA. The three exons encode 19, 222, and 534 codons, respectively. A RING finger domain characteristic of E3 ubiquitin ligases has been identified in the domain encoded by exon 2. These properties of ICP0 have led us to investigate the possibility that ICP0 expresses this function. In addition, it has been reported that foci of ICP0 observed in infected and transfected cells contain enhanced levels of conjugated ubiquitin, which is consistent with E3 ligase activity (21, 22).
Relevant to this report are the following: (i) In the R7914 recombinant virus, the aspartate 199 of ICP0 was replaced with alanine. This substitution abolished the binding cyclin D3 and precluded the colocalization of cyclin D3 with promyelocytic leukemia protein and the transport of ICP0 to the cytoplasm but did not preclude the degradation of ND10 structures (23). The R7914 mutant virus carrying the ICP0 D199A substitution exhibits a 10-fold reduction in viral yields from quiescent HEL fibroblasts and reduced neuroinvasiveness (24).
(ii) Ubiquitin is activated by the E1 ubiquitin-activating enzyme in an ATP-dependent manner to form a thioester with a conserved active site cysteine. The major E1 in both yeast and humans is Uba1. Activated ubiquitin is transesterified to a conserved cysteine of an E2 ubiquitin-conjugating enzyme. There are 13 E2s identified in yeast, and mammals have at least that number. The E3 ubiquitin ligase binds both the E2 and substrate and functions to assemble ubiquitin onto the substrate. E3s are by far the most diverse components of the system (reviewed in ref. 25). A recently described ubiquitination enzyme is the E4 multiubiquitin chain assembly factor, which binds ubiquitin moieties conjugated to the substrate and drives the polymerization of long polyubiquitin chains (26). This mechanism facilitates targeting of the substrate for degradation by the 26S proteasome.
(iii) A substrate-independent in vitro ubiquitination system can be constructed by addition of recombinant ubiquitin-activating (E1) enzyme, recombinant ubiquitin-conjugating (E2 or Ubc) enzyme, recombinant ubiquitin ligase (E3) enzyme, ubiquitin, and ATP. Here we report that a domain of the ICP0 encoded by exon 3 substitutes for and acts as an E3 ubiquitin ligase, whereas the domain encoded by exon 2 containing the RING finger binds a E2 protein.
Materials and Methods
Low passage HEL fibroblasts grown in 150-cm2 flasks were exposed to 10 plaque-forming units (pfu) of HSV-1(F) (27) or to the R7914 recombinant virus described elsewhere (24, 28). After 2 h of exposure, the inoculum was replaced with conditioned spent medium and incubated at 37°C.
Immunoprecipitation and Pulldown Experiments.
Infected or mock-infected cells were harvested, pelleted by centrifugation, and solubilized at 4°C in modified proteasome immunoprecipitation buffer [0.5% Nonidet P-40/50 mM Tris, pH 7.5/12% glycerol/0.1% Na orthovanadate/10 mM NaF/0.5 mM DTT/2.5 mM MgCl2/l-1-tosylamido-2-phenylethyl chloromethyl ketone/7-amino-1-chloro-3-tosylamido-2-heptanone/PMSF/an ATP regenerating system consisting of 0.2 mM ATP/1 mM creatine phosphate/15 units of creatine phosphokinase (29)]. After a brief sonication, lysates were clarified by centrifugation at 2,000 × g at 4°C and reacted with either normal mouse or rabbit sera for 1 h, precleared by mixing with 5 mg each of protein-G and protein-A Sepharose, and then reacted overnight at 4°C with either 2 μg of mouse monoclonal antiserum to proteasome subunit XAPC7 (α4, Affiniti catalogue no. PW8120), 5 μl of rabbit polyclonal anti-HSV thymidine kinase or 2 μl of rabbit anti-ICP0 exon 2 antibody (10). Each sample was then reacted with a mixture of protein-G Sepharose and protein-A Sepharose (5 mg each) for 1 h. The Sepharose pellet was collected, rinsed extensively with proteasome immunoprecipitation buffer, and then solubilized in disruption buffer (2% SDS/50 mM Tris, pH 6.8/3% sucrose/5% β-mercaptoethanol/bromophenol blue).
In Vitro Substrate-Independent Polyubiquitination.
Thirty microliters of in vitro reactions were performed in ubiquitination buffer [50 mM Tris, pH 7.5/2.5 mM MgCl/0.5 mM DTT] and contained 40 ng of recombinant E1 (Calbiochem catalogue no. 662070), 40 ng [His6]-UbcH3 (Affiniti, Mamhead, Exeter, U.K., catalogue no. UW8730), 2 μg of biotinylated ubiquitin (Affiniti catalogue no. UW8705), and 0.2 mM ATP along with the ATP regenerating system described above where indicated. Reaction mixtures also contained 5 μg of purified glutathione S-transferase (GST), GST-ICP0 exon 2 (pRB4994), or GST-ICP0 exon 3 (pRB4995), previously described (9). The mixtures were allowed to react at 37°C for 90 min. The reaction was either stopped by the addition of disruption buffer or subjected to affinity capture.
Affinity capture was done with either glutathione-Sepharose (Sigma) or Talon metal affinity resin (CLONTECH). Each sample (15 μl) from in vitro ubiquitination reactions described above was diluted on wet ice in 500 μl of ubiquitination buffer, mixed with either 1 mg of glutathione-Sepharose or 20 μl of Talon resin, and kept for 1 h at 4°C. The pellets were rinsed extensively with ubiquitination buffer before addition of disruption buffer.
Immunoblotting.
Cells were rinsed in PBS containing an EDTA-free protease inhibitor mixture (Roche Diagnostics) and lysed in PBS-A* (1% Nonidet P-40, 1% deoxycholate in PBS containing protease inhibitor). Cell lysates, affinity captured proteins, or immune precipitates were solubilized in disruption buffer, boiled for 5 min, subjected to electrophoresis on 10% N,N′-diallyltartardiamide-acrylamide gels, and transferred to nitrocellulose sheets. For detection of biotinylated ubiquitin, the nitrocellulose sheets were blocked for 1 h with PBS supplemented with 0.1% BSA and 0.1% Tween-20. After blocking, the membrane was reacted for 1 h with streptavidin-peroxidase (1:1,000 dilution, Bio-Rad) in blocking buffer, extensively rinsed, and developed by enhanced chemiluminescence according to instructions supplied by the manufacturer (Pierce). For all other immunoblots, the membranes were blocked for 1 h with 5% nonfat dry milk and reacted with the appropriate primary antibody overnight at 4°C. Monoclonal antibody to proteasome, subunit α7 (HC8) (Affiniti catalogue no. PW8110), was diluted 1:1,000 in PBS supplemented with 1% BSA and 0.1% Tween-20. Mouse monoclonal antibody against ICP0, Goodwin 1112 (Goodwin Institute, Plantation, FL; ref. 30), was diluted at 1:2,000. Rabbit polyclonal antiserum was diluted in antibody diluent as follows: ICP0 exon 2 (10) 1:1,000, Ccdc34 (Neomarkers Freemont catalogue no. RB-043-P1) 1:500, and GST-ORF P (31) 1:2,000. Immunoblots for ICP0 and GST were rinsed in PBS containing 0.1% Tween-20 and both reacted to secondary antibody [alkaline phosphatase-conjugated goat anti-rabbit (1:3,000), Sigma] for 2 h. Immunoblots for cdc34 and HC8 were rinsed and reacted to secondary antibody [peroxidase-conjugated goat anti-rabbit and anti-mouse (1:1,000), Sigma], respectively, for 2 h. Immunoblots were developed by either enhanced chemiluminescence (SuperSignal West Pico chemiluminescent substrate, Pierce) or 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium substrate (Sigma), according to instructions supplied by the manufacturer.
Results
Wild-Type ICP0 Associates with Proteasome Complexes in Infected Cells.
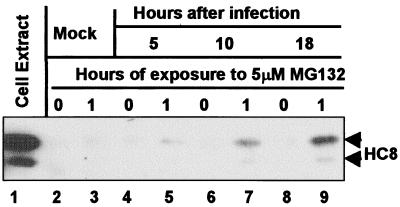
In these experiments, ICP0 was immune precipitated from lysates of HEL fibroblasts that were mock-treated or treated with MG132 at various times after exposure to 10 pfu of HSV-1(F) per cell. Electrophoretically separated immune precipitates were then probed with antibody to the proteasome subunit HC8 (α7). The procedures were as described in Materials and Methods. The results shown in Fig. 1 indicate that HC8 coprecipitated with ICP0 from lysates of MG132-treated infected cells but not from untreated infected cells (compare lanes 4 and 5, 6 and 7, and 8 and 9). The amount of HC8 that coprecipitated with ICP0 increased as infection progressed (compare lanes 5, 7, and 9). In other experiments, proteasome subunits HC3 (α2) and XAPC7 (α4) also coprecipitated with ICP0 (Fig. 3B and data not shown).
Figure 1.
Antibody to ICP0 pulls down the HC8 proteasome subunit from cells infected with wild-type virus and treated with MG132. HEL fibroblasts infected with 10 pfu of HSV-1(F) per cell were mock-treated or treated for 1 h before harvest at 5, 10, or 18 h after infection, lysed, and reacted with polyclonal rabbit antibody against ICP0-exon 2. The precipitate was solubilized, subjected to electrophoresis in a denaturing gel, and reacted with mouse monoclonal antibody against HC8.
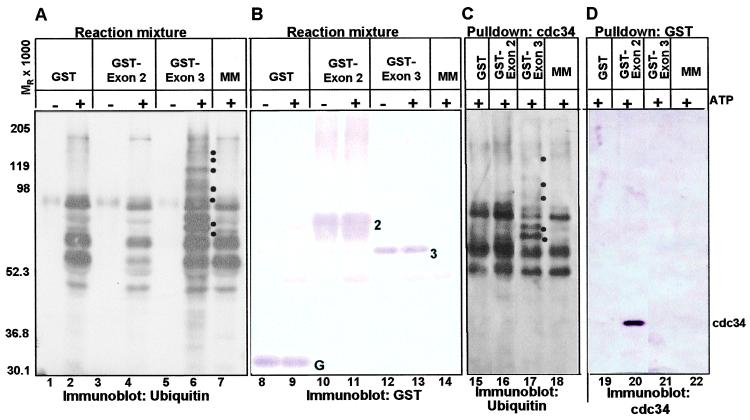
Figure 3.
The domain encoded by the carboxyl terminus of exon 3 of ICP0 acts as an E3 ligase, whereas the sequences encoded by exon 2 bind cdc34 ubiquitin-conjugating enzyme. (A) Immunoblots of electrophoretically separated products of substrate-independent in vitro ubiquitination reactions. GST (lanes 1 and 2), GST-exon 2 (lanes 3 and 4), GST-exon 3 (lanes 5 and 6), and no additional protein (lane 7) were added to the substrate-independent in vitro ubiquitination reaction master mix (MM) containing recombinant Uba1 (E1), recombinant cdc34, biotinylated ubiquitin, and ubiquitination buffer in the presence and absence of ATP and an ATP regenerating system, as described in Materials and Methods. The reaction was stopped after 90 min, and the reaction mixture was electrophoretically separated in a denaturing polyacrylamide gel and probed with streptavidin. (B) Electrophoretically separated reaction mixtures containing the indicated GST fusion protein in addition to the master mix (lanes 8–13) or the master mix alone (lane 14) in the presence and absence of ATP and an ATP regenerating system were probed with a rabbit polyclonal antibody directed against GST. (C) cdc34 was precipitated from reactions containing the indicated GST fusion protein in addition to the master mix (lanes 15–17) or the master mix alone (lane 18) in the presence of ATP and an ATP regenerating system. The precipitate was electrophoretically separated in a denaturing polyacrylamide gel and probed with streptavidin. (D) GST or GST fusion proteins were precipitated from reactions containing the indicated GST fusion protein in addition to the master mix (lanes 19–21) or the master mix alone (lane 22) in the presence of ATP and an ATP regenerating system by using glutathione Sepharose beads. The precipitate was electrophoretically separated in a denaturing polyacrylamide gel and probed with a rabbit polyclonal antibody directed against cdc34. The dots to the right of high molecular weight bands in lanes 6 and 17 identify ubiquitinated proteins; G, GST; 2, GST-exon 2 chimeric protein; 3, GST-carboxyl-terminal domain of exon 3 fusion protein.
To control for nonspecific association of ICP0 with immune complexes, we precipitated viral thymidine kinase protein with polyclonal antibody to the protein from lysates of HEL cells harvested at 5 h after infection, as described above. The electrophoretically separated precipitates were probed with antibody to ICP0. The results were that ICP0 was present in cell lysates but was not coprecipitated with the viral thymidine kinase (data not shown).
Inasmuch as ICP0 interacted with three different proteasomal subunits in the presence of MG132, but not in the absence, of the drug, we conclude that ICP0 interacts dynamically with proteasomes and that, in the presence of MG132, ICP0 remains associated with proteasomes.
ICP0 Enhances a Dynamic Association of Ubiquitinated cdc34 with Proteasomes.
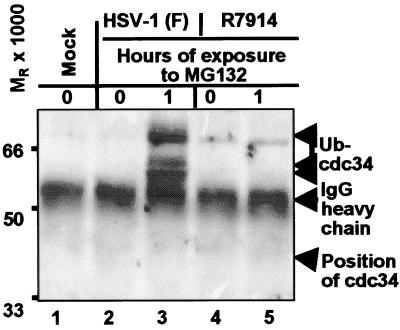
HEL cells were exposed to 10 pfu of HSV-1(F) or R7914 per cell and were either untreated or treated with 5 μm MG132 for 1 h prior to harvest at 9 h after infection. Lysates prepared in proteasome precipitation buffer supplemented with ATP were reacted with antibody to XAPC7. The immune precipitate was electrophoretically separated in a denaturing polyacrylamide gel and probed with antibody to cdc34 (UbcH3), the E2 enzyme that interacts with Skp1-Cdc53-F-box protein (SCF) (reviewed in ref. 25). As shown in Fig. 2, lane 3, the immune precipitate from lysates of MG132 treated wild-type virus-infected cells contained several cdc34 isoforms that formed bands with a MR > 50,000. These isoforms did not precipitate with proteasomal components from lysates of untreated wild-type virus-infected cells or of cells infected with R7914 recombinant virus, although small amounts of high molecular weight protein were present in all precipitates.
Figure 2.
Monoclonal antibody to XAPC7 pulls down cdc34 from lysates of HEL fibroblasts infected with wild-type HSV-1(F) and treated with MG132. Replicate cultures of HEL fibroblasts were infected with 10 pfu or HSV-1(F) or R7914 mutant virus per cell and either left untreated or treated with MG132 for 1 h before harvest at 9 h after infection. The immune precipitates obtained with monoclonal antibody against XAPC7 proteasome subunit were electrophoretically separated in denaturing gels and reacted with polyclonal rabbit serum made against cdc34.
The observation that cdc34 [MR of 32,000 (32)] coprecipitating with proteasome complexes migrated more slowly than expected suggests that it was covalently modified with numerous ubiquitin adducts. This conclusion is likely to be correct, inasmuch as, in addition to being charged at cysteine, yeast cdc34 is ubiquitinated at multiple lysines (33). Furthermore, because the higher MR cdc34 isoforms were present in significantly higher amounts in proteasomes precipitated from lysates of wild-type virus-infected cells treated with MG132 but not from those of mock-treated infected cells or cells infected with R7914 mutant, the results suggest that the enhanced interaction of ubiquitinated cdc34 with proteasomes is dynamic and mediated by wild-type ICP0.
ICP0 Expresses an E3 Ubiquitin Ligase Function.
In current models of the ubiquitin-proteasomal pathway, E3 ubiquitin ligases couple with E2 to bind substrate and facilitate the assembly of a polyubiquitin chain on the substrate (reviewed in ref. 25). Because ICP0 possesses a RING finger characteristic of many E3 enzymes, we used an in vitro ubiquitination system to determine whether the ICP0 chimeric proteins GST-exon 2 (ICP0 amino acids 111–240) or GST-exon 3 (ICP0 amino acids 568–773) promoted substrate-independent ubiquitin-protein ligation in the system described in Materials and Methods. In the experiment, the results of which are shown in Fig. 3A, the reaction mixture was electrophoretically separated in a denaturing polyacrylamide gel and reacted with streptavidin to identified ubiquitinated species. Fig. 3B shows an identical copy of the electrophoretically separated polypeptides reacted with antibody to GST. In a parallel experiment, the His6-cdc34 was collected by affinity chromatography, electrophoretically separated in a replicate denaturing polyacrylamide gel, and reacted with streptavidin. This procedure enabled the detection of ubiquitinated species interacting with cdc34 or ubiquitinated cdc34.
The results were as follows: (i) Significantly higher levels of ubiquitinated products and higher molecular weight ubiquitinated forms were observed in reactions containing GST-exon 3 in the presence of ATP (Fig. 3A, lane 6) than in any other reactions, suggesting that ICP0-exon 3 promotes ubiquitin-protein ligation. The observation that some bands in the ladder differ by approximately MR of 9,000 is consistent with the ligation of integral numbers of biotinylated ubiquitin molecules. This property is characteristic of E3 activity in substrate-independent in vitro systems (reviewed in ref. 25).
(ii) Comparison of lanes 5 and 6 in Fig. 3A indicates that the ubiquitination reaction and especially the appearance of higher molecular weight ubiquitinated proteins observed in lane 6 of Fig. 3A were ATP-dependent. This would be expected, because the ubiquitin-protein ligation is ATP dependent. Therefore, the ubiquitinated products are the consequence of the cascade of ubiquitinating enzymes and not an artifact of the system, because charging of ubiquitin by E1 is ATP dependent (34).
(iii) GST (Fig. 3A, lane 2), other GST fusion proteins (data not shown), or E1 and E2 without a putative E3 (Fig. 3A lane 7) failed to promote ubiquitin polymerization. Although ATP-dependent ubiquitin ligation was observed in the in vitro system in the absence of GST-exon 3, it occurred at much lower levels (compare lanes 2, 4, and 7 with lane 6), and high molecular weight ubiquitinated forms were not observed. This observation can be explained by basal ubiquitin-protein ligation activity in the in vitro system mediated by the E2 in the absence of stimulation by an E3.
(iv) Fig. 3B shows that the chimeric proteins GST-exon 2 and GST-exon 3 were comparable in amount and of predicted molecular weight. Therefore the difference in E3 activity between GST-exon 3 and GST-exon 2 cannot be attributed to differential addition of the GST fusion proteins to the reactions.
(v) Multiple ubiquitinated forms of cdc34 were observed in reactions containing GST-exon 3 but not in reactions containing GST, GST-exon 2, or no GST fusion protein (Fig. 3C). Observations that some bands in the ladder differed by approximately Mr of 9,000 and that there were protein bands with an Mr below the expected size (Mr of 41,000) for monoubiquitinated cdc34 are consistent with the ligation of integral numbers of ubiquitin molecules to cdc34. This observation suggests that ICP0-exon 3 promotes autoubiquitination of the E2, cdc34. Several bands in this ladder (Fig. 3C, lane 17) correspond to bands in the blot showing total ubiquitinated species (Fig. 3A, lane 6), suggesting that autoubiquitination of cdc34 accounts for a significant amount of the ubiquitin-protein ligation promoted by ICP0 exon 3 in the substrate independent in vitro ubiquitination system. In such systems, E3 activity often promotes autouibiquitination of the E2 (reviewed in ref. 25).
(vi) In substrate-independent in vitro ubiquitination systems, some RING finger E3 ligases were ubiquitinated (35). In this system, ICP0 exon 3 was not ubiquitinated to a significant extent (data not shown).
These results indicate that ICP0 has E3 ubiquitin ligase activity that maps to the region between amino acids 568 and 773 in the C-terminal portion of exon 3.
ICP0 Interacts with the E2 Ubiquitin-Conjugating Enzyme cdc34.
The results presented in Fig. 2 indicated a dynamic interaction of cdc34 with proteasomal subunits and that this interaction was mediated in infected cells by wild-type ICP0 and was apparent only when proteasomal function was inhibited by MG132. Also, all known classes of E3 ligases interact with E2 enzymes in some manner (reviewed in ref. 25). To test the hypothesis that ICP0 interacts with cdc34, GST or GST-ICP0 chimeric proteins were precipitated from the reaction mixture with glutathione Sepharose beads. The precipitates were electrophoretically separated in a denaturing polyacrylamide gel and reacted with an antibody directed against cdc34 (Fig. 3D). The results show that cdc34 coprecipitates from the reaction with GST-exon 2 but not with GST-exon 3 or GST (compare lane 20 with lanes 19 and 21). Thus, cdc34 specifically interacts with ICP0-exon 2, which contains the RING finger domain. Because GST-exon 3 promotes ubiquitination, it has to interact with cdc34 transiently or with low affinity. In many RING finger E3 ligases, the E3 RING finger domain binds the E2 (reviewed in ref. 25).
ICP0 Is Not Degraded in a Proteasome-Dependent Manner in Infected Cells.
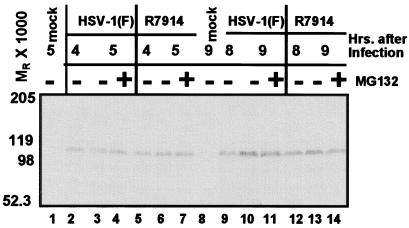
Inasmuch as both proteins targeted for degradation and components of the ubiquitin-proteasome degradation system are known to physically interact with proteasome complexes (reviewed in ref. 36) and that some RING finger E3 ligases such as TRAF2 target themselves for proteasome-dependent degradation (37), we examined the effect of MG132 treatment on ICP0 levels during infection of HEL cells exposed to 10 pfu of HSV-1(F) or R7914 recombinant virus. If ICP0 were degraded in a proteasome-dependent manner, the levels of ICP0 would be expected to be higher in drug-treated than in untreated infected cells. The procedures were as described in Materials and Methods. Electrophoretically separated proteins were probed with a rabbit polyclonal antibody directed against ICP0 exon 2 (Fig. 4) or monoclonal antibody to exon 3 (data not shown). Significant levels of ICP0 degradation products were observed in neither the presence nor the absence of MG132 in cells infected with HSV-1(F) and R7914 at any time point. We conclude that ICP0 was not degraded to a significant degree in a proteasome-dependent manner during the course of viral infection.
Figure 4.
ICP0 does not undergo proteasome-dependent degradation. HEL fibroblasts infected with 10 pfu of HSV-1(F) (lanes 2–7) or R7914 (lanes 9–14) per cell were mock-treated or treated with MG132 for 1 h before harvest at indicated times. The lysates were solubilized and subjected to electrophoresis in denaturing gels and reacted with a mouse monoclonal antibody against ICP0.
Discussion
This report sheds significant light on the involvement of ICP0 with the ubiquitin-proteasomal pathway. It is convenient to discuss the significance of key findings separately.
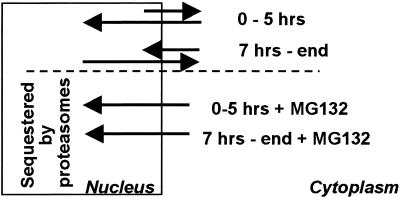
(i) ICP0 is dynamically associated with proteasomes and remains bound to proteasomes in the presence of proteasomal inhibitor MG132. This observation is consistent with the report published earlier that ICP0 colocalizes in dense spheroid-like structures with proteasomal subunits in the cytoplasm of cells infected with the d120 mutant (23). These results also shed light on the observation that at the midpoint of infection, i.e., after ICP0 is translocated to the cytoplasm, addition of MG132 causes the protein to be relocated to the nucleus (28). The results support the hypothesis that at later stages of infection, ICP0 shuttles between nucleus and cytoplasm and that in the presence of MG132, the shift in equilibrium to proteasome association causes a retention of ICP0 in the nucleus (Fig. 5). The location of ICP0 at the moment of capture, i.e., whether at the time of harvest and fixation of cells ICP0 is present in the nucleus or cytoplasm may well depend on the composition of proteasomes, and this may explain why in HEL fibroblasts ICP0 is found primarily in the cytoplasm after 5 h of infection, whereas in several human tumor cell lines the accumulation in the cytoplasm begins at a much later time.
Figure 5.
Model of the shuttling of ICP0 between nucleus and cytoplasm. As discussed, the results of this and preceding studies from this laboratory suggest that ICP0 shuttles between nucleus and cytoplasm depending on the nature of the dynamic association of the protein with proteasomes. In the presence of MG132, ICP0 is sequestered by proteasomes in nuclei either early or late in infection. In the absence of the drug, ICP0 is retained primarily in the nucleus early in infection and in cytoplasm at midpoint and later times after infection.
(ii) The association of the E2 protein cdc34 with proteasomes was significantly increased in cells infected with wild-type virus but not in cells infected with the R7914 mutant and treated with MG132. Furthermore, cdc34 formed several high molecular weight bands consistent with ubiquitinated forms. The results indicate that in wild-type virus-infected cells, cdc34 is dynamically associated with proteasomes in concert with and dependent on wild-type ICP0 but not on ICP0 carrying the D199A substitution in the sequences encoded in exon 2. The latter observation suggested that this interaction is mediated by the sequences encoding exon 2 of ICP0. Indeed, as shown in of Fig. 3D, cdc34 binds the sequences encoded by ICP0-exon 2 in the presence of ATP.
The E2 family is large and diverse, and the true range of E2 proteins capable of interacting with ICP0 remains to be investigated.
(iii) In the substrate independent in vitro ubiquitination system used in these studies, ICP0 acted as an E3 ubiquitin ligase, and this activity is encoded between codons 568 and 773 of exon 3. A caveat of this system is that it may not fully recapitulate the activity necessary to ubiquitinate a physiological substrate. Conceivably, a nonphysiologic function could be activated in vitro at high nonphysiologic concentrations. The results nevertheless are significant in that they point to two key features of ICP0. First, exon 2 bound the E2 cdc34 but did not exhibit E3 ubiquitin ligase activity. Second, the E3 ubiquitin ligase activity was independent of the RING finger domain encoded in exon 2.
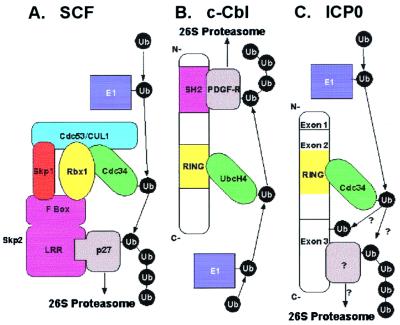
E3 ubiquitin ligases containing RING finger domains form two groups. The first (Fig. 6A) consists of E3 complexes such as SCF, which include a subunit with a RING finger domain that interacts with the cullin and E2 and a F-box protein that interacts with a specific substrate. The second (Fig. 6B) is a unimolecular class of E3 enzymes, which contain a RING finger domain that binds the E2, whereas the substrate interacts with a different domain. c-Cbl is the prototype of this class. In contrast to HECT domain E3 enzymes, both unimolecular and complex RING finger containing E3 enzymes do not appear to form a thioester intermediate with ubiquitin (reviewed in ref. 25). They bind substrates and E2 enzymes and are thought to have a catalytic activity that activates E2 enzymes. By binding both the substrate and E2, the RING finger E3 functions primarily to tether and present the substrate to the activated E2 protein, thus facilitating the transfer of ubiquitin to the substrate. We propose that ICP0 is a unimolecular E3 ubiquitin ligase as it promotes ubiquitin-protein ligation and binds the E2 cdc34 (Fig. 6C).
Figure 6.
Models for E3 ubiquitin ligase function. E1 ubiquitin-activating enzymes are shown in dark blue, E2 ubiquitin-conjugating enzymes in green, RING finger subunits and domains in yellow, substrate-binding subunits and domains in magenta, and substrates in gray. (A) Multicomponent E3 complex modeled after SCF (adapted from ref. 40). The substrate is recruited to the complex by a specific substrate-binding domain within a protein that contains an F-box motif. The F-box interacts with SCF components Skp1 and Rbx1. Recruitment of substrate p27 to SCF by the leucine-rich repeat (LRR) of the F-box protein Skp2 is depicted here. Other F-box proteins recruit different substrates (reviewed in ref. 41). SCF contains cullin 1 (CUL1), whereas other cullins are components of other multicomponent E3s. The cullin serves as a scaffold to bind Skp1, the RING finger protein Rbx1, and the E2 cdc34, which also interacts the RING finger. (B) RING finger unimolecular E3 ubiquitin ligase modeled after c-Cbl (adapted from ref. 25). The RING finger interacts with and allosterically activates the E2, UbcH4 (35). A Src-homology 2 (SH2) domain binds phosphotyrosine, such as platelet-derived growth factor receptor β (PDGF-Rβ). (C) Proposed model for ICP0 unimolecular E3 ubiquitin ligase activity. Cdc34 binds the RING finger domain in ICP0-exon 2. Unidentified substrates may bind exon 3, facilitating the transfer of ubiquitin from E2 to the substrate. Also, it is possible that ICP0 E3 activity catalyzes regulatory self-ubiquitination, which does not target ICP0 for degradation.
The fundamental difference between c-Cbl and ICP0 E3 ubiquitin ligase activities is that, whereas c-Cbl RING finger domain has intrinsic E3 activity (35), the RING finger domain in ICP0-exon 2 does not. Moreover, mutations in the RING finger domains of E3 enzymes Brca1, Siah-1, TRC8, NF-X1, kf-1, and Praja1 or addition of zinc chelators that disrupt the RING finger domain block E2 binding and ubiquitin polymerization promoted by these enzymes, suggesting interaction between the E3 RING finger domain and E2 is the seminal event in catalysis (38). Although Ubr1 and Rbx1 binding to E2s is not disrupted by RING finger mutations, E3 activity is (reviewed in ref. 25). Thus, the RING finger might mediate E3 function by promoting E2 catalytic activity, and the E2 might bind the E3 via other subunits and/or domains. In contrast, ICP0 exon 3 fragment promotes ubiquitin-protein ligation in the complete absence of the RING finger domain, showing that ICP0 intrinsic E3 activity is independent of the RING finger domain. This is unlike any RING finger E3 hitherto described, as the RING finger domain is required for E3 activity in all known RING finger E3 ligases (reviewed in refs. 25 and 38). Known RING finger domains of proteins such as HdmX/MdmX, which do not promote ubiquitin ligation, also do not have E3 ubiquitin ligase activity (39). Thus, the catalytic mechanism of the ICP0 E3 ligase must be distinct from that of the other RING finger E3 ligases.
The model we propose (Fig. 6C) is that E2 binding to the ICP0 RING finger domain in exon 2 serves to tether the E2 to the ICP0 E3 catalytic domain in exon 3, which stimulates E2 ubiquitin conjugation activity. This would function to increase the efficiency of ICP0 E3 ligase activity, because it would bring the E2 into the vicinity of the E3 catalytic domain, allowing it to promote transfer of ubiquitin from the E2. In the substrate-independent in vitro ubiquitination system, the high concentration of reagents could ensure that random diffusion is sufficient for GST-exon 3 to promote ubiquitin ligation from cdc34. We would expect a GST-fusion protein containing both domains to have increased E3 activity over GST-exon 3 in the in vitro system. The colocalization of conjugated ubiquitin with ICP0 requires the RING finger domain (21). Thus, although it is not required for in vitro E3 activity, the augmentation of E3 catalysis mediated by the RING finger domain may be required for meaningful E3 activity in vivo.
All RING finger E3 ubiquitin ligases known to date have both E2-binding and ubiquitin ligation promotion activities, In some E3 ligases, the two activities reside in the RING finger domains, whereas in others, exemplified in this case by ICP0, only one of these activities is localized in the RING finger domains, even though the protein or complex expresses both activities.
Acknowledgments
These studies were aided by grants from the National Cancer Institute (CA87761, CA83939, CA71933, and CA78766) of the United States Public Health Service. R.H. is a Howard Hughes Medical Institute Predoctoral Fellow. P.L. is a postdoctoral fellow of L'Association pour la Recherche sur le Cancer (ARC, France).
Abbreviations
- ICP0
infected cell protein 0
- HSV-1
herpes simplex virus 1
- HEL
human embryonic lung
- pfu
plaque-forming unit
- GST
glutathione S-transferase
- SCF
Skp1-Cdc53-F-box protein
References
- 1.Roizman B, Knipe D M. In: Herpes Simplex Viruses and Their Replication, in Fields Virology. 4th Ed. Howley P, Knipe D M, editors. Philadelphia: Lippincott; 2001. , in press. [Google Scholar]
- 2.Sacks W R, Schaffer P A. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stow N D, Stow E C. J Gen Virol. 1986;67:2571–2585. doi: 10.1099/0022-1317-67-12-2571. [DOI] [PubMed] [Google Scholar]
- 4.Maul G G, Guldner H H, Spivack J G. J Gen Virol. 1993;74:2679–2690. doi: 10.1099/0022-1317-74-12-2679. [DOI] [PubMed] [Google Scholar]
- 5.Everett R D, Maul G G. EMBO J. 1994;13:5062–5069. doi: 10.1002/j.1460-2075.1994.tb06835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maul G G, Everett R D. J Gen Virol. 1994;75:1223–1233. doi: 10.1099/0022-1317-75-6-1223. [DOI] [PubMed] [Google Scholar]
- 7.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaguchi Y, Tanaka M, Yokoymama A, Matsuda G, Kato K, Kagawa H, Hirai K, Roizman B. Proc Natl Acad Sci USA. 2001;98:1877–1882. doi: 10.1073/pnas.041592598. . (First Published February 6, 2001; 10.1073/pnas.041592598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawaguchi Y, Bruni R, Roizman B. J Virol. 1997;71:1019–1024. doi: 10.1128/jvi.71.2.1019-1024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawaguchi Y, Van Sant C, Roizman B. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett R D, Meredith M R, Orr A, Cross A, Kathoria M, Parkinson J. EMBO J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meredith M R, Orr A, Everett R D. Virology. 1994;200:457–469. doi: 10.1006/viro.1994.1209. [DOI] [PubMed] [Google Scholar]
- 13.Meredith M R, Orr A, Elliott M, Everett R D. Virology. 1995;209:174–187. doi: 10.1006/viro.1995.1241. [DOI] [PubMed] [Google Scholar]
- 14.Everett R D, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. J Virol. 1998;72:6581–6591. doi: 10.1128/jvi.72.8.6581-6591.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lees-Miller S P, Long M C, Kilvert M A, Lam V, Rice S A, Spencer C A. J Virol. 1996;70:7471–7477. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson J, Lees-Miller S P, Everett R D. J Virol. 1999;73:650–657. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett R D, Earnshaw W C, Findlay J, Lomonte P. EMBO J. 1999;18:1526–1538. doi: 10.1093/emboj/18.6.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lomonte P, Sullivan K F, Everett R D. J Biol Chem. 2001;276:5829–5835. doi: 10.1074/jbc.M008547200. [DOI] [PubMed] [Google Scholar]
- 19.Chelbi-Alix K M, de The H. Oncogene. 1999;18:935–941. doi: 10.1038/sj.onc.1202366. [DOI] [PubMed] [Google Scholar]
- 20.Parkinson J, Everett R D. J Virol. 2000;74:10006–10017. doi: 10.1128/jvi.74.21.10006-10017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Everett R D. J Virol. 2000;74:9994–10005. doi: 10.1128/jvi.74.21.9994-10005.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkinson J, Everett R D. J Virol. 2001;75:5357–5362. doi: 10.1128/JVI.75.11.5357-5362.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez P, Van Sant C, Roizman B. J Virol. 2001;75:3832–3840. doi: 10.1128/JVI.75.8.3832-3840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Sant C, Kawaguchi Y, Roizman B. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jackson P K, Eldridge A G, Freed E, Furstenthal L, Hsu J Y, Kaiser B K, Reimann J D R. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 26.Koegl M, Hoppe T, Schlenker S, Ulrich H D, Mayer T U, Jentsch S. Cell. 1999;96:635–644. doi: 10.1016/s0092-8674(00)80574-7. [DOI] [PubMed] [Google Scholar]
- 27.Ejercito P M, Kieff E D, Roizman B. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 28.Van Sant C, Lopez P, Advani S J, Roizman B. J Virol. 2001;75:1888–1898. doi: 10.1128/JVI.75.4.1888-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brandimarti R, Roizman B. Proc Natl Acad Sci USA. 1997;94:13973–13978. doi: 10.1073/pnas.94.25.13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ackermann M, Braun D K, Pereira L, Roizman B. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagunoff M, Randall G, Roizman B. J Virol. 1996;70:1810–1817. doi: 10.1128/jvi.70.3.1810-1817.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plon S E, Leppig K A, Do H N, Groudine M. Proc Natl Acad Sci USA. 1993;90:10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee A, Gregori L, Xu Y, Chau V. J Biol Chem. 1993;268:5668–5675. [PubMed] [Google Scholar]
- 34.Ciechanover A, Heller H, Katz-Etzion R, Hershko A. Proc Natl Acad Sci USA. 1981;78:761–765. doi: 10.1073/pnas.78.2.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joaziero C A P, Wing S S, Huang H, Leverson J D, Hunter T, Liu Y-C. Science. 1999;286:309–312. doi: 10.1126/science.286.5438.309. [DOI] [PubMed] [Google Scholar]
- 36.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 37.Duckett C S, Thompson C B. Genes Dev. 1997;11:2810–2821. doi: 10.1101/gad.11.21.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorick K L, Jensen J P, Fang S, Ong A M, Hatakeyama S, Weissman A M. Proc Natl Acad Sci USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp D A, Kratowicz S A, Sank M J, George D L. J Biol Chem. 1999;274:38189–38196. doi: 10.1074/jbc.274.53.38189. [DOI] [PubMed] [Google Scholar]
- 40.Tyers M, Willems A R. Science. 1999;284:601–604. doi: 10.1126/science.284.5414.601. [DOI] [PubMed] [Google Scholar]
- 41.Tyers M, Jorgensen P. Curr Opin Genet Dev. 2000;10:54–64. doi: 10.1016/s0959-437x(99)00049-0. [DOI] [PubMed] [Google Scholar]