Abstract
Available methods for differentiating human embryonic (ES) and induced pluripotent stem (iPS) cells into neurons are often cumbersome, slow and variable. Alternatively, human fibroblasts can be directly converted into induced neuronal (iN) cells. However, with present techniques conversion is inefficient, synapse formation is limited, and only small amounts of neurons can be generated. Here, we show that human ES and iPS cells can be converted into functional iN cells with nearly 100% yield and purity in less than two weeks by forced expression of a single transcription factor. The resulting ES-iN or iPS-iN cells exhibit quantitatively reproducible properties independent of the cell line of origin, form mature pre- and postsynaptic specializations, and integrate into existing synaptic networks when transplanted into mouse brain. As illustrated by selected examples, our approach enables large-scale studies of human neurons for questions such as analyses of human diseases, examination of human-specific genes, and drug screening.
INTRODUCTION
The generation of human embryonic stem cells (ES cells) and induced pluripotent stem cells (iPS cells) and their in vitro differentiation into potentially any desired cell type holds great promise, and may revolutionize the study of human disease (Hanna et al., 2010; Okita and Yamanaka, 2011; Blanpain et al., 2012). Given the lack of alternative sources, a major effort has been directed towards the development of differentiation protocols that convert pluripotent stem cells into neurons to allow examination of healthy human neurons and of neurons derived from patients with a variety of neurological diseases. In this approach, fibroblasts from patients with poorly understood diseases – such as schizophrenia or Alzheimer’s disease – are converted into iPS cells that are then differentiated into neurons to study the pathogenesis of these diseases (reviewed in Han et al., 2011; Ming et al., 2011; Brennand et al., 2012; Marchetto and Gage 2012). Moreover, elegant studies have described differentiation protocols that produce distinct types of neurons in vitro, although the number and properties of different types of human neurons in situ are largely unknown and are only now beginning to be defined. Overall, these studies suggest that derivation of neurons from human stem-cells may allow scientists to examine specific subtypes of neurons, to generate human neurons for regenerative medicine, and to investigate changes in human neurons in neuropsychiatric disorders (e.g., see Cho et al., 2008; Fasano et al., 2010; Kriks et al., 2011; Shi et al., 2012; Chambers et al., 2012; Ma et al., 2012). However, this approach of studying human neurons at present suffers from two major limitations.
The first limitation is based on characteristic differences between particular pluripotent cell lines (Osafune et al., 2008; Hu et al., 2009; Bock et al., 2011). These differences influence the properties of the neurons that are derived from these lines. For example, neurons derived by the same protocol from two different ES cell lines exhibited quite distinct properties (Wu et al., 2007). Moreover, ES and iPS cell lines may change as a function of time in culture (Mekhoubad et al., 2012). A systematic comparison of the neural differentiation potential of different ES and iPS cell lines revealed a large variation in conversion efficiency, and it is likely that maturation stages and functional properties of the resulting neurons are also variable (Hu et al., 2009).
The second limitation is related to the cumbersome, variable, and slow procedures needed for deriving neurons with functional properties from ES or iPS cells. Generating neurons by differentiation of ES or iPS cells requires months of tissue culture procedures, and renders large-scale studies difficult (Johnson et al., 2007). Moreover, differentiation of ES and iPS cells into neurons depends on specific environmental factors such as pharmacological agents and bioactive proteins that may be difficult to obtain with a consistent composition, injecting a further element of variation (Soldner and Jaenisch, 2012).
The two major limitations of current technologies for generating human neurons outlined above motivated us and others to develop methods for direct conversion of human fibroblasts into induced neurons, referred to as iN cells (Pang et al., 2011; Ambasudhan et al., 2011; Qiang et al., 2011; Pfisterer et al., 2011a and 2011b; Yoo et al., 2011; Caiazzo et al., 2011; Son et al., 2011). Although these efforts were successful and allow rapid production of human iN cells, all of the currently available protocols for generating human iN cells (as opposed to mouse iN cells) suffer from relatively low yields and low efficiency, and are further hampered by the limited availability and renewability of fibroblasts as starting materials. Moreover, the resulting iN cells often exhibited decreased competence for synapse formation. Specifically, we (Pang et al., 2011) and others (Pfisterer et al., 2011; Son et al., 2011) found that the same three transcription factors that convert mouse fibroblasts into iN cells (Brn2, Ascl1, and Myt1L; Vierbuchen et al., 2010) also trans-differentiate human fibroblasts into iN cells when combined with a fourth transcription factor (NeuroD1), a process that may be additionally facilitated by co-expression of microRNAs (Yoo et al., 2011; Ambasudhan et al., 2011). However, apart from the limited capabilities of iN cells produced by these procedures, these experiments did not clarify the minimal requirement of defined factors for trans-differentiating human non-neuronal cells into neurons, and suggested that ancillary factors, such as specific culture conditions, may introduce further variability into these trans-differentiation protocols. Together, these features make analysis of disease-related phenotypes using human iN cells difficult, especially since these protocols do not generate large amounts of iN cells that are fully competent to form synapses.
To address these problems, we here developed approaches that allow rapid and reproducible production of human iN cells from ES or iPS cells. The experiments we describe utilize a renewable resource and are scalable, and result in iN cells with reproducible properties that are independent of the starting ES or iPS cell line. Strikingly, our approach requires only a single transcription factor, and generates large amounts of human iN cells with robust synapse formation capabilities. Moreover, we demonstrate that the resulting iN cells can be used for analysis of human neuronal short-term plasticity, large-scale Ca2+-imaging, or analysis of loss-of-function states mimicking a human genetic disorder. Thus, the approach we describe may be generally useful not only to explore the cellular phenotype associated with neuropsychiatric disorders, but also for drug screening endeavors and for mechanistic studies.
RESULTS
A single transcription factor efficiently converts human ES and iPS cells into neurons
Following our initial observation that the combined expression of Brn2, Ascl1, and Myt1l induces functional neurons from human ES cells (Pang et al., 2011), we examined whether forced expression of a series of single transcription factors in ES and iPS cells might initiate iN cell differentiation. As in previous studies (Vierbuchen et al., 2010; Pang et al., 2011), we used lentiviral delivery for constitutive expression of rtTA (Urlinger et al., 2000), and tetracycline-inducible expression of exogenous proteins driven by a tetO promoter. Surprisingly, we found that overexpressing either neurogenin-2 (Ngn2) or NeuroD1 alone rapidly converted ES and iPS cells into neuronal cells (Figs. 1 and S1). Since this conversion was based on forced expression of a lineage-specific transcription factor and appears to be a direct lineage conversion similar to lineage conversion between somatic cells, we refer to the resulting neurons as iN cells as previously (Vierbuchen et al., 2010; Pang et al., 2011).
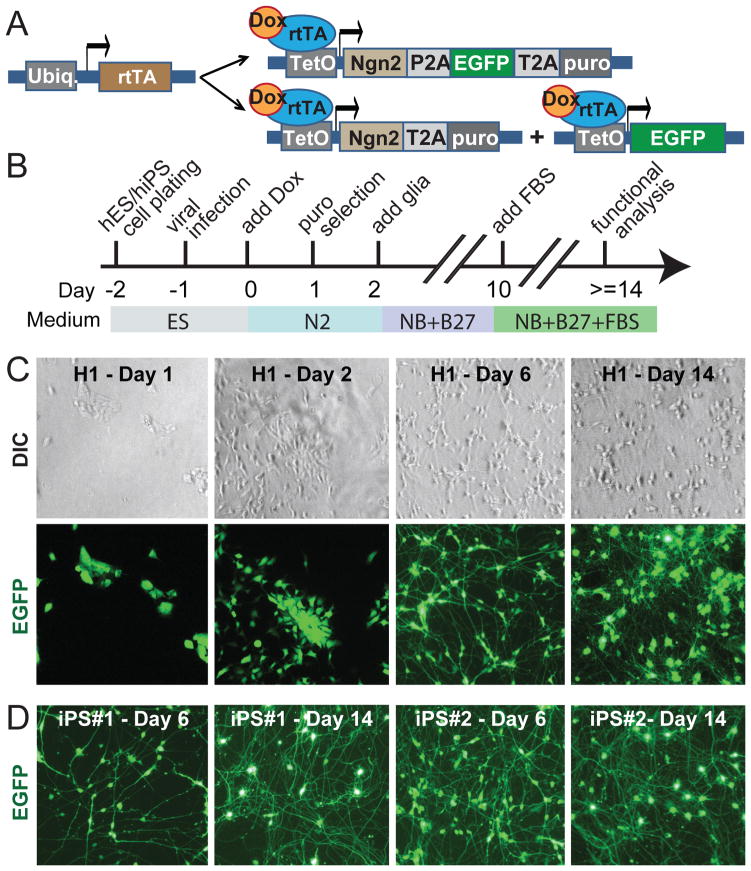
Figure 1. Rapid single-step generation of human iN cells.
A, Design of lentiviral vectors for Ngn2-mediated conversion of ES and iPS cells to iN cells. Cells are transduced with (i) a virus expressing rtTA and (ii) either a single additional virus expressing an Ngn2/EGFP/puromycin resistance gene as a fusion protein linked by P2A and T2A sequences, or with two viruses that separately express Ngn2/puromycin resistance gene and EGFP.
B, Flow diagram of iN cell generation.
C, Representative images illustrating the time course of the conversion of H1 ES cells into iN cells. Corresponding differential interference contrast (DIC) and GFP fluorescence pictures are shown on top and bottom.
D, Representative images of converted iN cells from two different iPS cells lines at day 6 and day 14. Note that iN cells are clearly identifiable already on day 6. For iN cells generated with NeuroD1 expression, see Fig. S1.
Because the effects of NeuroD1 and Ngn2 were similar, we decided to focus only on one factor and chose Ngn2. To selectively culture only cells expressing the transcription factor, we co-expressed a puromycin resistance gene with Ngn2 (allowing us to select for cells expressing Ngn2), and we additionally co-expressed EGFP (allowing us to identify lentivirally transduced cells). In the standard protocol (Fig. 1A), ES or iPS cells were plated on day −2, infected with lentiviruses on day −1, and Ngn2 expression was induced with doxycyclin on day 0. A 24 hr puromycin selection period was started on day 1, and mouse glia (primarily astrocytes) were added on day 2 to enhance synapse formation (Fig. 1B; Vierbuchen et al., 2010). Strikingly, forced Ngn2 expression converted ES and iPS cells into neuron-like cells in less than one week, and produced an apparently mature neuronal morphology in less than two weeks (Figs. 1C and 1D). This is faster than any currently available method for generating neurons from human ES or iPS cells (Table 1).
TABLE 1.
Comparison of the rapid iN cell generation method presented here with previously reported methods of generating neurons from ES/iPS cells
| targeted cell type | time to Tuj1/MAP2+ cells | time to functional synapses | yield of neurons | neuronal purity | neuron subtype purity | Reference | PMID |
|---|---|---|---|---|---|---|---|
| Neurons | ~ 6 weeks | 12 weeks | ND | 15–79%* | mixed | Hu et al., 2010, | 20160098 |
| NPCs | 4–5 weeks | ND | ND | ND | mixed | Zhang et al., 2001 | 11731781 |
| Neurons | 4 weeks | 7 weeks | ND | 89 ± 3%* | mixed | Johnson et al.,2007 | 17376968 |
| NPCs | ~3 weeks | ND | ND | 82%* | mixed | Chambers et al., 2009 | 19252484 |
| NPCs | ~3–4 weeks | ND | ND | ND | mixed | Elkabetz et al., 2008 | 18198334 |
| nociceptive sensory neurons | ~2 weeks | ND | ND | > 75%* | >60% ** | Chambers et al.,2012 | 22750882 |
| midbrain dopamine neurons | ~3–4 weeks | >7 weeks | ND | ND | 60–80%** | Kriks et al., 2011 | 22056989 |
| NPCs | 4 weeks | ND | ND | 40–70%* | mixed | Koch et al., 2009 | 19218428 |
| cortical excitatory neurons | ~3–4 weeks | > 8 weeks | ND | 40% * | <20%** | Espuny-Camacho et al., 2013 | 23395372 |
| neurons | 4 weeks | ~5 weeks with glia coculture | ND | 70–80%* | mixed | Wu et al., 2007 | 17693548 |
| inhibitory forebrain neurons | ~3–4 weeks | ~6 weeks | ND | ~70% ** | 80%** | Maroof et al., 2013 | 23642365 |
| inhibitory forebrain neurons | ~3–4 weeks | 8 weeks with glia coculture | ND | 92% ** | 76%** | Nicholas et al., 2013 | 23642366 |
| Neurons | ~3–4 weeks | ~4–5 weeks | ND | 90% ** | mixed | Israel et al., 2012 | 22278060 |
| excitatory neurons | ~1 week | 2 weeks | ~100%** | 100%** | ~100%*** | this study | n.a. |
based on 1 marker expression,
based on multiple markers,
based on functional data
All time periods listed are from the undifferentiated ES cell state; ND: not determined
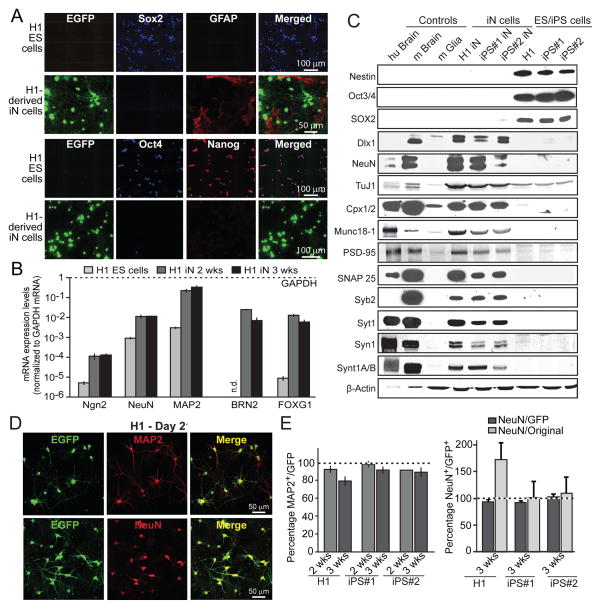
We stained H1 ES cells and H1-derived iN cells at 21 days after induction for EGFP (to identify cells with viral transduction), the stem-cell markers Sox2, Oct4, and Nanog, and the glial cell marker GFAP (to mark mouse astrocytes added to the iN cell culture for promotion of synapse formation). At the level of immunolabeling, expression of stem-cell markers was abolished in iN cells, consistent with a conversion of H1 ES cells into iN cells (Fig. 2A). Quantitative RT-PCR analyses revealed that iN cells expressed increased levels of endogenous Ngn2 as well as of two neuronal markers, NeuN and MAP2, whose levels were elevated ~100-fold (Fig. 2B). In addition, we observed an even larger induction of the expression of the transcription factors Brn2 and FoxG1, which are markers for excitatory cortical neurons (Fig. 2B).
Figure 2. Characterization of the properties and yield of Ngn2-generated iN cells.
A, Immunofluorescence images of H1 ES cells and H1 ES cell-derived iN cells (3 weeks after induction). H1 ES but not iN cells are positive for the ES cell markers Nanog, Sox2 and Oct4, while GFAP is only present in co-cultured astrocytes but not iN or ES cells.
B, Quantification of selected mRNA levels in H1 ES cells and in H1 ES-cell derived iN cells after 2 and 3 weeks of lentiviral infection. Levels are normalized for GAPDH mRNA levels as an internal control, and are shown on a logarithmic scale. Note that endogenous Ngn2 is induced ~20-fold, and endogenous Brain-2 and FOXG1 expression is induced >1,000-fold. Data are means ± SEM (n=3 independent experiments).
C, Immunoblot analyses of proteins extracted from human postmortem cortex (hu Brain), mouse brain (m Brain), and cultured mouse glia cells (m Glia), and of proteins solubilized from iN cells that were derived from H1 ES and two different iPS cell lines (3 weeks after induction) and from the starting ES and iPS cells as indicated. Proteins are identified on the left (Cpx1/2, complexin-1 and −2; Syb2, synaptobrevin-2; Syt1, synaptotagmin-1; Syn1, synapsin-1; Synt1A/B, syntaxin-1A and -1B).
D, Representative images of H1 ES-cell derived iN cells visualized via their EGFP fluorescence and immunolabelled for MAP2 or NeuN as indicated.
E, Yield of iN cell conversion of H1 ES cells and two different iPS cell lines. The percentage of EGFP-positive cells that also express the neuronal marker MAP2 after 2 or 3 weeks of conversion is shown on the left, and the yield of NeuN-positive cells at the right. NeuN-positive cell yields are calculated both in terms of the percentage of EGFP-positive cells (dark bars) or in terms of starting cell numbers (light bars). For the latter, the yield exceeds 100% for H1-derived but not iPS-derived iN cells because H1 cells still proliferate after lentiviral infection, while iPS cells do not because they are more sensitive to lentivirally induced cell death. Data are means ± SEMs (n=3 independent experiments). For additional data, see Fig. S2.
Immunoblotting experiments showed that the neuronal precursor cell (NPC) markers nestin and Sox2 were only expressed in the ES and iPS cells, whereas a series of well-established synaptic genes were only expressed in 3-week old Ngn2 iN cells (Figs. 2C and S2A). Quantitative RT-PCR measurements of the expression of the NPC markers Sox2 and nestin in the first two weeks after Ngn2 induction revealed a transient brief increase in these markers immediately after induction, with a rapid decline in expression (Fig. S2B). Furthermore, upon co-culture with mouse astrocytes H1-cell derived iN cells formed synapses with each other and with co-cultured COS cells expressing neuroligin-1 (Figs. S2C–S2E). Thus, iN cell generation involves a switch from a stem cell to a neuronal gene expression phenotype with stimulation of endogenous Ngn2 expression.
Measurements of the yield of iN cell conversion in three stem cell lines, H1 ES cells and two different iPS cells lines, showed that nearly 100% of surviving lentivirally infected ES and iPS cells were converted into neurons, revealing an unprecedented efficiency of conversion (Fig. 2D). When we calculated the number of iN cells generated as a function of starting ES or iPS cells, we observed an apparent increase with H1 ES-cell derived iN cells but not with the two iPS cell line derived iN cells (Fig. 2D). The increase in cell numbers in H1 ES-cell derived iN cells is due to the continuing division of H1 cells after plating; iPS-cell derived iN cells do not show such increased cell numbers because they exhibit some cell death in response to culture splitting and lentiviral infection, resulting in a partial loss of the iPS cells as iN cells are being generated. Overall, these data demonstrate that forced expression of a single transcription factor –Ngn2– induces neuronal differentiation with high yield.
ES- and iPS-cell derived iN cells exhibit reproducible gene expression patterns
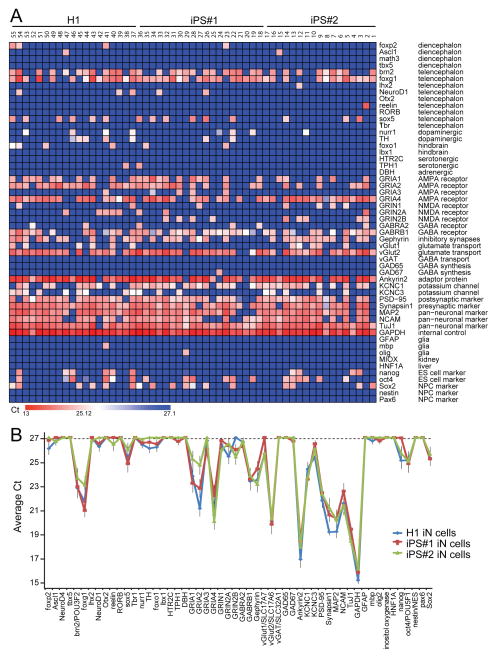
We next aimed to gain insight into the nature of the neurons generated and more importantly, to assess the reproducibility of Ngn2-induced production of iN cells from different ES and iPS cell lines. Towards this end, we quantitatively analyzed expression of 73 genes at the single-cell level using fluidigm-dependent mRNA measurements (Pang et al., 2011; Table S1). All fluidigm-mediated quantitative RT-PCR assays were validated using standard curves (Table S1). Analysis of more than 100 individual iN cells revealed a uniform but discrete pattern of gene expression in iN cells derived from H1 ES and two different iPS cell lines (Figs. 3 and S3). Specifically, Ngn2-iN cells expressed at high levels the telencephalic markers Brn-2, Cux1, and FoxG1 that are characteristic of layer 2/3 excitatory cortical neurons, but lacked other prominent forebrain transcription factors (e.g., Tbr1 and Fog2). iN cells consistently expressed AMPA-type glutamate receptors GluA1, A2, and A4, but lacked NMDA-type glutamate receptors 3 weeks after induction (Fig. 3A). Moreover, nearly all iN cells expressed vGlut2, and approximately 20% of iN cells expressed vGlut1. iN cells highly expressed GABAA-receptors but lacked the vesicular GABA-transporter vGAT or the GABA-synthetic enzyme glutamate decarboxylase (GAD). Ngn2 iN cells expressed all pan-neuronal markers tested, but lacked expression of markers for various glia cell types or for stem cells (Figs. 3A and S3B). These measurements show that Ngn2 iN cells are relatively homogeneous, and that they constitute excitatory neurons that express telencephalic markers suggestive of cortical layers 2/3.
Figure 3. iN cell generation involves reproducible changes in gene expression.
A, Single-cell quantitative RT-PCR analysis (Fluidigm) of the expression levels of the genes indicated on the right. Expression levels (expressed as Ct values) are color coded as shown on the bottom. mRNA levels were quantified in cytoplasm aspirated from individual iN cells using patch pipettes after 3 weeks of induction.
B, Comparison of gene expression profiles in iN cells differentiated from H1 ES and two different lines of iPS cells. The plot depicts average Ct values of the genes indicated at the bottom, with a cutoff of 27 cycles (on top of the 18 cycle pre-amplification).
For more extensive analyses of further marker genes, see Fig. S3.
Arguably the most important question in the production of iN cells – in fact, in the in vitro production of all human neurons – is reproducibility between lines. We therefore assessed this question for the Ngn2-based protocol in great detail. Comparison of the gene expression profiles between iN cells produced by forced differentiation of H1 ES cells and of two independent lines of iPS cells revealed a striking concordance in expression patterns (Figs. 3B and S3D). There was no major difference between stem cells in the expression of the genes tested. The highly similar transcriptional effects of Ngn2 indicate that forced expression of Ngn2 can override presumptive epigenetic differences between various pluripotent stem cell lines to induce differentiation of a single homogenuous population of excitatory forebrain neurons.
Human ES- and iPS-cell derived iN cells form synapses
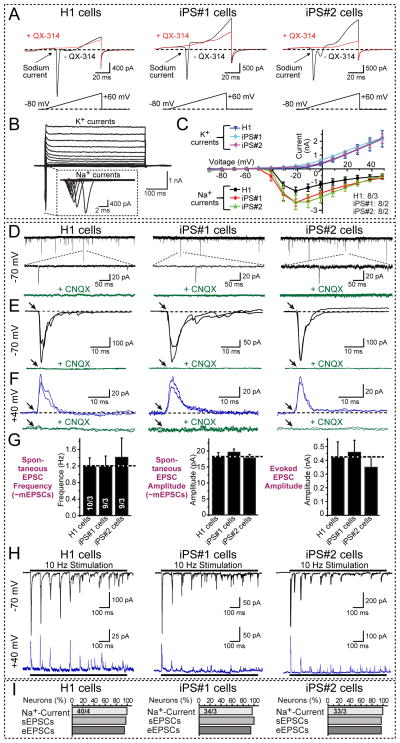
We next probed the ability of Ngn2-induced iN cells to differentiate into electrophysiologically active neurons and to form synapses. To promote synapse formation, we co-cultured iN cells with mouse glial cells (Pang et al., 2011). The iN cells reliably produced robust action potentials, and exhibited voltage-gated Na+- and K+-currents that were indistinguishable between iN cells derived from H1 ES and different iPS cell lines (Figs. 4A-4C and S4A). iN cells exhibited massive spontaneous synaptic activity that was blocked by the AMPA-receptor antagonist CNQX (Fig. 4D). Extracellular stimulation evoked EPSCs of large amplitudes, documenting abundant synapse formation (Figs. 4E and 4F). The kinetics of evoked EPSCs were identical at −70 mV and +40 mV holding potentials, and EPSCs were blocked by CNQX at both holding potentials. Thus, consistent with the gene expression profile described in Fig. 3A, EPSCs are entirely due to activation of AMPA-type and not of NMDA-type glutamate receptors, although we did observe small NMDA-receptor mediated synaptic currents in iN cells after more than 4 weeks of culture (Fig. S4B). When we quantified the frequency and amplitude of spontaneous EPSCs and the amplitude of evoked EPSCs, we found that they were indistinguishable between iN cells derived from H1 ES cells and two different iPS cell lines, and reproducible between experiments (Fig. 4G).
Figure 4. iN cells derived from ES and iPS cells form functional synapses.
A, Representative traces of membrane currents (upper panel) recorded following a ramp depolarization protocol (lower panel). Na+-currents were blocked by QX-314 (10 mM).
B, Representative traces of whole-cell voltage-clamp Na+- and K+-currents recorded in iN cells. iN cells were subjected to 10 mV step depolarizations from −90 mV to +50 mV at a −70 mV holding potential (pipette solution (in mM): 123 K-gluconate, 10 KCl, 1 MgCl2, 10 HEPES-KOH pH 7.2, 1 EGTA, 0.1 CaCl2, 1 K2ATP, 0.2 Na4GTP and 4 glucose).
C, Quantification of I/V curves of Na+- and K+-currents in iN cells derived from H1 ES cells and from two different iPS cell lines. Data are means ± SEMs; numbers of cells/cultures analyzed are shown on the right lower corner.
D, Representative traces of spontaneous EPSCs (likely mEPSCs based on size); middle panels depict expansions of selected events. The lower traces display block of spontaneous EPSCs by 50 μM CNQX.
E & F, Representative traces of evoked EPSCs monitored at −70 mV (E) and at +40 mV (F); lower panels show that CNQX completely blocks all EPSCs. Two superimposed traces are shown.
G, Quantification of the frequency (left panel) and amplitude of spontaneous EPSCs (middle panel) and of the amplitude of evoked EPSCs (right panel). Data are means ± SEM; numbers in the left bars indicate the number of cells/independent experiments performed, and apply to all panels. Note that iN cells derived from different ES/iPS cell lines exhibit quantitatively similar synaptic properties.
H, Representative traces of EPSCs evoked by a 10 Hz stimulus train applied for 1 s and monitored at −70 mV (top traces) and +40 mV (bottom traces).
I, Quantification of the rate of successful observations of voltage-gated Na+-currents, spontaneous EPSCs (sEPSCs), and evoked EPSCs (eEPSCs) in iN cells. Numbers in top bars indicate the number of cells/independent experiments performed.
Stimulus trains of 10 Hz revealed fast synaptic depression, showing that iN cell synapses exhibit short-term plasticity (Fig. 4H). No inhibitory synaptic events were observed when Ngn2-induced human iN cells were co-cultured with glia cells, but strong inhibitory synaptic inputs onto the iN cells were detected when we co-cultured iN cells with mouse cortical neurons (Figs. S4C–S4E). This experiment demonstrated that iN cells integrate into a synaptic network with the mouse cortical neurons, and that they are fully capable of forming inhibitory postsynaptic specializations. Quantifications showed that the vast majority of all iN cells, when co-cultured with mouse glia cells or cortical neurons, contained voltage-gated Na+- and K+-currents, exhibited spontaneous synaptic activity, and displayed evoked EPSCs (Fig. 4I).
Application of human iN cells to study normal and diseased human neurons
To explore the potential use of ES- or iPS-cell derived iN cells for monitoring drug activities, studying human synaptic plasticity, or modeling human disease states, we examined Ngn2 iN cells in a variety of paradigms. We first tested the use of optogenetics to directly probe the formation of presynaptic specializations of iN cells onto co-cultured mouse neurons (Figs. 5A–5C). When we selectively expressed the channelrhodopsin variant CHIEF in iN cells and co-cultured the iN cells with mouse neurons, we found that this approach led to an accurate definition of presynaptic function in the human iN cells that allows measurement of synaptic transmission between two connected neurons without the need to separately patch these neurons.
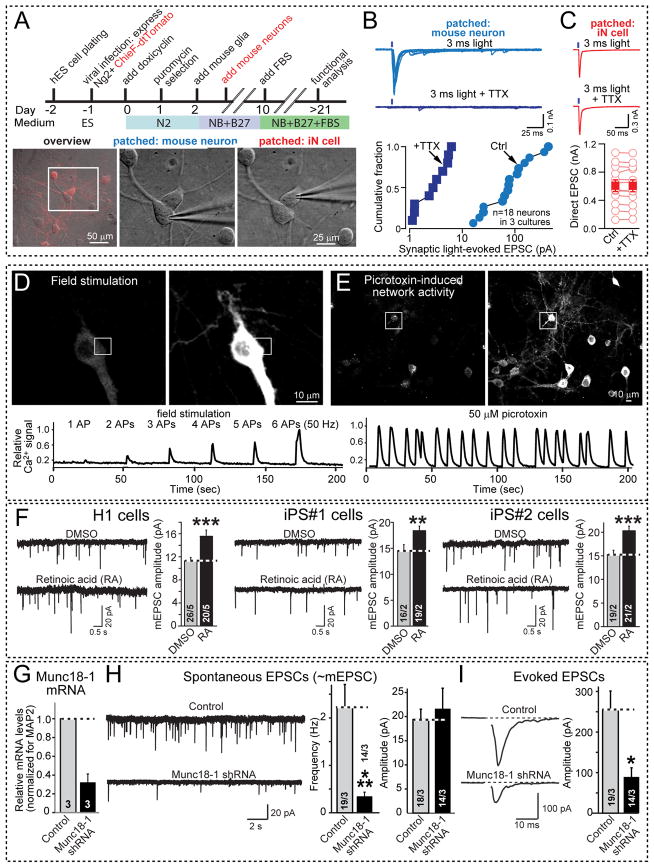
Figure 5. Use of rapidly induced human iN cells for characterizing human synapses and modeling human diseases.
A–C, Optogenetic mapping of functional presynaptic specializations formed by iN cells onto co-cultured cortical mouse neurons. A, Strategy for the generation of channelrhodopsin-expressing iN cells (top), and combined tdTomato fluorescence and DIC images (overview) or DIC images only (images of patched mouse neurons and iN cells) to illustrate the selectively patching human iN cells or mouse neurons (bottom). ES cells were co-infected with lentivirus expressing the channelrhodopsin variant CHIEF as a td-Tomato fusion protein at the time of Ngn2 transduction. B, Synaptic responses triggered by presynaptic optogenetic stimulation of iN cells and monitored in postsynaptic mouse neurons (top, representative traces; bottom, summary graph of the evoked EPSC amplitudes). Responses were triggered by 3 ms blue light pulses without or with 0.5 μM TTX (to block presynaptic action potentials induced by channelrhodopsin). C, Channelrhodopsin-mediated presynaptic depolarizations monitored in human iN cells (top, representative traces; bottom, summary graph of the light-evoked EPSC). As in B, responses were triggered by light pulses in the absence or presence of 0.5 μM TTX, but TTX has no effect because the recorded current is directly induced by channelrhodopsin activation which is not inhibited by TTX.
D & E, Ca2+-imaging of human iN cells. D, Representative images of GCaMP6M-expressing iN cells cultured alone (top) or E, of co-cultured with cortical primary neurons (top) in the absence (left panels) or presence of a Ca2+-signal (right panels). Traces of Ca2+-signals induced by field stimulation (D; indicated action potential (AP) inducing pulses were delivered at 50 Hz) or by network activity triggered by 0.1 mM picrotoxin (E) monitored in these iN cells are shown on the bottom.
F, iN cells derived from H1 ES cells or two different iPS cell lines exhibit retinoic acid (RA) dependent increases in synaptic strength as a model of homeostatic plasticity. iN cells were incubated with 1 μM RA for 45 min, and the amplitude of spontaneous miniature EPSCs (mEPSCs) was recorded in TTX (T-test,). Data shown are means ± SEMs (n = 12 cells/3 independent experiments for DMSO, and 14/3 for RA treatments; statistical significance was assessed by Student’s t-test; **P<0.01; ***P<0.001).
G–I, Effect of a Munc18-1 loss-of-function on synaptic transmission in human iN cells. G, quantification of the Munc18-1 knockdown (KD) efficiency in iN cells. Munc18-1 mRNA levels were measured by quantitative RT-PCR in conrol iN cells and iN cells infected with Munc18-1 KD lentivirus, and normalized to MAP2 as an endogenous control (n=3 independent experiments). H, Representative traces of spontaneous EPSCs monitored in control and Munc18-1 KD iN cells from H1 ES cells (left), and quantifications of the frequency (center) and amplitudes of spontaneous EPSCs (right). Numbers of cells/ independent experiments performed are indicated; I, Representative traces of evoked EPSCs of control and Munc18-1 KD iN cells derived from H1 cells (left), and quantification of spontaneous EPSCs amplitudes (right). Numbers of cells/independent experiments performed are shown in the bars. Data are means ± SEMs; statistical significance was assessed by Student’s t-test (*P<0.05; ***P<0.001).
We then examined the possibility of monitoring activity-dependent Ca2+-transients in entire populations of iN cells using the genetically expressed Ca2+-sensor gCamp6M, which is an advanced version of gCamp5 (Akerboom et al., 2012). We found that Ca2+-transients induced even by single isolated action potentials could be detected in our iN cells (Fig. 5D). The amplitude of the Ca2+-signal correlated well with the number of action potentials elicited. Conversely, when we co-cultured iN cells with mouse neurons, we observed typical network activity in iN cells that was induced by addition of the GABA-receptor blocker picrotoxin (Fig. 5E). These Ca2+-imaging examples demonstrate that it is possible to use iN cells for monitoring network activity of iN cells over larger populations of cells, for example during drug screening projects.
In another experiment, we tested whether synapses in Ngn2 produced iN cells can be modulated. Recent studies revealed that retinoic acid rapidly upregulates postsynaptic AMPA-type glutamate receptors via a direct synaptic action, and that this signaling pathway is activated by activity blockade (Aoto et al., 2008; Wang et al., 2011). We thus explored the ability of exogenously applied retinoic acid to upregulate postsynaptic glutamate receptors (Fig. 5F). Indeed, acute treatment (~45 min) of iN cells with retinoic acid significantly enhanced the amplitude of postsynaptic mEPSCs that are mediated by AMPA-type glutamate receptors without changing the frequency of mEPSCs, demonstrating that the retinoic acid-dependent synaptic signaling pathway is operational in iN cells and thus also applies to humans. The effect was equally observed with iN cells derived from H1 ES cells and with iN cells derived from two different iPS cell lines (Figs. 5F and S5).
Finally, we examined whether iN cells can potentially be used to monitor a disease state. We produced a knockdown (KD) of Munc18-1, resulting in a 75% decrease in Munc18-1 mRNA levels (Fig. 5G). Heterozygous loss-of-function mutations of Munc18-1 (gene symbol STXBP1) have been associated not only with severe infantile epileptic encephalopathies (Ohtahara and West syndromes), but also with moderate to severe cognitive impairment and nonsyndromic epilepsy, suggesting that the functions of human neurons are very sensitive to Munc18-1 levels (Pavone et al., 2012). Strikingly, KD of Munc18-1 in human iN cells, such that Munc18-1 levels are decreased but not abolished, led to a major decrease in the frequency but not the amplitude of spontaneous EPSCs, which based on their size probably represent mEPSCs (Fig. 5H). Moreover, KD of Munc18-1 caused a >50% decrease in evoked EPSCs in iN cells (Fig. 5I). Thus, decreasing the Munc18-1 levels in human iN cells produces a major phenotype consistent with the deleterious phenotype observed in heterozygous loss-of-function mutations observed in Ohtahara syndrome.
Functional integration of human iN cells into mouse brain
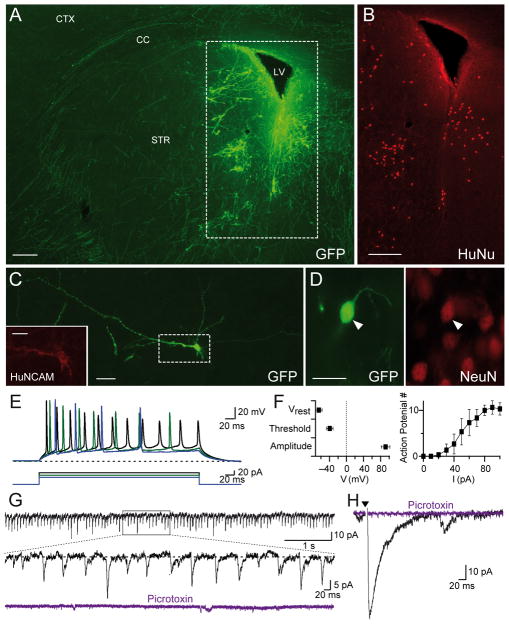
To probe the competence of Ngn2-induced iN cells to form synapses in vivo and not only in vitro, we injected EGFP-labeled iN cells on day 6 into the striatum of newborn mice (postnatal day 2), and analyzed the mouse brains six weeks later.
Immunofluorescence staining revealed that the injected iN cells had dispersed throughout the striatum, and formed extensive dendritic arborizations (Fig. 6A). Numerous EGFP-positive processes were found throughout the striatum and extending through the corpus callosum into the non-transplanted hemisphere. The human iN cells were selectively labeled by antibodies to human nuclei (Fig. 6B), human NCAM (Fig. 6C), and NeuN (Fig. 6D). Electrophysiological recordings from acute slices in current-clamp mode showed that the transplanted iN cells exhibited a resting potential of ~−60 mV, fired trains of action potentials when injected with current, and displayed a near physiological action-potential firing threshold and action potential amplitude (Figs. 6E and 6F). Moreover, recordings in voltage-clamp mode demonstrated that the transplanted neurons received highly active spontaneous inhibitory synaptic inputs as documented by the blockade of the synaptic events by picrotoxin (Fig. 6G). Inhibitory synaptic events would be expected given the preponderance of inhibitory medium spiny neurons in the striatum. Accordingly, inhibitory postsynaptic currents could also be elicited by extracellular stimulation, confirming that the transplanted neurons received abundant inhibitory synaptic inputs from the surrounding neurons in the striatum (Fig. 6H).
Figure 6. Functional integration of transplanted H1-derived iN cells into mouse brain.
A, Representative image of EGFP-expressing iN cells in mouse striatum. Note dispersion of the iN cells into the parenchyma (CC, corpus callosum; CTX, cortex; LV, lateral ventricle; STR, striatum; scale bar: 200 μm).
B, Labeling of the area in A. with antibodies to the marker protein HuNu specific for human neurons (scale bar: 200 μm).
C, Morphology of a single EGFP- and human NCAM- positive iN cells after transplantation (scale bars: 20 μm and 10 μm (inset)).
D, Example of an EGFP-positive human iN cell (left panel) that is co-labeled with NeuN (right panel; scale bar: 20 μm).
E, Example traces showing action-potential generation (upper traces) in response to current pulses (lower traces) applied with 10 pA step sizes. For the recording configuration from transplanted iN cells, see Fig. S6.
F, Average resting membrane potential (Vrest), action-potential threshold (Threshold), and action-potential height (Height) measured in transplanted iN cells (n = 4, left panel). Number of action-potentials (AP #) plotted against current pulse values (n = 4, right panel).
G, Representative traces of spontaneous postsynaptic inhibitory currents (IPSCs) monitored in transplanted iN cells; currents are blocked by picrotoxin.
I, Evoked IPSCs elicited by extracellular stimulation (arrowhead) and blocked by addition of picrotoxin in transplanted iN cells.
DISCUSSION
Previous studies demonstrated the principal feasibility of converting non-neuronal human cells into iN cells, but also described a low conversion efficiency and a diminished capacity of the resulting iN cells for synapse formation (Pang et al., 2011; Ambasudhan et al., 2011; Qiang et al., 2011; Pfisterer et al., 2011a and 2011b; Yoo et al., 2011; Caiazzo et al., 2011; Son et al., 2011). However, realization of the potential of iN cells for studying the pathogenesis of neurological diseases, for developing drug screening systems, and for producing neurons for regenerative medicine requires the capability of producing human iN cells at a large scale and high yield, and necessitates the generation of iN cells that readily form synapses. Moreover, such goals would be facilitated by a high degree of reproducibility of iN cell generation independent of the starting cell line, and by production of a relatively homogeneous population of functional iN cells for experiments.
In the present study, we describe a new, highly effective method that generates a homogeneous population of iN cells by forced expression of a single transcription factor in ES or iPS cells. We demonstrate that the new method results in the reproducible generation of the same type of neuron with quantitatively the same properties independent of the ES or iPS cell line used. The entire procedure generates iN cells in only a few weeks, allowing a rapid turnaround of experiments, and the resulting iN cells exhibit short-term plasticity, are modulated at the level of their synapses, and integrate into neuronal networks when transplanted into the mouse brain. Moreover, the new iN cells can be used for studying synaptic properties including plasticity, for large-scale Ca2+-imaging for example for drug screening purposes, and for disease modeling as exemplified in our Munc18-1 KD experiments. Thus, we believe that the approach described here has the potential to enable mechanistic and translational studies on human neurons that exceed currently existing capabilities, and hope that the simplicity of the approach will allow its wide dissemination. Table 1 shows a comparison of the properties of the method described here with selected other widely used methods to illustrate the advantages and disadvantages of the various approaches that have been described.
It is surprising that forced expression of a single transcription factor can produce iN cells from human ES and iPS cells, suggesting that this transcription factor alone initiates a specific neuronal differentiation process. Previous experiments showed that in mouse ES cells, expression of single transcription factors facilitates neuronal differentiation (e.g., see Hamada et al., 2006; Kondo et al., 2008; Sugimoto et al., 2009), but the synaptic competence of the resulting neuron-like mouse cells was not assessed. Moreover, obtaining human iN cells has historically been more challenging than obtaining mouse iN cells (Vierbuchen et al., 2010; Pang et al., 2011). It has been reported that forced overexpression of Sox2 alone can convert mouse and human fibroblasts into NPCs (Ring et al., 2012), although our own experiments required additional transcription factors besides Sox2 for generation of NPCs (Lujan et al., 2012).
Exogenous Ngn2 probably induces neural differentiation by activating a transcription factor cascade, including endogenous Ngn2. Consistent with its embryonic expression in the developing dorsal forebrain, Ngn2 produced one particular type of neuron that is characterized by specific markers, such as the presence of vGlut2, Cux1, and Brn2, suggesting that it may represent a relatively immature excitatory layer2/3 cortical neuron. It is possible that single use of another transcription factor, or addition of other transcription factors to Ngn2 in our protocol, will result in other types of neurons as shown previously for the BAM factor combination (Pfisterer et al., 2011; Caiuzzi et al., 2011; Son et al., 2011).
We do not at present understand what determines the ability of a neuron to form synapses. Most neuron-like cells can produce action potentials and elaborate postsynaptic specializations – even non-neuronal cells can be made to generate postsynaptic specializations by simple expression of postsynaptic cell-adhesion molecules (Biederer et al., 2002) – but the ability to form presynaptic specializations seems to be specific for a ‘real’ neuron (Yang et al., 2011). Ngn2-induced iN cells form robust synapses among themselves when cultured in the presence of mouse astrocytes (which supply unknown synaptogenic factors) in a manner that was not previously observed for any human iN cells, and occurs much faster than during conventional guided ES/iPS cell differentiation approaches. The synapses that are thus generated exhibit full function and are capable of short-term plasticity and direct modulation by retinoic acid, suggesting that Ngn2 iN cells provide a useful system for studies in which the effects of mutations or pharmacological agents on synaptic transmission in human neurons is investigated.
For the purpose of studying neurological diseases, the relatively homogeneous nature of the iN cells we here generated (which differs from the properties of neurons generated by most other approaches; Table 1) is an advantage as well as a disadvantage. It is advantageous in that phenotypes will not be occluded by heterogeneous properties of different types of neurons, but it might be disadvantageous if phenotypes that are specific to a particular subset of neurons will be examined. Thus, this system will be generally applicable to analysis of mutations in genes with a general neuronal action, for example FMRP, neuroligins, and MECP2. However, even for analysis of diseases that manifest in specific types of neurons, such as Parkinson’s disease, Ngn2 iN cells may be useful because in many neurological diseases the pathological processes are not restricted to the specfiic types of neurons in which the disease becomes manifest. Specifically, even if disease such as Parkinson’s or Huntington’s disease manifest in a dysfunction of dopaminergic or striatal neurons, respectively, this manifestation likely represents a particular vulnerability of specific types of neurons to a general disease process, and not a disease process that is restricted to these types of neurons. Thus, even for such diseases it may not only be feasible, but actually be productive to examine Ngn2-generated iN cells as a homogeneous population of glutamatergic neurons, especially in co-culture with mouse neurons or after transplantation into the mouse brain.
EXPERIMENTAL PROCEDURES
Cell culture
H1 ES cells were obtained from WiCell Research Resources (Wicell, WI); the iPS#1 line was derived from dermal fibroblasts of a Dystrophic epidermolysis bullosa patient carrying homozygous mutations in COL7A1, while the iPS#2 line was derived from dermal fibroblast of a sickle cell anemia patient and genetically corrected by homologous recombination (Sebastiano et al., 2011). This type VII collagen gene is not expressed in neurons, patients with mutations have no brain phenotypes, and our study demonstrates that the mutation of this gene does not affect the molecular and functional properties of Ngn2-mediated iN cells. Both iPS lines were generated by infecting with a floxed polycistronic lentiviral reprogramming vector followed by Cre-mediated loop-out of the reprogramming factors (Sommer et al., 2009). ES and iPS cells were maintained as feeder-free cells in mTeSR™1 medium (Stem Cell Technologies; Xu et al., 2010). Mouse glial cells were cultured from the forebrain of newborn wildtype CD1 mice (Franke et al., 1998). Briefly, newborn mouse forebrain homogenates were digested with papain and EDTA for 30 min, cells were dissociated by harsh trituration to avoid growing of neurons, and plated onto T75 flasks in DMEM supplemented with 10% FBS. Upon reaching confluence, glial cells were trypsinized and replated at lower density a total of three times to remove potential trace amounts of mouse neurons before the glia cell cultures were used for co-culture experiment with iN cells. Mouse cortical neurons were cultured as described (Pang et al., 2011), added to iN cells 4–5 days after infection, and co-cultured for an additional 2 weeks.
Virus generation
Lentiviruses were produced as described (Pang et al., 2010) in HEK293T cells (ATCC, VA) by co-transfection with three helper plasmids (pRSV-REV, pMDLg/pRRE and vesicular stomatitis virus G protein expression vector) (12 μg of lentiviral vector DNA and 6 μg of each of the helper plasmid DNA per 75 cm2 culture area) using calcium phosphate (Chen and Okayama, 1987). Lentiviruses were harvested with the medium 46 hr after transfection, pelleted by centrifugation (49,000 × g for 90 min), resuspended in MEM, aliquoted, and snap-frozen in liquid N2. Only virus preparations with >90% infection efficiency as assessed by EGFP expression or puromycin resistance were used for experiments. For details of lentiviral constructs, see Supplementary Methods.
Generation of iN cells from human ES and iPS cells
ES and iPS cells were treated with Accutase (Innovative Cell Technologies) and plated as dissociated cells in 24 well plates (H1: 1×104 cells/well; iPS cells: 1.5×104 cells/well) on day −2 (Fig. 1B). Cells were plated on matrigel- (BD Biosciences) -coated coverslips in mTeSR™1 containing 2 μM thiazovivin (Bio Vision). On day −1, lentivirus prepared as described above (0.3 μl/well of 24 well plate) was added in fresh mTeSR™1 medium containing polybrene (8 μg/μl, Sigma). On day 0, the culture medium was replaced with N2/DMEM/F12/NEAA (Invitrogen) containing human BDNF (10 μg/l, PeproTech), human NT-3 (10 μg/l, PeproTech) and mouse laminin (0.2 mg/l, Invitrogen). Doxycycline (2 g/l, Clontech) was added on day 0 to induce TetO gene expression, and retained in the medium until the end of the experiment. On day 1, a 24 h puromycin selection (1 mg/l) period was started. On day 2, mouse glia cells were added in Neurobasal medium supplemented with B27/Glutamax (Invitrogen) containing BDNF, and NT3; Ara-C (2 g/l, Sigma) was added to the medium to inhibit astrocyte proliferation. After day 2, 50% of the medium in each well was exchanged every 2 days. FBS (2.5%) was added to the culture medium on day 10 to support astrocyte viability, and iN cells were assayed on day 14 or 21 in most experiments. The efficiency of conversion of ES and iPS cells into iN cells was calculated by two approaches from counts of cell densities in four random fields on each coverslip (Fig. 2D): (1) as the percentage of EGFP-positive lentivirally-transduced cells that also express MAP2 or NeuN; (2) as the percentage of starting cells that become NeuN-positive.
Immunofluorescence experiments were performed essentially as described (Pang et al., 2011). Briefly, cultured iN cells were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature, washed three times with PBS, and incubated in 0.2% Triton X-100 in PBS for 10 min at room temperature. Cells were blocked in PBS containing 10% goat serum for 1 hr at room temperature. Primary antibodies were applied overnight, cells were washed in PBS for three times and blocked with 10% goat serum for 15 min, secondary antibodies were applied for 1 hr. Transplanted iN cells in mouse striatum were analyzed by immunofluorescence staining after mice were transcardially perfused with saline followed by 4% paraformaldehyde. After overnight post-fixation, brains were removed, cryoprotected in 30% sucrose, and cut in 40 μm thick coronal sections. Free-floating sections were washed in PBS, incubated with PBS containing 0.25% Triton X-100 and 5% FBS for 1hr and stained overnight with primary antibodies. Following washes, sections were incubated with secondary antibodies for 2 hrs, washed, mounted on glass slides and coverslipped. For antibody details, see Supplementary Methods.
Gene expression analyses
For quantitative RT–PCR analyses of pooled cultured cells, RNA was isolated using the RNAqueous Kit (Applied Biosystems), treated with DNase (Applied Biosystems), and reverse-transcribed with Superscript III (Invitrogen). mRNA levels were quantified by real-time PCR assay using the Applied Biosystems 7900HT Fast real-time PCR system and RQ analysis software. For quantitative RT–PCR analyses of single cells, cytoplasm from individual cells was aspirated with a patch pipette, and mRNA levels were measured in the cytoplasm using the Fluidigm Biomark dynamic array system as described (Pang et al., 2011). For all quantitative RT-PCR assays, titrations of total human brain RNA were included in each experiment, and only primers that demonstrated a linear amplification with R2 values of >0.98 were included (see Supplemental Materials and Table S1 for details).
RNAi experiments
Oligonucleotides containing the human Munc18-1 shRNA sequence (GGCACAGATGCTGAGGGAGAG) were cloned into the XhoI/XbaI cloning site downstream of the human H1 promoter in the L309-mCherry lentiviral vector (Yang et al 2010). Lentiviruses for control (no shRNA) and the Munc18-1 KD were prepared as described above and used to infect H1-iN cells 5 days after doxycycline addition. iN cells were analyzed at 3 weeks after Ngn2 induction by determining the KD efficacy using RT-PCR and by electrophysiology.
Ca2+-imaging experiments were performed with iN cells that were infected with a lentivirus expressing GCaMP6M on day 3 after induction, co-cultured with mouse cortical neurons at day 6 after induction, and analyzed at day 21. See Supplemental Materials for details.
Channelrhodopsin experiments
H1 cells were co-infected with viruses expressing Ngn2 and ChiEF2-tdTomato on day 1, and mouse cortical neurons were added for co-culture on day 3. iN cells were analyzed at day 21 as described in detail in the Supplemental Methods.
Transplantation
H1-derived iN cells were dissociated using Enzyme-Free Cell Dissociation Buffer (Gibco) 7 days after infection (i.e., on day 6) without co-culture of astrocytes, and 105 cells were unilaterally injected under hypothermia-induced anesthesia into the striatum of postnatal day 2 NOD-SCID; IL2Rγ knock-out mice. Mice were processed for immunocytochemistry or slice electrophysiology six weeks after transplantation.
Electrophysiology of cultured iN cells was performed essentially as described (Maximov and Südhof, 2005; Pang et al., 2011). Stimulus artifacts for evoked synaptic responses were removed for graphic representation. For electrophysiological recordings of transplanted cells, we prepared acute coronal slices (140 μm) of striatum from mice 6 weeks after transplantation using a vibratome (Leica, VT1200 S). GFP-positive hES-iN cells in slices were visualized using an X-cite 120Q fluorescence lamp (Lumen Dynamics) and an Olympus BX51WI microscope equipped with a Rolera-XR camera (Qimaging). Whole-cell patches were established at room temperature using MPC-200 manipulators (Sutter Instrument) and Multiclamp 700B amplifier (Molecular Devices) controlled by Clampex 10 Data Acquisition Software (Molecular Devices). Cells were recorded in current-clamp mode for intrinsic firing properties and switched to voltage-clamp mode (−70 mV) for synaptic measurements. Evoked responses were generated using a concentric bipolar electrode (FHC) connected to Isolated Pulse Stimulator 2100 (A–M systems). Picrotoxin (50 μM, Tocris) was used to block inhibitory synaptic responses. For more details, see Supplementary Methods.
Data presentation and statistics
All data shown are means ± SEMs; all statistical analyses were performed using Student’s t-test comparing the test sample to the control sample examined in the same experiments.
Supplementary Material
Research highlights.
Neurogenin-2 overexpression rapidly transforms ES and iPS cells into neurons
Neurogenin-2 induced human neurons form spontaneous excitatory synaptic networks
Synapses formed by neurogenin-2 induced human neurons exhibit short-term plasticity
After transplantation, neurogenin-2 induced human neurons are synaptically integrated
Acknowledgments
We would like to thank Drs. V. Sebastiano, B. Haddad, and B. Berninger for advice and reagents. This study was supported by grants from the Ellison Medical Foundation (AG-NS-0709-10 M.W.), the NIH (P50 AG010770-18A1 and R01 MH092931 to M.W. and T.C.S., and P50 MH086403 to L.C. and T.C.S.), the California Institute for Regenerative Medicine (RT2-02061 to M.W. and T.C.S.), and the Department of Defense (PR100175P1 to M.W.). M.W. is a New York Stem Cell Foundation-Robertson Investigator, N.Y. N.Y. was supported by a fellowship from the Berry Foundation, H.A. by a fellowship from the Swedish Research Council and the Swedish Society for Medical Research, and C.P. by a fellowship from the Deutsche Forschungsgemeinschaft.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Daley GQ, Hochedlinger K, Passegué E, Rossant J, Yamanaka S. Stem cells assessed. Nat Rev Mol Cell Biol. 2012;13:471–476. doi: 10.1038/nrm3371. [DOI] [PubMed] [Google Scholar]
- Brennand KJ, Simone A, Tran N, Gage FH. Modeling psychiatric disorders at the cellular and network levels. Mol Psychiatry. 2012 doi: 10.1038/mp.2012.20. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27:275–80. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi SH, Studer L. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol. 2012;30:715–20. doi: 10.1038/nbt.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MS, Lee YE, Kim JY, et al. Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:3392–3397. doi: 10.1073/pnas.0712359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008;22:152–65. doi: 10.1101/gad.1616208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espuny-Camacho I, Michelsen KA, Gall D, Linaro D, Hasche A, Bonnefont J, Bali C, Orduz D, Bilheu A, Herpoel A, Lambert N, Gaspard N, Péron S, Schiffmann SN, Giugliano M, Gaillard A, Vanderhaeghen P. Pyramidal neurons derived from human pluripotent stem cells integrate efficiently into mouse brain circuits in vivo. Neuron. 2013;77:440–56. doi: 10.1016/j.neuron.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Fasano CA, Chambers SM, Lee G, Tomishima MJ, Studer L. Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell. 2010;6:336–347. doi: 10.1016/j.stem.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Franke B, Figiel M, Engele J. CNS glia are targets for GDNF and neurturin. Histochem Cell Biol. 1998;110:595–601. doi: 10.1007/s004180050322. [DOI] [PubMed] [Google Scholar]
- Hamada M, Yoshikawa H, Ueda Y, Kurokawa MS, Watanabe K, Sakakibara M, Tadokoro M, Akashi K, Aoki H, Suzuki N. Introduction of the MASH1 gene into mouse embryonic stem cells leads to differentiation of motoneuron precursors lacking Nogo receptor expression that can be applicable for transplantation to spinal cord injury. Neurobiol Dis. 2006;22:509–522. doi: 10.1016/j.nbd.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Han SS, Williams LA, Eggan KC. Constructing and deconstructing stem cell models of neurological disease. Neuron. 2011;70:626–644. doi: 10.1016/j.neuron.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BY, Weick JP, Yu J, Ma LX, Zhang XQ, Thomson JA, Zhang SC. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc Natl Acad Sci U S A. 2010;107:4335–40. doi: 10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, Goldstein LS. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–20. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Weick JP, Pearce RA, Zhang SC. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J Neurosci. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P, Opitz T, Steinbeck JA, Ladewig J, Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc Natl Acad Sci U S A. 2009;106:3225–30. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Sheets PL, Zopf DA, Aloor HL, Cummins TR, Chan RJ, Hashino E. Tlx3 exerts context-dependent transcriptional regulation and promotes neuronal differentiation from embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:5780–5785. doi: 10.1073/pnas.0708704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim JW, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, Yang L, Beal MF, Surmeier DJ, Kordower JH, Tabar V, Studer L. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Lujan E, Chanda S, Ahlenius H, Südhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci U S A. 2012;109:2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Hu B, Liu Y, Vermilyea SC, Liu H, Gao L, Sun Y, Zhang X, Zhang SC. Human embryonic stem cell-derived GABA neurons correct locomotion deficits in quinolinic acid-lesioned mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Gage FH. Modeling brain disease in a dish: really? Cell Stem Cell. 2012;10:642–645. doi: 10.1016/j.stem.2012.05.008. [DOI] [PubMed] [Google Scholar]
- Maroof AM, Keros S, Tyson JA, Ying SW, Ganat YM, Merkle FT, Liu B, Goulburn A, Stanley EG, Elefanty AG, Widmer HR, Eggan K, Goldstein PA, Anderson SA, Studer L. Directed differentiation and functional maturation of cortical interneurons from human embryonic stem cells. Cell Stem Cell 2013. 2013;12:559–72. doi: 10.1016/j.stem.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Brüstle O, Muotri A, Studer L, Wernig M, Christian KM. Cellular reprogramming: recent advances in modeling neurological diseases. J Neurosci. 2011;31:16070–16075. doi: 10.1523/JNEUROSCI.4218-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas CR, Chen J, Tang Y, Southwell DG, Chalmers N, Vogt D, Arnold CM, Chen YJ, Stanley EG, Elefanty AG, Sasai Y, Alvarez-Buylla A, Rubenstein JL, Kriegstein AR. Functional Maturation of hPSC-Derived Forebrain Interneurons Requires an Extended Timeline and Mimics Human Neural Development. Cell Stem Cell. 2013;12:573–86. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci. 2011;366:2198–2207. doi: 10.1098/rstb.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafune K, Caron L, Borowiak M, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nature Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- Pang ZP, Cao P, Xu W, Südhof TC. Calmodulin controls synaptic strength via presynaptic activation of calmodulin kinase II. J Neurosci. 2010;30:4132–4142. doi: 10.1523/JNEUROSCI.3129-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Südhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavone P, Spalice A, Polizzi A, Parisi P, Ruggieri M. Ohtahara syndrome with emphasis on recent genetic discovery. Brain Dev. 2012 Jun;34(6):459–468. doi: 10.1016/j.braindev.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Pfisterer U, Wood J, Nihlberg K, Hallgren O, Bjermer L, Westergren-Thorsson G, Lindvall O, Parmar M. Efficient induction of functional neurons from adult human fibroblasts. Cell Cycle. 2011a;10:3311–3316. doi: 10.4161/cc.10.19.17584. [DOI] [PubMed] [Google Scholar]
- Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Björklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011b;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L, Fujita R, Yamashita T, Angulo S, Rhinn H, Rhee D, Doege C, Chau L, Aubry L, Vanti WB, Moreno H, Abeliovich A. Directed conversion of Alzheimer’s disease patient skin fibroblasts into functional neurons. Cell. 2011;146:359–371. doi: 10.1016/j.cell.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HP, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldner F, Jaenisch R. Medicine. iPSC disease modeling. Science. 2012;338:1155–1156. doi: 10.1126/science.1227682. [DOI] [PubMed] [Google Scholar]
- Sommer CA, Sommer AG, Longmire TA, Christodoulou C, Thomas DD, Gostissa M, Alt FW, Murphy GJ, Kotton DN, Mostoslavsky G. Excision of reprogramming transgenes improves the differentiation potential of iPS cells generated with a single excisable vector. Stem Cells. 2010;28:64–74. doi: 10.1002/stem.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Furuno T, Nakanishi M. Effect of NeuroD2 expression on neuronal differentiation in mouse embryonic stem cells. Cell Biol Int. 2009;33:174–179. doi: 10.1016/j.cellbi.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc Natl Acad Sci USA. 2000;97:7963–7968. doi: 10.1073/pnas.130192197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Xu J, Pang ZP, Ge W, Kim KJ, Blanchi B, Chen C, Südhof TC, Sun YE. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci U S A. 2007;104:13821–138266. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhu XW, Hahm HS, Wei WG, Hao EG, Hayek G, Ding S. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci USA. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Morishita W, Buckmaster PS, Pang ZP, Malenka RC, Südhof TC. Distinct Neuronal Coding Schemes in Memory Revealed by Selective Erasure of Fast Synchronous Synaptic Transmission. Neuron. 2012;73:990–1001. doi: 10.1016/j.neuron.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Ng YH, Pang ZP, Südhof TC, Wernig M. Induced Neuronal (iN) Cells: How to Make and Define a Neuron. Cell Stem Cell. 2011;9:517–525. doi: 10.1016/j.stem.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SC, Wernig M, Duncan ID, Brüstle O, Thomson JA. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat Biotechnol. 2001;19:1129–33. doi: 10.1038/nbt1201-1129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.