Abstract
The aim of this work was to identify Lactobacillus johnsonii NCC533 (La1) surface molecules mediating attachment to intestinal epithelial cells and mucins. Incubation of Caco-2 intestinal epithelial cells with an L. johnsonii La1 cell wall extract led to the recognition of elongation factor Tu (EF-Tu) as a novel La1 adhesin-like factor. The presence of EF-Tu at the surface of La1 was confirmed by analysis of purified outer surface protein extract by immunoblotting experiments, by electron microscopy, and by enzyme-linked immunosorbent assays of live bacteria. Furthermore, tandem mass spectrometry analysis proved that EF-TU was expressed at the La1 surface as an intact molecule. Using recombinant La1 EF-Tu protein, we were able to determine that its binding to intestinal cells and to mucins is pH dependent. Competition experiments suggested that EF-Tu has an important role in La1 mucin binding capacity. In addition, immunomodulation studies performed on HT29 cells showed that EF-Tu recombinant protein can induce a proinflammatory response in the presence of soluble CD14. Our in vitro results indicate that EF-Tu, through its binding to the intestinal mucosa, might participate in gut homeostasis.
Probiotic bacteria, mainly lactic acid bacteria and bifidobacteria, have been shown to have beneficial effects on the immune defenses and to alleviate or prevent diverse intestinal disorders (3, 4, 16, 25, 27, 30, 39, 42, 47). Several in vitro studies have shown that one of them, Lactobacillus johnsonii NCC533 (La1), is able to bind to epithelial cell lines (5, 8, 9, 21) and can induce the secretion of different cytokines in coculture systems (9, 24). Furthermore, human and animal studies have demonstrated that La1 has immune adjuvant effects (40, 44) and can also act as a modulator of nonspecific immune responses (6, 14, 29, 48, 53, 62).
The mechanisms underlying these beneficial effects are not completely understood, but it is believed that the maximum probiotic effects can be achieved if the organisms adhere to mucus and/or intestinal epithelial cells (31, 62). It has recently been shown that lipoteichoic acid (LTA), a molecule associated with the surface of La1 bacteria, participates in their adhesion to intestinal cells (21) and has an immunomodulatory effect on gut homeostasis (64). However, competition experiments indicated that LTA is not the only surface molecule mediating La1 binding to epithelial cells (21). Indeed, it had already been suggested by Bernet et al. (5) that proteinaceous compounds are involved in the attachment of bacteria to these cells. This observation is in accordance with recent studies showing that surface proteins of other lactobacilli participate in adhesion to epithelial cell lines, gastrointestinal mucins, or extracellular matrix proteins (1, 26, 58, 60).
In this work, therefore, we have investigated the ability of La1 surface proteins to attach to intestinal epithelial cells and mucoproteins. We have identified the elongation factor Tu as a novel surface protein possessing the characteristics of an adhesion factor. Using the recombinant His-tagged La1 EF-Tu protein purified from Escherichia coli, we have demonstrated an adhesin-like role of this molecule and shown that it is able to induce a proinflammatory response.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. johnsonii NCC533 (La1) and NCC 1657, Lactobacillus acidophilus NCC 90 and NCC 12, Lactobacillus paracasei NCC 2461, Lactobacillus gasseri NCC 907, Lactobacillus reuteri NCC 2458, Lactobacillus helveticus NCC 1643, and Bifidobacterium sp. strains NCC 362 and NCC 490 were obtained from the Nestlé Culture Collection (Lausanne, Switzerland). L. johnsonii ATCC 33200 was obtained from the American Type Culture Collection (Manassas, Va.). Lactobacilli were grown overnight at 37°C in DE Man-Rogosa-Sharpe broth (Difco), and bifidobacteria were grown overnight in brain heart infusion- 0.5% cysteine (Difco) under anaerobic conditions.
Cell lines and culture conditions.
Caco-2 and HT29 human intestinal cells (American Type Culture Collection) were used between passages 40 and 90 and cultured as previously described (21, 64). HT29-MTX (methotrexate-treated) cells were grown according to the method of Lesuffleur et al. (38).
Isolation of bacterial outer surface proteins.
Bacterial pellets were incubated in 5 M guanidine-HCl (pH 7.0) (10 mg [wet weight]/ml) or 0.5 M lithium chloride as described previously (41). Contamination of extracts with other bacterial fractions was assessed by sodium dodecyl sulfate-4 to 20% polyacrylamide gel electrophoresis (SDS-4 to 20% PAGE) (Bio-Rad Laboratories, Hercules, Calif.) under nonreducing conditions and by Western blotting, using a rabbit antibody against enzyme I from the phosphotransferase system (50).
Preparation of bacterial cell wall extract.
Cell wall proteins were prepared using a slightly modified protocol (7, 12, 35). Bacterial pellets (500-ml cultures) were washed twice with cold phosphate-buffered saline (PBS), suspended in a solution containing 30 mM ammonium carbonate- 1 mM phenylmethylsulfonyl fluoride- 5 mM EDTA- 10% sucrose at pH 8.0, and incubated for 30 to 60 min at 37°C in the presence of 2,000 U of mutanolysin (Sigma Chemical Co., St. Louis, Mo.) and 20 mg of lysozyme (109,000 U/mg; Sigma). Protoplast formation was followed by monitoring the decrease in optical density at 590 nm (optical density decreases until protoplast formation is finished). The suspensions were then centrifuged at 10,000 × g for 10 min at 4°C, the supernatants were dialyzed against 50 mM ammonium bicarbonate (pH 7.0), and aliquots were stored at −20°C.
Preparation of bacterial cytoplasm.
The bacterial pellets (250-ml cultures) were washed twice in PBS and then suspended in 30 mM ammonium hydrogen carbonate, pH 8.0, containing 1 mM phenylmethylsulfonyl fluoride. After two passages through a cell disrupter (One Shot cell disrupter version V4-53-5/97; Constant System Ltd., Warwick, England) at a pressure of 40,000 lb/in2, the cells were centrifuged at 15,000 × g for 20 min at 4°C, and the resulting supernatants were further centrifuged at 45,000 rpm for 1 h at 4°C in a Ti60 rotor. The supernatant containing the cytoplasmic extract was passed through a 0.22-μm-pore-size Millipore filter and stored at −20°C.
Mucin purification from HT29-MTX cell line.
The cell culture medium of 3-week-old HT29-MTX cells was replaced by serum-free medium for 16 to 24 h. Purification of the secreted mucins was performed by size exclusion chromatography using a Sepharose Cl-4B column (Amersham) as described previously (18, 19). The total protein concentration was determined using the bicinchoninic acid kit (Pierce, Perbio Science, Lausanne, Switzerland) or the Bio-Rad protein assay according to the manufacturer's instructions. Total sugars were assessed as described previously (15). Mucins were eluted in the void volume, thus indicating a molecular mass of >2 × 106 kDa. Mucin purity was controlled by SDS-4 to 20% PAGE. Mucin preparations were reduced and alkylated before gel loading. Total proteins were revealed by silver staining (Invitrogen Life Technologies, Rockville, Md.), and sugars were revealed by periodic-acid- Schiff staining (Merck AG, Geneva, Switzerland) as previously described (18). Gel analysis showed an apparent molecular mass of >300 kDa. The preparation contained other proteins entrapped in the mucins which could not be eliminated by Flavourzyme treatment (18) without affecting the mucins themselves. Due to their high sugar content (60 to 80%), mucins were preferentially visualized by periodic-acid-Schiff staining. The yield was 6 to 7 μg of pure mucin/T75 flask. For comparison with the concentration of mucins isolated from the normal human colon, the concentration of HT29 MTX mucins was expressed as micrograms of total sugar per milliliter.
Isolation of normal human gut mucins.
The mucus layer was scraped from a resected piece of a normal human colon, placed in cold 0.05 M Tris-HCl, pH 7.5, and homogenized with a Polytron P T 3100 (Kinematica AG, Littau, Switzerland) at 6,500 rpm for 30 s at 4°C. The suspension was incubated in an ice bath for 1 h with 1% Triton X-100 (Sigma) with agitation and with a cocktail of inhibitors (Complete protease inhibitor cocktail; Roche Diagnostics, Mannheim, Germany). After centrifugation, mucin preparations were purified by size exclusion chromatography, and their purities were assessed as described above. Identification of the mucins was performed by Western blot analysis using commercial anti-human MUC2 and MUC3 antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.) according to the manufacturer's instructions. This mucin preparation was almost devoid of other contaminating proteins and was shown to represent mostly MUC2.
Preparation of anti-La1 surface antibodies.
Rabbit serum against live La1 (109 bacteria/injection) was prepared by EUROGENTEC (Herstal, Belgium). The purified immunoglobulin G (IgG) fraction was incubated for 2 h at 4°C with a washed suspension of an overnight La1 culture in PBS. After the washing, bound antibodies were eluted at pH 2.3, immediately neutralized, and purified on a protein A-Sepharose column (Amersham Biosciences, Otelfingen, Switzerland) according to the manufacturer's instructions and controlled by enzyme-linked immunosorbent assay (ELISA) on whole live La1 or by Western blot analysis of La1 cell wall extracts run on SDS-4 to 20% acrylamide gels.
Detection of La1 surface adhesins.
T75 flasks containing differentiated Caco-2 cells or undifferentiated HT29 cell monolayers were washed two times with either PBS (pH 7.2) or 0.05 M sodium acetate buffer (pH 5.0) containing 0.1 M NaCl and then incubated for 1 h at room temperature with 12 ml of PBS or acetate buffer containing 100 μg of La1 cell wall extract/ml. Negative controls were performed by incubating cells in the respective buffers alone. After three washes with the respective buffers, cell monolayers were harvested with a rubber policeman and transferred into 50-ml Falcon tubes, washed once more, and then transferred into-1.5 ml Eppendorf tubes. To elute La1 adhesins, the pellets were gently suspended in 1 ml of 0.1 M glycine-HCl, pH 3.0, and centrifuged for 5 min at 1,500 × g in a Sorvall Biofuge 13. Supernatants were neutralized with 0.2 ml of 1 M Tris-HCl buffer, pH 8.0. After dialysis against 33% PBS overnight at 4°C, the extracts were concentrated threefold. Samples were run on an SDS-4 to 20% polyacrylamide gel under nonreducing conditions and further analyzed by Western blotting on Immunoblot polyvinylidene difluoride (PVDF) membranes (Bio-Rad) using rabbit anti-La1 surface and anti-ovalbumin antibodies (2 μg/ml), followed by goat anti-rabbit Ig conjugated to alkaline phosphatase (Sigma) at a dilution of 1/2,000. La1 proteins that had been bound to the cells were revealed by the 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate (Zymed Laboratories, San Francisco, Calif.) according to the manufacturer's instructions. The same samples were run on a separate gel adjacent to an La1 cell wall extract, blotted on a Sequi PVDF membrane (Bio-Rad), and stained with Coomassie blue as described by the manufacturer. For subsequent control experiments, the same eluates were migrated together with 0.03 μg of the recombinant EF-Tu protein and 0.1 μg of cell wall extract and revealed with rabbit anti-La1 surface antibody (2 μg/ml), mouse anti-EF-Tu antibodies (1/5,000), or a monoclonal antibody hybridoma supernatant, 9C2c2, made upon immunization with a guanidine-HCl extract of La1 bacteria (21). Anti-mouse Ig coupled to alkaline phosphatase (Santa Cruz) was used as a second antibody at a dilution of 1/2,000.
Protein sequencing.
The band corresponding to the La1 50-kDa (P50) protein band detected with the anti-La1 surface antibody was excised from the PVDF membrane stained with Coomassie blue, and the sequence of the first 12 amino acids was determined (Analytical Research and Services, Departement für Chemie und Biochemie, Universität Bern, Switzerland). The major P50 peptide sequence was compared to the total predicted translated proteins of NCC533 La1 using the program BlastP (2).
Expression cloning.
The EF-Tu gene was amplified from NCC533 chromosomal DNA with the primers 5′ATATATTCATGACAGAAAAAGAACATTACG3′ and 5′ATATATGGATCCTCAAGGATTTCAGTAACTTGACC3′, introducing BspHI and BamHI sites, respectively (underlined). Amplification was performed using the Pwo polymerase (Roche Molecular Biochemicals) on an Applied Biosystems 9700 under the following conditions: incubation for 5 min at 95°C followed by 30 cycles of 30 s at 95°C, 30 s at 50°C, and 3 min at 68°C, and finally incubation for 7 min at 68°C before holding at 4°C. The amplicon was digested with BspHI and BamHI and ligated into the expression plasmid pET24d (Novagen Inc., Madison, Wis.) digested with NcoI and BamHI. The cloning fuses the carboxyl terminus of the EF-Tu protein to the linker and His tag of pET24d, giving plasmid pDP649. Plasmid pDP649 was transformed into the expression host BL21(DE3) RIL (Stratagene Inc., La Jolla, Calif.), protein expression was induced with isopropyl-β-d-thiogalactopyranoside, and the recombinant protein was purified by Ni2+-nitrilotriacetic acid affinity chromatography (Qiagen AG, Basel, Switzerland).
Preparation of polyclonal antibodies against recombinant EF-Tu and normal human gut mucins.
Immunization of BALB/c mice with EF-Tu and normal gut mucins was done as described previously (22). Serum samples from mice were tested by ELISA or Western blot analysis for reactivity against the antigen. Normal mouse serum was obtained from nonimmunized mice from the same lot.
ELISA with bacteria.
Nunc Maxisorb polyvinyl wells (Fisher Scientific AG, Wohlen, Switzerland) were coated overnight at 4°C with 100 μl of a concentration of 0.3 × 108 La1/ml in PBS, pH 7.2. To control for bacterial lysis, supernatants were carefully removed from the wells and tested for the presence of DNA (Ribogreen quantitation kit; Molecular Probes, Eugene, Oreg.) and of the intracellular marker aldolase (35). After quenching of nonspecific binding with 10 mg of gelatin/ml in PBS (gelatin Goldruck; Schweizerhall Chemie AG, Basel, Switzerland), the bacteria were incubated for 2 h at 4°C with increasing dilutions of anti-EF-Tu mouse antibodies, anti-La1 LTA monoclonal antibody (21), anti-La1 surface rabbit antibodies, or normal mouse serum and washed three times with PBS-0.05% Tween 20, followed by an incubation with rabbit anti-mouse or goat anti-rabbit Ig antibodies coupled to horseradish peroxidase (Zymed) at a dilution of 1/2,000 in PBS-0.05% Tween 20. After the washing, the enzymatic activity was revealed with the ImmunoPure TMB substrate kit (Pierce) and measured at 450 nm with a Dynatech MR 5000 microtiter plate reader.
Assays of EF-Tu binding to intestinal cell lines and mucins.
HT29 cells (10,000/well) or Caco-2 cells (20,000/well) were cultivated in 96-well NUNCLON surface flat-bottom cell culture plates (Nunc) in their respective media for 1 week. They were then washed twice with PBS or 0.05 M sodium acetate- 0.1 M NaCl buffer, pH 5.0. Dilutions (100 μl) of La1 cell wall extract (100 μg/ml) or recombinant EF-Tu (100 pmol/ml) in the same buffers were added, and the cells were incubated for 1 h at room temperature. After three washings with their respective buffers, the cells were fixed for 10 min in the dark with a solution of 2.5% paraformaldehyde in PBS and washed twice with PBS. The cells were then incubated with mouse anti-EF-Tu antibodies (1/2,000) or a mouse monoclonal antibody against the His tag epitope (0.1 μg/ml) (Qiagen) for 1 h at the same temperature, followed by 100 μl of a 1/2,000 dilution of rabbit anti-mouse Ig conjugated to horseradish peroxidase (Zymed). The enzymatic activity bound to the wells was determined as described above.
Mucin-binding studies used Nunc Maxisorb 96 polyvinyl well plates coated with 50 μl of a solution of HT29-MTX mucins (0.5 μg of total sugar/ml) or of a normal human gut mucin preparation in 0.05 M carbonate-bicarbonate buffer (pH 9.6) overnight at 4°C. After saturation with 200 μl of a solution of 10 mg of gelatin/ml in PBS, the plates were treated as described above.
Binding of normal human gut mucins to L. johnsonii La1.
Maxisorb polyvinyl wells were coated overnight with 100 μl of a solution of 0.3 × 108 La1/ml in PBS, pH 7.2. After saturation with gelatin, they were washed two times with acetate buffer (pH 5.0) and incubated with serial dilutions of normal human gut mucins in the same buffer containing 2 mg of gelatin/ml and 0.05% Tween 20 for 2 h at 4°C. After three washings with the same buffer, the bacteria were fixed with 2.5% paraformaldehyde and incubated with normal mouse serum or mouse anti-mucin antibodies at a dilution of 1/2,000 and then with rabbit anti-mouse Ig conjugated to horseradish peroxidase for 1 h at the same temperature described above. Enzymatic activity was determined as described above. For competition studies, human gut mucins (1.5 μg/ml) were added to the same bacteria in the presence of serial dilutions of EF-Tu recombinant protein starting at 5 μg/ml and incubated under the same conditions described above.
Electron microscopy.
Stationary-phase bacteria were treated as previously described (21), and ultrathin sections were incubated with mouse anti-EF-Tu recombinant protein antibodies and/or normal mouse serum at a dilution of 1/50 for 3 h at room temperature, followed by a goat anti-mouse Ig-gold (10-nm diameter) conjugate (Chemie-Brunswig AG) for 3 h at 4°C. Sections were finally examined in a Philips CM 12 electron microscope at an acceleration voltage of 60 kV.
Tandem mass spectrometry.
A guanidine-HCl extract of L. johnsonii La1 (2 μg/slot) was run on an SDS-12% acrylamide gel under nonreducing conditions. The gel was stained with Coomassie brilliant blue G-250 (57). The band corresponding to the protein recognized by anti-EF-Tu recombinant antibodies was excised from the gel, in-gel digested with trypsin, and analyzed by nano-electrospray ionization-mass spectrometry as previously described (45).
Stimulation assay with HT29 cells.
The stimulation assay with HT29 cells was done as described previously (64). Briefly, HT29 cells were plated at 104/well in 96-well flat-bottom plates. After incubation for 5 days, the HT29 cells were washed twice with serum-free medium before the addition of the La1 EF-Tu recombinant protein, native or heated at 100°C for 20 min in the presence or absence of 2% human milk (HM) in Dulbecco's modified Eagle's medium. In some wells, MY4, murine anti-CD14 monoclonal antibody (Coulter Instrumentation Laboratory, Schlieren, Switzerland), and/or control mouse IgG2b immunoglobulins (Sigma) were also added at a final concentration of 20 μg/ml. Native or heated lipopolysaccharide (LPS) from E. coli O55:B5 (Sigma) was used as a positive control at different dilutions. The cell viability was examined using a cytotoxicity kit (Roche Diagnostic, Rotkreuz, Switzerland). Detection of LPS contamination was done using the Limulus amebocyte lysate endochrome test (Charles River Endosafe, Charleston, S.C.). Detection of HT29 cell interleukin-8 (IL-8) release was done essentially as described previously (64).
RESULTS
Detection of La1 cell wall proteins binding to differentiated Caco-2 cells and undifferentiated HT29 cells.
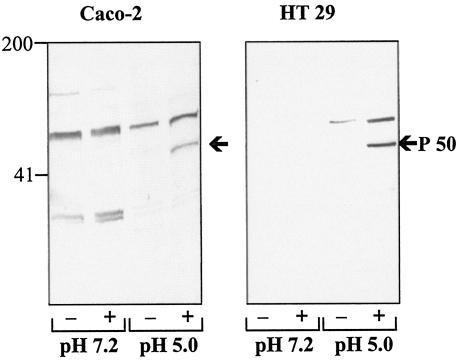
To identify La1 proteins mediating adhesion to epithelial cells, La1 cell wall extracts were incubated with two different cell lines at different stages of differentiation. Bound La1 proteins eluted from differentiated Caco-2 cells were analyzed by Western blotting and revealed with anti-La1 surface antibodies. Eluates of Caco-2 cells incubated with La1 cell wall extracts at pH 5.0, but not at pH 7.2, showed one specific band with an apparent molecular mass of 50 kDa (Fig. 1). The other bands detected were due to Caco-2 cell proteins that cross-react with the anti-La1 surface antibodies, as they were observed even in the absence of La1 cell wall extracts. Furthermore, these bands were absent from the blots incubated with the control anti-ovalbumin antibodies (data not shown).
FIG. 1.
Detection of La1 cell wall proteins binding to differentiated Caco-2 cells and HT29 cells. Caco-2 or HT29 cells were incubated with buffer alone (−) or with an La1 cell wall extract (+) at pH 7.2 or 5.0. Bound proteins were eluted as described in the text, submitted to SDS-PAGE, and analyzed by Western blotting using anti-La1 surface antibodies. The results of one representative of three independent experiments are shown for each cell line.
A protein band exhibiting a similar apparent molecular mass was also observed when undifferentiated HT29 cells were used (Fig. 1), suggesting that the same La1 protein could bind to two different intestinal cell lines.
Identification of the La1 protein bound to Caco-2 and HT29 cells.
The band with an apparent molecular mass of 50 kDa (P50) was excised from the membrane, and the sequence of the first 12 amino acids was determined. The result gave Ala-Glu-Lys-Gln-Val-Tyr-Glu-Arg-Thr-Lys-Ala-Leu as the major sequence. This sequence was compared to the total predicted translated proteins of L. johnsonii NCC533 using the bioinformatics program BlastP, which returned a strong match to the gene LJ 1009 (accession number AE017198) (56). The translated protein of LJ 1009 was compared to the current protein databases using the program BlastP, which returned a significant (84%) identity to EF-Tu of Lactobacillus plantarum WCSF1 (33), involved in protein translation (Fig. 2). For further studies, the recombinant His-tagged EF-Tu protein was prepared in E. coli. The purified recombinant protein, analyzed by SDS-PAGE, migrated with an apparent molecular mass of 50 kDa (data not shown).
FIG. 2.
Identification of the protein bound to Caco-2 and HT29 cells. AA seq., major sequence of the first 12 amino acids of the protein with an apparent molecular mass of 50 kDa. The L. johnsonii and L. plantarum EF-Tu proteins show 84% sequence identity. Asterisks and dots represent identical and similar amino acids, respectively. Amino acids that are identical in the AA sequence and NCC533 EF-Tu sequence are underlined.
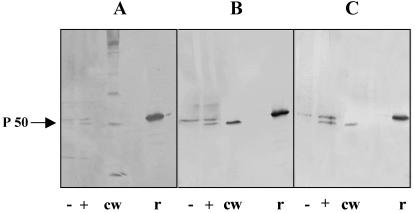
As the La1 EF-Tu sequence showed 57% similarity to the Homo sapiens EF-Tu mitochondrial precursor (66), further experiments were performed to verify that the identified EF-Tu was of La1 origin and to exclude the possibility of a false-positive result due to cross-reaction with a protein of human origin. The eluates from Caco-2 cells preincubated with La1 cell wall extracts at pH 5.0 were analyzed by Western blotting using three different antibodies (Fig. 3). Two protein bands were detected with either anti-La1 surface antibodies (Fig. 3A), anti-EF-Tu antibodies (Fig. 3B), or the monoclonal antibody 9C2c2 produced upon immunization with La1 guanidine-HCl extract (Fig. 3C). The major band, with a lower apparent molecular weight, was also detected in La1 cell wall extracts but not in the eluates of Caco-2 cells obtained in the absence of La1 cell wall extract, indicating that this protein is of La1 origin. In contrast, the minor band, with a higher apparent molecular weight, was also detected in the eluates of Caco-2 cells obtained in the absence of La1 cell wall extract but not in La1 cell wall extracts, suggesting that some human mitochondrial EF-Tu (MW = 49,541) was released during elution at pH 3 and reacted with our antibodies. In addition, as observed in Fig. 1, this minor band was not detected in all preparations, indicating that the amounts of mitochrondrial EF-Tu released from Caco-2 cells during acid treatment were variable. All three antibodies also recognized the La1 recombinant EF-Tu protein. This recombinant protein exhibited a higher apparent molecular weight than the native protein due to the six histidine residues and a linker added to the molecule for production in E. coli. Indeed, the predicted molecular weights for the recombinant and the native protein are 46,629.5 and 43,664.0, respectively. Similar results were obtained with HT29 cell eluates (data not shown).
FIG. 3.
Verification of the La1 origin of the identified EF-Tu molecule. Western blots of eluates from Caco-2 cells incubated with La1 cell wall extract (+) or buffer alone (−) were revealed with anti-La1 surface antibodies (A), mouse anti-recombinant EF-Tu polyclonal antibodies (B), and a monoclonal antibody against an unknown La1 outer surface protein (C). As controls, 0.03 μg of EF-Tu recombinant protein (r) and 0.1 μg of cell wall extract (cw) were included in the Western blots. The results of one representative of three independent experiments are shown.
In another set of experiments, the La1 protein band recognized by the anti-EF-Tu antibodies in a guanidine-HCl surface protein extract was analyzed by nano-electrospray ionization-mass spectrometry (data not shown). The peptides generated from the La1 protein perfectly matched the predicted EF-Tu protein sequence, with no detectable contamination.
All these results unequivocally demonstrate that the unknown protein is EF-Tu and also confirm both its bacterial origin and its surface localization.
Binding properties of EF-Tu.
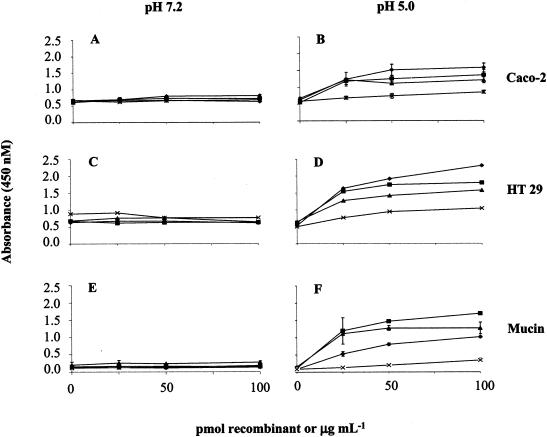
To characterize the binding properties of La1 EF-Tu, different experiments were performed using the recombinant protein or the La1 cell wall extract in combination with an anti-His tag monoclonal antibody or anti-recombinant EF-Tu polyclonal antibodies (Fig. 4). The recombinant EF-Tu was also able to bind to undifferentiated Caco-2 cells (Fig. 4B), as well as to HT29 cells (Fig. 4D), at pH 5.0. No specific binding was observed when the incubations were performed at pH 7.2 (Fig. 4A and C). These results are in accordance with the experiments aimed at identifying the La1 surface proteins involved in attachment to intestinal cells (Fig. 1), even though the Caco-2 cells used in the former experiments were already differentiated.
FIG. 4.
Binding properties of EF-Tu. (A to D) Caco-2 cells (20,000/well) and HT29 cells (10,000/well) were cultivated for 1 week and incubated at pH 7.2 or 5.0, as indicated, with serial dilutions of the recombinant EF-Tu or La1 cell wall extract. Bound EF-Tu recombinant molecules were revealed by anti-His tag antibody (♦) and anti-EF-Tu polyclonal antibodies (▪). La1 cell wall-associated EF-Tu was revealed by anti-EF-Tu antibodies (▴). Binding of an unrelated His-tagged La1 recombinant molecule was revealed by anti-His-tag antibody (×). (E and F) Binding of the same molecules to HT29-MTX mucins. The standard deviations are indicated by the error bars. The results are representative of at least five independent experiments.
The capacity of EF-Tu recombinant protein and/or the La1 cell wall extract to bind to mucins was also investigated using mucins purified from HT29-MTX cells (Fig. 4E and F). The binding pattern of EF-Tu to mucins was similar to that observed with the cell lines, i.e., specific binding was observed only at pH 5.0. To exclude the possibility of nonspecific binding mediated by the His tag epitope, a cell wall extract of La1 was incubated at different dilutions with the cells or mucins. The presence of cell-bound EF-Tu at pH 5.0 was again detected by the polyclonal antibodies (Fig. 4B, D, and F). Furthermore, another His-tagged La1 recombinant protein used as a control did not exhibit any specific binding.
As the most predominant mucin secreted by HT29-MTX cells is MUC 5AC, mostly present in the gastric mucosa (38), the ability of the EF-Tu recombinant protein to bind to the purified preparation of normal human colonic mucins was also tested (data not shown). Even though MUC 2 was the most abundant colonic mucin (data not shown), similar results were obtained, suggesting that common binding sites for EF-Tu exist in the different mucins.
Competition assays between EF-Tu and La1 bacteria for binding to normal gut mucins.
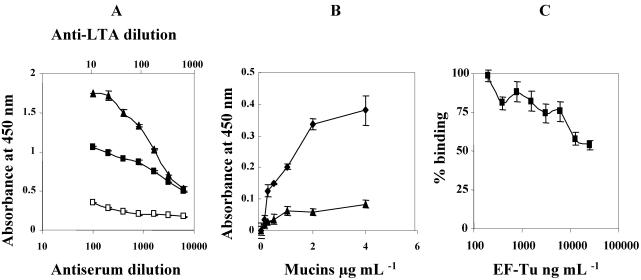
To evaluate the contribution of the EF-Tu molecule to the binding of La1 bacteria to mucins, we set up another binding test. As a first step, the presence of EF-Tu on the surface of La1 was demonstrated by comparing the binding of mouse EF-Tu antibodies with the binding of normal mouse serum (negative control) (Fig. 5A). A monoclonal antibody against LTA, another La1 surface molecule, was used as a positive control. The supernatants of the wells coated with La1 did not contain DNA, aldolase, or any other protein, indicating that the bacteria maintained their integrity on the ELISA plate wells and also that no release of EF-Tu protein from the bacteria occurred during the incubation period.
FIG. 5.
(A) Detection of EF-Tu on live La1. Polyvinyl wells were coated with bacteria, which were incubated with anti-EF-Tu polyclonal antibodies (▪) and anti-La1 LTA monoclonal antibody supernatant (▴). The negative control was normal mouse serum (□). (B) Detection of mucin binding to La1. The same bacteria were incubated with normal human gut mucins at pH 5.0 and fixed with paraformaldehyde. La1-bound mucins were revealed with mouse anti-mucin polyclonal antibodies (♦). The control was normal mouse serum (▴). (C) Competition experiment between EF-Tu and La1 bacteria for mucin binding. The same bacteria were incubated with a constant amount (1.5 μg/ml) of mucins in the presence of serial dilutions of EF-Tu. La1-bound mucins were revealed with mouse anti-mucin polyclonal antibodies. Mucin binding was expressed as the absorbance at 450 nm. Residual binding of mucins to La1 bacteria in the presence of EF-Tu is expressed relative to the binding in the absence of EF-Tu, which was considered 100%. The standard deviations are indicated by the error bars. The results are representative of three independent experiments for each panel.
To determine the correct amount of mucins needed to perform the competition assay, bacteria were incubated with increasing amounts of normal gut mucins at pH 5.0 (Fig. 5B). The binding of mucins increased to reach a plateau level at ∼4 μg/ml. The specificity of the binding was assessed with normal mouse serum.
Based on these previous results, competition studies were performed with a constant mucin concentration of 1.5 μg/ml in the presence of serial dilutions of recombinant EF-Tu protein. The results clearly showed that EF-Tu has an important role in the mucin binding activity of La1, as it could prevent up to 40% of the binding of mucins to La1 bacteria (Fig. 5C).
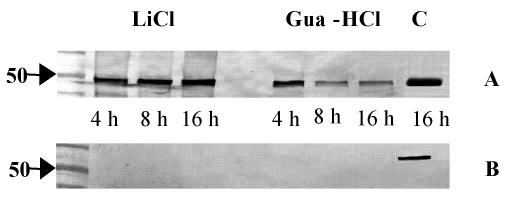
Detection of EF-Tu at the surfaces of La1 bacteria.
To confirm the presence of EF-Tu on the La1 surface, outer surface proteins were extracted with LiCl or guanidine-HCl at different times of culture and analyzed by Western blotting using anti-EF-Tu antibodies. EF-Tu was detected after different times of culture (Fig. 6A). As depicted in Fig. 6A, EF-Tu was very abundant in the cytoplasmic fraction. Therefore, to exclude the possibility of contamination of the outer surface protein preparation by cytoplasmic EF-Tu due to cell lysis, blots were also probed with an anti-enzyme I antibody. Indeed, enzyme I, a protein of a phosphotransferase system which is not present on the surfaces of bacteria (33, 54), was detected only in La1 cytoplasm (Fig. 6B).
FIG. 6.
Localization of EF-Tu in La1 bacterial compartments. Western blots were made with outer surface proteins of La1 extracted with LiCl or guanidine-HCl at different times of cuture and with cytoplasm after 16 h of culture (C). They were incubated with anti-EF-Tu antibodies (A) or with anti-enzyme I antibodies (B) as a control for cytoplasmic contamination. The results are representative of three independent experiments.
The primary structure of La1 EF-Tu present at the surface was confirmed by nano-electrospray tandem mass spectrometry. The sequence of the peptides obtained by in-gel tryptic digestion of the EF-Tu band, when compared to the La1 EF-Tu theoretical protein by sequence tag searching (data not shown), confirmed that EF-Tu was present at the La1 surface as an intact molecule.
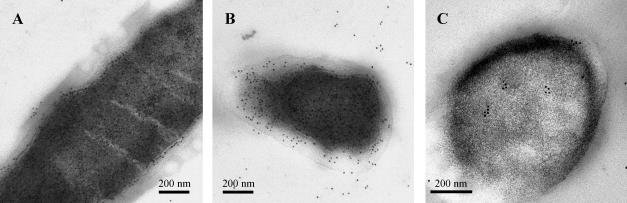
The surface localization of EF-Tu was also analyzed by electron microscopy (Fig. 7A and B). Positive immunostaining was observed on the outer surface of the bacteria, in the external layer of the bacterial cell wall, and in the cytoplasm. Figure 7C represents a section treated with normal mouse serum as a negative control.
FIG. 7.
Immunogold staining of EF-Tu on La1 thin sections. (A and B) Positive EF-Tu labeling on cytoplasm, membrane, cell wall, and outer surface. (C) La1 treated with normal mouse serum.
Detection of EF-Tu in outer surface protein extracts of other bacteria.
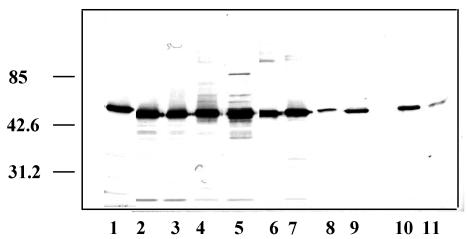
To investigate whether the presence of EF-Tu on the surface of La1 was a rare event, we looked for its presence in the outer surface proteins of other lactobacilli and bifidobacteria (Fig. 8). EF-Tu was detected in all strains tested, although with a weaker intensity in L. paracasei, L. helveticus, and the two bifidobacteria, which may imply a different expression level at the surface or a lower cross-reaction with the anti-La1 EF-Tu antibodies.
FIG. 8.
Detection of EF-Tu in guanidine-HCl extracts of other bacteria. Western blots were made with 1 μg of each bacterial outer surface protein extract and revealed with anti-EF-Tu polyclonal antibodies. Lanes: 1, L. johnsonii La1 (NCC533); 2, L. johnsonii NCC 1657; 3, L. johnsonii ATCC 33200; 4, L. acidophilus NCC 90; 5, L. acidophilus NCC 12; 6, L. reuteri NCC 2458; 7, L. gasseri NCC 907; 8, L. paracasei NCC 2461; 9, L. helveticus; 10, bifidobacterium Bb 12 NCC 362; 11, bifidobacterium Bl 29 NCC 490. Experiments were repeated at least twice. Molecular mass standards (in kilodaltons) are indicated on the left.
Stimulation assays with HT29 cells.
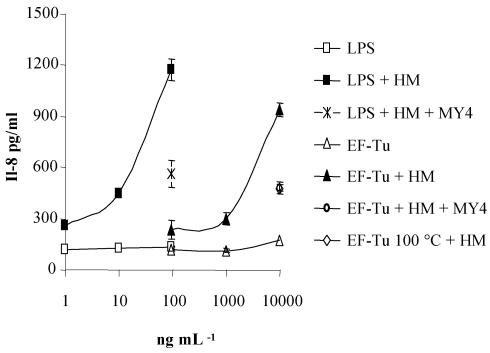
It was recently demonstrated that the human intestinal epithelial cell line HT29 secretes IL-8 in response to gram-negative bacteria or LPS and that this stimulation is soluble CD14 (sCD14) dependent (36, 64). Since sCD14 is known to interact with components of gram-positive bacteria (17), we examined whether the EF-Tu molecule was able to stimulate IL-8 release from HT29 cells. Addition of 10 μg of recombinant EF-Tu protein/ml resulted in an important increase in IL-8 secretion only in the presence of HM used as a source of sCD14 (Fig. 9). The involvement of sCD14 was confirmed by the fact that this increase was greatly diminished when EF-Tu was incubated in the presence of MY4 anti-CD14 antibodies. When the recombinant EF-Tu protein was denatured by heat treatment at 100°C, IL-8 release was diminished by 60%, indicating that the stimulation was not due to LPS contamination. Indeed, the amount of LPS of E. coli origin present in the recombinant-protein preparation used was 1 ng per 10 μg of protein, which is not sufficient to induce the IL-8 secretion observed. For comparison and validation of the system, HT29 cells were also incubated in the presence of LPS and HM, which revealed that the stimulatory potential of EF-Tu was ∼100 times lower than that of LPS. In contrast to that observed with EF-Tu, heating of LPS only resulted in a 10% decrease in IL-8 secretion.
FIG. 9.
IL-8 release by HT29 cells upon stimulation with EF-Tu recombinant molecule. IL-8 production was measured by ELISA in supernatants of HT29 cells incubated for 24 h with various amounts of the EF-Tu recombinant molecule in the presence (▴) or absence (▵) of HM (2%) as a source of sCD14. IL-8 release was measured when EF-Tu was incubated in the presence of anti-CD14 antibody (○) or heated at 100°C for 20 min (⋄). Controls of HT29 cell activation were done by incubating various amounts of LPS from E. coli in the presence (▪) or absence (□) of HM, or by incubation in the presence of MY4 anti-CD14 monoclonal antibody (×). The standard deviations are indicated by the error bars. The results are representative of three independent experiments.
DISCUSSION
The aim of this work was to identify La1 surface proteins mediating the attachment to intestinal epithelial cell lines and/or mucins and to further study whether they were able to trigger an immunomodulatory response in the gut. The identification of one of these proteins as EF-Tu was rather unexpected. Indeed, this molecule is a guanosine nucleotide binding protein that plays a central role in protein synthesis in the cytoplasm. The predicted L. johnsonii NCC533 EF-Tu protein does not possess an identifiable signal sequence (61) (the determined amino-terminal sequence lacks the first methionine), an LPXTG peptidoglycan anchor motif, or a lipoprotein motif (PROSITE PS00013) to explain its localization on the surface of La1. However, the presence of EF-Tu in the different compartments of microorganisms has been well documented. Actually, since 1976, the EF-Tu molecule, originally thought to be restricted to the cytoplasm of bacteria, has been shown to also be associated with the membrane of E. coli (32) under laboratory culture conditions and with the periplasm of Neisseria gonorrhoeae (55). Recently, Nakamura et al. (49), studying sake spoilage lactobacilli, declared EF-Tu to be “an envelope-associated protein that can be released from the cell by osmotic shock.” Finally, EF-Tu was also identified as a major cell wall-associated component of Mycobacterium leprae (43) and Mycoplasma pneumoniae (13). Interestingly, while M. pneumoniae EF-Tu binds to fibronectin, a component of the extracellular matrix (13), La1 EF-Tu does not have this property. Indeed, it does not bind to fibronectin or to collagen (data not shown).
Using several methods and control experiments, we have shown unequivocally that this molecule is also localized at the surfaces of La1 bacteria. The analyses we have performed on other lactobacilli and bifidobacteria demonstrate that the presence of EF-Tu associated with the surfaces of La1 bacteria is not an exceptional fact. This observation is in accordance with the results of Nakamura et al. (49), who described the protein as a common antigen of many lactobacilli These findings suggest that EF-Tu can be added to a growing list of enzymes that one would expect to see only in the cytoplasm but which have been detected on the surface and are thought to bind through charge and/or hydrophobic interactions (20).
Binding of the EF-Tu protein present in La1 cell wall extracts or of the recombinant protein to intestinal epithelial cell lines and to human mucins was performed at pH 5.0 and 7.2. Three facts supported this choice. First, bacterial-adhesion studies (8) showed that La1 adhesion to mucus and Caco-2 cells was pH dependent and was higher at pH 5.0 than at pH 7.2. Second, it has been postulated that the pH of the gut lumen is neutral but becomes gradually more acidic at the mucus-covered surface, due to the sialic acid residues and sulfated content of the mucin. Binding at pH 5.0 would thus be more representative of physiological conditions (8). Finally, the pH dependency of this process had already been observed for other mucus binding proteins (60) or whole bacteria binding to epithelial cells or mucins (8, 21, 23). The binding of the recombinant EF-Tu protein to gastric or intestinal mucin preparations was similar, implying that oligosaccharide determinants, common to both types of mucin, may be involved in this binding. Binding of mucins to La1 could be inhibited up to 40% by EF-Tu, which supports an important role of this protein in the attachment of La1 bacteria to the gastrointestinal mucus layer.
Our results demonstrate clearly that EF-Tu can have two functions, depending on its cytoplasmic or surface location. Indeed, it has already been shown that α-enolase, a cytoplasmic enzyme, also functions as a plasmin(ogen) binding protein (51) when present on the surfaces of several organisms (52). The same is true for ornithine carbamoyltransferase, which is also a putative adhesin of Staphylococcus epidermidis (28).
The immunomodulatory properties of EF-Tu have been demonstrated using HT29 cells. This cell line has already been used to study the immune response triggered by components of gram-negative and gram-positive bacteria in the presence of sCD14 (36, 64, 65). Indeed, it has been shown that this molecule can bind to numerous components of gram-positive bacteria, including peptidoglycan, LTA, lipoarabinomannan, and mannuronic acid polymers (17, 26). Even though we had first determined that EF-Tu alone was not able to bind to epithelial cells at pH 7.2, we have further demonstrated that the EF-Tu recombinant molecule does bind to HT29 cells at neutral pH by using sCD14 as a ligand. Furthermore, once bound to HT29 cells, EF-Tu elicits a CD14-dependent proinflammatory response.
Up to now, studies of immunomodulation by isolated lactobacillus components have been principally focused on their signaling on peripheral blood mononuclear cells (10, 11, 24, 63). Using HT29 cells as a model, we showed that La1 EF-Tu binds to intestinal epithelial cells and stimulates proinflammatory reactions in the absence of other immune cells normally present in the lamina propria.
Using the same system, in vitro experiments have shown that intact La1 bacteria are not able to stimulate IL-8 secretion from these cells even in the presence of sCD14 (64) or peripheral blood mononuclear cells (9). This discrepancy may arise from the presence of LTA, another La1 surface-associated molecule mediating binding to epithelial cells (21) and exhibiting anti-inflammatory properties (64). Furthermore, these two molecules may be effective only when they are released as metabolites under acidic conditions (data not shown). Experiments are now under way to determine whether cell triggering by La1 EF-Tu involves a signal transduction pathway(s) similar to those induced by LPS or peptidoglycan and/or contributes, like LTA (37, 46), to nonimmunological mechanisms of the gastrointestinal tract defenses.
Acknowledgments
We thank F. Schuepbach, A.-C. Pittet, and J. Horlbeck for their excellent technical assistance and Harald Nothaft and Fritz Titgemeyer for the generous gift of the anti-enzyme I antibody. We are also indebted to K. Vidal for helpful suggestions and to C. Cherbut for critical review of the manuscript.
Editor: F. C. Fang
REFERENCES
- 1.Aleljung, P., W. Shen, B. Rozalska, U. Hellman, A. Ljungh, and T. Wadstrom. 1994. Purification of collagen-binding proteins of Lactobacillus reuteri NCIB 11951. Curr. Microbiol. 28:231-236. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Apostolou, E., P. V. Kirjavainen, M. Saxelin, H. Rautelin, V. Valtonen, S. J. Salminen, and A. C. Ouwehand. 2001. Good adhesion properties of probiotics: a potential risk for bacteremia? FEMS Immunol. Med. Microbiol. 31:35-39. [DOI] [PubMed] [Google Scholar]
- 4.Arvola, T., K. Laiho, S. Torkkeli, H. Mykkanen, S. Salminen, L. Maunula, and E. Isolauri. 1999. Prophylactic Lactobacillus GG reduces antibiotic-associated diarrhea in children with respiratory infections: a randomized study. Pediatrics 104:e64. (Online.) [DOI] [PubMed] [Google Scholar]
- 5.Bernet, M. F., D. Brassart, J. R. Neeser, and A. L. Servin. 1994. Lactobacillus acidophilus LA 1 binds to cultured human intestinal cell lines and inhibits cell attachment and cell invasion by enterovirulent bacteria. Gut 35:483-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernet-Camard, M. F., V. Lievin, D. Brassart, J. R. Neeser, A. L. Servin, and S. Hudault. 1997. The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl. Environ. Microbiol. 63:2747-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bican, P., and A. Spahni. 1988. Reversion of Lactobacillus lactis protoplasts. Experientia 44:348-351. [DOI] [PubMed] [Google Scholar]
- 8.Blum, S., R. Reniero, E. J. Schiffrin, R. Crittenden, T. Mattila-Sandholm, A. C. Ouwehand, S. Salminen, A. von Wright, M. Saarela, M. Saxelin, K. Collins, and L. Morelli. 2000. Adhesion studies for probiotics: need for validation and refinement. Trends Food Sci. Technol. 10:405-410. [Google Scholar]
- 9.Blum, S., D. Haller, A. Pfeifer, and E. J. Schiffrin. 2002. Probiotics and immune response. Clin. Rev. Allergy Immunol. 22:287-309. [DOI] [PubMed] [Google Scholar]
- 10.Chen, T., P. Isomaki, M. Rimpilainen, and P. Toivanen. 1999. Human cytokine responses induced by gram-positive cell walls of normal intestinal microbiota. Clin. Exp. Immunol. 118:261-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, T., P. Toivanen, and O. Vainio. 2002. Suppression of antigen-specific lymphocyte proliferation by Gram-positive bacterial cell walls. APMIS 110:490-498. [DOI] [PubMed] [Google Scholar]
- 12.Cull, M., and C. S. McHenry. 1990. Preparation of extracts from prokaryotes. Methods Enzymol. 182:147-153. [DOI] [PubMed] [Google Scholar]
- 13.Dallo, S. F., T. R. Kannan, M. W. Blaylock, and J. B. Baseman. 2002. Elongation factor Tu and E1 β subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46:1041-1051. [DOI] [PubMed] [Google Scholar]
- 14.Donnet-Hughes, A., F. Rochat, P. Serrant, J. M. Aeschlimann, and E. J. Schiffrin. 1999. Modulation of nonspecific mechanisms of defense by lactic acid bacteria: effective dose. J. Dairy Sci. 82:863-869. [DOI] [PubMed] [Google Scholar]
- 15.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 68:350-356. [Google Scholar]
- 16.Dunne, C., L. Murphy, S. Flynn, L. O'Mahony, S. O'Halloran, M. Feeney, D. Morrissey, G. Thornton, G. Fitzgerald, C. Daly, B. Kiely, E. M. Quigley, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 1999. Probiotics: from myth to reality. Demonstration of functionality in animal models of disease and in human clinical trials. Antonie Leeuwenhoek 76:279-292. [PubMed] [Google Scholar]
- 17.Dziarski, R., A. J. Ulmer, and D. Gupta. 2000. Interactions of CD14 with components of gram-positive bacteria. Chem. Immunol. 74:83-107. [DOI] [PubMed] [Google Scholar]
- 18.Faure, M., D. Moennoz, F. Montigon, L. B. Fay, D. Breuille, P. A. Finot, O. Ballevre, and J. Boza. 2002. Development of a rapid and convenient method to purify mucins and determine their in vivo synthesis rate in rats. Anal. Biochem. 307:244-251. [DOI] [PubMed] [Google Scholar]
- 19.Finnie, I. A., A. D. Dwarakanath, B. A. Taylor, and J. M. Rhodes. 1995. Colonic mucin synthesis is increased by sodium butyrate. Gut 36:93-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischetti, V. A. 2000. Surface protein on Gram-positive bacteria, p. 11-23. In V. A. Fischetti (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 21.Granato, D., F. Perotti, I. Masserey, M. Rouvet, M. Golliard, A. Servin, and D. Brassart. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 65:1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Granato, D. A., and P. F. Piguet. 1986. A mouse monoclonal IgE antibody anti-bovine milk beta-lactoglobulin allows studies of allergy in the gastrointestinal tract. Clin. Exp. Immunol. 63:703-710. [PMC free article] [PubMed] [Google Scholar]
- 23.Greene, J. D., and T. R. Klaenhammer. 1994. Factors involved in adherence of lactobacilli to human Caco-2 cells. Appl. Environ. Microbiol. 60:4487-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haller, D., P. Serrant, D. Granato, E. J. Schiffrin, and S. Blum. 2002. Activation of human NK cells by staphylococci and lactobacilli requires cell contact-dependent costimulation by autologous monocytes. Clin. Diagn. Lab. Immunol. 9:649-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamann, L., V. El Samalouti, A. J. Ulmer, H. D. Flad, and E. T. Rietschel. 1998. Components of gut bacteria as immunomodulators. Int. J. Food Microbiol. 41:141-154. [DOI] [PubMed] [Google Scholar]
- 26.Heinemann, C., J. E. Hylckama Vlieg, D. B. Janssen, H. J. Busscher, H. C. van der Mei, and G. Reid. 2000. Purification and characterization of a surface-binding protein from Lactobacillus fermentum RC-14 that inhibits adhesion of Enterococcus faecalis 1131. FEMS Microbiol. Lett. 190:177-180. [DOI] [PubMed] [Google Scholar]
- 27.Hoyos, A. B. 1999. Reduced incidence of necrotizing enterocolitis associated with enteral administration of Lactobacillus acidophilus and Bifidobacterium infantis to neonates in an intensive care unit. Int. J. Infect. Dis. 3:197-202. [DOI] [PubMed] [Google Scholar]
- 28.Hussain, M., G. Peters, G. S. Chhatwal, and M. Herrmann. 1999. A lithium chloride-extracted, broad-spectrum-adhesive 42-kilodalton protein of Staphylococcus epidermidis is ornithine carbamoyltransferase. Infect. Immun. 67:6688-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibnou-Zekri, N., S. Blum, E. J. Schiffrin, and T. von der Weid. 2003. Divergent patterns of colonization and immune response elicited from two intestinal Lactobacillus strains that display similar properties in vitro. Infect. Immun. 71:428-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ichikawa, H., T. Kuroiwa, A. Inagaki, R. Shineha, T. Nishihira, S. Satomi, and T. Sakata. 1999. Probiotic bacteria stimulate gut epithelial cell proliferation in rat. Dig. Dis. Sci. 44:2119-2123. [DOI] [PubMed] [Google Scholar]
- 31.Isolauri, E., S. Salminen, and T. Mattila-Sandholm. 1999. New functional foods in the treatment of food allergy. Ann. Med. 31:299-302. [DOI] [PubMed] [Google Scholar]
- 32.Jacobson, G. R., and J. P. Rosenbusch. 1976. Abundance and membrane association of elongation factor Tu in E. coli. Nature 261:23-26. [DOI] [PubMed] [Google Scholar]
- 33.Kamionka, A., S. Parche, H. Nothaft, J. Siepelmeyer, K. Jahreis, and F. Titgemeyer. 2002. The phosphotransferase system of Streptomyces coelicolor. Eur. J. Biochem. 269:2143-2150. [DOI] [PubMed] [Google Scholar]
- 34.Kleerebezem, M., J. Boekhorst, R. Van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. De Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA. 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kling, D. E., L. C. Madoff, and J. L. Michel. 1999. Subcellular fractionation of group B Streptococcus. BioTechniques 27:24-26, 28. [DOI] [PubMed] [Google Scholar]
- 36.Labeta, M. O., K. Vidal, J. E. Nores, M. Arias, N. Vita, B. P. Morgan, J. C. Guillemot, D. Loyaux, P. Ferrara, D. Schmid, M. Affolter, L. K. Borysiewicz, A. Donnet-Hughes, and E. J. Schiffrin. 2000. Innate recognition of bacteria in human milk is mediated by a milk-derived highly expressed pattern recognition receptor, soluble CD14. J. Exp. Med. 191:1807-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lemjabbar, H., and C. Basbaum. 2002. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat. Med. 8:41-46. [DOI] [PubMed] [Google Scholar]
- 38.Lesuffleur, T., F. Roche, A. S. Hill, M. Lacasa, M. Fox, D. M. Swallow, A. Zweibaum, and F. X. Real. 1995. Characterization of a mucin cDNA clone isolated from HT-29 mucus-secreting cells. The 3′ end of MUC5AC? J. Biol. Chem. 270:13665-13673. [DOI] [PubMed] [Google Scholar]
- 39.Lin, M. Y., and F. J. Chang. 2000. Antioxidative effect of intestinal bacteria Bifidobacterium longum ATCC 15708 and Lactobacillus acidophilus ATCC 4356. Dig. Dis. Sci. 45:1617-1622. [DOI] [PubMed] [Google Scholar]
- 40.Link-Amster, H., F. Rochat, K. Y. Saudan, O. Mignot, and J. M. Aeschlimann. 1994. Modulation of a specific humoral immune response and changes in intestinal flora mediated through fermented milk intake. FEMS Immunol. Med. Microbiol. 10:55-63. [DOI] [PubMed] [Google Scholar]
- 41.Lortal, S., J. van Heijenoort, K. Gruber, and U. B. Sleytr. 1992. S-layer of Lactobacillus helveticus ATCC 12046: isolation, chemical characterization and re-formation after extraction with lithium chloride. J. Gen. Microbiol. 138:611-618. [Google Scholar]
- 42.Mangiante, G., P. Canepari, G. Colucci, P. Marinello, C. Signoretto, N. Nicoli, and S. Bengmark. 1999. A probiotic as an antagonist of bacterial translocation in experimental pancreatitis. Chir. Ital. 51:221-226. [PubMed] [Google Scholar]
- 43.Marques, M. A., S. Chitale, P. J. Brennan, and M. C. Pessolani. 1998. Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect. Immun. 66:2625-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marteau, P., J. P. Vaerman, J. P. Dehennin, S. Bord, D. Brassart, P. Pochart, J. F. Desjeux, and J. C. Rambaud. 1997. Effects of intrajejunal perfusion and chronic ingestion of Lactobacillus johnsonii strain La1 on serum concentrations and jejunal secretions of immunoglobulins and serum proteins in healthy humans. Gastroenterol. Clin. Biol. 21:293-298. [PubMed] [Google Scholar]
- 45.Marvin, L. F., V. Parisod, L. B. Fay, and P. A. Guy. 2002. Characterization of lactosylated proteins of infant formula powders using two-dimensional gel electrophoresis and nanoelectrospray mass spectrometry. Electrophoresis 23:2505-2512. [DOI] [PubMed] [Google Scholar]
- 46.Mattar, A. F., D. H. Teitelbaum, R. A. Drongowski, F. Yongyi, C. M. Harmon, and A. G. Coran. 2002. Probiotics up-regulate MUC-2 mucin gene expression in a Caco-2 cell-culture model. Pediatr. Surg. Int. 18:586-590. [DOI] [PubMed] [Google Scholar]
- 47.McIntosh, G. H., P. J. Royle, and M. J. Playne. 1999. A probiotic strain of L. acidophilus reduces DMH-induced large intestinal tumors in male Sprague-Dawley rats. Nutr. Cancer 35:153-159. [DOI] [PubMed] [Google Scholar]
- 48.Michetti, P., G. Dorta, P. H. Wiesel, D. Brassart, E. Verdu, M. Herranz, C. Felley, N. Porta, M. Rouvet, A. L. Blum, and I. Corthesy-Theulaz. 1999. Effect of whey-based culture supernatant of Lactobacillus acidophilus (johnsonii) La1 on Helicobacter pylori infection in humans. Digestion 60:203-209. [DOI] [PubMed] [Google Scholar]
- 49.Nakamura, J., D. Ito, K. Nagai, Y. Umehara, M. Hamachi, and C. Kumagai. 1997. Rapid and sensitive detection of hiochi bacteria by amplification of hiochi bacterial common antigen gene by PCR method and characterization of the antigen. J. Ferment. Bioeng. 83:161-167. [Google Scholar]
- 50.Nothaft, H., D. Dresel, A. Willimek, K. Mahr, M. Niederweis, and F. Titgemeyer. 2003. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. J. Bacteriol. 185:7019-7023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pancholi, V., and V. A. Fischetti. 1997. A novel plasminogen/plasmin binding protein on the surface of group A streptococci. Adv. Exp. Med. Biol. 418:597-599. [DOI] [PubMed] [Google Scholar]
- 52.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 53.Pantoflickova, D., I. Corthésy-Theulaz, G. Dorta, M. Stole, P. Isler, F. Rochat, M. Enslen, and A. L. Blum. 2003. Favourable effect of regular intake of fermented milk containing Lactobacillus johnsonii on Helicobacter pylori associated gastritis. Aliment. Pharmacol. Ther. 18:1-9. [DOI] [PubMed] [Google Scholar]
- 54.Parche, S., H. Nothaft, A. Kamionka, and F. Titgemeyer. 2000. Sugar uptake and utilisation in Streptomyces coelicolor: a PTS view to the genome. Antonie Leeuwenhoek 78:243-251. [DOI] [PubMed] [Google Scholar]
- 55.Porcella, S. F., R. J. Belland, and R. C. Judd. 1996. Identification of an EF-Tu protein that is periplasm-associated and processed in Neisseria gonorrhoeae. Microbiology 142:2481-2489. [DOI] [PubMed] [Google Scholar]
- 56.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Baretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed]
- 57.Rabilloud, T. 2002. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics 2:3-10. [PubMed] [Google Scholar]
- 58.Rojas, M., F. Ascencio, and P. L. Conway. 2002. Purification and characterization of a surface protein from Lactobacillus fermentum 104R that binds to porcine small intestinal mucus and gastric mucin. Appl. Environ. Microbiol. 68:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roos, S., P. Aleljung, N. Robert, B. Lee, T. Wadstrom, M. Lindberg, and H. Jonsson. 1996. A collagen binding protein from Lactobacillus reuteri is part of an ABC transporter system? FEMS Microbiol. Lett. 144:33-38. [DOI] [PubMed] [Google Scholar]
- 60.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148:433-442. [DOI] [PubMed] [Google Scholar]
- 61.Schatz, G., and B. Dobberstein. 1996. Common principles of protein translocation across membranes. Science 271:1519-1526. [DOI] [PubMed] [Google Scholar]
- 62.Schiffrin, E. J., F. Rochat, H. Link-Amster, J. M. Aeschlimann, and A. Donnet-Hughes. 1995. Immunomodulation of human blood cells following the ingestion of lactic acid bacteria. J. Dairy Sci. 78:491-497. [DOI] [PubMed] [Google Scholar]
- 63.Tejada-Simon, M. V., and J. J. Pestka. 1999. Proinflammatory cytokine and nitric oxide induction in murine macrophages by cell wall and cytoplasmic extracts of lactic acid bacteria. J. Food Prot. 62:1435-1444. [DOI] [PubMed] [Google Scholar]
- 64.Vidal, K., A. Donnet-Hughes, and D. Granato. 2002. Lipoteichoic acids from Lactobacillus johnsonii strain La1 and Lactobacillus acidophilus strain La10 antagonize the responsiveness of human intestinal epithelial HT29 cells to lipopolysaccharide and gram-negative bacteria. Infect. Immun. 70:2057-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vidal, K., M. O. Labeta, E. J. Schiffrin, and A. Donnet-Hughes. 2001. Soluble CD14 in human breast milk and its role in innate immune responses. Acta Odontol. Scand. 59:330-334. [DOI] [PubMed] [Google Scholar]
- 66.Woriax, V. L., W. Burkhart, and L. L. Spremulli. 1995. Cloning, sequence analysis and expression of mammalian mitochondrial protein synthesis elongation factor Tu. Biochim. Biophys. Acta 1264:347-356. [DOI] [PubMed] [Google Scholar]