Abstract
Enteropathogenic Escherichia coli (EPEC) produces a lesion on epithelial cells called the attaching and effacing (AE) lesion. All genes necessary for AE are encoded within the locus of enterocyte effacement (LEE). EPEC also adheres in a characteristic pattern to epithelial cells by forming microcolonies, usually referred to as localized adherence (LA). LA is mediated by the bundle-forming pilus and flagella. The LEE genes are directly activated by the LEE-encoded regulator (Ler). Transcription of Ler is under the control of Per, integration host factor, Fis, BipA, and quorum sensing (QS), specifically through the luxS system. QS activates expression of the LEE genes in EPEC, with QseA activating transcription of ler. Here we report that transcription of the LEE genes and type III secretion are diminished in both luxS and qseA mutants. Transcription of the LEE genes is affected in both mutants mostly during the mid-exponential phase of growth. Transcription of qseA itself is diminished throughout growth in a luxS mutant and is under autorepression. Furthermore, QS activation of type III secretion is independent of per, given that QseA still activates type III secretion in a per mutant strain. Both mutants are deficient in adherence to epithelial cells and form smaller microcolonies. Several factors may contribute to this abnormal behavior: transcription of LEE genes and type III secretion are diminished, and expression of flagella and Per is altered in both mutants. These results suggest that QS is involved in modulating the regulation of the EPEC virulence genes.
Enteropathogenic Escherichia coli (EPEC) is a major cause of diarrhea in children in developing countries (29). EPEC is part of a group of pathogens that includes enterohemorrhagic E. coli (EHEC), Citrobacter rodentium, and Hafnia alvei, all of which are able to cause a lesion on the intestinal epithelial cells named the attaching and effacing (AE) lesion. This lesion is characterized by the destruction of the microvilli and rearrangement of the cytoskeleton to form a pedestal-like structure which cups the bacteria individually (21, 28, 48). The genes involved in the formation of the AE lesion are encoded within a chromosomal pathogenicity island named the locus of enterocyte effacement (LEE) (25). The LEE region contains five major operons, i.e., LEE1, LEE2, LEE3, tir (LEE5), and LEE4 (6, 26), which encode a type III secretion system (TTSS) (18), an adhesin (intimin) (19), and the intimin adhesin receptor (Tir) (20), which is translocated to the epithelial cell through the bacterial TTSS. The LEE genes are directly activated by the LEE-encoded regulator (Ler), which is encoded by the first gene in the LEE1 operon (1, 7, 26, 33, 39). Mutations in ler abolished the ability of EPEC and EHEC to produce AE lesions (7, 9). Bustamante et al. (1) reported that Ler acts as an antirepressor for H-NS, probably by competing with H-NS for binding to the regulatory regions of LEE2 and LEE3. Ler and H-NS also compete to activate transcription of LEE5 (16, 33). Taken together, these studies demonstrated that Ler directly activates transcription of the LEE genes by competing for binding with H-NS, which represses their expression in the absence of Ler.
EPEC produces a type IV pilus called bundle-forming pilus (BFP) (11), which is associated with bacterial clustering and formation of tight microcolonies on tissue culture and human intestinal cells, a phenotype referred to as the localized adherence pattern (3, 34). Recent work by Giron et al. (12) suggests that the EPEC flagellum also functions as an adhesin and is essential for microcolony formation. EPEC contains a large plasmid, referred to as the EPEC adherence factor (EAF) plasmid. The EAF plasmid encodes a regulator of virulence genes called Per (plasmid-encoded regulator), consisting of the products of the three open reading frames perA, perB, and perC. PerA is an AraC homologue (14) and activates the expression of the bfp operon, encoding the BFP (47). The per loci also activate the expression of ler, which activates expression of the LEE2, LEE3, LEE5, and LEE4 operons in EPEC in a regulatory cascade (26). Transcription of ler is also regulated by integration host factor (9), Fis (13), and BipA (15). Transcription of per is autoactivated by Per (24) and downregulated by GadX (37).
EPEC LEE genes are also regulated through quorum sensing (40). Quorum sensing is a mechanism of cell-to-cell signaling via the production of compounds, known as autoinducers, that allow a bacterium to “sense” its own population as well as the population of other bacteria in a given environment. Bacterial intercellular communication provides a mechanism for the regulation of gene expression resulting in coordinated population behavior. The functions controlled by quorum sensing are varied and reflect the needs of a particular species of bacteria inhabiting a given niche. Quorum sensing was first described as being involved in the regulation of luminescence in Vibrio fischeri (30) and since then has been shown to be a widespread gene regulation mechanism present in both gram-negative and gram-positive bacteria. Quorum-sensing-controlled processes include bioluminescence, virulence factor expression, biofilm development, and conjugation, among others (4, 10). One of the most widely distributed systems is the luxS system, first described for Vibrio harveyi (44, 46). The autoinducer (referred to as autoinducer-2 [AI-2]) is a furanosyl-borate-diester (2), and the enzyme responsible for its synthesis is encoded by the luxS gene (46). LuxS is an enzyme involved in the detoxification of S-adenosylmethionine, and it converts ribose-homocysteine into homocysteine and 4,5-dihydroxy-2,3-pentanedione, which is the precursor of AI-2 (35). We recently reported that the presence of luxS is necessary for the production of another autoinducer, AI-3, which is the actual autoinducer involved in quorum-sensing regulation of the LEE and flagellar genes (42)
In EPEC, quorum sensing activates transcription of a LysR-like regulator, the quorum-sensing E. coli regulator A (QseA) (38), which activates transcription of Ler (the activator of the LEE genes). Furthermore, quorum sensing also activates expression of the flagellar regulon through the QseBC two-component system (41, 43).
Here we report the effects of a luxS mutation and a qseA mutation on the transcription of the LEE genes, qseA, per, and bfp. We also report that both mutants showed decreased type III secretion, decreased adhesion to cultured epithelial cells, and altered flagellation and motility. Our results suggest that EPEC pathogenesis is a temporally controlled process and that quorum sensing plays an important role in the regulation of these virulence traits.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in this study are listed in Table 1. The prototype EPEC strain E2348/69 was obtained from James B. Kaper. Strain VS102 is a luxS isogenic mutant of strain E2348/69. We initially constructed plasmid pVS68 by amplifying the luxS gene with Pwo polymerase (Boehringer Manheim) by using primers K1663 (5′-GTCGACGCCGCTGATACCGAACCG-3′) and K1664 (5′-GTCGACGCGGTGCGCACTAAGTACAA-3′) and cloning into the EcoRV site of pBluescript KSII (Stratagene) (40). We cloned a chloramphenicol resistance cassette derived from pACYC184 into an EcoRV site in the middle of luxS; this construct was then cloned into the suicide vector pCVD442, which contains an R6K origin of replication (5), generating plasmid pVS98. The EPEC luxS mutant strain, named VS102, was generated by allelic exchange of the luxS gene in pVS98 by using chloramphenicol and sucrose selection as previously described (5). VS104 is VS102 complemented with pVS84, which is wild-type luxS cloned into pACYC177 (Table 1). The qseA mutant strain VS193 has been previously described (38). Strain MPS150 is VS193 complemented with plasmid pVS150, which is wild-type qseA cloned into pACYC177. Plasmid pVS241 was constructed by amplifying the qseA gene with Pfx polymerase (Invitrogen) by using primers qseAF and qseAR (Table 2) and cloning this amplicon into pBADMycHisA (Invitrogen) digested with XhoI and HindIII. All E. coli strains were grown aerobically at 37°C in Luria-Bertani (LB) medium or Dulbecco's modified Eagle's medium (DMEM) (Invitrogen). Selective antibiotics were added at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; nalidixic acid, 100 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Reference or source |
|---|---|---|
| Strains | ||
| E2348/69 | Wild-type EPEC strain; Nalr | James B. Kaper |
| VS102 | E2348 luxS | This study |
| VS104 | VS102 with plasmid pVS84 | This study |
| VS193 | E2348 qseA | 38 |
| MPS150 | VS193 with plasmid pVS150 | This study |
| Plasmids | ||
| pACYC177 | Cloning vector | New England Biolabs |
| pCVD442 | Suicide vector | 5 |
| pBADMycHisA | C-terminal Myc-His tag vector | Invitrogen |
| pVS84 | luxS cloned in pACYC177 | 40 |
| pVS150 | qseA cloned in pACYC177 | 38 |
| pVS160 | qseA::lacZ in PRS551 | 38 |
| pVS241 | qseA into pBADMycHisA | This study |
TABLE 2.
Primers used in this study
| Primer name | Sequence | Gene |
|---|---|---|
| eaeF2 | 5′-CGGAATTCCGATCTGATGCCAATGACT AAT-3′ | eae (LEE5) |
| eaeRL | 5′-CGGGATCCAGCTTATGCTTGTGCCGGGT-3′ | eae (LEE5) |
| PerAF | 5′-CACAATAGAATCCAACTCCT-3′ | perA |
| PerAR | 5′-ACCCAACCAAACAAGATATTT-3′ | perA |
| 23SF | 5′-GGATGTTGGCTTAGAAGCAG-3′ | 23S RNA |
| 23SR | 5′-CAGCTGGTATCTTCGACTGA-3′ | 23S RNA |
| BFPF | 5′-ATGGTGCTTGCGCCTTGCTGC-3′ | bfpA |
| BFPR | 5′-CCTCCCATATAATACGCCC-3′ | bfpA |
| qseAF | 5′-CTCGAGGGAACGACTAAAACGCATGT CGG-3′ | qseA |
| qseAR | 5′-AAGCTTCTTCTCTTTCCCGCGCCCGT-3′ | qseA |
Recombinant DNA techniques.
Plasmid purification, PCR, restriction, ligation, transformation, and DNA gel electrophoresis were performed by standard methods (32). DNA sequence analysis was performed at the University of Texas Southwestern Medical Center Sequencing Core Facility on an ABI automated sequencer with DNA purified by use of Qiagen midi-columns.
RNA dot blotting.
RNA was isolated according to standard procedures (32) from strains E2348/69, VS102, VS104, VS193, and MPS150 grown at 37°C to early exponential phase (optical density at 600 nm [OD600], 0.2), mid-exponential phase (OD600, 0.5), late exponential phase (OD600, 1.0), and stationary phase (OD600, 2.0) in DMEM (Invitrogen). RNA slot blotting was performed with 1 μg of total RNA in triplicate. The RNA was denatured for 60 min in 1 M glyoxal-0.1 M MOPS (morpholinepropanesulfonic acid) (pH 6.8) at 65°C and applied to a nylon membrane under vacuum by using a Bio-Rad dot blot apparatus. The wells were then washed with 500 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membranes were cross-linked, hybridized with different DNA probes by using UltraHyb from Ambion at 42°C, washed first with 2× SSC-0.1% sodium dodecyl sulfate (SDS) and then with 0.2× SSC-0.1% SDS at 42°C, and exposed to X-ray film. Densitometry was performed with a Bio-Rad imager. DNA probes for LEE5, bfpA, perA, and 23S RNAs were generating by PCR with Taq DNA polymerase, amplifying these genes or genes within these operons by using the oligonucleotide primers described in Table 2. These probes were labeled by random primer extension with Ready-to-go DNA labeling beads from Amersham and [α-32P]CTP according to the manufacturer's instructions.
β-Galactosidase assays.
The strains containing the qseA::lacZ fusion (pVS160) were grown to early exponential phase (OD600, 0.2), mid-exponential phase (OD600, 0.5), late exponential phase (OD600, 1.0), and stationary phase (OD600, 2.0) in DMEM at 37°C. These cultures were diluted 1:10 in Z buffer (Na2HPO4, 0.06 M; NaH2PO4, 0.04 M; KCl, 0.01 M; MgSO4, 0.001 M; β-mercaptoethanol, 0.05 M) and assayed for β-galactosidase activity by using o-nitrophenyl-β-d-galactopyranoside as a substrate as previously described (27).
SDS-PAGE and Western blotting.
Total proteins were extracted from strains E2348/69, VS102, VS104, VS193, and MPS150 grown in LB medium or DMEM to the same OD600. Briefly, 1 ml of culture was pelleted (12,000 × g for 5 min at 4°C), and suspended in 400 μl of phosphate-buffered saline (PBS) and 100 μl of 5× sample buffer (20% SDS, 20% glycerol, 200 mM Tris base [pH 6.8], 0.001% bromophenol blue). Protein concentration was measured by using the Lowry assay (32). Equal amounts of total proteins were subjected to SDS-12% PAGE (32). The membrane was later stained with amido black to ensure that the same amounts of proteins had been loaded in each well. Western blotting procedures were performed as previously described (32), and blots were probed with polyclonal antisera directed against BfpA (11) and flagellin (12). Secreted proteins were extracted from strains E2348/69, VS102, VS104, VS193, and MPS150 grown in DMEM at 37°C to an OD600 of 1.0 as described by Jarvis et al. (18), subjected to SDS-12% PAGE, and stained with Coomassie brilliant blue. For detection of Tir, secreted proteins were transferred to a polyvinylidene difluoride membrane and blotted against polyclonal anti-Tir antiserum (6).
FAS test.
Fluorescent actin staining (FAS) tests were performed as initially described by Knutton et al. (22). Briefly, bacterial strains were grown for 18 h in LB medium at 37°C. From the uninduced overnight cultures, 105 CFU (equivalent to an OD600 of 0.05) was added to HeLa cells and incubated for 3 h at 37°C 5% CO2, after which epithelial cells were permeabilized with 0.2% Triton and treated with fluorescein isothicyanate-phalloidin to visualize the accumulation of actin beneath and around the bacteria attached to the HeLa cells (which is the hallmark of the AE lesions). The bacteria were stained with propidium iodide.
Adherence assays.
Bacterial strains were grown for 18 h in LB medium at 37°C. From the uninduced overnight cultures, 105 CFU (equivalent to an OD600 of 0.05) was added to HeLa cells, incubated for 3 h at 37°C with 5% CO2, washed with PBS, fixed with methanol, and stained with Giemsa stain. For quantification of adherence, after a 3-h incubation, the nonadherent bacteria were removed by washing with PBS and the HeLa cells were lysed with 1% Triton. Serial dilutions of the bacterial cells were plated in LB agar plates, and CFU were counted.
Motility assays.
Motility assays were performed at 37°C on 0.3% LB or DMEM agar. The motility halo was observed at 16, 24, and 48 h.
RESULTS
Regulation of transcription of the LEE genes by quorum sensing.
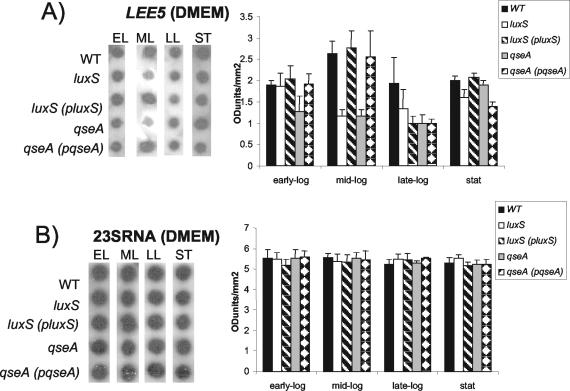
We have previously reported that the LEE genes are activated by quorum sensing in both EHEC and EPEC through the luxS quorum-sensing system. In a previous study we have examined transcriptional activation of LEE::lacZ fusions in an E. coli K-12 background with preconditioned medium (40). We also previously reported that QseA activates transcription of ler in response to quorum sensing in both EPEC and EHEC (38). In those studies (38, 40), we investigated primarily the effect of quorum sensing in the EHEC LEE promoters. The investigation of quorum sensing in EPEC LEE promoters was performed only with preconditioned medium within an E. coli K-12 background (40). In EPEC there is an additional level of regulation of the LEE genes through Per, which is absent in E. coli K-12 (14, 26). Given that we also mapped different promoters for the LEE operons in EHEC than in EPEC (26, 40), we decided to investigate quorum-sensing regulation of the EPEC LEE promoters in depth within an EPEC background. In order to have a more complete picture of the role of quorum sensing in the expression of the LEE genes in EPEC, we measured transcription of these genes by using RNA slot blotting throughout growth (Fig. 1). All of these tests were performed with two RNA samples spotted in triplicate, and Fig. 1 shows the results obtained for the RNA slot blotting and densitometry readings of the three tests. These blots were stripped and then hybridized with a probe to 23S rRNA to ensure equal loading of RNA in the wells (Fig. 1B).
FIG. 1.
Quorum-sensing regulation of transcription of the LEE genes throughout growth. (A) RNA dot blot hybridized with the LEE5 probe. RNAs were extracted from the wild-type (WT) (E2348/69), luxS mutant (VS102), luxS complemented (VS104), qseA mutant (VS193), and qseA complemented (MPS150) strains grown in DMEM to the early exponential (EL), mid-exponential (ML), late exponential (LL), and stationary (ST) phases of growth. The graph corresponds to densitometry measurements of three spots. Error bars indicate standard deviations. (B) RNA dot blot hybridized with the 23S RNA probe. RNAs were extracted from strains grown in DMEM.
We observed that transcription of LEE5 in DMEM was downregulated in both the luxS and qseA mutants during mid-exponential-phase growth and was restored to wild-type levels upon complementation of these mutations (Fig. 1A). The luxS quorum-sensing system has been reported to be involved in gene regulation during mid-exponential-phase growth (44, 45). Our results concerning LEE5 differential regulation in the luxS mutant are in agreement with this expression profile. Transcription of LEE5 is mildly reduced only in the qseA mutant in early exponential phase. The different expression profiles for the luxS and qseA mutants suggest that this regulation is multifactorial and that qseA is one of several transcriptional factors involved in the regulation of the LEE genes.
Regulation of qseA transcription.
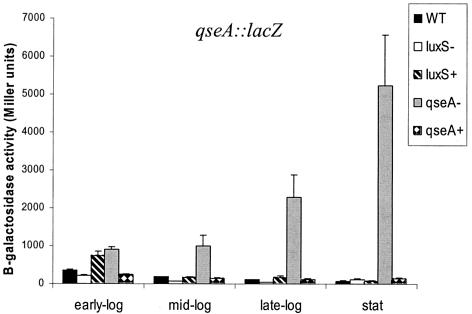
The differential regulation in a qseA and a luxS mutant could also be the result of autoregulation of qseA expression. Given that qseA belongs to the family of LysR transcriptional factors and that these regulators are known to autorepress their own expression (36), we also investigated autoregulation of qseA transcription. We monitored transcriptional regulation of qseA throughout growth in DMEM by using a qseA::lacZ fusion (Fig. 2). Transcription of qseA was diminished in the luxS mutant 2.5-fold throughout growth and was restored upon complementation of this mutation. There was no expression of qseA and no effect of a luxS mutation on qseA transcription during stationary-phase growth (transcription of qseA::lacZ was 50 Miller units, which is the same as the transcriptional background of the vector alone [data not shown]).
FIG. 2.
Quorum-sensing regulation of qseA transcription throughout growth in DMEM. β-Galactosidase assays of pVS160 (qseA::lacZ in pRS551) in the wild-type (WT) (E2348/69), luxS mutant (VS102), luxS complemented (VS104), qseA mutant (VS193), and qseA complemented (MPS150) strains were performed. Error bars indicate standard deviations.
Transcription of qseA was upregulated throughout growth in a qseA mutant (Fig. 2) and was restored upon complementation of this mutation. QseA expression was upregulated 2.5-fold in early-exponential-phase growth, 5.5-fold in mid-exponential-phase growth, 18-fold in late-exponential-phase growth, and 84-fold in stationary-phase growth in the qseA mutant. These results suggest that qseA activation of the LEE genes is restricted to early- and mid-exponential-phase growth (Fig. 1), because this is when its expression is repressed the least. During late-exponential- and stationary-phase growth, repression of qseA transcription is more pronounced. The opposite effects of a luxS mutation and a qseA mutation in the transcription of qseA suggest that fine-tuning of the amount of QseA throughout growth is important in modulating LEE gene expression.
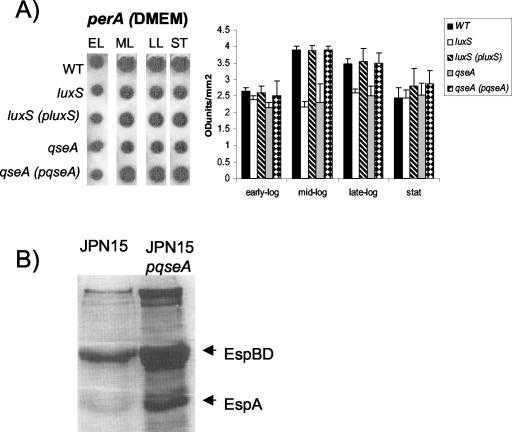
QseA activates type III secretion independent of Per.
Transcription of the LEE genes has been shown to be activated in a cascade fashion, with Per activating transcription of ler (26). We observed decreased transcription of per in both the luxS and qseA mutants in mid- and late-exponential-phase growth (Fig. 3A), confirming our previous observation, with a perA::lacZ fusion, that transcription of the per operon was positively modulated by quorum sensing (40). Given that QseA activates transcription of ler in both EPEC and EHEC (38) and that there is no Per in EHEC, we investigated whether QseA can activate type III secretion in the absence of Per. We introduced plasmid pVS241, which has the qseA gene under the control of a pBAD promoter, into the JPN15 strain (cured of the EAF plasmid and, consequently, of the per genes). We then investigated protein secretion in JPN15 in the presence and absence of pVS241, both with induction with 0.2% arabinose. Figure 3B shows that type III secretion is highly increased by QseA in the absence of Per, suggesting that QseA acts independently of Per to activate the LEE genes in EPEC.
FIG. 3.
(A) RNA slot blot hybridized with the perA probe. RNAs were harvested from the wild-type (WT) (E2348/69), luxS mutant (VS102), luxS complemented (VS104), qseA mutant (VS193), and qseA complemented (MPS150) strains grown in DMEM to the early exponential (EL), mid-exponential (ML), late exponential (LL), and stationary (ST) phases of growth. The graph corresponds to densitometry measurements of three slot blots. Error bars indicate standard deviations. (B) Coomassie blue-stained SDS-PAGE from secreted protein preparations from JPN15 and JPN15 containing plasmid pVS241 (QseA-His under control of an arabinose-inducible promoter) induced with 0.2% arabinose.
Type III secretion and AE lesion formation.
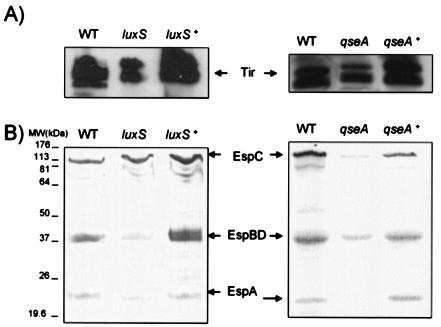
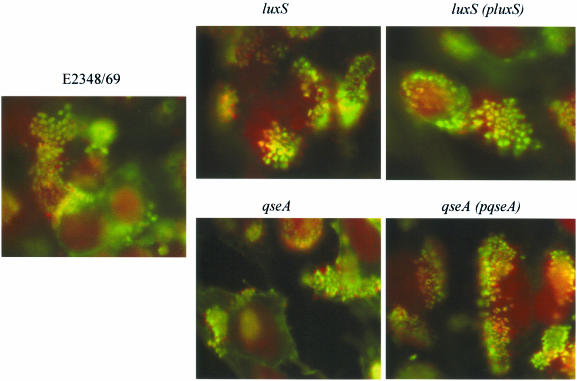
Type III secretion was diminished in both the luxS and qseA mutants and was restored upon complementation of these mutations (Fig. 4). This phenotype was expected, given that transcription of the LEE genes was decreased in both of these mutants. However, even though these mutants have decreased type III secretion, it is not eliminated, and consequently both mutants are still able to produce AE lesions on cultured epithelial cells (Fig. 5). AE lesion formation was observed by using the FAS test (22), which allows visualization of the accumulation of actin beneath and around the bacteria attached to the HeLa cells (which is a hallmark of AE lesion formation). These tests are of a qualitative nature, allowing observation only of whether a mutant is still able to produce AE lesions.
FIG. 4.
(A) Western blot analysis with Tir antiserum to probe secreted proteins prepared from the wild-type (WT), luxS mutant, luxS complemented, qseA mutant, and qseA complemented strains grown in DMEM to an OD600 of 1.0. (B) Coomassie blue-stained SDS-PAGE from secreted protein preparations from the WT, luxS mutant, luxS complemented, qseA mutant, and qseA complemented strains grown in DMEM to an OD600 of 1.0.
FIG. 5.
FAS test of the wild-type, luxS mutant, luxS complemented, qseA mutant, and qseA complemented strains incubated with HeLa cells for 3 h. The bacteria were stained red with propidium iodide, and the cytoskeleton was stained green with fluorescein isothiocyanate-phalloidin.
Localized adhesion in epithelial cells.
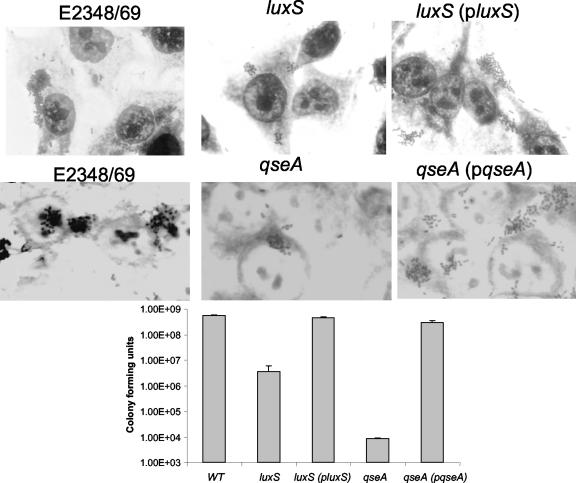
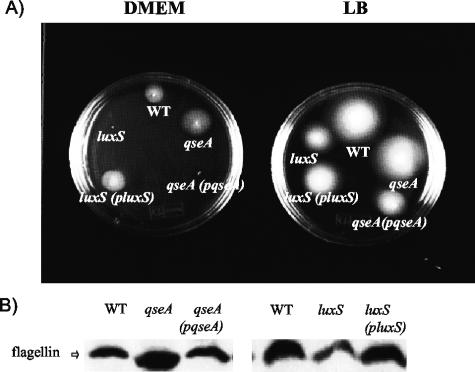
Besides AE lesion formation, one of the hallmarks of EPEC infection is its pattern of adherence to epithelial cells forming microcolonies (34). Efficient microcolony formation is multifactorial and involves expression of the LEE genes (most importantly, those for intimin and Tir), BFP, and flagella (11, 12, 17, 20). Both the luxS and qseA mutants formed smaller microcolonies than the wild type and complemented strains on cultured HeLa cells (Fig. 6). Adherence of the luxS mutant was two orders of magnitude lower than those of the wild-type and complemented strains. The qseA mutant had an even more pronounced defect in adherence, with adherence four orders of magnitude less than those of the wild-type and complemented strains (Fig. 6). These results are consistent with the lower transcription of the LEE genes (LEE5 encodes both Tir and intimin) (Fig. 1) and the diminished Tir secretion in these mutants (Fig. 4). We did not observe any difference in transcription of bfpA or in BFP expression in either of the mutants (data not shown). Expression of flagella was reduced in a luxS mutant and increased in a qseA mutant (Fig. 7B). In agreement with this phenotype, the luxS mutant showed reduced motility and the qseA mutant showed increased motility in both DMEM and LB medium (Fig. 7A). Increased motility in LB in the qseA mutant is not very apparent because of saturation; the qseA mutant and the wild-type strain halos are already touching each other. These phenotypes suggest that successful microcolony formation and adhesion are dependent in the correct timing and dosage of flagellar and LEE gene expression.
FIG. 6.
Adherence of the wild-type (WT), luxS mutant, luxS complemented, qseA mutant, and qseA complemented strains to HeLa cells after 3 h of incubation. The graph represent CFU of adherent bacteria. Error bars indicate standard deviations.
FIG. 7.
(A) Motility of the wild-type (WT), luxS mutant, luxS complemented, qseA mutant, and qseA complemented strains in DMEM (48 h) and LB medium (16 h). (B) Western blot analysis of whole-cell lysates grown in LB medium to an OD600 of 1.0 and probed with polyclonal antiflagellum antiserum.
DISCUSSION
EPEC colonizes the proximal small intestine and causes profuse and persistent watery diarrhea lasting up to 120 days (8, 31). EPEC pathogenesis has several steps. First the bacteria adhere to the intestinal epithelial cells, probably through the EspA filament, which is secreted by the LEE-encoded TTSS (17, 23). Tir then is translocated through the LEE-encoded TTSS and inserts itself into the mammalian cell membrane, where it serves as the intimin receptor allowing the intimate attachment characteristic of AE lesion formation (20). Other EPEC cells then interact with each other, forming large microcolonies (17). The successful formation of these microcolonies requires BFP and flagella (11, 12). The present knowledge about EPEC pathogenesis suggests that expression of EPEC virulence genes is dependent on the concerted action of several regulatory factors.
In this paper we describe a detailed study of quorum-sensing regulation of virulence gene expression in EPEC. We have previously reported that transcription of the LEE genes from EPEC within an E. coli K-12 background was activated through quorum sensing when preconditioned medium was used (40). Here we monitored transcriptional regulation of the LEE genes, per, qseA, and bfpA in an EPEC background throughout growth. Our results demonstrated that transcription of the LEE5 operon was diminished only in mid-exponential phase in the luxS mutant (Fig. 1) and in the early- and mid-exponential growth phases in the qseA mutant. One explanation for this differential regulation can be inferred from the observations that QseA autorepresses its own transcription (Fig. 2) and that this autorepression is more pronounced during late-exponential- and stationary-phase growth. Furthermore, there is also activation of the LEE genes through Per (1, 24, 26).
The differential patterns of gene expression observed for a luxS mutant and a qseA mutant suggest that there are additional transcriptional factors involved in this regulation. Thus, this regulation is multifactorial. QseA is one of several transcriptional factors involved in the fine-tuning of LEE gene expression, and we have preliminary data suggesting the involvement of at least two other novel transcriptional factors in this regulation. The concerted action of several transcriptional factors is what one observes in a luxS mutant.
In contrast to that for EHEC, type III secretion in EPEC is diminished but never eliminated in a luxS mutant (Fig. 4) (42). This differential regulation can be explained by the additional control of the LEE genes through Per, which is absent in EHEC (26). In EPEC, GadX also represses transcription of the LEE genes through Per at acidic pHs (possibly when EPEC is crossing the stomach) and activates their transcription at alkaline pHs (possibly when EPEC reaches the small intestine) (37). EPEC has to coordinate transcription of the LEE genes with microcolony formation. This is when quorum-sensing regulation may play an active role. Transcription of the LEE genes, as well as flagellation and motility, is altered in both luxS and qseA mutants (Fig. 1 and 6). These mutants exhibit opposite phenotypes with regard to flagellar regulation. Flagellation and motility are diminished in a luxS mutant, while they are augmented in a qseA mutant. We have recently described another regulator in the quorum-sensing cascade, QseBC, which is involved in activation of the flagellar regulon (43). QseBC activates transcription of flhDC (the master flagellar regulator) and, consequently, of the whole regulon. The regulatory region of flhDC is identical in EHEC and EPEC, suggesting that this regulation will be conserved. Therefore, the opposite flagellar regulation phenotypes of the luxS and qseA mutants may again be the result of multifactorial genetic control. Disruption of quorum-sensing signaling affects expression of the LEE genes and the flagellar regulon, thereby interfering with microcolony formation and adherence to epithelial cells (Fig. 6 and 8).
FIG. 8.
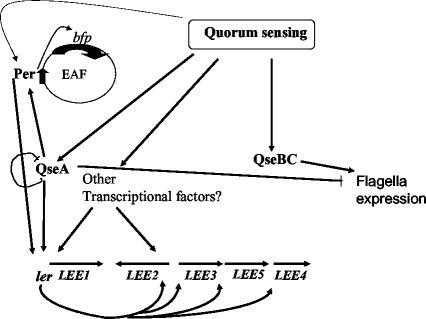
Schematic representation of virulence gene regulation by quorum sensing in EPEC. Ler activates transcription of the LEE genes. Transcription of ler is activated by quorum sensing through QseA and other, yet-unidentified transcriptional factors. QseA autorepresses its own transcription. Transcription of bfp is activated by Per, and transcription of per is autoactivated by Per and positively modulated by QseA. Quorum-sensing regulation of the flagellar regulon is activated by QseBC and repressed by QseA.
Quorum-sensing regulation is different in EPEC and EHEC. EPEC colonizes the proximal small intestine, which is though to have very little or no resident flora. Therefore, while quorum sensing is primarily an interspecies signaling system during EHEC infection, it seems to be used for intraspecies signaling during EPEC infection. Another remarkable difference between EPEC and EHEC regarding quorum-sensing regulation involves QseA. In EHEC, QseA activates transcription of the LEE genes but is not involved in the regulation of the flagellar regulon (38). In EPEC, QseA clearly represses flagellation and motility (Fig. 7). This difference may also be reflective of the differential role played by flagella in EPEC and EHEC pathogenesis. While flagella in EHEC are used mostly for swimming, they are involved in adherence and microcolony formation in EPEC (12), thus causing the need to coordinate transcription of the LEE genes with flagellation.
These studies suggest that disruption of quorum-sensing regulation can have pleiotropic effects on EPEC pathogenesis, altering adherence, type III secretion, and flagellation. A better understanding of quorum-sensing regulation in EPEC will provide invaluable insights into the disease caused by this bacterium.
Acknowledgments
We thank Jorge Giron (University of Arizona Medical College) for providing anti-EspA, anti-BFP, and anti-FliC polyclonal antisera. We thank Kevin McIver (University of Texas Southwestern Medical Center) for technical assistance with the RNA slot blotting and critical review of the manuscript. We also thank Eric Hansen (University of Texas Southwestern Medical Center) and Alfredo Torres (University of Texas Medical Branch) for critical review of the manuscript.
This work has been supported by start-up funds from the University of Texas Southwestern Medical Center and NIH grants AI054468 and AI053067. M.P.S. was supported by a fellowship from Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (FAPESP) grant 00/02844-1.
Editor: V. J. DiRita
REFERENCES
- 1.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 2.Chen, X., S. Schauder, N. Potier, A. Van Dorssealaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 3.Cravioto, A., R. J. Gross, S. M. Scotland, and B. Rowe. 1979. An adhesive factor found in strains of Escherichia coli belonging to the traditional infantile enteropathogenic serotypes. Curr. Microbiol. 3:95-99. [Google Scholar]
- 4.de Kievit, T. R., and B. H. Iglewski. 2000. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 68:4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1189. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagundes-Neto, U. 1996. Enteropathogenic Escherichia coli infection in infants: clinical aspects and small bowel morphological alterations. Rev. Microbiol. 27:117-119. [Google Scholar]
- 9.Friedberg, D., T. Umanski, Y. Fang, and I. Rosenshine. 1999. Hierarchy in the expression of the locus of enterocyte effacement genes of enteropathogenic Escherichia coli. Mol. Microbiol. 34:941-952. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 11.Giron, J. A., A. S. Ho, and G. K. Schoolnik. 1991. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science 254:710-713. [DOI] [PubMed] [Google Scholar]
- 12.Giron, J. A., A. G. Torres, E. Freer, and J. B. Kaper. 2002. The flagella of enteropathogenic Escherichia coli mediate adherence to epithelial cells. Mol. Microbiol. 44:361-379. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg, M. D., M. Johnson, J. C. Hinton, and P. H. Williams. 2001. Role of the nucleoid-associated protein Fis in the regulation of virulence properties of enteropathogenic Escherichia coli. Mol. Microbiol. 41:549-559. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, A. J., M. Farris, P. Alefounder, P. H. Williams, M. J. Woodward, and C. D. O'Connor. 2003. Co-ordination of pathogenicity island expression by the BipA GTPase in enteropathogenic Escherichia coli (EPEC). Mol. Microbiol. 48:507-521. [DOI] [PubMed] [Google Scholar]
- 16.Haack, K. R., C. L. Robinson, K. J. Miller, J. W. Fowlkes, and J. L. Mellies. 2003. Interaction of Ler at the LEE5 (tir) operon of enteropathogenic Escherichia coli. Infect. Immun. 71:384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hicks, S., G. Frankel, J. B. Kaper, G. Dougan, and A. D. Phillips. 1998. Role of intimin and bundle-forming pili in enteropathogenic Escherichia coli adhesion to pediatric intestinal tissue in vitro. Infect. Immun. 66:1570-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 21.Knutton, S., M. M. Baldini, J. B. Kaper, and A. S. McNeish. 1987. Role of plasmid-encoded adherence factors in adhesion of enteropathogenic Escherichia coli to HEp-2 cells. Infect. Immun. 55:78-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Laguna, Y., E. Calva, and J. L. Puente. 1999. Autoactivation and environmental regulation of bfpT expression, the gene coding for the transcriptional activator of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 33:153-166. [DOI] [PubMed] [Google Scholar]
- 25.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nealson, K. H., T. Platt, and J. W. Hastings. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothbaum, R., A. J. McAdams, R. Giannella, and J. C. Partin. 1982. A clinicopathologic study of enterocyte-adherent Escherichia coli: a cause of protracted diarrhea in infants. Gastroenterology 83:441-454. [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Sanchez-SanMartin, C., V. H. Bustamante, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of the orf19 gene and the tir-cesT-eae operon of enteropathogenic Escherichia coli. J. Bacteriol. 183:2823-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scaletsky, I. C., M. L. Silva, and L. R. Trabulsi. 1984. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 45:534-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 36.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 37.Shin, S., M. P. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 38.Sperandio, V., C. C. Li, and J. B. Kaper. 2002. Quorum-sensing Escherichia coli regulator A (QseA): a regulator of the LysR family involved in the regulation of the LEE pathogenicity island in enterohemorrhagic Escherichia coli. Infect. Immun. 70:3085-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sperandio, V., J. L. Mellies, R. M. Delahay, G. Frankel, J. A. Crawford, W. Nguyen, and J. B. Kaper. 2000. Activation of enteropathogenic Escherichia coli (EPEC) LEE2 and LEE3 operons by Ler. Mol. Microbiol. 38:781-793. [DOI] [PubMed] [Google Scholar]
- 40.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 100:8951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sperandio, V., A. G. Torres, and J. B. Kaper. 2002. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol. Microbiol. 43:809-821. [DOI] [PubMed] [Google Scholar]
- 44.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 46.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 48.Tzipori, S., I. K. Wachsmuth, C. Chapman, R. Birden, J. Brittingham, C. Jackson, and J. Hogg. 1986. The pathogenesis of hemorrhagic colitis caused by Escherichia coli O157:H7 in gnotobiotic piglets. J. Infect. Dis. 154:712-716. [DOI] [PubMed] [Google Scholar]