Abstract
Introduction
The molecular links between shock-response and adaptation remain poorly understood, particularly for extremophiles. This has hindered rational engineering of solvent tolerance and correlated traits (e.g., productivity) in extremophiles. To untangle such molecular links, here we established a model that tracked the microevolution from shock to adaptation in thermophilic bacteria.
Method
Temporal dynamics of genomes and transcriptomes was tracked for Thermoanaerobacter sp. X514 which under increasing exogenous ethanol evolved from ethanol-sensitive wild-type (Strain X) to tolerance of 2%- (XI) and eventually 6%-ethanol (XII). Based on the reconstructed transcriptional network underlying stress tolerance, genetic engineering was employed to improve ethanol tolerance and production in Thermoanaerobacter.
Results
The spontaneous genome mutation rate (μg) of Thermoanaerobacter sp. X514, calculated at 0.045, suggested a higher mutation rate in thermophile than previously thought. Transcriptomic comparison revealed that shock-response and adaptation were distinct in nature, whereas the transcriptomes of XII resembled those of the extendedly shocked X. To respond to ethanol shock, X employed fructose-specific phosphotransferase system (PTS), Arginine Deiminase (ADI) pathway, alcohol dehydrogenase (Adh) and a distinct mechanism of V-type ATPase. As an adaptation to exogenous ethanol, XI mobilized resistance-nodulation-cell division (RND) efflux system and Adh, whereas XII, which produced higher ethanol than XI, employed ECF-type ϭ24, an alcohol catabolism operon and phase-specific heat-shock proteins (Hsps), modulated hexose/pentose-transport operon structure and reinforced membrane rigidity. Exploiting these findings, we further showed that ethanol productivity and tolerance can be improved simultaneously by overexpressing adh or ϭ24 in X.
Conclusion
Our work revealed thermophilic-bacteria specific features of adaptive evolution and demonstrated a rational strategy to engineer co-evolving industrial traits. As improvements of shock-response, stress tolerance and productivity have been crucial aims in industrial applications employing thermophiles, our findings should be valuable not just to the production of ethanol but also to a wide variety of biofuels and biochemicals.
Keywords: Shock, Adaptation, Ethanol, Microevolution, Thermophile
Introduction
Adaptive evolution is a universal theme of life on our planet [1-3]. It is also a widely practiced strategy for selecting and engineering economically valuable traits [4-6]. Adaptive evolution typically starts from an environmental change and results in genetically inheritable adaptation [3,7]. The initial cellular response, which usually includes a transient reprogramming of cellular activities, is termed “shock”, while the subsequent cellular state that involves inheritable traits resulted from long-term exposure and selection (i.e., after generations) is termed “adaptation”. Numerous studies have investigated the cellular programs underlying shock (e.g., Escherichia coli[5,8,9]; Saccharomyces cerevisiae[10,11] and Clostridium acetobutylicum[12,13]) or adaptation [6,11,14-21], but few have attempted to test their links by tracking the temporal development from shock-response to the eventual adaptation [10,22,23]. The vast numbers of genetic variables (e.g., strains used) and environmental variables (e.g., culture conditions) across the studies hampered meaningful comparisons between shock- and adaptation-responses. Thus the molecular links between shock and adaptation are not yet well established, and the temporal characteristics and genetic mechanisms defining the shock-to-adaptation process remain elusive [10,11]. Moreover, whether and how the process was shaped by ecological parameters such as temperature is largely unknown.
Although most contemporary life forms are found at a narrow range of 24-40°C [24], the thermophiles thrive under optimal temperature of >50°C [24]. These organisms play a profound role in evolution and ecology of our biosphere (primordial life on earth is believed by many to be thermophilic [2]). They have also found wide applications in biotechnology. For example, thermophilic gram-positive anaerobes (TGPAs) such as certain Thermoanaerobacter and Clostridium species are of interest in producing solvents (e.g., ethanol, butanol and isopropanol) from lignocelluloses under a Consolidated Bioprocessing (CBP) scheme [25,26], due to their thermophilic nature (60-65°C), wide spectrum of carbon-sources [27,28] and co-utilization of pentose and hexose [25]. However, as is the case in many mesophiles, TGPAs are generally sensitive to excessive concentrations of their own solvent products [27], which reduce cell vitality, impair membrane integrity, inhibit enzymes and/or perturb intracellular pH balance [29,30]. Cellular tolerance to solvents can be derived by adaptive evolution of the wild-type strains via exposure to exogenous solvents for months or even years [30,31]. However, solvent-tolerant strains derived via such a strategy usually exhibit lower productivity of the solvent (although this reduction in yield was not proportional; [4,31]); this negative correlation between tolerance and productivity represents a major hurdle in strain development. In fact, although for many organisms genetic engineering of either solvent tolerance (e.g., [4,6]) or solvent productivity (e.g., [25,27,32]) has been accomplished, simultaneous improvement of solvent tolerance and productivity has not been demonstrated in thermophiles [4,33]. Devising a rational strategy to counter this hurdle might require a mechanistic understanding of the co-evolution of such linked traits.
Here we developed a model of adaptive evolution for thermophiles by evolving, under increasing concentrations of exogenous ethanol, a Thermoanaerobacter sp. X514 (NCBI Taxonomy ID: 399726) lineage from the ethanol-sensitive wild-type to tolerance of 2% ethanol, and eventually to 6%-ethanol tolerance. The genomes, transcriptomes and gene networks were traced and compared on a temporal scale, which unveiled the molecular links between shock and adaptation and revealed unusual features of “thermophilic” adaptive evolution. Furthermore, these findings enabled us to demonstrate that for wild-type TGPA strains, the two linked and co-evolving traits of ethanol productivity and ethanol tolerance could be simultaneously improved by genetic approaches (overexpressing an iron-containing Adh enzyme (Teth5140145-0146) or an ECF subfamily RNA polymerase sigma-24 factor regulator (ϭ24, Teth5141847-1848)).
Results
An experimental model of adaptive evolution from shock to adaptation in thermophilic bacteria
Thermoanaerobacter sp. X514 is a TGPA we previously used for dissecting the mechanism of pentose-hexose co-utilization [25] and ethanol production [34]. We first designed a trackable experimental model of adaptive evolution for thermophiles (Figure 1A and 1B) by exploiting the observed ethanol-sensitivity of wild-type X514 (designated “X”; Additional file 1A) to the mutant strain XI (“low ethanol tolerance”, i.e., tolerating 2% ethanol; Additional file 2A; Methods) that was one isolate of Xp (a mixed culture of mutants that were developed from X via sequential transfer) and eventually to another mutant strain XII (“high-ethanol tolerance”, i.e., tolerating 6% ethanol; Additional file 2A; Methods). The three developmental phases of ethanol tolerances represented by X, XI (and Xp) and XII were interrogated under three “Views” (Figure 1A): (i) Phenotypic adaptation (in X, XI and XII) (View I; Additional file 2). (ii) Transcriptomic responses that respectively defined four time-points under ethanol shock and the two phases of ethanol adaptation (View II; Figures 2 and Figure 3). Under the particular culture medium, the ethanol concentration that caused stress but not significant cell death in X was 0.15% (v/v), which was selected for the ethanol shock assay (Additional file 1A). Thus, subsequent shock responses were examined by comparing X-0.15% (X cells cultured at defined medium with 0.15% exogenous ethanol; View IIA) to X-0% (X cells cultured without exogenous ethanol); moreover, dynamics of shock responses was tested by sampling X transcriptomes at four time points (0.5 h, 1 h, 2 h and 4 h) upon ethanol exposure. On the other hand, mechanism of ethanol tolerance was revealed by the transcriptomes of XI (XI-0% and XI-2%; XI cells cultured under 0% and 2% ethanol respectively) and those of XII (XII-0% and XII-6%; XII cells cultured under 0% and 6% ethanol respectively; View IIB). In addition to microarray-based expression profiling, RNA-Seq was employed so as to detect all structural and sequence changes of transcripts during the microevolution. (iii) Genome sequence mutations (for X, Xp, XI and XII) (View III; Figure 2). To precisely identify every genotypic change along the microevolution, we resequenced the X, XI and XII genomes and the metagenome of Xp, using the previously finished wild-type X514 genome [34] as reference.
Figure 1.
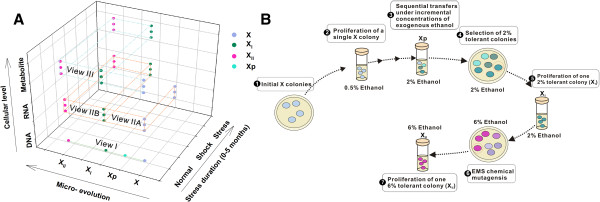
Experimental strategy tracking the genotypes and phenotypes along the microevolution from ethanol shock to tolerance. (A) Overview of experimental design. The three biological replicates for each sampled condition were indicated as dots. (B) Experiments to derive Xp (XI) and XII that tolerated 2% and 6% (v/v) ethanol respectively. EMS: ethylmethane sulfonate. Xp: the mixed culture which grew under 2% ethanol and from which XI was isolated.
Figure 2.
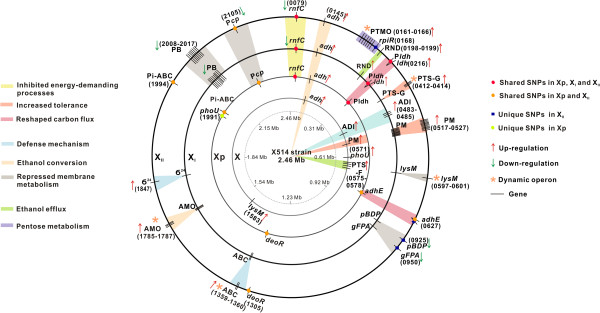
Genomic and transcriptomic events defining each developmental phases over the complete time-course. Key mutated genes, gene with significantly changed expression levels and dynamic operons were illustrated. Pldh: the promoter of lactate dehydrogenase; PTMO: pentose transport and metabolism operon; PM: purine metabolic genes; PTS-G/F: glucose/fructose specific PTS system; pBDP: peptidoglycan binding domain-containing protein; gFPA: glucosamine--fructose-6-phosphate aminotransferase; AMO: alcohol metabolism operon; Pi-ABC: phosphate ABC transporter; PB: peptidoglycan biosynthesis; Pcp: the promoter of Serine-type D-Ala-D-Alacarboxypeptidase. IDs of the corresponding genes in X514 were shown.
Figure 3.
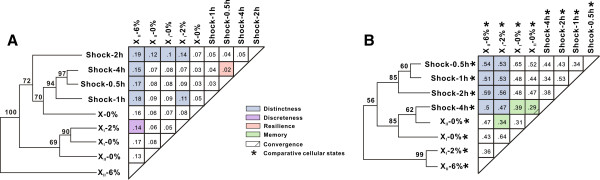
Links among the cellular states of shock and tolerance as defined by transcriptome. (A) Links among the nine cellular states. (B) Links among the eight relative cellular states. The number in each square represented the distance between each pair-wise comparison as calculated based on Pearson Correlation. Hierarchical clustering of the nine cellular states and the eight relative cellular states were shown respectively. Numbers on the branches represent bootstrap values as percentages of 1000 resampling efforts of the dataset. The clustering analysis was performed by TM4 software [57].
Higher concentrations of exogenous ethanol slowed down growth in all strains (Additional file 2A), with reduction in cell length correlated with improvements in tolerance (Additional file 3). Glycolysis in the tolerant mutants was as robust as that in the wild-type yet the carbohydrate metabolism was altered, as in the absence of exogenous ethanol, XI and XII consumed more glucose (100:120:115 for X: XI: XII) yet produced less ethanol (100:45:70 for X: XI: XII) than X (Additional file 2B).
In Xp, XI and XII genomes (in reference to X; Methods), 76 (74 single-nucleotide polymorphisms (SNPs) and two single-base indels), 20 (19 SNPs and one single-base indel) and 45 (43 SNPs and two single-base indels) mutations were respectively found (Figure 2; Additional file 4). Along the microevolution, the ratio between non-coding sequence mutations and those in coding sequence stayed largely unchanged (from 17.6% in XI to 15.4% in XII); moreover the functional profile of non-synonymous mutations shifted from regulatory functions to structural proteins (e.g., membrane and sugar phosphate metabolisms; Additional file 4). On the other hand, in X, XI and XII transcriptomes, expression-altered genes were mostly concentrated in amino acid metabolism, cell motility, coenzyme metabolism, carbohydrate metabolism and energy production (Additional file 5). Along the microevolution, the functional profile of expression-altered genes shifted from one mainly featuring amino acid metabolism and cell motility to one primarily of energy production, suggesting ethanol adaptation but not shock compromised energy generation. Together, the microevolution featured decreasing numbers of expression-altered genes (314 genes under XI-0% vs X-0% and 189 genes under XII-0% vs X-0% were significantly changed) yet increasing numbers of DNA-sequence changes (20 mutations in XI and 45 in XII), suggesting a temporal shift from changes in gene expression to changes in genome sequence (Figure 2, Additional file 5 and Additional file 4). Interestingly, expression alteration and DNA-sequence changes exhibited complementary functional landscapes. In XI, the former were mainly found in amino acid, coenzyme and carbohydrate metabolisms, yet the latter mainly involved regulatory functions (transcription factors (TFs) and cis-regulatory elements) (Figure 2 and Additional file 5). In XII, expression alteration mainly involved carbohydrate metabolism, energy production and amino acid metabolism, while DNA-sequence changes mainly involved membrane and sugar phosphate metabolism genes and TFs (Figure 2 and Additional file 5).
Comparison between XI and X genomes revealed a spontaneous mutation rate per genome per generation (μg) of 0.045 given 440 generations. For another thermophile Thermusthermophilus, under its optimal growth condition, a μg of 0.00093 was proposed based on mutation reporter gene pyrEF[35], which was nearly 10-fold lower than the mesophile E.coli (0.0048); the much lower μg in thermophiles (mean μg 0.00079, range 1.4-fold) than mesophiles (mean μg 0.0040, range 2.9-fold) was interpreted as due to the rapid accumulation of deleterious mutations at a temperature only 5-10oC higher [35]. However, under ethanol stress, our experimentally estimated μg of 0.045 for X514 appeared to be two orders of magnitude higher than that of optimal-growth Thermusthermophilus and actually slightly higher than that of E.coli under isobutanol stress (0.026) [5], contradicting with the current notion [35].
Relationship among transcriptomic programs that respectively underlie shock, low-tolerance and high-tolerance
Relationship among the four shock-stages (X-0.15% at 0.5h, 1h, 2h and 4h upon exogenous-ethanol exposure) and the four adaptation states (XI-0%, XI-2%, XII-0% and XII-6%) were unveiled by pair-wisely comparing the nine transcriptomic states (including X-0%; each state in biological triplicates; Figure 3A; Methods). Three observations were apparent. (i) Shock-response and adaptation were distinct in nature (i.e., “Distinctness”), as mutant states (XI and XII) and X states formed two separate clusters, with large pairwise distances between them (mostly over 0.1). (ii) Adaptation was phased (i.e., “Discreteness”), as XII-6% formed an independent clad with a large pairwise distance (0.14) between XII-6% and XI-2%. (iii) For both shock-response and adaptation, their temporal development exhibited a certain degree of “Resilience”. From the onset of environmental change, the transcriptomes were first altered to a distinct state but then returned to one that was closer to the original state (Figure 3A, Additional file 1B and Additional file 5): for example, in shock, X-4 h was closer than both X-1 h and X-2 h to X-0.5 h; similarly, in adaptation, XII-0% was closer to X-0% than XI-0%.
Furthermore, to distinguish between environmental and genetic effects, the relative transcriptomic changes that included the six “normalized” transcriptomes due to environmental perturbation (X-0.15% vs X-0% at each of 0.5 h, 1 h, 2 h and 4 h; XI -2% vs XI-0%; XII-6% vs XII-0%) and the other two caused by genetic changes (XI -0% vs X-0%; XII-0% vs X-0%) were pair-wisely compared (Figure 3B; Methods). Two findings emerged. (i) Changes of the cellular states corresponding to the evolving tolerance exhibited a degree of “Memory” left by a priori exposure to the stimuli, as demonstrated by a) the similarity between XI-0% (vs X-0%) and X-0.15%-4h (vs X-0%-4h), and b) the similarity between XII-0% (vs X-0%) and XI-2% (vs XI-0%) (Figure 3B). (ii) The shock and adaptation programs appeared progressing towards a shared destiny of cellular state, as the shortest pair-wise distance (at 0.296) among the 28 such distances was actually found between the high-tolerance stage (XII-0% (vs X-0%)) and the wild-type under the most extended shock (X-0.15%-4h (vs X-0%-4 h)) (Figure 3B). This suggested “Convergence”, where the high-tolerance cells, in the absence of stimuli, retained certain transcriptomic features of those under extended shock.
A dynamic yet coordinated response to ethanol shock (View IIA)
Ethanol-shock networks
How gene networks of thermophiles respond to environmental stimuli has been poorly understood [36]. For X, two genome-wide gene co-expression networks respectively characterizing ethanol-shock cells (ES+) and control cells (ES-) were constructed via co-expression analysis and then compared (Methods; Additional file 6). ES+ (216 nodes; 45 of them were hypothetical proteins representing new components of ethanol-shock response; Part I of Additional file 7; Additional file 6) was 23.7% smaller than ES- (283 nodes), with eleven modules each of at least five nodes found in each network (module sizes vary substantially both within and between networks, ranging from 5 to 93 nodes). There were 30 ES+–specific (e.g., small multi-drug export, channel protein, cell wall hydrolase and ethanolamine utilization protein EutN) and 97 ES-–specific nodes (electron complex and ribosomal proteins), suggesting repression of energy metabolism and protein translation and activation of detoxification under shock.
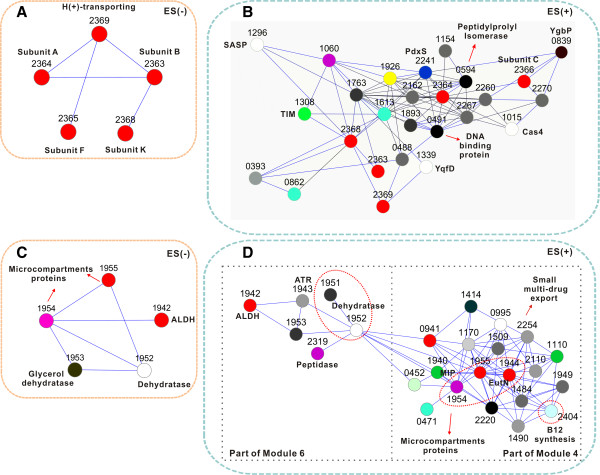
Moreover, for the 186 nodes shared between ES+ and ES-, inter-node relationships were distinct. Among the top twenty such nodes with the most connections in ES+, several were known to play pivotal roles in ethanol shock. The first group was V-type ATPase, which maintains intracellular pH homeostasis in S. cerevisiae upon ethanol shock [11]. In ES-, these genes (Teth5142363-2365 and Teth5142368-2369) constituted a module (Module 10) free of inter-module links (Figure 4A and Additional file 6; also observed in our recently reported Thermoanaerobacter glycobiome network [25]). However, in ES+, these genes became connected with other genes; not only were they found in the largest module (Module 1; with 59 nodes) (Additional file 6), but also directly linked to genes protecting cells from organic solvent damages or related to membrane structure (Figure 4B; Part I of Additional file 7).
Figure 4.
Gene co-expression network of the wild-type strain under ethanol shock.(A) The sub-module of V-type ATPase under control (ES-). (B) The sub-module of V-type ATPase under ethanol shock (ES+). Only the first neighbors (genes directly connected to the V-type ATPase genes) were shown. (C) The sub-module of genes involved in defense mechanism under control (ES-). (D) The sub-module of genes involved in defense mechanism under ethanol shock (ES+). Color code was as in Additional file 6. Blue lines indicated positive correlation.
The second group consisted of genes that metabolize or exclude toxic compounds. In ES-, Module 11 (5 genes) was free of connections with other modules, indicating these five genes (aldehyde dehydrogenase (Teth5141942, aldh), microcompartment proteins (Teth5141954-1955) and dehydratases (Teth5141952-1953) (Figure 4C and Additional file 6) formed a single functional unit to metabolize toxic intermediates [37]. Interestingly, in ES+, the unit expanded to include 25 genes organized into two modules (Module 4 and 6) (Figure 4D and Additional file 6): Aldh converts acetaldehyde (the toxic product of ethanol oxidation) to acetate; Teth5142404 participates in vitamin B12 synthesis; Teth5141943 (ATP-cobalaminadenosyltransferase; converting B12 to coenzyme B12) provides the essential coenzyme to B12-dependent microcompartment protein (Teth5141944); the latter and additional such proteins (Teth5141954-1955) remove extra aldehyde through major intrinsic protein (MIP) channel (Teth5141940); multi-drug exporter (Teth5142254) excludes toxic chemicals; peptidase S51 (Teth5142319) degrades misfolded proteins and prevents their accumulation. Several (Teth5141940, Teth5141943-1944 and Teth5142254) of these genes were absent in ES-.
Dynamics of ethanol-shock networks
Transcriptomes respectively sampled at 0.5 h, 1 h, 2 h and 4 h upon ethanol-shock revealed a total of 520 genes differentially expressed (X-0.15% vs X-0%) in at least one time-point (|log2R| ≥1; Additional file 8). Their expression patterns formed ten temporal clusters (Additional file 9) that include both stimulatory and inhibitory responses. An iron-containing adh (Teth5140145; one of the nine adh genes in X514 genome) and ADI Pathway (Cluster 9) [25,38] were the earliest tide, peaking at 0.5h and then quickly subduing. The second (Cluster 7) surged at 1h, among which was the small acid-soluble spore protein gene (Teth5141739) that protects DNA backbone from chemical and enzymatic cleavage. The third (Cluster 3) was induced at 2 h and included lysM (Teth5141583) which encodes a general peptidoglycan binding function and resists ethanol damage [39]. Cluster 1, consisting of purine metabolism, fatty acid metabolism and fructose PTS genes, maintained upregulation within 2 h. On the other hand, several clusters were downregulated. Amino acid metabolism genes (valine, arginine and tyrosine; Cluster 2) represented the earliest inhibited genes (from 0.5 h to 4 h), followed by histidine metabolism and carbohydrate transport regulators (Cluster 5; inhibited from 1 h to 4 h). Subsequently, dipeptide transport, ion transport, carbohydrate transport and flagella synthesis fell during 2 h to 4 h (Clusters 4 and 6; although Cluster 4 later restored to the control level), followed by ribosomal proteins, DNA replication and carbohydrate metabolism genes (Clusters 8 and 10) that were repressed at 4 h. Prolonged ethanol exposure extending from 0.5 h to 4 h resulted in dramatic increase of downregulated genes (from 37 to 350 genes; 14.1% of genome) and decrease of upregulated genes (from 40 to 4 genes) (Additional file 1B).
Therefore, upon ethanol shock, X mobilized a highly dynamic yet coordinated program organized in ten temporal clusters and involved dozens of upregulated genes (Additional file 9), however it was not clear whether any among them contributed to ethanol tolerance. Thus we next examined the 2%- and 6%-ethanol-tolerant mutants.
Adaptation strategy of the low-tolerance mutant
Genome mutations in XI and Xp
Mutations can be beneficial, neutral or deleterious [1]. In XI, 20 SNPs were identified that included 17 SNPs (6 nonsense and 11 missense) in coding and 3 in non-coding sequences (Additional file 4). Among them, six SNPs in coding regions were synonymous and thus likely neutral. To distinguish beneficial SNPs in XI, we sequenced the metagenome of Xp to a depth equivalent to 140 sequence coverage of X514 genome, the pool of mutants that tolerated 2% ethanol, reasoning that non-synonymous mutations with higher mutation frequency (percentage of mutated reads to all reads at a single-base locus in the X514 mutant community) were more beneficial (Additional file 10B).
Three such XI-mutations were present in Xp (Part II of Additional file 7) with high frequency (>80%), implicating energy conversion (COG C, electron transport complex I (Teth5140079) and lactate dehydrogenase (Ldh, Teth5140216)) and ion transport process (COG P, TrkH family potassium uptake protein (Teth5140140)) (Figure 2, Additional file 10C, Additional file 11 and Additional file 12; Part II of Additional file 7). Among them, one insertion mutation was found between the -10 box and the -35 box of the predicted promoter of an ldh (Teth5140216) that catalyzes lactate formation (Figure 2 and Additional file 11). The distance (but not the sequence) between these two conserved elements is crucial for regulating gene expression [40]; thus this regulatory adaptation likely led to ldh upregulation which was consistent with the increased ldh expression level (Figure 2) and then elevated lactate production in XI (Additional file 2B).
A priori ethanol stress reshaped gene network of the cell
Comparison between XI-0% and X-0% revealed that a priori long-term ethanol exposure reshaped metabolisms, even in the absence of exogenous ethanol (Additional file 13A and Additional file 14B). First, central carbon metabolisms were transcriptionally altered. Corresponding to the one insertion mutation in promoter, one ldh (Teth5140216; the only ldh in X514 genome) was induced, whereas solvent formation genes were inhibited that included butyrate kinase (Teth5140936), phosphate butyryltransferase (Teth5140937), and iron-containing adh and its associated NADH oxidase (Teth5140145-0146) (Additional file 15A). In addition, several glycolysis and pentose phosphate pathway (PPP) genes (Teth5140161, Teth5140163-0165, Teth5140575-0576 and Teth5141896; Additional file 15A) were induced. Thus glycolysis in tolerant mutants was robust with carbon flux shifting from ethanol- to lactate-deriving, explaining the increased glucose consumption and elevated lactate production (Additional file 2B). Second, additional solvent formation genes were suppressed in mutants. For example, B12 dependent euts (Teth5141943-1946), whose expression level positively correlates with ethanol production in X [25], were downregulated in XI, likely contributing to the decrease in ethanol titer (Additional file 2B). Third, coenzyme B metabolism was either up- (B1, B2 and B5) or down-regulated (B12; Additional file 15A). B12 biosynthesis (Teth5140298-0320) and related cobalt transporters (Teth5141931-1934 and Teth5140297-0326), which provide coenzyme B12 to ethanol formation genes [25], were all downregulated, leading to the lower ethanol production (Part III of Additional file 7). Fourth, genes in several stress-response pathways [10,41] were upregulated in XI (Part III of Additional file 7). Among them was up-regulation of NAD/NADP octopine dehydrogenase (Teth5142108; Additional file 15A). This multifunctional enzyme catalyzed the reversible reductive condensation of arginine and pyruvic acid to D-octopine [42]. The arginine can protect cells against ethanol damage [25]. On the other hand, the octopine dehydrogenase activity was significantly correlated with the ability to buffer the acidic end products of anaerobic metabolism in the marine invertebrate cephalopods [42]. As more lactate acid was produced in XI (Additional file 2B), higher activity of this gene probably led to acid-damage resistance. Finally, nitrogen metabolism and cell wall/membrane metabolism were perturbed in XI (Part III of Additional file 7).
Long-term ethanol stress changed the cellular stress-response program
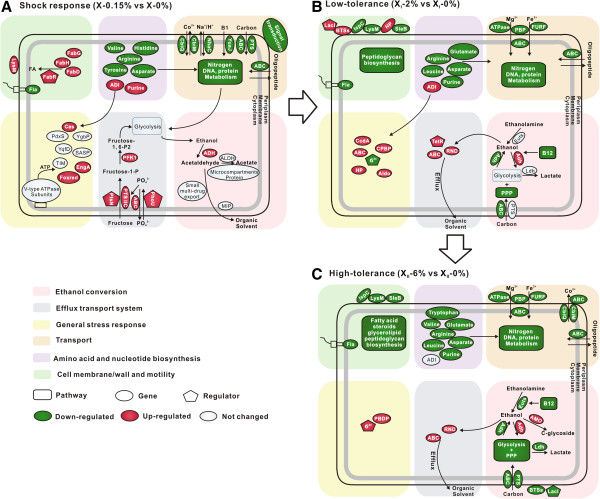
Comparison between XI-2% and XI-0% revealed how long-term stress altered the stress-response program (View IIB; Figure 5B and Additional file 14A). Overall, XI-2% featured an inhibited metabolism, including carbon metabolism (PPP pathways), energy conversion (e.g., acetate kinase (ak), adh, and 3-isopropylmalate dehydrogenase (ipmdh)), cell membrane metabolism, DNA metabolism, coenzyme biosynthesis (B1, B2, and B12), amino acid synthesis and cell motility (Additional file 16A). However, several genes were upregulated. (i) Extracellular solute-binding protein and binding-protein-dependent transport systems inner membrane component in COG G (Teth5142194-2202, Teth5140534 and Teth5141044) were upregulated (Figure 5B and Additional file 14A), however the associated carbohydrate transport systems were not induced, suggesting changed cell surface interactions but not increased carbohydrate transports. (ii) Efflux systems were elevated that likely removed intracellular ethanol, including tetracycline repressor (TetR) family transcription factor [43], RND family efflux transporter [44] and capsule polysaccharide biosynthesis [45] (Additional file 16A). (iii) An iron-containing adh (Teth5140145) and the ADI pathway (Teth5140483-0485) were activated.
Figure 5.
Molecular events underpinning development of ethanol tolerance in thermopiles. The transition from the shock response in X (A) to the low-tolerance in XI(B) and eventually to the high-tolerance in XII(C) was shown. Key transport, metabolic and regulatory genes and pathways were illustrated. Fla: flagellar biosynthesis; BTSs: binding-protein-dependent transport systems; Aldo: aldo/ketoreductase; PBD: peptidoglycan binding domain-containing protein; CcdA: cytochrome c biogenesis protein; TSPP: transport system permeaseprotein.PBP: periplasmic binding protein; IspC: cell wall hydrolase/autolysin; CPBP: capsule polysaccharide biosynthesis protein; PBDP: peptidoglycan binding domain-containing protein; FURF: ferric uptake regulator family protein; HP: hypothetical protein.
Therefore in both X and XI, ethanol inhibited carbohydrate, amino acid and flagellar synthesis. The defense mechanisms shared between shock and low-tolerance included the adh operon that controls intracellular ethanol [25] and ADI pathway that maintains pH balance [25,38]. On the other hand, X appeared to employ fructose-specific PTS systems for extruding ethanol [46], whereas XI mobilized RND and TetR presumably to activate efflux pumps for ethanol [43] and extracellular solute-binding protein to tune cell-surface interactions (Figure 2, Figure 5A-B and Additional file 14A).
Adaptation strategy of the high-tolerance mutant
Genome mutations in XII
The three apparently beneficial mutations of XI, found at electron transport complex I, ldh promoter and TrkH family potassium uptake protein respectively, were also found in XII (Figure 2 and Additional file 10D), consistent with XII being derived from XI. Moreover, 12.1% of non-synonymous mutations in XII were found in Xp but not in XI, indicating these mutations might further enhance ethanol tolerance (Figure 2 and Additional file 10D; Part IV of Additional file 7). In both Xp and XII, Leu598?→?Gln was found in the Fe-ADH domain of AdhE (bifunctional aldehyde/alcohol dehydrogenase; Additional file 17B). Furthermore, in XII, one more SNP (His748?→?Arg) was identified in this domain (near the active-site iron and the cofactor-binding sites), likely altering the interaction between the ADH-domain and NADP (Additional file 17B; Methods) [4]. In addition, in XII, analogous to the mutated RodA (Teth5142127) in Xp, another rod shape-determining protein, MreC, was mutated (Val132?→?Gly, Teth5142133). MreC plays a role in determining cell shape (e.g., the rod-shape in E. coli[47]), likely underlying the shortened length of XII cells (Additional file 3).
XII harbored additional SNPs that were absent in both Xp and XI. They were mostly in two categories: ribose metabolism (e.g., Thr94?→?Ala in RpiR) and cell membrane metabolism (Figure 2 and Additional file 10C; Part IV of Additional file 7). Thus the altered ribose and membrane metabolism in XII might contribute to the enhanced tolerance.
Features of the cellular state of XII
Transcriptomic comparison of XII-0% versus X-0% revealed characteristics of XII state, including altered carbon-metabolisms (inhibited adh cluster (Teth5140145-0146) in glycolysis and induced oxaloacetate decarboxylase (oad, Teth5141582; converting oxaloacetate into pyruvate)) in pyruvate metabolism, induced stress response, repressed nitrogen metabolism and inhibited cell wall/membrane metabolism (Additional file 13B and Additional file 18; Part IV of Additional file 7). Notably, fabR (Teth5141728), a global regulator of membrane lipid biosynthesis in many Gram-positive bacteria [48], was up-regulated in XII-0%, together with teth5141723-1727. FabG (Teth5141723) and fabD (Teth5141724) were involved in long chain fatty acid biosynthesis, whereas fabH (Teth5141726) was the determining factor in branched-chain fatty acid biosynthesis [49]. Therefore, XII reinforced membrane rigidity against ethanol damage [30]. Such expression patterns were also observed under ethanol shock in X at 1h and 2h, revealing a link between shock response and adaptation that likely underlay the observed “Memory” effect in this adaptive evolution (Figure 3B). In addition, the induction of heat shock proteins (Hsps, which were a universal response to ethanol stress in mesophiles) was not observed under either the shock to X (X-0.15% vs X-0%) or the stress to the mutants (XI-2% vs XI-0% and XII-6% vs XII-0%) (Part V of Additional file 7), suggesting one TGPA-specific feature of adaptive evolution.
Adaptation from low-tolerance to high-tolerance
Comparison of XII-0% and XI-0% transcriptomes explained the increased tolerance yet higher ethanol productivity in XII. Higher ethanol-tolerance generally correlated with lower ethanol-productivity, as XI and XII produced less ethanol than X (Additional file 2B), which was due to rewired glycolysis (XI and XII), suppressed solvent formation genes (XI and XII) and inhibited co-enzyme B biosynthesis (XI). Surprisingly, XII both tolerated and produced higher ethanol than XI (Additional file 2B; p = 0.01), suggesting positive co-evolution of the two traits under certain circumstances (Additional file 8 and Additional file 14B).
In XII-0% (vs XI-0%), 21.6% (535) of the genes were upregulated, while 1.3% (33 genes) downregulated. Upregulation was mostly in three categories. First, solvent formation genes (6.54%) were induced, which included euts (Teth5141940-1946), de novo B12 biosynthesis (Teth5140299-0320) and butyrate biosynthesis (Teth5140936-0944). These likely underpinned the higher-than-XI ethanol-production of XII.
Second, among the carbon/ion transporters repressed in XI and XII (X-0% as baseline), 51 genes (10% of all upregulated genes) were expressed higher in XII than XI (e.g., Teth5140323-0326 for cobalt transport in B12 biosynthesis), indicating a lesser degree of inhibition in XII. Notably, dynamically regulated operon structures were observed in these upregulated carbon transporters between XI and XII, including pentose transport (Teth5140161-0166) and glucose transport (Teth5140412-0414) (Figure 2 and Additional file 19). In XI, genes involved in pentose transport were clustered in a single operon (Additional file 19A), as was the case for glucose transport genes (Additional file 19C). However, in XII, pentose transport genes were transcribed in three suboperons (Additional file 19B), whereas glucose transport genes formed two suboperons (Additional file 19D). Such dynamic operon structures might be a mechanism to precisely tune the ratios of enzyme-encoding transcripts as an adaptation strategy specific at the high-tolerance phase.
Third, general stress-response (5.2% of all upregulated genes; [36,41]) was specifically induced in XII-0% (vs XI-0%). It included ABC transporters, Hsps (e.g., DnaK, GrpE, and Hsp20), peptidases, CRISPRs-associated (Cas) immunity system, ADI pathway, and oxidoreductases. It also included several genes induced in XI-2% but not in XI -0% (vs X-0%; i.e., the “Memory” of XII-0%), such as the TetR regulator in efflux systems and the aldo/ketoreductase in oxidoreduction (Additional file 12B). Interestingly, Teth5141359-1361, an ABC transporter operon in COG V (defense mechanism) was “dynamic”. In XI, Teth5141359-1361 constituted a single operon (Additional file 19E). However, in XII, they formed two suboperons, with the first (Teth5141359-1360, encoding hypothetical protein and ABC transporter) up-regulated in XII-0% (vs XI-0%) while the second (Teth5141361, a hypothetical protein) not induced (Additional file 19F), suggesting dynamic operon as a mechanism for gene-specific upregulation within an operon.
The distinct stress-response program of XII and its link to XI
Comparison between XII-6% and XII-0% demonstrated how long-term stress shaped the stress-response program in the high-tolerance phase (Figure 5C). In XII-6%, despite the downregulation of 1583 genes, 16 were upregulated (Additional file 16B). (i) One extracytoplasmic function (ECF) subfamily ϭ24 locus (Teth5141847-1848; Figure 5C and Additional file 14A) was induced, which encodes RNA polymerase subunits that regulate intracellular responses to various extracellular stimuli [41]. In Bacillus subtilis, this ECF ϭ (SigM) was activated under ethanol shock and salt stress [50]. (ii) RND family efflux system operon (Teth5140198-0200) was upregulated, which extrudes ethanol [44]. (iii) The iron-containing adh (Teth5140145-0146) operon and the alcohol catabolism operon (Teth5141785-1787), which convert ethanol into other intermediate metabolites, was expressed higher in XII-6% than XII-0%. The Teth5141786 activated nucleotide sugar and then Teth5141787 transferred glycosyl from nucleotide sugar to alcohol, forming C-glycoside to reduce the intracellular alcohol concentration. Interestingly, in XII-6%, due to functional correlation, they constituted one operon, instead of two suboperons (Teth5141785-1786 and Teth5141787) in XII-0% (Additional file 20), suggesting dynamic operons can be condition-dependent. (iv) Peptidoglycan binding domain-containing protein (Teth5140954) in COG V (defense mechanism) was induced, which can protect cell wall from autolysis.
Comparison between XII-6% and XI-2% revealed 72 (3%) upregulated genes and 725 (29%) downregulated genes (Figure 2 and Additional file 14A; Part VI of Additional file 7), which are key to the 2%-to-6% tolerance improvement. The 72 upregulated genes include, (i) iron-containing adh (Teth5140145-0146) operon, which suggested positive correlation between ethanol conversion and tolerance; (ii) purine metabolism operons (Teth5140517-0525 and Teth5142354-2355), which was also upregulated in X under ethanol shock and thus was a shock-adaptation link (Additional file 14A); (iii) cell wall and membrane biosynthesis genes (Teth5141784-1789, Teth5140797-0799 and Teth5141976); (iv) hsps(including dnaK, dnaJ, grpE and hrcA; Teth5142078-2081), which were upregulated under 6%-ethanol stress (XII-0% vs XI-0%; XII-6% vs XI-2%) but not under shock or 2%-ethanol stress (X-0.15% vs X-0%; XI-2% vs XI-0%), suggesting their phase-specific functioning; (v) the ϭ24 (Teth5141847) which was also a unique feature of XII upon ethanol stress; (vi) ribosome protein genes (Teth5140864-0881), which suggested a potential contribution of translation machineries in XII tolerance; (vii) transporter genes including ABC transporter (Teth5140253-0254, COG P), Na+/H+ exchanger (Teth5141320, COG P) and cellobiose specific PTS IIA (Teth5140265), which were repressed by ethanol in both XI and XII (X as baseline) yet the inhibition was alleviated in XII-6% (vs XI-2%), potentially explaining the growth of XII but not XI under 6% ethanol.
Thus XI and XII mobilized linked yet distinct defense mechanisms (Figure 2 and Figure 5). The former included RND efflux system and adh operon, both controlling intracellular ethanol. For the latter, XI mobilized TetR presumably to activate efflux pumps for ethanol [43] and ADI pathway to maintain pH balance [25], whereas XII employed ϭ24, alcohol metabolism operon and peptidoglycan binding domain-containing protein (Teth5140954).
Exploiting the molecular links between shock response and adaptation for simultaneous improvement of ethanol tolerance and titer
The higher ethanol titer and higher tolerance in XII than XI suggested ethanol production level was not necessarily negatively correlated with ethanol tolerance. The unraveled links and distinctions among the stress-response programs in X, XI and XII provided rational genetic strategies to engineer the two co-evolving traits.
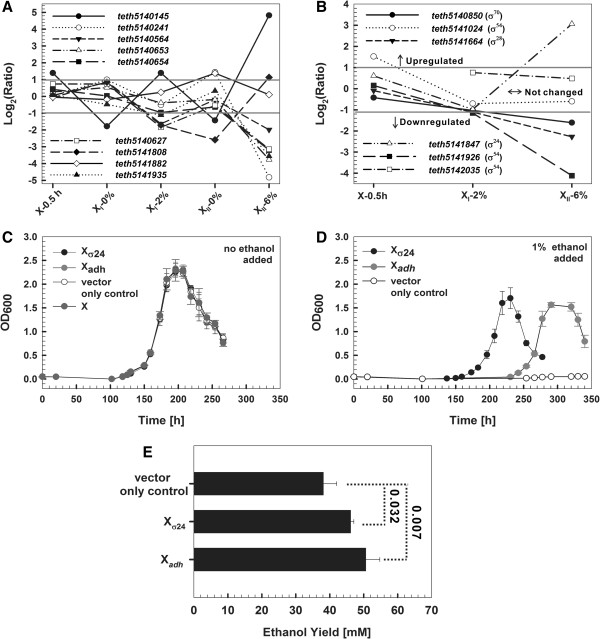
First, adhs were involved in both ethanol production and tolerance [4,51]; however the sheer number and apparent redundancy of this metabolic enzyme (nine such loci/operons in X514) in most bacterial genomes confounded rational engineering. In our microevolution model, an iron-containing adh (Teth5140145) exhibited an unique transcriptional choreography (Figure 6A): upon exposure to ethanol, its transcription was induced (by 2.6 folds) in X and XI and dramatically (by 28.2 folds) induced in XII, yet was inhibited in XI and XII (which produced lower ethanol) when grown without ethanol (Additional file 2B). Such a transcriptional pattern was highly distinct from any of the other eight adhs, whose expression were either unaltered (8 in X, 6 in XI and 2 in XII) or severely inhibited (2 in XI and 6 in XII, by 3-48 folds; Figure 6A). Thus Teth5140145 might be a key junction between the co-evolving tolerance and titer. To test this hypothesis, the Teth5140145-0146 locus was cloned into a replicating plasmid pIKM1, and transformed into X. This adh-overexpressing strain (Xadh) showed improvement in both titer and tolerance: Xadh produced 33% more ethanol than plasmid control strain Xvector (p = 0.007; Figure 6E); moreover, growth (as measured by OD600; Methods) under 0.25%, 0.5% and 1% exogenous ethanol were all enhanced, e.g., by 31.8 folds under 1% (Figure 6D and Additional file 21A-B), suggesting greatly improved tolerance.
Figure 6.
Genetic approaches that improved both ethanol production and tolerance. (A-B) Transcriptional programs of nine adhs and six ϭ factors from ethanol shock to tolerance. (C-D) Growth curves of Thermoanaerobacter sp. X514 wild-type strain and wild-type strains that carried on the pIKM1 plasmid: a vector-only control, an adh locus (Teth5140145-0146) or a ϭ24 cluster (Teth5141847-1848) under 0% (C) and 1% (D) exogenous ethanol. Strains were grown at 45oC in QRCM medium. (E) Ethanol production of these engineering strains (Xvector, XadhE and Xϭ24) at stationary phase, which were grown at 45oC in QRCM medium.
Second, sigma factors are an evolutionarily conserved group of RNA-polymerase subunits that plays regulatory roles [52]. X514 harbors five types of sigma factors: ϭ70, ϭ54, ϭ24, ϭ28 and ϭ29. Interestingly, in our microevolution model, ϭ24 (Teth5141847) was unique in that it was the only ϭ factor up-regulated under 6% ethanol (Figure 6B). In E.coli ϭ24 was previously recognized as a heat-shock-specific ϭ factor [41]. Our engineered strain overexpressing Teth5141847-1848 (Xϭ24) showed dramatic improvement in growth, e.g., 102-fold enhancement of the control (Xvector; as measured by OD600) and 26% faster than Xadh under 1%-ethanol (Figure 6D and Additional file 21A-B). Furthermore, Xϭ24 produced 21% higher ethanol than Xvector (p = 0.032; Figure 6E).
Finally, we found that such two co-evolving traits of ethanol productivity and ethanol tolerance can also be improved by non-genetic approaches (e.g., adaptive evolution and medium supplementation of vitamin B12, Additional file 21C; Part VII of Additional file 7).
Discussion
Solvent tolerance and productivity are both crucial traits in a CBP scheme of biofuel production from cellulose, where cellulase production, cellulose hydrolysis, pentose-hexose co-utilization (one crucial feature of X514 [25]) and solvent production take place in a single bioreactor for maximal energy- and cost-efficiency [4,27,30]. Simultaneous improvement of ethanol tolerance and ethanol titer has been successfully reported in mesophiles (e.g., S. cerevisiae and C. acetobutylicum), but not yet in thermophilic bacteria [4,33]. In mesophiles such as S. cerevisiae and C. acetobutylicum, improvement in both ethanol tolerance and titer were achieved by screening strain libraries overexpressing mutant genes [14,53] or via genomic shuffling [33], yet these approaches required a set of pre determined candidate genes (e.g., two TFs, spt15 and taf25 were selected as the targets to generate mutation libraries by gTME [14]) or laborious mutant selection steps [14,33,53], limiting them to a narrow range of hosts (e.g., well studied model organisms). However, as all adaptation started from shock, delineating the temporal characteristics of the adaptation process and testing the mechanistic links between shock and adaptation should serve as essential foundation for modulating genome evolution, including the rational engineering of co-evolving traits.
First, our experimental model revealed the links and distinctions, both global and local, between shock response and adaptation. The “discreteness” of microevolution suggested the feasibility of phase-specific modulation of microevolution, which might carry certain advantages. This was validated by our experiments where overexpressing ϭ24 (a gene induced specifically at the high-tolerance phase) improved tolerance more dramatically (102 folds vs 31.8 folds) than overexpressing iron-containing adh (Teth5140145; a gene consistently induced along shock-to-adaptation development). Moreover, “Convergence” raised the possibility of modeling and engineering tolerance based on shock-responses. This hypothesis was validated by our experiment that overexpression of iron-containing adh locus (Teth5140145-0146), which were the earliest responders in shock-response, improved tolerance (Figure 6D).
Second, our study unveiled TGPA-specific features of adaptive evolution, compared to mesophiles (e.g., E. coli and S. cerevisiae). (i) The higher μg (0.045) in Thermoanaerobacter sp. X514 than that in E. coli (0.026; [5]) under solvent stress does not support the theoretically postulated much lower μg in thermophiles than in mesophiles [35]. Our finding thus tentatively suggested that TGPAs balance between the deleterious effect of the average mutation and the cost of further reducing mutation rate not by reducing μg but likely by lowering deleteriousness of mutations under stress. (ii) TGPA-specific features of ethanol-shock were unveiled, such as the most vulnerable amino acid metabolism in X514 versus the activated tryptophan biosynthesis in S. cerevisiae, specific activated ADI pathway (e.g., arginine deiminase) yet without activating transcription of Hsps (one key feature of the general shock-response in mesophiles [8,10]), and different mechanisms of V-type-ATPase in resisting ethanol (Figure 4B). (iii) TGPA-specific features of ethanol-tolerance were also revealed: distinct mechanisms in membrane metabolism [29,30], specific ADI pathway [38] and its related NAD/NADP octopine (Part III of Additional file 7), and a different solvent-response role of Hsps [11,13,31] (absent induction under either shock or stress; Part V of Additional file 7). Noticeably, Hsps do not seem to play a prominent role in solvent response in thermophiles and their high levels sustaining in thermophiles in the absence of stress was possibly a consequence of long-term evolution under high temperature.
Finally, by elucidating the molecular choreograph underlying an adaptive evolution under solvents, this study demonstrated a strategy to rationally identify the gene targets for engineering the tolerance-productivity relationship. In addition, our experiments showed that simultaneous improvement of ethanol tolerance and productivity is feasible in ethanogenic thermophilic bacteria, and it can be accomplished via genetic routes (e.g., metabolic enzymes (an adh loci; Teth5140145-0146) or transcriptional regulators (a ϭ24 cluster; Teth5141847-1848)). Furthermore, it is conceivable that these novel gene targets identified by our approach can serve as the foundation for rational protein-engineering or mutant protein screening to further improve ethanol tolerance and productivity in this and related thermophiles.
Conclusions
In adaptive evolution, the molecular links between shock-response and adaptation remain poorly understood, which hinders rational engineering of solvent tolerance and correlated traits (e.g., productivity). In this study an experimental model was established to track the shock-to-adaptation microevolution in thermophiles. Under ethanol stress, the spontaneous genome mutation rate (μg) in Thermoanaerobacter, at 0.045, appears to be equivalent to that in mesophiles (e.g. E.coli) [5]. Shock-response and adaptation were distinct in nature, yet both temporally phased and resilient. In the absence of stimuli, transcriptomic states of tolerance mutants resembled their stressed parental strains, while that of high-tolerance mutants resembled the extendedly shocked wild-type. Interestingly, responses to ethanol stress were phase-specific. Upon ethanol shock, X employed fructose-specific PTS, ADI pathway, Adh and a distinct mechanism of V-type ATPase. As an adaptation to ethanol, XI mobilized RND efflux system and Adh, whereas XII, which produced higher ethanol than XI, employed ϭ24, an alcohol catabolism operon and phase-specific Hsps, modulated the operon structures of hexose/pentose transport and reinforced membrane rigidity. Exploiting these links and distinctions between shock-response and adaptation, we showed ethanol productivity and tolerance can be simultaneously improved by genetic approaches (overexpressing iron-containing adh or ϭ24).
Therefore, this study revealed thermophilic-bacteria specific features of adaptive evolution and demonstrated a rational strategy to engineer the co-evolution of industrial traits. As improvements of shock-response, stress tolerance and productivity have been crucial and shared aims in industrial applications employing thermophiles [26], our findings should be valuable not just to the production of ethanol but also to a wide variety of biofuels and biochemicals.
Methods
Adaptive evolution for improved ethanol tolerance
Thermoanaerobacter sp. X514 was cultured anaerobically in QRCM medium (1.5 g/L KH2PO4, 4.2 g/L Na2HPO4.12H2O, 3 g/L NaCl, 0.5 g/L NH4Cl, 0.2 g/L MgCl2, 5 g/L yeast extract, 10 g/L tryptone, 2 mg/L resazurin and 0.05g/L and Cys-HCl) supplemented with 50 mM glucose [54] at 60°C without shaking. For adaptation evolution under exogenous ethanol, sequential transfer was employed. The wild type strain (X) was initially inoculated into QRCM containing 0.5% (v/v) ethanol. When OD600 reached the maximum, cultures were immediately transferred into fresh 0.5%-ethanol medium on a 1:10 volume ratio. The transfer was repeated until OD600 reached a reproducible maximum value, cells were inoculated into 1%-ethanol medium. The cycle was repeated with increasing ethanol concentrations (until 2% ethanol) for approximately 440 generations over five months. A single clone that grew under 2% ethanol, XI, was isolated from the mixed cultures of mutant pools (Xp) (Figure 1B and Additional file 2A). XI was mutagenized with ethyl methanesulfonate (EMS) [55] and the mutant pool screened on 6%-ethanol agar QRCM plate and subsequently in liquid QRCM (Figure 1B and Additional file 2A). One clone that grew the fastest in liquid QRCM (termed XII) was isolated. Further experiments confirmed that the ethanol tolerance phenotypes of both XI and XII were inheritable and stable after culturing for at least 60 generations in ethanol-free medium. OD600 improvement was measured by OD600 of the treatment divided by that of the control when the former reached the maximum.
In summary, X was not exposed a priori to exogenous ethanol and thus represented a phase of “ethanol sensitivity”, where growth was inhibited at defined medium in the mid-log with 0.15% exogenous ethanol (Additional file 1A). XI represented the “low-tolerance” phase while XII represented the “high-tolerance” phase, as they tolerated 2% and 6% ethanol respectively.
Ethanol shock
To determine the effect of ethanol on X growth, a wide range of ethanol concentrations (0.1%?~?2% (v/v)) were tested first (data not shown) and then narrowed down to 0.15% (v/v) that caused stress but not significant cell death in defined medium (0.08 g/L CaCl2∙2H2O, 1.0 g/L NH4Cl, 0.2 g/L MgCl2∙6H2O, 1.0 g/L NaCl, 7.2 g/L HEPES, 2.52 g/L NaHCO3, 0.05 g/L L-cysteine-HCl, 1 ml 1000× trace element stock and 1 ml 1000× vitamin stock solution) supplemented with 50 mM glucose [8,25] (Additional file 1A). Thus all subsequent ethanol shock assays were conducted at 0.15% (v/v) ethanol in defined medium. In triplicate experiments, ethanol was added to the medium for an exogenous concentration of 0.15% when X was grown to exponential phase (OD600 = 0.12). An identical volume of water was added to the controls. After 0.5 h, 1 h, 2 h and 4 h of the addition, cells were harvested and cell pellets frozen immediately in liquid N2 and stored at -80°C prior to RNA extraction for microarray experiments.
Ethanol stress
The mutant strains were grown at 60°C in triplicates in defined medium without ethanol (XI and XII), with 2% ethanol (XI) and with 6% ethanol (XII) respectively. Cells were harvested at mid-exponential phase followed by microarray experiments as described above. For sugars and metabolites quantification, samples were harvested at stationary phase and analyzed using HPLC [25].
Effects of exogenous vitamin B12 on ethanol production for Thermoanaerobacter sp. X514 (X, XI and XII)
To test the effect of co-enzyme B12 on ethanol productivity, 0×, 1×, 2× to 4× B12 (0.1 mg/L as 1×) was added to the defined medium at 60°C, followed by inoculation of X, XI and XII, respectively. Ethanol concentration at stationary phase was measured by HPLC [25].
Microarray experiments and data analysis
Thermoanaerobacter sp. X514 whole-genome oligonucleotide (70mer) microarray [25] was used in this study. Total cellular RNA and genomic DNA were isolated, labeled and then hybridization and data analysis performed as previously described [25]. For ethanol shock, the trasncriptomes of X-0.15% (X cells cultured at defined medium with 0.15% exogenous ethanol; Additional file 1A) were compared to that of X-0% (X cells cultured without exogenous ethanol). On the other hand, the transcriptomes of XI under 0% and 2% ethanol and those of XII under 0% and 6% ethanol were compared respectively to reveal the mechanism of ethanol adaptation. Cutoffs of mean |log2 (Rtreatment/Rcontrol)| ≥1.0 and |Z score|?≥?2.0 were used to determine significant expression changes [25,34]. The totally 39 microarray datasets were deposited as NCBI GEOGSE32630.
Gene co-expression networks for shock response
Twelve microarray datasets for the ethanol shock (three replicates for each of the four time points) and the corresponding twelve control datasets were respectively generated and used via co-expression analysis to construct the two co-expression networks with random matrix theory approach [25,56] (Additional file 6). For each spot on microarray, a normalized Cy5/Cy3 ratio (R) was calculated and its logarithmic transformation performed. Pearson correlation coefficient cut-off was 0.98 (both for ES+ and ES-) between each gene-pair. The modules were separated by fast greedy modularity optimization [56]. Hierarchical clustering analysis was performed using K-Means/K-Medians Clustering to identify expression patterns each shared by a sub-set of genes throughout the duration of the ethanol shock response (Additional file 9).
Hierarchical clustering analysis of transcriptomes
Hierarchical clustering analysis (support tress) among the nine transcriptional profiles from shock (X-0.15% at each of the four time points of 0.5 h, 1 h, 2 h and 4 h) to adaptation (XI-0%, XI-2%, XII-0%, XII-6%, each in triplicates) was performed with TM4 software [57] based on Pearson Correlation. Similarly, the clustering analysis among the eight “relative” transcriptomic changes (using genes with more than 2 fold changes) from shock (X-0.15% vs X-0% at each of the four time points of 0.5 h, 1 h, 2 h and 4 h) to adaptation (XI-0% vs X-0%, XI-2% vs XI-0%, XII-0% vs X-0%, XII-6% vs XII-0%, each in triplicates) was performed via the same method.
RNA-Seq for detecting structural variation of transcripts
For X, XI and XII, 10 μg of the same total RNA samples from ethanol stress and normal growth condition (in the absence of ethanol) in triplicates were used for high-throughput RNA-Sequencing. The cDNA libraries (X-0%, XI-0%, XI-2%, XII-0% and XII-6%) were constructed as previously described [58]. The samples were quantified spectrophotometrically using Nanodrop (Thermo, USA) and sequenced in a Solexa GA-IIx (Illumina, USA). The raw 2 × 100bp reads, after quality screening, were mapped to the Thermoanaerobacter sp. X514 reference genome sequence [34] (NCBI accession number: NC_010320.1) using SOAP [59], allowing for 2nt mismatches. These uniquely mapped sequences were further analyzed to calculate transcript coverage map based on the number of uniquely mapped reads per locus. The RNA-Seq datasets were deposited as NCBI accession number SRA046273.1.
Detection of dynamic operon structures
Genes within an operon were defined based on continuous read coverage, transcript abundance and detection of pair-end reads among these genes in all of the triplicates. A new operon was defined when two requirements were met in each of the triplicate samples: 1) uniquely mapped pair-end reads were detected; 2) a significant change (greater than two-fold) of read coverage was found between genes in one predicted polycistron.
Genome-wide mutation profiling via whole-genome sequencing
Genomic DNA of X, Xp, XI and XII were isolated [25] and then shot gun libraries constructed [58] and sequenced on Solexa GA-IIx (Illumina, USA). MAQ [60], Samtools [61] and GATK [62] were used respectively for read alignment to Thermoanaerobacter sp. X514 reference genome (NC_010320.1; [34]) and SNP calling. Those predicted SNPs shared among the three were then manually examined (Additional file 10) and then validated by sequencing a selected set of mutated genes (five in XI and six in XII) using gene-specific primer pairs (Additional file 12A). For each gene, ten clones were randomly picked for Sanger sequencing (Invitrogen, USA). The results were consistent with Solexa sequencing (Additional file 12B). All sequences were deposited under NCBI accession number SRA046273.1.
Plasmid and strain construction
A 3.4 kb PCR fragment encoding the 3.1 kb iron-containing adh and NADH oxidase gene (Teth5140145-0146) flanked by its 197 bp promoter and 48 bp transcription terminator region was amplified from the genome of Thermoanaerobacter sp. X514 and subsequently ligated into plasmid pIKM1 [54] through EcoRI and BamHI. Similarly, 1.7 kb PCR fragment encoding the 1.4 kb ϭ24 factor and hypothetical protein gene (Teth5141847-1848) with its 245 bp promoter and 80 bp transcription terminator region was amplified and subcloned into pIKM1 through XbaI and KpnI. Plasmids were then transformed into Thermoanaerobacter sp. X514 wild type strain (X) based on our published protocol [54]. These plasmid containing strains were cultured in QRCM medium at 45°C and cell samples were collected at stationary phase for ethanol titer quantification.
Homology modeling of the structure of mutant proteins
For the X514 wild-type and mutant protein sequences (DeoR family transcriptional factor (Teth5141305) and the ADH domain of AdhE (Teth5140627), queries were aligned to structural templates in PDB using NCBI protein BLAST. The homology models of 3D protein structures that included cofactors and ligands were constructed via MODELLER [63].
Abbreviations
AK: Acetate kinase; Aldh (Teth5141942): Aldehyde dehydrogenase; Adh (Teth5140145): Alcohol dehydrogenase; ADI pathway (Teth5140483-0485): Arginine deiminase pathway; AdhE (Teth5140627): Bifunctional aldehyde/alcohol dehydrogenase; Cas immunity system: CRISPRs-associated immunity system; Euts (Teth5141943-1946): Ethanolamine utilization protein genes; XII: 6% ethanol tolerant mutant; EMS: Ethyl methanesulfonate; μg: Genome mutation rate; Hsps: Heat shock proteins; ipmdh: 3-isopropylmalate dehydrogenase; Ldh (Teth5140216): Lactate dehydrogenase; XI: One single colony from Xp; Oad (Teth5141582): Oxaloacetate decarboxylase; PTS: Phosphotransferase system; Xp: Pool of 2% ethanol tolerant spontaneous mutants of Thermoanaerobacter sp. X514; MreC (Teth5142133): Rod shape-determining protein; RodA (Teth5142127): Rod shape-determining protein; RND efflux system: Resistance-nodulation-cell division family efflux transporter; TetR: Tetracycline repressor family transcription factor; TGPAs: Thermophilic gram-positive anaerobes; X: Wild-type Thermoanaerobacter sp. X514.
Competing interests
The authors declare that they have no conflict of interest.
Authors’ contributions
LL and JX conceived and designed the study. LL, YJ, HS, XZ, LT and KW performed experiments. JZ and ZH assisted microarray experiment design and analysis. QT constructed co-expression gene networks. RH and QZ assisted RNA-Seq data analysis. YL and QC assisted protein structure modeling. JX and LL analyzed all data and wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Impact of ethanol shock on the growth and gene expression of Thermoanaerobacter sp. X514 (wild type). A) Ethanol of different concentrations was added to cultures in early-mid exponential phase, and growth was subsequently monitored as OD600. Data were averaged from triplicate cultures, with error bars indicating standard deviations. B) Numbers of the significantly up- or down-regulated genes under each condition. Each column represented the number of genes with significant expression changes (|log2R|?≥?1 and |Z score| ≥2|) at the corresponding time points.
Thermoanaerobic growth conditions and glucose fermentation by Thermoanaerobacter sp. X514 wild type and ethanol tolerant mutants. A) Growth curves of X, XI and XII in defined medium with or without exogenous ethanol. B) Substrate utilization and product profiles of X, XI and XII in defined medium.
Scanning electron microscopy images of the Thermoanaerobacter sp. X514 wild-type and mutants.
Statistical analysis of the mutations in X, Xp, XI and XII Genomes.
Functional patterns of significantly expression-altered Genes in XI (A) and XII (B) under ethanol-supplemented and ethanol-free media respectively. Proportions of differentially expressed genes under each COG among the total number of differentially expressed genes were indicated.
Gene co-expression networks of Thermoanaerobacter under ethanol shock and normal growth condition. A) A global view of the network under normal growth condition (ES-). B) A global view of the network under ethanol shock (ES+). Number in bracket represents the number of genes in each module. Each node represented a gene, which was color-coded using its predicted COG-based functional classification (http://www.ncbi.nlm.nih.gov/nuccore/NC_010320; NC_010320.1).
Supplemental Materials.
The List of differentially expressed genes by X for at least one time points in each cluster under 0.15% (v/v) ethanol shock.
Clustering results of gene-expression patterns in the X under ethanol shock. Ten clusters were obtained by the HCL method (TM4 software). Each panel displayed the expression dynamics of one such cluster. The horizontal axis indicated the time points of the data, and vertical axis was log (base 2) expression ratio.
Genomic mutations shared among the three mutations-lists identified by MAQ, Samtools and GATK respectively. A) X; B) Xp; C) XI; D) XII.
The SNPs in non-coding region. Red: the mutated base; base in brackets: reference base at mutated point; dash: single base deletion. Promoters were predicted by Berkeley Drosophila Genome Project (prokaryote organism; http://www.fruitfly.org/seq_tools/promoter.html).
Experimental validation of SNPs in XI and XII. A) The primer sequences used for PCR and then Sanger sequencing. B) The SNPs validated by Sanger sequencing.
Priori ethanol stress rewired cellular networks (XI and XII). Key transport, metabolic and regulatory genes and pathways were illustrated. OD: NAD/NADP octopine dehydrogenase; FITP: ferrous iron transport protein B. Other abbreviations were as in Figure 5.
Developmental model of solvent-tolerance traits in thermopiles. A) Model of cellular responses upon environmental stimuli: the transition from shock response (wild type) to low-tolerance (XI) and eventually to high-tolerance (XII). B) Model of the cellular states after a priori long-term exposure to exogenous ethanol. Boxes represented genes/pathways, whereas box size indicated the number of differentially expressed genes in the pathway. Differential-expression ratios (log2R) for the genes/pathways were represented by colors. Asterisks indicated non-synonymous mutated genes in the pathways or the SNPs in predicted promoters of genes. Tra (P): ABC transporter related in COG P; MM: membrane metabolism; AAM: amino acid metabolism; LFA: long-chain fatty acid.
Differentially expressed genes in mutant. A) XI under normal growth condition (XI-0%). B) XII under normal growth condition (XII-0%).
Differentially expressed genes in mutants under ethanol stress. A) XI under 2% ethanol (XI-2%). B) XII under 6% ethanol (XII-6%).
Homology models of the mutant protein structures DeoR (A) and AdhE (B) in Thermoanaerobacter sp. X514. Mutation sites, cofactors and ligands were respectively labeled.
The Dynamic operons between X and XII (Teth5140597-0601). Gray and black areas respectively represented the coverage of mRNA in X and XII in ethanol-free media. The scale with the relative score indicated, for each library and at a given nucleotide (among the three biological replicates), the mean number of uniquely mapped reads normalized to the total number of such reads.
The Dynamic operons between XI and XII in the absence of ethanol. A-B) Teth5140161-0166; C-D) Teth5140412-0414; E-F) Teth5141359-1361. The scale was as in Additional file 18.
The Dynamic operon between XII-0% and XII-6% (Teth5141785-1787). The scale was as in Additional file 18.
Genetic and non-genetic approaches that improved both ethanol production and tolerance.A-B) Growth curves of Thermoanaerobacter sp. X514 wild type strains that carried different plasmids (a vector-only control, an adh cluster (Teth5140145-0146) or a ϭ24 cluster (Teth5141847-1848)). Strains were grown at 45°C in QRCM medium with 0.25% and 0.5% added ethanol respectively. C) Fermentation experiments testing effects of vitamin B12 on ethanol production of tolerant mutants in defined medium at 60°C. Vitamin B12 concentrations in defined medium were indicated on x-axis. The means (and standard deviation) for the three biological replicates were shown for each sample. Differences in means were evaluated using one-tailed paired t-test (those with p<0.05 were shown).
Contributor Information
Lu Lin, Email: linlu@qibebt.ac.cn.
Yuetong Ji, Email: jiyt@qibebt.ac.cn.
Qichao Tu, Email: qichao@ou.edu.
Ranran Huang, Email: huangrr@qibebt.ac.cn.
Lin Teng, Email: tenglin@qibebt.ac.cn.
Xiaowei Zeng, Email: zengxw@qibebt.ac.cn.
Houhui Song, Email: songhh@qibebt.ac.cn.
Kun Wang, Email: wangwei@qibebt.ac.cn.
Qian Zhou, Email: zhouqian@qibebt.ac.cn.
Yifei Li, Email: liyf@qibebt.ac.cn.
Qiu Cui, Email: cuiqiu@qibebt.ac.cn.
Zhili He, Email: zhe@rccc.ou.edu.
Jizhong Zhou, Email: jzhou@ou.edu.
Jian Xu, Email: xujian@qibebt.ac.cn.
Acknowledgements
This work was supported by Grants 2011CB707404 and 2011BAD22B02 from MoST of China, 91231205 and 30870572 from NNSF of China, JQ200822 and BS2009SW022 from NSF of Shandong and KSCX1-YW-11C2 from CAS, and by the US NSF EPSCoR Grant EPS-0814361 and the Oklahoma Bioenergy Center.
References
- Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Wiegel J, Adams MWW. Thermophiles: The Keys to the Molecular Evolution and the Origin of Life. Philadelphia: Taylor & Francis e-Library; 2003. [Google Scholar]
- Lenski RE, Ofria C, Pennock RT, Adami C. The evolutionary origin of complex features. Nature. 2003;423:139–144. doi: 10.1038/nature01568. [DOI] [PubMed] [Google Scholar]
- Brown SD, Guss AM, Karpinets TV, Parks JM, Smolin N, Yang S, Land ML, Klingeman DM, Bhandiwad A, Rodriguez M Jr. et al. Mutant alcohol dehydrogenase leads to improved ethanol tolerance in Clostridium thermocellum. Proc Natl Acad Sci USA. 2011;108:13752–13757. doi: 10.1073/pnas.1102444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minty JJ, Lesnefsky AA, Lin F, Chen Y, Zaroff TA, Veloso AB, Xie B, McConnell CA, Ward RJ, Schwartz DR. et al. Evolution combined with genomic study elucidates genetic bases of isobutanol tolerance in Escherichia coli. Microb Cell Fact. 2011;10:18. doi: 10.1186/1475-2859-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S, Wu TY, Machado IM, Huang WC, Chen PY, Pellegrini M, Liao JC. Evolution, genomic analysis, and reconstruction of isobutanol tolerance in Escherichia coli. Mol Syst Biol. 2010;6:449. doi: 10.1038/msb.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford BJ, Dahl RH, Price RE, Szmidt HL, Benke PI, Mukhopadhyay A, Keasling JD. Functional genomic study of exogenous n-butanol stress in Escherichia coli. Appl Environ Microbiol. 2010;76:1935–1945. doi: 10.1128/AEM.02323-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brynildsen MP, Liao JC. An integrated network approach identifies the isobutanol response network of Escherichia coli. Mol Syst Biol. 2009;5:277. doi: 10.1038/msb.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley D, Bandara A, Fraser S, Chambers PJ, Stanley GA. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J Appl Microbiol. 2010;109:13–24. doi: 10.1111/j.1365-2672.2009.04657.x. [DOI] [PubMed] [Google Scholar]
- Ma M, Liu ZL. Mechanisms of ethanol tolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2010;87:829–845. doi: 10.1007/s00253-010-2594-3. [DOI] [PubMed] [Google Scholar]
- Tomas CA, Welker NE, Papoutsakis ET. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl Environ Microbiol. 2003;69:4951–4965. doi: 10.1128/AEM.69.8.4951-4965.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Beamish J, Papoutsakis ET. Transcriptional analysis of butanol stress and tolerance in Clostridium acetobutylicum. J Bacteriol. 2004;186:2006–2018. doi: 10.1128/JB.186.7.2006-2018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- Gill RT, Wildt S, Yang YT, Ziesman S, Stephanopoulos G. Genome-wide screening for trait conferring genes using DNA microarrays. Proc Natl Acad Sci USA. 2002;99:7033–7038. doi: 10.1073/pnas.102154799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnecke TE, Lynch MD, Karimpour-Fard A, Lipscomb ML, Handke P, Mills T, Ramey CJ, Hoang T, Gill RT. Rapid dissection of a complex phenotype through genomic-scale mapping of fitness altering genes. Metab Eng. 2010;12:241–250. doi: 10.1016/j.ymben.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Winkler J, Kao KC. Harnessing recombination to speed adaptive evolution in Escherichia coli. Metab Eng. 2012;14:487–495. doi: 10.1016/j.ymben.2012.07.004. [DOI] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes LH, Almario MP, Kao KC. Genomic library screens for genes involved in n-butanol tolerance in Escherichia coli. PLoS One. 2011;6:e17678. doi: 10.1371/journal.pone.0017678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff LB, Gill RT. Engineering genomes in multiplex. Curr Opin Biotechnol. 2011;22:576–583. doi: 10.1016/j.copbio.2011.04.019. [DOI] [PubMed] [Google Scholar]
- Borden JR, Papoutsakis ET. Dynamics of genomic-library enrichment and identification of solvent tolerance genes for Clostridium acetobutylicum. Appl Environ Microbiol. 2007;73:3061–3068. doi: 10.1128/AEM.02296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou SA, Gaida SM, Papoutsakis ET. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab Eng. 2010;12:307–331. doi: 10.1016/j.ymben.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LBA, Gill RT. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotech. 2010;28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- Brock TD, editor. In thermophiles: general, molecular and applied microbiology. New York: John Wiley & Sons; 1986. [Google Scholar]
- Lin L, Song H, Tu Q, Qin Y, Zhou A, Liu W, He Z, Zhou J, Xu J. The Thermoanaerobacter glycobiome reveals mechanisms of pentose and hexose co-utilization in bacteria. PLoS Genet. 2011;7:e1002318. doi: 10.1371/journal.pgen.1002318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Xu J. Dissecting and engineering metabolic and regulatory networks of thermophilic bacteria for biofuel production. Biotechnol Adv. 2013. [DOI] [PubMed]
- Taylor MP, Eley KL, Martin S, Tuffin MI, Burton SG, Cowan DA. Thermophilic ethanologenesis: future prospects for second-generation bioethanol production. Trends Biotechnol. 2009;27:398–405. doi: 10.1016/j.tibtech.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Shaw AJ, Podkaminer KK, Desai SG, Bardsley JS, Rogers SR, Thorne PG, Hogsett DA, Lynd LR. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci USA. 2008;105:13769–13774. doi: 10.1073/pnas.0801266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M, Tuffin M, Burton S, Eley K, Cowan D. Microbial responses to solvent and alcohol stress. Biotechnol J. 2008;3:1388–1397. doi: 10.1002/biot.200800158. [DOI] [PubMed] [Google Scholar]
- Timmons MD, Knutson BL, Nokes SE, Strobel HJ, Lynn BC. Analysis of composition and structure of Clostridium thermocellum membranes from wild-type and ethanol-adapted strains. Appl Microbiol Biotechnol. 2009;82:929–939. doi: 10.1007/s00253-009-1891-1. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol. 2010;6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond-Watts BB, Bellerose RJ, Chang MCY. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat Chem Biol. 2011;7:222–227. doi: 10.1038/nchembio.537. [DOI] [PubMed] [Google Scholar]
- Mao S, Luo Y, Zhang T, Li J, Bao G, Zhu Y, Chen Z, Zhang Y, Li Y, Ma Y. Proteome reference map and comparative proteomic analysis between a wild type Clostridium acetobutylicum DSM 1731 and its mutant with enhanced butanol tolerance and butanol yield. J Proteome Res. 2010;9:3046–3061. doi: 10.1021/pr9012078. [DOI] [PubMed] [Google Scholar]
- Hemme CL, Fields MW, He Q, Deng Y, Lin L, Tu Q, Mouttaki H, Zhou A, Feng X, Zuo Z. et al. Correlation of genomic and physiological traits of thermoanaerobacter species with biofuel yields. Appl Environ Microbiol. 2011;77:7998–8008. doi: 10.1128/AEM.05677-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JW. Avoiding dangerous missense: thermophiles display especially low mutation rates. PLoS Genet. 2009;5:e1000520. doi: 10.1371/journal.pgen.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moat AG, Foster JW, Spector MP. Microbial stress responses. 4. New York: Wiley-Liss, Inc; 2002. (Microbial Physiology). [Google Scholar]
- Penrod JT, Roth JR. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J Bacteriol. 2006;188:2865–2874. doi: 10.1128/JB.188.8.2865-2874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelis MD, Mariotti L, Rossi J, Servili M, Fox PF, Rollan G, Gobbetti M. Arginine catabolism by sourdough lactic acid bacteria: Purification and characterization of the arginine deiminase pathway enzymes from Lactobacillus sanfranciscensis CB1. Appl Environ Microbiol. 2002;68:6193–6201. doi: 10.1128/AEM.68.12.6193-6201.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Turner PC, Mclennan AG, Bates AD, White MRH. Instant Notes in Molecular Biology. 3. Liverpool, UK: University of Liverpool; 2001. [Google Scholar]
- Boor KJ. Bacterial stress responses: what doesn't kill them can make then stronger. PLoS Biol. 2006;4:e23. doi: 10.1371/journal.pbio.0040023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibel BA, Thuesen EV, Childress JJ. Light-limitation on predator-prey interactions: consequences for metabolism and locomotion of deep-sea cephalopods. Biol Bull. 2000;198:284–298. doi: 10.2307/1542531. [DOI] [PubMed] [Google Scholar]
- Hillerich B, Westpheling J. A new TetR family transcriptional regulator required for morphogenesis in Streptomyces coelicolor. J Bacteriol. 2008;190:61–67. doi: 10.1128/JB.01316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop MJ. Engineering microbes for tolerance to next-generation biofuels. Biotechnol Biofuels. 2011;4:32. doi: 10.1186/1754-6834-4-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts IS. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu Rev Microbiol. 1996;50:285–315. doi: 10.1146/annurev.micro.50.1.285. [DOI] [PubMed] [Google Scholar]
- Okochi M, Kurimoto M, Shimizu K, Honda H. Increase of organic solvent tolerance by overexpression of manXYZ in Escherichia coli. Appl Microbiol Biotechnol. 2007;73:1394–1399. doi: 10.1007/s00253-006-0624-y. [DOI] [PubMed] [Google Scholar]
- Cabeen MT, Jacobs-Wagner C. Bacterial cell shape. Nat Rev Microbiol. 2005;3:601–610. doi: 10.1038/nrmicro1205. [DOI] [PubMed] [Google Scholar]
- Schujman GE, Paoletti L, Grossman AD, de Mendoza D. FapR, a bacterial transcription factor involved in global regulation of membrane lipid biosynthesis. Dev Cell. 2003;4:663–672. doi: 10.1016/S1534-5807(03)00123-0. [DOI] [PubMed] [Google Scholar]
- Alsaker KV, Spitzer TR, Papoutsakis ET. Transcriptional analysis of spo0A overexpression in Clostridium acetobutylicum and its effect on the cell's response to butanol stress. J Bacteriol. 2004;186:1959–1971. doi: 10.1128/JB.186.7.1959-1971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh MJ, Moir A. Sigma M, an ECF RNA polymerase sigma factor of Bacillus subtilis 168, is essential for growth and survival in high concentrations of salt. Mol Microbiol. 1999;32:41–50. doi: 10.1046/j.1365-2958.1999.01323.x. [DOI] [PubMed] [Google Scholar]
- Peng H, Wu G, Shao W. The aldehyde/alcohol dehydrogenase (AdhE) in relation to the ethanol formation in Thermoanaerobacter ethanolicus JW200. Anaerobe. 2008;14:125–127. doi: 10.1016/j.anaerobe.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Vorholt JA, Francez-Charlot A, Frunzke J, Reichen C, Ebneter JZ, Gourion B. Sigma factor mimicry involved in regulation of general stress response. Proc Natl Acad Sci USA. 2009;106:3467–3472. doi: 10.1073/pnas.0810291106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong ME, Lee KS, Yu BJ, Sung YJ, Park SM, Koo HM, Kweon DH, Park JC, Jin YS. Identification of gene targets eliciting improved alcohol tolerance in Saccharomyces cerevisiae through inverse metabolic engineering. J Biotechnol. 2010;149:52–59. doi: 10.1016/j.jbiotec.2010.06.006. [DOI] [PubMed] [Google Scholar]
- Lin L, Song H, Ji Y, He Z, Pu Y, Zhou J, Xu J. Ultrasound-mediated DNA transformation in thermophilic Gram-positive anaerobes. PLoS One. 2010;5(9):e12582. doi: 10.1371/journal.pone.0012582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto JJ, Fernandezcastillo R, Megias M, Ruizberraquero F. Ethyl Methanesulfonate Mutagenesis in Extremely Halophilic Archaebacteria-Isolation of Auxotrophic Mutants of Haloferax-Mediterranei and Haloferax-Gibbonsii. Curr Microbiol. 1992;24:41–47. doi: 10.1007/BF01570098. [DOI] [Google Scholar]
- Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. Functional molecular ecological networks. MBio. 2010;1(4):e00169–00110. doi: 10.1128/mBio.00169-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol. 2006;411:134–193. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- Camarena L, Bruno V, Euskirchen G, Poggio S, Snyder M. Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 2010;6:e1000834. doi: 10.1371/journal.ppat.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Angel GD, Rivas MA, Hanna M. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Impact of ethanol shock on the growth and gene expression of Thermoanaerobacter sp. X514 (wild type). A) Ethanol of different concentrations was added to cultures in early-mid exponential phase, and growth was subsequently monitored as OD600. Data were averaged from triplicate cultures, with error bars indicating standard deviations. B) Numbers of the significantly up- or down-regulated genes under each condition. Each column represented the number of genes with significant expression changes (|log2R|?≥?1 and |Z score| ≥2|) at the corresponding time points.
Thermoanaerobic growth conditions and glucose fermentation by Thermoanaerobacter sp. X514 wild type and ethanol tolerant mutants. A) Growth curves of X, XI and XII in defined medium with or without exogenous ethanol. B) Substrate utilization and product profiles of X, XI and XII in defined medium.
Scanning electron microscopy images of the Thermoanaerobacter sp. X514 wild-type and mutants.
Statistical analysis of the mutations in X, Xp, XI and XII Genomes.
Functional patterns of significantly expression-altered Genes in XI (A) and XII (B) under ethanol-supplemented and ethanol-free media respectively. Proportions of differentially expressed genes under each COG among the total number of differentially expressed genes were indicated.
Gene co-expression networks of Thermoanaerobacter under ethanol shock and normal growth condition. A) A global view of the network under normal growth condition (ES-). B) A global view of the network under ethanol shock (ES+). Number in bracket represents the number of genes in each module. Each node represented a gene, which was color-coded using its predicted COG-based functional classification (http://www.ncbi.nlm.nih.gov/nuccore/NC_010320; NC_010320.1).
Supplemental Materials.
The List of differentially expressed genes by X for at least one time points in each cluster under 0.15% (v/v) ethanol shock.
Clustering results of gene-expression patterns in the X under ethanol shock. Ten clusters were obtained by the HCL method (TM4 software). Each panel displayed the expression dynamics of one such cluster. The horizontal axis indicated the time points of the data, and vertical axis was log (base 2) expression ratio.
Genomic mutations shared among the three mutations-lists identified by MAQ, Samtools and GATK respectively. A) X; B) Xp; C) XI; D) XII.
The SNPs in non-coding region. Red: the mutated base; base in brackets: reference base at mutated point; dash: single base deletion. Promoters were predicted by Berkeley Drosophila Genome Project (prokaryote organism; http://www.fruitfly.org/seq_tools/promoter.html).
Experimental validation of SNPs in XI and XII. A) The primer sequences used for PCR and then Sanger sequencing. B) The SNPs validated by Sanger sequencing.
Priori ethanol stress rewired cellular networks (XI and XII). Key transport, metabolic and regulatory genes and pathways were illustrated. OD: NAD/NADP octopine dehydrogenase; FITP: ferrous iron transport protein B. Other abbreviations were as in Figure 5.
Developmental model of solvent-tolerance traits in thermopiles. A) Model of cellular responses upon environmental stimuli: the transition from shock response (wild type) to low-tolerance (XI) and eventually to high-tolerance (XII). B) Model of the cellular states after a priori long-term exposure to exogenous ethanol. Boxes represented genes/pathways, whereas box size indicated the number of differentially expressed genes in the pathway. Differential-expression ratios (log2R) for the genes/pathways were represented by colors. Asterisks indicated non-synonymous mutated genes in the pathways or the SNPs in predicted promoters of genes. Tra (P): ABC transporter related in COG P; MM: membrane metabolism; AAM: amino acid metabolism; LFA: long-chain fatty acid.
Differentially expressed genes in mutant. A) XI under normal growth condition (XI-0%). B) XII under normal growth condition (XII-0%).
Differentially expressed genes in mutants under ethanol stress. A) XI under 2% ethanol (XI-2%). B) XII under 6% ethanol (XII-6%).
Homology models of the mutant protein structures DeoR (A) and AdhE (B) in Thermoanaerobacter sp. X514. Mutation sites, cofactors and ligands were respectively labeled.
The Dynamic operons between X and XII (Teth5140597-0601). Gray and black areas respectively represented the coverage of mRNA in X and XII in ethanol-free media. The scale with the relative score indicated, for each library and at a given nucleotide (among the three biological replicates), the mean number of uniquely mapped reads normalized to the total number of such reads.
The Dynamic operons between XI and XII in the absence of ethanol. A-B) Teth5140161-0166; C-D) Teth5140412-0414; E-F) Teth5141359-1361. The scale was as in Additional file 18.
The Dynamic operon between XII-0% and XII-6% (Teth5141785-1787). The scale was as in Additional file 18.
Genetic and non-genetic approaches that improved both ethanol production and tolerance.A-B) Growth curves of Thermoanaerobacter sp. X514 wild type strains that carried different plasmids (a vector-only control, an adh cluster (Teth5140145-0146) or a ϭ24 cluster (Teth5141847-1848)). Strains were grown at 45°C in QRCM medium with 0.25% and 0.5% added ethanol respectively. C) Fermentation experiments testing effects of vitamin B12 on ethanol production of tolerant mutants in defined medium at 60°C. Vitamin B12 concentrations in defined medium were indicated on x-axis. The means (and standard deviation) for the three biological replicates were shown for each sample. Differences in means were evaluated using one-tailed paired t-test (those with p<0.05 were shown).