Abstract
Background
Phyllodes tumors are uncommon breast tumors that account for less than 0.5% of all breast malignancies. After metastases develop, the prognosis is poor, with very few patients living more than 1 year. The biology of this unusual cancer is not understood and, consequently, no potential targets for treatments are currently available. There has been an exponential increase in the number of commercially available tumor profiling services. Herein, we report a case of metastatic malignant phyllodes tumor for which a comprehensive molecular analysis was performed by using Clinical Laboratory Improvement Amendments (CLIA)-certified labs, providing new insights into the potential opportunities for molecularly targeted therapies for this extremely rare disease.
Methods
Next-generation sequencing was performed by using the FoundationOne™ platform (Foundation Medicine, Cambridge, MA). Whole-genome array-based comparative genomic hybridization (array CGH) was performed by using the DNAarray™ (CombiMatrix Diagnostics, Irvine, CA). Immunohistochemical and morphoproteomics analysis were performed at Consultative proteomics®, The University of Texas, UT Health Medical School, Houston,TX (Robert E Brown Lab); Clarient Diagnostics, Aliso Viejo, CA; and Caris Life Sciences Target one, Irving, TX, USA.
Results
Next-generation sequencing showed 3 aberrant genes: activating mutation Q61L on NRAS; inactivating mutations Q504* and K740* on RB1; and TP53 loss. Whole-genome array-based comparative genomic hybridization (array CGH) revealed amplifications of chromosome (chr.) 1 (CKS1B gene), chr. 8 (MYC gene), and chr. 9 (CDKN2A gene) Deletions of chr. 17 (TP53), chr. 10 (GATA3), chr. 11 (FGF4 and CCND1 genes), and chr.22 (PDGFβ). Immunohistochemical analysis for relevant markers showed a positive staining for transducing-like enhancer of split (TLE) 3; secreted protein acidic and rich in cysteine (SPARC) was expressed at 2-3+ in the cytoplasm of the tumors cells, whereas mammalian target of rapamycin (mTOR) was expressed up to 2+ in the nuclei of the tumor cells.
Conclusions
We describe for the first time an NRAS mutation with concomitant activation of PI3K/Akt/mTOR in phyllodes tumor. We also found markers for sensitivity to taxane-based therapies, especially albumin-bound paclitaxel. Exploring the biology of rare malignancies by CLIA certified labs may be reasonable strategy for the development of targeted treatments.
Keywords: Phyllodes tumor, Phyllodes sarcoma, Sarcoma, Targeted therapy, Chemotherapy, SPARC, TLE3, NRAS, P53, P13K, Mutation, Rare disease, Breast cancer, Breast sarcoma, Next generation sequencing, CLIA, Immunohistochemistry, Morphoproteomics, Proteomics, Whole genome sequencing
Background
Phyllodes tumors are uncommon breast tumors that account for less than 0.5% of all breast malignancies [1]. Histologically, a phyllodes tumor is a fibroepithelial neoplasia subdivided into benign, borderline, and malignant subtypes. Malignant phyllodes are the most aggressive subtype in this spectrum and correspond to about 20% of all phyllodes tumors [2]. They are characterized by high mitotic rates, marked stromal cellularity and atypia, and infiltrative margins [3]. In contrast to benign and borderline lesions, malignant tumors can metastasize in up to 22% of cases [1,4-6]. After metastases develop, the prognosis is poor, with very few patients living more than 1 year [7,8]. The biology of this unusual cancer is not understood and consequently, no potential targets for treatments are currently available.
Herein we report a case of metastatic malignant phyllodes tumor for which a comprehensive molecular analysis was performed by using Clinical Laboratory Improvement Amendments (CLIA)-certified labs, providing new insights into the potential opportunities for molecularly targeted therapies for this extremely rare disease.
Methods
We reviewed the medical chart of a patient with phyllodes tumor that presented to the Department of Investigational cancer therapeutics for targeted therapy options. The patient requested a commercially available comprehensive molecular analysis by CLIA certified labs.
Next-generation sequencing
Next-generation sequencing was performed by using the Clinical Laboratory Improvement Amendments (CLIA)-approved FoundationOne™ platform (Foundation Medicine, Cambridge, MA, USA). FoundationOne™ is a targeted assay utilizing next generation sequencing in routine cancer specimens. The test simultaneously sequences the entire coding sequence of 236 cancer-related genes (3,769 exons) plus 48 introns from 20 genes often rearranged or altered in cancer to an average depth of coverage of >250X. It detects all class of genomic alterations (including base substitutions, insertions and deletions, copy number alterations and rearrangements) using routine FFPE tissue samples that may be as small as 40 μm.
Whole-genome array-based comparative genomic hybridization (array CGH)
Whole-genome array-based comparative genomic hybridization (array CGH) was performed by using the DNAarray™ (CombiMatrix Diagnostics, Irvine, CA). Whole-genome array-based comparative genomic hybridization (array CGH) was performed using the DNAarray™ - Breast Profile designed to detect genome-wide copy number variations. The array contains 3000+ unique large-insert clones covering coding and non-coding human genome sequences with content sourced from the UCSC hg18 human genome (NCBI build 36) and an average probe spatial resolution of ~800 Kb. DNA copy number in the patient sample was evaluated in relation to a reference diploid DNA sample, and evaluation of copy number changes was performed in all covered regions of the genome including pericentromeric, subtelomeric, and loci representative of malignant genomic changes.
Immunohistochemical and morphoproteomics analysis
Immunohistochemical and morphoproteomics analysis were performed at Consultative proteomics®, The University of Texas, UT Health Medical school, Houston,TX, USA (Robert E Brown Lab); Clarient Diagnostics, Aliso Viejo, CA, USA; and Caris Life Sciences Target one, Irving, TX, USA.
Fluorescence in situ hybridization (FISH)
FISH was performed at Clarient Diagnostics, Aliso Viejo, CA, USA. HER-2 gene amplification was assessed utilizing the PathVysion assay (Vysis Corp., Downers Grove, Illinois, USA). The identification probes for the HER-2 (SpectrumOrange) and alphasatellite DNA sequence at the centromeric region of chromosome 17 (SpectrumGreen) were hybridized according to the manufacturer’s guidelines. At least twenty non-overlapping nucleicontaining at least one orange and one green signal were enumerated. The ratio of orange signals (HER-2 gene) to green signals (chromosome 17) was calculated. A ratio greater thanor equal to 2.0 is considered as amplified based on the FDA approval in this kit. The College of American Pathologists (CAP) HER-2 consensus conference 2002 suggested that a ratio of 1.8-2.2 be considered as borderline.
Results and discussion
Case history
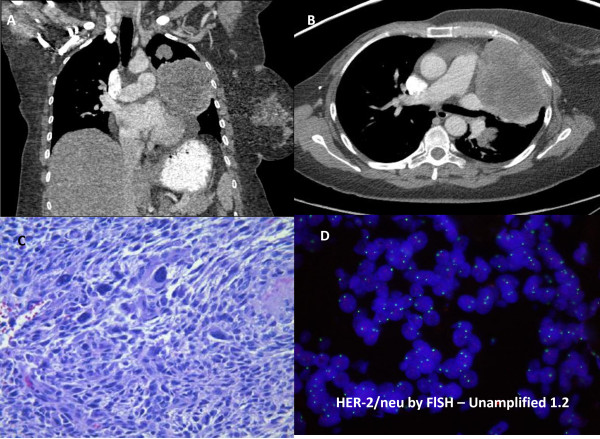
A 55-year-old woman with a metastatic malignant phyllodes tumor presented to our clinic to discuss treatment options. Fifteen months earlier, a mass had been detected in her right breast for which a biopsy was consistent with metaplastic carcinoma/sarcoma. She underwent total mastectomy, and the diagnosis washigh-grade sarcoma arising in a phyllodes tumor with no positive lymph nodes (0/14). Immunohistochemical analysis showed that the lesion was negative for estrogen and progesterone receptor and HER-2/neu. During a follow-up examination 12 months later, computed tomography (CT) of the chest revealed five lung masses with the dominant mass measuring 6 × 4.5 × 4.5 cm within the lingula. No other sites of metastatic disease were detected. A repeated chest CT also showed multiple masses in the left lung, which is consistent with metastatic disease. The largest mass measured 10.2 × 7.6 × 8.1 cm (Figure 1A and B). Pathology review of the mastectomy product at our institution confirmed the diagnosis of a malignant phyllodes tumor characterized by stromal overgrowth of a high-grade spindle cell sarcoma with marked stromal pleomorphism, a high mitotic rate, focal cystic change, and areas of necrosis (Figure 1C). Considering the rarity of this disease and the lack of standard care options, the patient requested a commercially available comprehensive molecular analysis (Tables 1 and 2) including a whole-genome array-based comparative genomic hybridization study (array CGH) (Figures 2) and immunohistochemistry and morphoproteomics studies. (Figure 3).
Figure 1.
Imaging studies, histopathology and FISH studies: UA and B: CT of the chest showing metastatic masses in the lung. C Hematoxylin and Eosin stained section of the tumor from the right mastectomy shows a malignant sarcoma/malignant phyllodes tumor that has high stromal cellularity and a mitotic index of 11 per 10 high power fields. DHER-2/neu by Fluorescence in Situ Hybridization (FISH) HER-2 gene amplification was assessed utilizing the PathVysion assay (Vysis Corp., Downers Grove, Illinois). The identification probes for the HER-2 (SpectrumOrange) and alpha satellite DNA sequence at the centromeric region of chromosome 17 (SpectrumGreen) were hybridized according to the manufacturer’s guidelines. At least twenty non-overlapping nuclei containing at least one orange and one green signal were enumerated. The ratio of orange signals (HER-2 gene) to green signals (chromosome 17) was calculated. A ratio greater than or equal to 2.0 is considered as amplified based on the FDA approval in this kit. The CAP HER-2 consensus conference 2002 suggested that a ratio of 1.8-2.2 be considered as borderline.
Table 1.
Summary of molecular profiling analyses from various CLIA certified methods in a patient with malignant phyllodes sarcoma
| Marker | Result | Details |
|---|---|---|
|
Focused exome sequencing | ||
|
NRAS |
Mutated |
Q61L |
|
TP53 |
Deletion |
Also proved by aCGH1 |
|
RB1 |
Mutated |
Q504* and K740* |
|
Array comparative hybridization | ||
|
Amplifications |
Chr. 1, 8 and 9 |
CKS1B, MYC and CDKN2A genes |
|
Deletions |
Chr. 10, 11, 17 and 22 |
GATA3, CCND1, TP53 and PDGFB gene |
|
Immunohistochemistry | ||
|
p-AKT |
Positive |
Intensity of 2 in 40% of cells |
|
PDGFRβ |
Positive |
Intensity of 2 in 90% of cells |
|
PDGFRα |
Positive |
Intensity of 2 in 80% of cells |
|
EGFR |
Positive |
Intensity of 2 in 70% of cells |
|
TLE3 |
Positive |
Intensity of 2 in 35% of cells |
|
PTEN |
Present |
Intensity of 2 in 90% of cells |
|
Kit (CD117) |
Negative |
No staining |
|
Morphoproteomic immunohistochemistry | ||
|
mTOR |
Positive |
2+ morpho test |
|
SPARC |
Positive |
2-3+ morpho test |
| ER-α | Negative | Morpho test |
1aCGH: array-based comparative genomic hybridization.
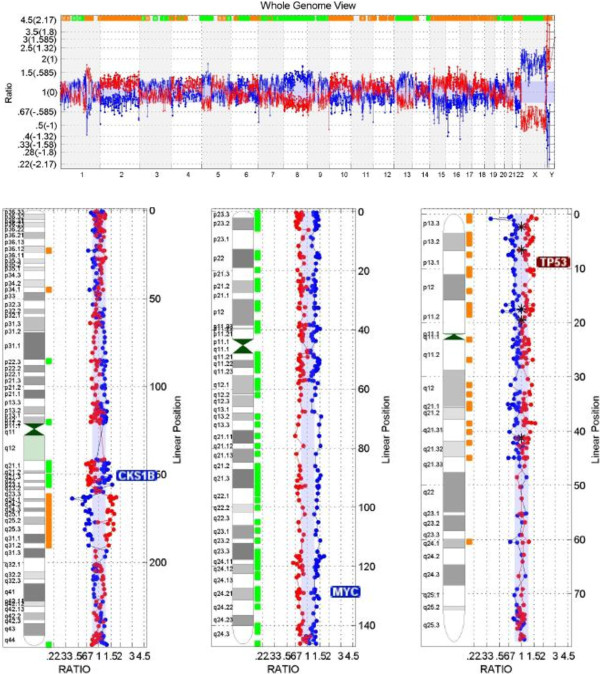
Figure 2.
Shows genome viewer as reported by Combimatrix positive for high level chromosomal instability (CIN). Amplification of chromosome 1 (CKS1B gene), chromosome 8 (MYC gene), and chromosome 9 (CDKN2A gene) were detected. Deletions of chromosome 17 (TP53 gene), chromosome 10 (GATA3), chromosome 11 (FGF4 and CCND1 genes) and chromosome 22 (PDGFB gene) were also detected.
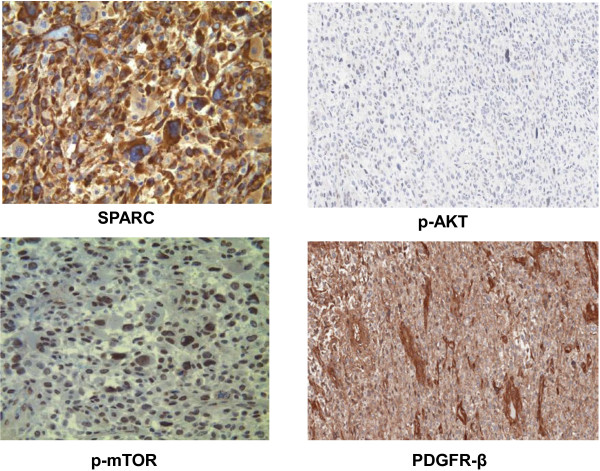
Figure 3.
Immunohistochemistry and morphoproteomic studies: SPARC (secreted protein acidic and rich in cysteine) is expressed at 2-3+ in the cytoplasm of the tumor cells. The mammalian target of rapamycin (mTOR) Ser 2448 is expressed up to 2+ in the nuclei of the tumor cells. P-AKT and PDGFR β IHC staining showing an intensity of 2+.
Next-generation exome sequencing and whole-genome array-based comparative genomic hybridization
Three genes were identified with significant abnormalities: activating mutation Q61L on NRAS; inactivating mutations Q504* and K740* on RB1; and TP53 loss. Whole-genome array-based comparative genomic hybridization (array CGH) was performed by using the CLIA-certified DNAarray™ (CombiMatrix Diagnostics, Irvine, CA). This analysis was highly positive for chromosomal instability, revealing amplifications of chromosome (chr.) 1 (CKS1B gene), chr. 8 (MYC gene), and chr.9 (CDKN2A gene) (Figure 2). Deletions of chr. 17 (TP53), chr. 10 (GATA3), chr. 11 (FGF4 and CCND1 genes), and chr. 22 (PDGFβ) were also detected (Figure 2). In addition, HER-2/neu was confirmed as negative by Fluorescence in Situ Hybridization (FISH) (Figure 1D).
Immunohistochemical and morphoproteomics analysis
Immunohistochemical analysis for relevant markers showed a positive staining for transducing-like enhancer of split (TLE) 3; secreted protein acidic and rich in cysteine (SPARC) was expressed at 2-3+ in the cytoplasm of the tumors cells, whereas mammalian target of rapamycin (mTOR) was expressed up to 2+ in the nuclei of the tumor cells (Figure 3). Other positive relevant markers were phospho-AKT, PTEN, VEGF, PDGFR β, PDGFR α, ERCC1, and EGFR. Negative markers included ER alpha, CD117, and MGMT. All of these analyses were performed at Consultative proteomics®, The University of Texas, UT Health Medical School Houston,TX (Robert E Brown Lab); Clarient Diagnostics, Aliso Viejo, CA; and Caris Life Sciences Target one, Irving, TX.
Discussion
We have reported the first complete characterization by genomic and proteomic analysis of a metastatic phyllodes tumor of the breast by commercially available profiling services (Table 2). This characterization was performed in CLIA laboratories and not in a research environment, meaning that clinical targeted therapy decisions could be made including enrollment of a patient into a clinical trial or off label use of an FDA approved agent (depending upon the molecular aberration) that could benefit this patient. With the recent increase in the number of commercially available molecular profiling services these tests are just a phone call away within the patients reach and no longer in the realm of just research publications in high impact journals. There has been an exponential increase in motivated patients with cancer who have the resources requesting these services and present to the clinic with these profiles. These present with a challenge and an opportunity for practicing oncologists. These have been useful in unraveling the biology of extremely complex and rare diseases that have no standard care therapy.
Table 2.
Summary of molecular aberrations in genes, receptors and pathways by various CLIA certified methods in a patient with phyllodes tumor
| RAS/RAF/MAPK | P13CK/AKT/mTOR | Cell cycle/Apoptosis/Tumor Suppressor/Other pathways | Chemosensitivity markers | ||
|---|---|---|---|---|---|
|
CLIA Method,Lab and location |
|
|
|
Positive markers |
Negative Markers |
|
Foundation Next Gen Sequencing, Cambridge, Boston MA |
NRASQ61L |
|
RB1 mutation and TP53 loss |
|
|
|
aCGHCombimatrix Array, Irvine, CA |
|
|
CKS1B amplification, TP53 loss |
|
|
|
IHC –Clarient,Aliso Viejo, CA |
|
p-AKT |
|
PDGFR-α and β PTEN |
Her-2 |
|
Morphoproteomics, UT Houston, TX |
|
p-MTOR, |
|
SPARC |
ER-alpha |
|
Caris Target Now, Irving, Tx |
|
|
|
TLE3, ERCC1 |
|
| Potential Targeted therapy options | MEK inhibitors | P13CK*/AKT*/mTOR inhibitors | MEK inhibitors | Taxanes, albumin-bound paclitaxel; PDGFR inhibitors | |
Potential targeted therapy options based on best pre-clinical and/or clinical evidence are listed in the last row.
The case reported herein has several clinical features typical of metastatic malignant phyllodes tumor. Previous published reports have reported a median age at diagnosis of 50 years, the time to development of metastatic lesions between 12 and 24 months after surgery [7], and a predominance of metastasis to the lungs [7,9,10]. Moreover, this patient presented with well-characterized risk factors for the development of metastatic disease, including the presence of stromal overgrowth, mastectomy at initial surgery, larger tumor size, and high mitotic index [11].
Metastatic malignant phyllodes tumor is associated with a dismal prognosis. Mean overall survival duration in this setting is 30 months according to some series [9]. The stromal component is accepted as being responsible for the metastatic behavior, and systemic treatment often is based on guidelines for soft tissue sarcoma. Previous series showed some activity of cisplatin combined with etoposide [12] or doxorubicin [13] and of ifosfamide [14]. Nonetheless, larger series evaluating the role of adjuvant chemotherapy suggested that this subtype of breast tumor presents low sensitivity to chemotherapy [15].
Some recent studies are describing genetic changes associated with this disease. Array CGH has determined that the most frequent changes were gain of 1q and loss of 3p [16]. Interestingly, in one study, gain of 1q material was significantly associated with histologically defined stromal overgrowth and a higher likelihood of recurrence [17]. Here we described a patient presenting stromal overgrowth and a metastatic recurrence with a genetic gain in 1q associated with CSK1B gene amplification, Amplification and over expression of the CSK1 gene inhibited apoptosis of cells through the MEK/ERK pathway and was associated with poor prognosis in breast cancer cells [18]. Other genetic imbalances described herein, such as gain in chr. 8 and loss in chr.10, have already been described, suggesting a high level of genomic instability in these tumors [19].
In fact, mutations in the tumor suppressor gene TP53 appear to lead to a high level of chromosomal instability and drive oncogenesis in soft tissue sarcomas [20]. Loss of TP53 in our patient might be associated with the higher level of chromosomal instability detected. There are no reports of p53 loss in phyllodes tumors in the Catalogue of Somatic Mutations in Cancer (COSMIC) database, although 2 of 30 patients (7%) presented with TP53 mutations. Previous reports suggested a relationship between TP53 expression and the malignant potential of phyllodes tumor [21,22] but the consequences of this genetic abnormality still needs to be clarified. Other genetic abnormalities in phyllodes tumor that have been described in the COSMIC database (as of February 2013) are CDKN2A mutation (1/25 ptes), KIT mutation (1/26 ptes), and PI3KCA mutation (1/1 pte).
Our genomic analysis reports for the first time a mutation in the NRAS gene in breast sarcomas. The mutation detected here (Q61L) has been shown to markedly attenuate GTP hydrolysis maintaining NRAS in an active GTP state [23]. Activation of this protein causes cell growth, differentiation, and survival mainly through the RAF/MAPK/ERK pathway [23]. Targeting this pathway with MEK inhibitors showed activity for patients with melanoma presenting with NRAS mutations [24]. Nonetheless, NRAS is believed to activate PI3K signaling in addition to the MAPK pathway and, indeed, we demonstrated strong expression of p-AKT and p-mTOR in this patient, suggesting concomitant activation of the PI3K pathway. This activation was not mediated by PI3KCA mutation or PTEN loss in this patient, indicating again a role for NRAS-mediated signaling. Recent evidence suggested that combining the targeting of both the MEK/ERK and PI3K/mTOR pathways might be a better strategy for the treatment of NRAS mutant tumors [25]. Considering both the presence of the NRAS mutation and CSK1B amplification, the use of a MEK inhibitor (especially with agents blocking thePI3K pathway) would be reasonable for this patient.
Other interesting findings in this patient were the expression of TLE3 and SPARC. The first acts downstream of beta-catenin influencing microtubule stability, and a previous study indicated that TLE3 expression was associated with improved response to taxane-based therapy in breast tumors [26]. The positivity of SPARC indicates that there may be greater delivery of albumin-bound paclitaxel to the malignant sarcoma, since SPARC is a facilitator that allows more chemotherapeutic agents to concentrate in the surrounding tumoral microenvironment [27]. Tumor responses to albumin-bound paclitaxel have already been linked to SPARC expression in some tumors [28].
The expression of estrogen receptor alpha was negative in most of the tumor, which is a well-described finding for the stromal component of phyllodes tumors [29]. Expression of both PDGFRα and β have already been described in phyllodes tumors and have been associated with high histologic grade and worse prognosis [30]. The therapeutic implication of this finding is not well understood, although a previous response to sunitinib, a known PDGFR inhibitor, in a metastatic phyllodes tumor was reported [31]. Of note, the patient who responded to sunitinib was also treated with paclitaxel, which our data showed may be active in this disease. A major limitation of this comprehensive molecular profiling is the assessment of response to molecularly targeted matched therapy. We made several treatment recommendations for this particular patient based on the discussion provided in the manuscript. These included combination of MEK inhibitors and PI3K inhibitors with or without a taxane based regimen. Unfortunately this patient came from a different country where these drugs are not available as clinical trials. In addition, due to insurance issues the patient could not be treated on our service. This is a common issue in clinic especially because insurance companies usually request a high level of evidence for allowing treatments in even rare diseases.
Conclusion
In summary, this is the first report of a whole genomic profile and proteomics analysis of a metastatic phyllodes tumor of the breast. We described an NRAS mutation with concomitant activation of PI3K/Akt/mTOR, suggesting a potential role for a combination of MEK and PI3K inhibitors. We also found markers for sensitivity to taxane-based therapies, especially albumin-bound paclitaxel. Exploring the biology of rare malignancies may be a reasonable strategy for the development of targeted treatments.
Competing interests
The authors declare no competing interests other than those mentioned in the acknowledgements section.
Authors’ contributions
All authors contributed to writing the manuscript. DLFJ, AC and VS conceived the manuscript. AC and VS provided clinical expertise. DLFJ, AC and VS analysed the data. DLFJ, AC, VS wrote the paper. AC and VS provided sarcoma expertise. DLFJ and VS provided cellular, molecular and targeted therapy expertise. All authors read and approved the final manuscript.
Contributor Information
Denis L Fontes Jardim, Email: jardimde@gmail.com.
Anthony Conley, Email: aconley@mdanderson.org.
Vivek Subbiah, Email: vsubbiah@mdanderson.org.
Acknowledgements
We thank the patient and the family for their motivation in seeking extensive profiling services. The molecular profiling services were requested by the patient and performed in the CLIA certified labs, Foundation Medicine, Cambridge, MA, USA; Caris target Now, Irving, Texas, USA; UT Houston Morphoproteomics, Houston Texas; Combimatrix CGH, Irvine, CA, USA; Clarient, Aliso Viejo, CA, USA through “N-of-One therapeutics”, Boston, MA, USA a fee for service resource for patients with cancer. We wish to acknowledge Dr. Robert Brown, MD for his extensive morphoproteomics and insightful discussions. We wish to acknowledge Dr. Funda Meric-Bernstam, MD for supporting the publication. The University of Texas MD Anderson cancer center is supported by National Institutes of Health grant CA016672. Vivek Subbiah, MD has clinical trial research support from Genentech/Roche, Glaskosmithkline and Northwest Biotherapeutics.
References
- Geisler DP, Boyle MJ, Malnar KF, McGee JM, Nolen MC, Fortner SM, Broughan TA. Phyllodes tumors of the breast: a review of 32 cases. Am Surg. 2000;66(4):360–366. [PubMed] [Google Scholar]
- Bellocq JP, Magro G. In: World Health Organization classification of tumours: Pathology and genetics tumours of the breast and female genital organs. Tavassoli FAD,P, editor. Lyon: IARC Press; 2003. Fibroepithelial tumours; pp. 99–103. [Google Scholar]
- Hawkins RE, Schofield JB, Fisher C, Wiltshaw E, McKinna JA. The clinical and histologic criteria that predict metastases from cystosarcoma phyllodes. Cancer. 1992;69(1):141–147. doi: 10.1002/1097-0142(19920101)69:1<141::AID-CNCR2820690125>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Moffat CJ, Pinder SE, Dixon AR, Elston CW, Blamey RW, Ellis IO. Phyllodes tumours of the breast: a clinicopathological review of thirty-two cases. Histopathology. 1995;27(3):205–218. doi: 10.1111/j.1365-2559.1995.tb00212.x. [DOI] [PubMed] [Google Scholar]
- Mokbel K, Price RK, Mostafa A, Wells CA, Carpenter R. Phyllodes tumour of the breast: a retrospective analysis of 30 cases. Breast. 1999;8(5):278–281. doi: 10.1054/brst.1999.0058. [DOI] [PubMed] [Google Scholar]
- Reinfuss M, Mitus J, Smolak K, Stelmach A. Malignant phyllodes tumours of the breast. A clinical and pathological analysis of 55 cases. Eur J Cancer. 1993;29A((9):1252–1256. doi: 10.1016/0959-8049(93)90067-p. [DOI] [PubMed] [Google Scholar]
- Suzuki-Uematsu S, Shiraishi K, Ito T, Adachi N, Inage Y, Taeda Y, Ueki H, Ohtani H. Malignant phyllodes tumor composed almost exclusively of a fibrosarcomatous component: a case report and review of malignant phyllodes tumors with metastases. Breast cancer (Tokyo, Japan) 2010;17(3):218–224. doi: 10.1007/s12282-009-0099-7. [DOI] [PubMed] [Google Scholar]
- Asoglu O, Ugurlu MM, Blanchard K, Grant CS, Reynolds C, Cha SS, Donohue JH. Risk factors for recurrence and death after primary surgical treatment of malignant phyllodes tumors. Ann Surg Oncol. 2004;11(11):1011–1017. doi: 10.1245/ASO.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Belkacemi Y, Bousquet G, Marsiglia H, Ray-Coquard I, Magne N, Malard Y, Lacroix M, Gutierrez C, Senkus E, Christie D. et al. Phyllodes tumor of the breast. Int J Radiat Oncol Biol Phys. 2008;70(2):492–500. doi: 10.1016/j.ijrobp.2007.06.059. [DOI] [PubMed] [Google Scholar]
- Macdonald OK, Lee CM, Tward JD, Chappel CD, Gaffney DK. Malignant phyllodes tumor of the female breast: association of primary therapy with cause-specific survival from the Surveillance, Epidemiology, and End Results (SEER) program. Cancer. 2006;107(9):2127–2133. doi: 10.1002/cncr.22228. [DOI] [PubMed] [Google Scholar]
- Fou A, Schnabel FR, Hamele-Bena D, Wei XJ, Cheng B, El Tamer M, Klein L, Joseph KA. Long-term outcomes of malignant phyllodes tumors patients: an institutional experience. Am J Surg. 2006;192(4):492–495. doi: 10.1016/j.amjsurg.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Burton GV, Hart LL, Leight GS Jr, Iglehart JD, McCarty KS Jr, Cox EB. Cystosarcoma phyllodes. Effective therapy with cisplatin and etoposide chemotherapy. Cancer. 1989;63(11):2088–2092. doi: 10.1002/1097-0142(19890601)63:11<2088::AID-CNCR2820631103>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Allen R, Nixon D, York M, Coleman J. Successful chemotherapy for cystosarcoma phyllodes in a young woman. Arch Intern Med. 1985;145(6):1127–1128. doi: 10.1001/archinte.1985.00360060195030. [DOI] [PubMed] [Google Scholar]
- Hawkins RE, Schofield JB, Wiltshaw E, Fisher C, McKinna JA. Ifosfamide is an active drug for chemotherapy of metastatic cystosarcoma phyllodes. Cancer. 1992;69(9):2271–2275. doi: 10.1002/1097-0142(19920501)69:9<2271::AID-CNCR2820690911>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Morales-Vasquez F, Gonzalez-Angulo AM, Broglio K, Lopez-Basave HN, Gallardo D, Hortobagyi GN, De La Garza JG. Adjuvant chemotherapy with doxorubicin and dacarbazine has no effect in recurrence-free survival of malignant phyllodes tumors of the breast. Breast J. 2007;13(6):551–556. doi: 10.1111/j.1524-4741.2007.00510.x. [DOI] [PubMed] [Google Scholar]
- Jee KJ, Gong G, Ahn SH, Park JM, Knuutila S. Gain in 1q is a common abnormality in phyllodes tumours of the breast. Anal cell pathol: J Euro Society for Anal Cell Pathol. 2003;25(2):89–93. doi: 10.1155/2003/803192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Birdsall S, Osin P, Gusterson B, Shipley J. Phyllodes tumors of the breast analyzed by comparative genomic hybridization and association of increased 1q copy number with stromal overgrowth and recurrence. Genes Chromosomes Cancer. 1997;20(3):275–281. doi: 10.1002/(SICI)1098-2264(199711)20:3<275::AID-GCC8>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Wang XC, Tian LL, Tian J, Wu HL, Meng AM. Overexpression of Cks1 is associated with poor survival by inhibiting apoptosis in breast cancer. J Cancer Res Clin Oncol. 2009;135(10):1393–1401. doi: 10.1007/s00432-009-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jones AM, Mitter R, Springall R, Graham T, Winter E, Gillett C, Hanby AM, Tomlinson IP, Sawyer EJ, Phyllodes Tumour C. A comprehensive genetic profile of phyllodes tumours of the breast detects important mutations, intra-tumoral genetic heterogeneity and new genetic changes on recurrence. J Pathol. 2008;214(5):533–544. doi: 10.1002/path.2320. [DOI] [PubMed] [Google Scholar]
- Perot G, Chibon F, Montero A, Lagarde P, De The H, Terrier P, Guillou L, Ranchere D, Coindre JM, Aurias A. Constant p53 pathway inactivation in a large series of soft tissue sarcomas with complex genetics. Am J Pathol. 2010;177(4):2080–2090. doi: 10.2353/ajpath.2010.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatalica Z, Finkelstein S, Lucio E, Tawfik O, Palazzo J, Hightower B, Eyzaguirre E. p53 protein expression and gene mutation in phyllodes tumors of the breast. Pathol Res Pract. 2001;197(3):183–187. doi: 10.1078/0344-0338-00031. [DOI] [PubMed] [Google Scholar]
- Korcheva VB, Levine J, Beadling C, Warrick A, Countryman G, Olson NR, Heinrich MC, Corless CL, Troxell ML. Immunohistochemical and molecular markers in breast phyllodes tumors. Appl Immunohistochem Mol Morphol: AIMM/official publication of the Society for Appl Immunohistochem. 2011;19(2):119–125. doi: 10.1097/PAI.0b013e3181f5349a. [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. RAS oncogenes: weaving a tumorigenic web. Nat Rev Cancer. 2011;11(11):761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascierto PA, Schadendorf D, Berking C, Agarwala SS, Van Herpen CM, Queirolo P, Blank CU, Hauschild A, Beck JT, St-Pierre A. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–256. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- Posch C, Moslehi H, Feeney L, Green GA, Ebaee A, Feichtenschlager V, Chong K, Peng L, Dimon MT, Phillips T, Combined targeting of MEK and PI3K/mTOR effector pathways is necessary to effectively inhibit NRAS mutant melanoma in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America. 2013. [DOI] [PMC free article] [PubMed]
- Kulkarni SA, Hicks DG, Watroba NL, Murekeyisoni C, Hwang H, Khoury T, Beck RA, Ring BZ, Estopinal NC, Schreeder MT. et al. TLE3 as a candidate biomarker of response to taxane therapy. Breast cancer res: BCR. 2009;11(2):R17. doi: 10.1186/bcr2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradishar WJ. Albumin-bound paclitaxel: a next-generation taxane. Expert Opin Pharmacother. 2006;7(8):1041–1053. doi: 10.1517/14656566.7.8.1041. [DOI] [PubMed] [Google Scholar]
- Desai N, Trieu V, Damascelli B, Soon-Shiong P. SPARC expression correlates with tumor response to albumin-bound paclitaxel in head and neck cancer patients. Transl Oncol. 2009;2(2):59–64. doi: 10.1593/tlo.09109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse GM, Lee CS, Kung FY, Scolyer RA, Law BK, Lau TS, Putti TC. Hormonal receptors expression in epithelial cells of mammary phyllodes tumors correlates with pathologic grade of the tumor: a multicenter study of 143 cases. Am J Clin Pathol. 2002;118(4):522–526. doi: 10.1309/D206-DLF8-WDNC-XJ8K. [DOI] [PubMed] [Google Scholar]
- Feakins RM, Wells CA, Young KA, Sheaff MT. Platelet-derived growth factor expression in phyllodes tumors and fibroadenomas of the breast. Hum Pathol. 2000;31(10):1214–1222. doi: 10.1053/hupa.2000.18481. [DOI] [PubMed] [Google Scholar]
- Park IH, Kwon Y, Kim EA, Lee KS, Ro J. Major response to sunitinib (Sutene) in metastatic malignant phyllodes tumor of breast. Invest New Drugs. 2009;27(4):387–388. doi: 10.1007/s10637-008-9195-3. [DOI] [PubMed] [Google Scholar]