Abstract
Background
Kashin-Beck disease is a kind of degenerative osteoarthropathy. Genetic factors may play an important role in the pathogenesis of KBD.
Objective
To investigate the association of the selenoprotein genes GPX1 (rs1050450, rs1800668, and rs3811699), TrxR2 (rs5748469), and DIO2 (rs225014) with Kashin-Beck disease (KBD) in a Tibetan population and to investigate the association of these SNPs with the serum iodine/selenium concentration in the Tibetan population.
Design
Five SNPs including rs1050450, rs1800668, and rs3811699 in the GPX1 gene, rs5748469 in the TrxR2 gene, and rs225014 in the DIO2 gene were analyzed in Tibetan KBD patients and controls using the SNaPshot method. P trend values of the SNPs were calculated using an additive model.
Results
None of the five SNPs in the three genes showed a significant association with KBD. Haplotypes TCC, TTC and TTT of rs1050450, rs1800668 and rs3811699 in GPX1 showed a significant association with KBD and controls with P value of 0.0421, 5.0E-4 and 0.0066, respectively. The GPX1 gene (rs1050450) showed a potential significant association with the iodine concentration in the Tibetan study population (P = 0.02726). However, no such association was detected with the selenium concentration (P = 0.2849).
Conclusion(s)
In this study, we showed that single SNPs in the genes GPX1 (rs1050450, rs1800668 and rs3811699), TrxR2 (rs5748469), and DIO2 (rs225014) may not be significantly associated with KBD in a Tibetan population. However, haplotype analysis of SNPs rs1050450, rs1800668 and rs3811699 in GPX1 gene showed a significant association with KBD. The results suggested that GPX1 gene play a protective role in the susceptivity of KBD in Tibetans. Furthermore, the GPX1 gene (rs1050450) may be significantly associated with the serum iodine concentration in Tibetans.
Introduction
Kashin–Beck Disease (KBD) is named after the two Russian Cossack doctors Nikolai Kashin and Evgeny Beck who first described bone deformities in patients in Russia in 1848 and 1906, respectively [1]. Today, KBD is known as an endemic, chronic, and degenerative osteoarthropathy, with the involvement of epiphyseal cartilage damage, joint damage, and gradual deformation of the bone and joints [2]–[4]. KBD is endemic in a crescent-shaped area encompassing South-Eastern Siberia to North China, Central China, and Chinese Tibet; it is also endemic in Mongolia and North Korea [5]. China has the most KBD patients in the world [1]. The most frequently involved joints are the ankles, knees, wrists, and elbows. The disease often occurs in children aged 5–15 years and is age related, and serious KBD is responsible for significant disability. In some KBD endemic regions in China, the incidence of KBD is about 8.3% (2.5 million of 30 million urban residents affected) [6].
The etiology of KBD is largely unknown. The risk factors are thought to include deficiency in trace elements, mainly selenium and iodine deficiency [7]–[14]. In addition, mycotoxins such as Trichothecene mycotoxin (T-2), which are produced by various fungi such as F. compactum, F. moniliforme, and F. oxysporum in contaminated storage grains, are also suspected factors in KBD susceptibility [15]–[17]. Organic substances such as humic acid and fulvic acid in drinking water have also been implicated in the disease [3], [18], [19]. Among all of these risk factors, selenium and iodine have been extensively studied. Recently we also confirmed that low selenium and iodine concentrations are associated with KBD [20].
Genetic factors also play an important role in the pathogenesis of KBD. Xiong et al. showed that the polymorphisms of the selenoprotein GPX1 gene (rs1050450 and Pro200Leu) were significantly different between patients with KBD and controls (P = 0.013) in a Han Chinese population [21]. To further investigate the potential relationship between selenoprotein genes and KBD susceptibility in Tibetans, we analyzed the association of the three selenoprotein genes GPX1 (rs1050450, rs1800668, and rs3811699), TrxR2 (rs5748469), and DIO2 (rs225014) with Tibetan KBD in this study. Moreover, we investigated the association between these SNPs and serum selenium and iodine concentrations in Tibetans.
Materials and Methods
Study population
KBD patients and matched normal controls in this study were recruited from Tibetan populations in the same endemic villages in Song Pan, Ruo Er Gai, and Hong Yuan counties in the Aba Tibetan Autonomous prefecture of Sichuan Province, China. Clinical examination was performed as the methods of Moreno-Reyes [7] and Greulich [22]. Patients show joint of fingers, toes, knees and ankles swelling (Figure 1). The KBD patients showed specific changes on X-ray photography; they did not have other arthritis diseases such as rheumatoid arthritis (RA), Osteoarthritis (OA), or local inflammation. Normal controls were individuals with a normal joint examination and no other bone or joint disease. Clinical information about the patients and the controls is listed in Table 1. Veinal bloods (5 ml) of the participants were collected for DNA extraction. The Institutional Review Boards of the Sichuan Academy of Medical Sciences & Sichuan Provincial People's Hospital approved this study. All of the participants received and signed the informed consent.
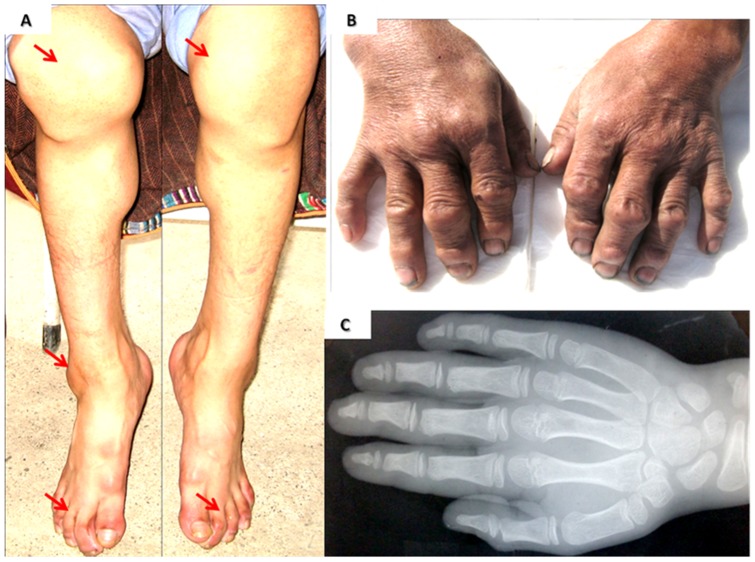
Figure 1. Clinical features of KBD patients.
A. A female KBD patient with her knee, ankle and toe joints deformity, the arrows show multiple joints of this patient are affected. B. A male KBD patient with his fingers joints deformity. C. The X-ray picture of a KBD patient's hand.
Table 1. Characteristics of the KBD cases and controls.
| Characteristics | Cases(n = 638) | Controls(n = 324) |
| Age, mean ± SD | 53.3±13.07 | 54.5±15.81 |
| Sex, male/female | 244/394 | 157/167 |
| Degree | ||
| I | 241 | – |
| II | 336 | – |
| III | 60 | – |
| IV | 1 | – |
Selection of SNPs
Following a review of the literature, we selected five single nucleotide polymorphisms (SNPs) in three selenoprotein genes to genotype: glutathione peroxidase 1 (GPX1, rs1050450), thioredoxin reductase 2 (TrxR2, rs5748469), and iodothyronine deiodinase type 2 (DIO2, rs225014) to genotype [21]. We also referenced the NCBI SNP database and selected rs1800668 and rs3811699 in the GPX1 gene for genotyping. Information pertaining to the five SNPs selected is shown in Table 2.
Table 2. Conditions used for genotyping assays.
| SNPs | Primers | Ta(°C) |
| GPX1(rs1050450) | FW:CGCCAAGAACGAAGAGATTC | 58 |
| missense CCC⇒ CTC Pro200Leu | RV:CTGACACCCGGCACTTTATT | |
| S30-C/T:CTGACATCGAAGCCCTGCTGTCTCAAGGGC | ||
| SV40-A/G:CCAAGCAGCCGGGGTAGGAGGGGCGCCCTAGGCACAGCTG | ||
| GPX1(rs1800668) | FW:ACAGGAGAGGAGGGCTGTTT | 63.5 |
| UTR-5 C/T | RV:AGAAGGCATACACCGACTGG | |
| S60-C/T:CCTGTGCCACGTGACCCGCCGCCGGCCAGTTAAAAGGAGGCGCCTGCTGGCCTCCCCTTA | ||
| GPX1(rs3811699) | FW:AGGACTTCCTGGCCTAGCTC | 63.3 |
| nearGene-5 C/T | RV:CCAGAGGGATCTAGGCTTCC | |
| S50C/T:GCCAAGGAAACGCTGCCGGAGTCCTCCCTCCCTGGCCTCCTCAGGCTGCA | ||
| DIO2(rs225014) | FW:TCGTGAAAGGAGGTCAAGT | 58 |
| missense C/T ACA⇒ GCA Thr92Ala;Thr128Ala | RV:GTGGCAATGTGTTTAATGTG | |
| S50-C/T:CTCAGCTATCTTCTCCTGGGTACCATTGCCACTGTTGTCACCTCCTTCTG | ||
| TrxR2(rs5748469) | FW:TCCCAAAGTGCTGTGAGT | 58 |
| missense A/C GCC⇒ TCC Ala66Ser | RV:ACCTGCTGCTCTCCTTATC | |
| S40-A/C:TGCCTACCTTGGGGAGAAGGTTCCACGTAGTCCACCACGG |
Genotyping
Genomic DNA was extracted using a Gentra Puregene Blood DNA kit (Minneapolis, MN). SNP genotyping was performed by the dye terminator-based SNaPshot method (Applied Biosystems, Foster City, CA). The SNP analysis was performed on the ABI 3130XL genetic analyzer (Applied Biosystems). The genotypes of the SNPs were determined by Genemapper software (Applied Biosystems). All of the SNPs reported in this manuscript had a genotyping success rate >96 percent and accuracy as judged by random regenotyping of 10 percent of the samples in the subject group. The PCR and SNaPshot primers are listed in Table 2.
Statistical analysis
The Hardy-Weinberg equilibrium (HWE) for each SNP polymorphism was tested by the χ2 test with df = 1. The P values of the SNPs were calculated using an additive model. Unadjusted odds ratios of the alleles and genotypes were estimated by the χ2 test. The LD block structure was examined by using software Haploview Vision 4.2. The D and r2 values for all pairs of SNPs were calculated, and the haplotype blocks were estimated by using software Haploview Vision 4.2.All of the statistical analyses were performed using the software SPSS version 10.0. P<0.05 was considered statistically significant.
Results
Genotype analysis of the KBD group and the controls
All of the five SNPs selected were successfully genotyped. However, the results showed that none of these five SNPs were significantly associated with KBD (Table 3). Although the SNP rs1050450 (Pro200Leu) in the GPX1 gene was reported to be significantly associated with KBD in previous studies in a Han Chinese population (P = 0.013), this SNP did not show a significant association with KBD in the Tibetan population in this study (P = 0.1031) [21]. The other two SNPs in the GPX1 gene (rs1800668 at 5′UTR of the gene and rs3811699 at 3′upstream of the gene) also showed no association with KBD (P = 0.7614 and P = 0.8351, respectively). In contrast to the results that Xiong et al. reported in the Han population, neither the DIO2 gene (rs225014) nor the TrxR2 gene (rs5748469) showed significant differences in the Tibetan KBD group compared with the control group (P = 0.7287 and P = 0.4426, respectively).
Table 3. Genotype and allele frequencies of polymorphisms across selenoprotein genes in Tibetan.
| SNPs | Controls | KBD | Trend | ||
| number | Freq | number | Freq. | p-value(OR) | |
| GPX1(rs1050450) | 0.1031(0.74) | ||||
| CC | 271 | 0.836 | 559 | 0.876 | |
| CT | 53 | 0.164 | 79 | 0.124 | |
| TT | 0 | 0 | 0 | 0 | |
| C-allele | 595 | 0.918 | 1197 | 0.938 | |
| T-allele | 53 | 0.082 | 7 | 0.062 | |
| P_HWE | 0.1089 | 0.0956 | |||
| GPX1(rs1800668) | 0.7614(1.06) | ||||
| CC | 183 | 0.806 | 197 | 0.804 | |
| CT | 40 | 0.176 | 41 | 0.167 | |
| TT | 4 | 0.018 | 7 | 0.029 | |
| C-allele | 406 | 0.894 | 435 | 0.883 | |
| T-allele | 48 | 0.106 | 55 | 0.112 | |
| P_HWE | 0.3046 | 0.012 | |||
| GPX1(rs3811699) | 0.8351(0.95) | ||||
| TT | 202 | 0.838 | 212 | 0.841 | |
| CT | 35 | 0.145 | 37 | 0.147 | |
| CC | 4 | 0.017 | 3 | 0.012 | |
| T-allele | 439 | 0.911 | 461 | 0.915 | |
| C-allele | 43 | 0.089 | 43 | 0.085 | |
| P_HWE | 0.0988 | 0.3467 | |||
| DIO2(rs225014) | 0.7287(1.04) | ||||
| TT | 94 | 0.388 | 158 | 0.356 | |
| CT | 113 | 0.467 | 228 | 0.514 | |
| CC | 35 | 0.145 | 58 | 0.131 | |
| T-allele | 301 | 0.622 | 544 | 0.613 | |
| C-allele | 183 | 0.378 | 344 | 0.387 | |
| P_HWE | 0.9121 | 0.0844 | |||
| TrxR2(rs5748469) | 0.4426(0.86) | ||||
| AA | 137 | 0.714 | 209 | 0.749 | |
| AC | 50 | 0.26 | 63 | 0.226 | |
| CC | 5 | 0.026 | 7 | 0.025 | |
| A-allele | 324 | 0.844 | 481 | 0.882 | |
| C-allele | 60 | 0.156 | 77 | 0.138 | |
| P_HWE | 0.8642 | 0.3958 | |||
Haplotype analysis of the three SNPs in the GPX1 gene
We examined the three SNPs in gene GPX1 (rs3811699, rs1050450, and rs1800668) in all tested samples using the program Haploview (Vision4.2). The haplotypes TCC, TTC and TTT generated from these three SNPs proved to be significantly different between the KBD cases and controls (P = 0.0421, 0.0005 and 0.0066, Table 4). These individuals showed protective feature in the susceptibility of KBD with the odds ratios of 0.69, 0.22 and 0.15 respectively.
Table 4. The haplotype association of gpx1 with KBD in this study.
| Haplotype | Frequencies | Chi Square | P value | Odds ratio (95% CI) |
| TCC | 0.848 | 4.132 | 0.0421 | 0.69(0.49–0.98) |
| CTT | 0.06 | 0.222 | 0.6376 | 1.14(0.67–1.91) |
| TTC | 0.032 | 12.183 | 5.00E-04 | 0.22(0.088–0.553) |
| CCT | 0.022 | 0.436 | 0.5092 | 1.34(0.57–3.14) |
| TCT | 0.016 | 0.296 | 0.5861 | 1.32(0.49–3.57) |
| TTT | 0.015 | 7.371 | 0.0066 | 0.15(0.03–0.724) |
Comparison of the Results of the Genotype Analysis Based on the Subjects' Iodine or Selenium Concentrations
Given that KBD is related to iodine or selenium deficiency, we analyzed the genotypes of the KBD group and the controls in relation to the subjects' serum iodine or selenium concentrations which were tested previously [23]. The results are shown in Table 5 and Table 6 for iodine and selenium, respectively. We found no significant differences between the KBD group and the controls, providing additional evidence for the absence of an association between the five SNPs and KBD in the Tibetan population. The power calculation results are shown in Table 7.
Table 5. Genotype and allele frequencies of polymorphisms across selenoprotein genes by serum iodine concentration in Tibetan population.
| SNPs | Controls | KBD | Controls | KBD | Trend | Higher group | Lower group | Higher group | Lower group | Trend |
| number | number | Freq. | Freq. | p-value | number | number | Freq. | Freq. | p-value | |
| GPX1(rs1050450) | 0.9129 | *0.02726 | ||||||||
| CC | 146 | 155 | 0.811 | 0.816 | 154 | 147 | 0.77 | 0.865 | ||
| CT | 34 | 35 | 0.189 | 0.184 | 46 | 23 | 0.23 | 0.135 | ||
| TT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| C-allele | 326 | 345 | 0.906 | 0.908 | 354 | 317 | 0.885 | 0.932 | ||
| T-allele | 34 | 35 | 0.094 | 0.092 | 46 | 23 | 0.115 | 0.068 | ||
| P_HWE | 0.1617 | 0.162 | 0.0661 | 0.14013 | 0.3441 | |||||
| GPX1(rs1800668) | 0.1401 | 0.3982 | ||||||||
| CC | 205 | 207 | 0.804 | 0.881 | 187 | 181 | 0.379 | 0.823 | ||
| CT | 49 | 22 | 0.192 | 0.094 | 48 | 33 | 0.2 | 0.15 | ||
| TT | 1 | 6 | 0.004 | 0.026 | 5 | 6 | 0.021 | 0.027 | ||
| C-allele | 459 | 436 | 0.9 | 0.928 | 422 | 395 | 0.879 | 0.898 | ||
| T-allele | 51 | 34 | 0.1 | 0.072 | 58 | 45 | 0.121 | 0.102 | ||
| P_HWE | 0.2808 | 0 | 0.3634 | 0.0066 | ||||||
| GPX1(rs3811699) | 0.8763 | 0.14013 | ||||||||
| TT | 200 | 212 | 0.84 | 0.841 | 205 | 207 | 0.804 | 0.881 | ||
| CT | 34 | 37 | 0.143 | 0.147 | 49 | 22 | 0.192 | 0.094 | ||
| CC | 4 | 3 | 0.017 | 0.012 | 1 | 6 | 0.004 | 0.026 | ||
| T-allele | 434 | 461 | 0.912 | 0.915 | 459 | 436 | 0.9 | 0.928 | ||
| C-allele | 42 | 43 | 0.088 | 0.085 | 51 | 34 | 0.1 | 0.072 | ||
| P_HWE | 0.08364 | 0.3467 | 0.2808 | 0 | ||||||
| DIO2(rs225014) | 0.6574 | 0.4836 | ||||||||
| TT | 170 | 175 | 0.73 | 0.742 | 70 | 81 | 0.348 | 0.382 | ||
| CT | 56 | 56 | 0.24 | 0.237 | 105 | 106 | 0.522 | 0.5 | ||
| CC | 7 | 5 | 0.03 | 0.021 | 26 | 25 | 0.129 | 0.118 | ||
| T-allele | 396 | 406 | 0.85 | 0.86 | 245 | 268 | 0.609 | 0.632 | ||
| C-allele | 70 | 66 | 0.15 | 0.14 | 157 | 158 | 0.391 | 0.368 | ||
| P_HWE | 0.3712 | 0.8347 | 0.16748 | 0.2748 | ||||||
| TrxR2(rs5748469) | 0.4689 | 0.6574 | ||||||||
| AA | 136 | 209 | 0.716 | 0.749 | 170 | 175 | 0.73 | 0.742 | ||
| AC | 49 | 63 | 0.258 | 0.226 | 56 | 56 | 0.24 | 0.237 | ||
| CC | 5 | 7 | 0.026 | 0.025 | 7 | 5 | 0.03 | 0.021 | ||
| A-allele | 321 | 481 | 0.845 | 0.862 | 396 | 406 | 0.85 | 0.86 | ||
| C-allele | 59 | 77 | 0.155 | 0.138 | 70 | 66 | 0.15 | 0.14 | ||
| P_HWE | 0.8164 | 0.3958 | 0.3712 | 0.8347 |
Table 6. Genotype and allele frequencies of polymorphisms across selenoprotein genes by serum selenium concentration in Tibetan population.
| SNPs | Controls | KBD | Controls | KBD | Trend | Higher group | Lower group | Higher group | Lower group | Trend |
| number | number | Freq. | Freq. | p-value | number | number | Freq. | Freq. | p-value | |
| GPX1(rs1050450) | 0.932 | 0.2849 | ||||||||
| CC | 147 | 155 | 0.812 | 0.816 | 174 | 125 | 0.798 | 0.839 | ||
| CT | 34 | 35 | 0.188 | 0.184 | 44 | 24 | 0.202 | 0.161 | ||
| TT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| C-allele | 328 | 345 | 0.906 | 0.908 | 392 | 274 | 0.899 | 0.919 | ||
| T-allele | 34 | 35 | 0.094 | 0.092 | 44 | 24 | 0.101 | 0.081 | ||
| P_HWE | 0.1631 | 0.162 | 0.2849 | 0.0975 | ||||||
| GPX1(rs1800668) | 0.701 | 0.924 | ||||||||
| CC | 182 | 197 | 0.809 | 0.804 | 173 | 206 | 0.797 | 0.811 | ||
| CT | 39 | 41 | 0.173 | 0.167 | 41 | 40 | 0.189 | 0.157 | ||
| TT | 4 | 7 | 0.018 | 0.029 | 3 | 8 | 0.014 | 0.031 | ||
| C-allele | 403 | 435 | 0.896 | 0.888 | 387 | 452 | 0.892 | 0.89 | ||
| T-allele | 47 | 55 | 0.104 | 0.112 | 47 | 56 | 0.108 | 0.11 | ||
| P_HWE | 0.2706 | 0.0121 | 0.749 | 0.002 | ||||||
| GPX1(rs3811699) | 0.887 | 0.8969 | ||||||||
| TT | 201 | 212 | 0.841 | 0.841 | 185 | 228 | 0.833 | 0.848 | ||
| CT | 34 | 37 | 0.142 | 0.147 | 35 | 36 | 0.158 | 0.134 | ||
| CC | 4 | 3 | 0.017 | 0.012 | 2 | 5 | 0.009 | 0.019 | ||
| T-allele | 436 | 461 | 0.912 | 0.915 | 405 | 492 | 0.912 | 0.914 | ||
| C-allele | 42 | 43 | 0.088 | 0.085 | 39 | 46 | 0.088 | 0.086 | ||
| P_HWE | 0.082 | 0.347 | 0.018 | 0.8099 | ||||||
| DIO2(rs225014) | 0.19 | 0.6357 | ||||||||
| TT | 57 | 94 | 0.425 | 0.337 | 63 | 88 | 0.389 | 0.351 | ||
| CT | 61 | 150 | 0.455 | 0.538 | 72 | 139 | 0.444 | 0.554 | ||
| CC | 16 | 35 | 0.119 | 0.125 | 27 | 24 | 0.167 | 0.096 | ||
| T-allele | 175 | 338 | 0.653 | 0.606 | 198 | 315 | 0.611 | 0.627 | ||
| C-allele | 93 | 220 | 0.347 | 0.394 | 126 | 502 | 0.389 | 0.373 | ||
| P_HWE | 0.9585 | 0.0359 | 0.0035 | 0.4085 | ||||||
| TrxR2(rs5748469) | 0.461 | 0.6092 | ||||||||
| AA | 136 | 209 | 0.716 | 0.749 | 154 | 191 | 0.748 | 0.726 | ||
| AC | 49 | 63 | 0.258 | 0.226 | 47 | 65 | 0.228 | 0.247 | ||
| CC | 5 | 7 | 0.026 | 0.025 | 5 | 7 | 0.024 | 0.027 | ||
| A-allele | 321 | 481 | 0.845 | 0.862 | 355 | 447 | 0.862 | 0.85 | ||
| C-allele | 59 | 77 | 0.155 | 0.138 | 57 | 79 | 0.138 | 0.15 | ||
| P_HWE | 0.8164 | 0.3958 | 0.5367 | 0.606 |
Table 7. Statistical Power Calculations for a Paired t Test and Their Effect on Desired Sample Size.
| iodine(case/control) | selenium(case/control) | iodine(high/low) | selenium(high/low) | |
| α | 0.05 | 0.05 | 0.05 | 0.05 |
| 1-β | 0.8 | 0.8 | 0.8 | 0.8 |
| n | 90 | 1337 | 3 | 56 |
α, (alpha)the threshold value below which statistical significance will be declared.
1-β, (one minus beta) the statistical power.
n, the sample size.
Comparison of the Results of the Genotype Analysis Based on the Higher and Lower Serum Iodine or Selenium Concentration Groups
To further investigate the association of the five SNPs with the serum iodine or selenium concentration, we analyzed the genotype of those in the higher group and lower group of serum iodine or selenium concentration in the Tibetan population (subjects combined the patients and the controls). The mean serum iodine concentration of the combined samples including both cases and controls was 37.33 μg/L. The mean serum iodine concentration of the combined samples including both cases and controls was 37.33 μg/L. The mean serum iodine concentration in the lower group was 24.35 μg/L and 51.64 μg/L in the higher group. The mean serum selenium concentration was 28.65 μg/L. In the lower group, it was 18.18 μg/L, and in the higher group it was 44.40μg/L. The results are presented in the right column of Table 5 and Table 6 for iodine and selenium, respectively. The power calculation results are shown in Table 7. The GPX1 gene (rs1050450) showed a potential significant association with the iodine concentration, with trend p value of 0.02726. However, rs1050450 was not associated with the selenium concentration (trend p value = 0.2849). These might be because the sample size is two less than the power calculated number size 1337 (Table 7). Other SNPs showed no significant association with either iodine or the selenium concentration.
Discussion
KBD is believed to be a complex disease involving genetic factors, as well as environmental factors such as selenium and iodine deficiency [1]. Based on a candidate gene approach, Xiong et al. reported that SNP rs1050450 in the selenoprotein Gpx1 gene was significantly associated with KBD in a Han Chinese population [21], potentially linked this genetic variant to selenium and iodine deficiency in patients. In China, Tibetans who live in the plateau region are one of the most susceptible populations to KBD. Thus, in this study, we examined the genotype of five selenoprotein SNPs including rs1050450, rs1800668, and rs3811699 in the Gpx1gene, rs225014 in the Dio2 gene, and rs5748569 in the TrxR2 gene in KBD patients and controls in a Tibetan population. Our results provided no support for any one of the five SNPs being significantly associated with KBD in the Tibetan population. However, haplotype analysis of SNPs rs1050450, rs1800668 and rs3811699 in GPX1 gene showed a significant association of KBD. Our previous study indicated that the concentrations of serum selenium and iodine in KBD patients were significantly lower than that of controls living in the same village [23], suggesting that genetic variants may affect selenium and/or iodine metabolism. In this study, by haplotype analysis, we found haplotypes TCC, TTC and TTT generated from three SNPs rs1050450, rs1800668, and rs3811699 of gpx1 gene proved to be significantly associated with KBD and play a protective role from the disease. This is consistent with the fact that supplement of selenium is beneficial for KBD children [13].
In this study, for the first time, we observed that the GPX1 gene rs1050450 is significantly associated with the serum iodine concentration (P trend = 0.027). The GPX1 gene encodes a member of the glutathione peroxidase family; SNP rs1050450 in this gene is polymorphic at codon 200, resulting in either a proline or a leucine at that position. Previous studies have suggested that the GPX1-200Leu variant has about 10% lower GPX activity than the wild-type enzyme [24] and that the frequency of the Leu allele is strongly associated with the risk of cancer, such as lung cancer, breast cancer, meningioma, and prostate cancer [25]–[29]. In the present study, we observed that the frequency of the Leu allele of rs1050450 is associated with a relatively higher iodine concentration.
Iodine is an essential component of the hormones produced by the thyroid gland. Therefore, iodine is essential for mammalian life [30]. In mammals, after iodine is absorbed and distributed in the serum, most of it will be transported by the sodium/iodide symporter (NIS) symporter into the thyrocyte where iodine is oxidated in a reaction catalyzed by the hemoprotein thyroid peroxidase (TPO) and then incorporated into the thyroglobulin molecule (Tg-I) [30]–[32] (Fig. 2). In this reaction, H2O2 is generated by the NADPH-dependent thyroxidase (ThOx) and is required as a substrate by TPO for the iodination in Tg. The thyroid hormones triiodothyronine (T3) and tetraiodothyronine (T4) are then released into the bloodstream. When the iodine supply is sufficient, this H2O2 generation is the limiting step for thyroid hormone synthesis. Therefore, if the KM of TPO for H2O2 is very high, the higher amounts of H2O2 are produced than be consumed by the iodination process [34], the more likely potentially exposing the thyroid gland to free radical damage can be produced. Thus, to prevent organ damage, H2O2, H2O2 should be reduced to H2O immediately after the iodination process. Several selenoproteins participate in the protection of thyrocytes and the prevention of damage to the thyroid gland of H2O2 by catalyzing glutathione and H2O2 to glutathione disulfide and H2O [30]. Selenium-dependent GPX gene is one of the first and most important antioxidant enzymes identified in humans and one of only a few proteins known in higher vertebrates to contain selenocysteine. GPX1 can remove H2O2 from many tissues and cells to decrease oxidative damage. By searching human genes' expression data, we observed that GPX1 is very highly expressed in thyroid tissue (http://www.genecards.org/cgi-bin/carddisp.pl?gene=GPX1&search=GPX1). This suggests that this gene is an important H2O2 remover in the thyroid gland. When iodine is deficient, the thyroid would increase the generation of H2O2 to maintain the balance of thyroid hormone synthesis. Therefore, the gland requires a higher level of GPX activity to clear the H2O2 and prevent H2O2-induced damage. In contrast, when iodine is sufficient, less H2O2 and GPX are needed to maintain the hormone balance. Thus, we suspect that the iodine concentration and GPX1 genetic variant might have undergone adaptive evolution in the distant past.
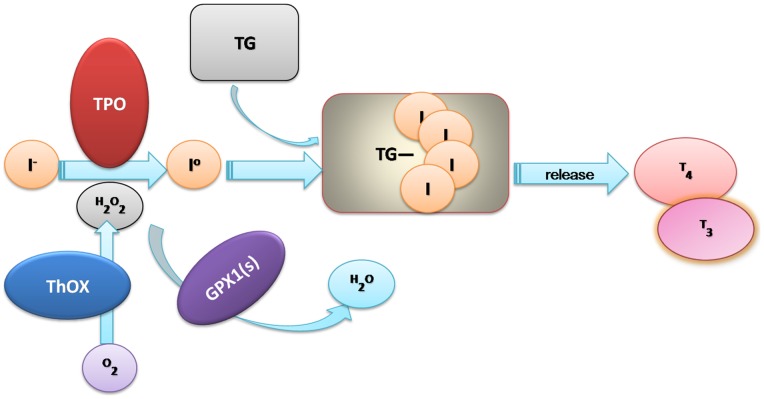
Figure 2. Iodine and GPXs involved in Thyroid Hormones Biosynthesis.
The serum sodium iodide is transported into the thyrocyte and then iodine is incorporated into the thyroglobulin molecule (Tg) in a reaction catalyzed by the hemoprotein thyroid peroxidase (TPO). In this reaction, H2O2 generated by the NADPH-dependent thyroxidase (ThOx) is required as substrate by TPO for the iodination and coupling of tyrosyl residues in Tg. Then, thyroid hormones triiodothyronine (T3) and tetraiodothyronine (T4) are released into the bloodstream. H2O2 used in this reaction decreases the amount of H2O2 that would otherwise be available for damaging oxidation reactions. Selenium-dependent glutathione peroxidase 1 (GPX 1) and other GPX s remove H2O2 from the tissues, also decreasing oxidative damage. (Modified from: J. Köhrle et al. Selenium, the thyroid, and the endocrine system. Endocrine Reviews, December 2005, 26(7):944–984; Lyn Patrick, ND. Iodine: deficiency and therapeutic considerations. Alternative Medicine Review, 2008, 13(2):116–127).
The biosynthesis of selenoproteins is highly regulated by its upstream effectors [33], [34] and by the supply of selenium, which acts as a substrate for the first step in the biosynthesis of Sec-containing proteins. Yet we found no association between the genetic variant Sec-containing coding gene GPX1 and the selenium concentration in this study. This might because the variant of rs1050450 does not affect the yield of GPX, although it does affect the activity of GPX1. As the selenium concentration can affect the mole yield of Sec-containing GPX, it can, therefore, influence the activity of the enzymes. Thus, iodine deficiency increases H2O2 generation, whereas selenium deficiency decreases H2O2 disposal [30]. This results in oxidative cell damage causing chronic inflammation or autoimmune diseases. This linkage may explain why selenium and iodine deficiency are associated with many diseases, such as KBD and other impaired immune function–related disease [14], [32].
Funding Statement
Supported by Department of Science and Technology of China (2007BAI25B02) and Department of Science and Technology of Sichuan Province (08ZC0479) (to Z. Yang); Health Department of Sichuan Province (110196) and China's Post-doctoral Scientific Fund of the 49 Projects funded On-oriented (Serial No.1612) (to L. Huang). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Stone R (2009) Diseases. A medical mystery in middle China. Science 324: 1378–1381. [DOI] [PubMed] [Google Scholar]
- 2. Kolsteren P (1992) Kashin-Beck disease. Ann Soc Belg Med Trop 72: 81–91. [PubMed] [Google Scholar]
- 3. Peng A, Yang C, Rui H, Li H (1992) Study on the pathogenic factors of Kashin-Beck disease. J Toxicol Environ Health 35: 79–90. [DOI] [PubMed] [Google Scholar]
- 4. Pasteels JL, Liu FD, Hinsenkamp M, Rooze M, Mathieu F, et al. (2001) Histology of Kashin-Beck lesions. Int Orthop 25: 151–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hinsenkamp M, Ryppens F, Begaux F, Mathieu F, De Maertelaer V, et al. (2001) The anatomical distribution of radiological abnormalities in Kashin-Beck disease in Tibet. Int Orthop 25: 142–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Y, Guo X, Ping Z, Yu M, Shi X, et al. (2009) Main source of drinking water and familial aggregation of Kashin-Beck disease: a population based on case-control family study. Ann Epidemiol 19: 560–566. [DOI] [PubMed] [Google Scholar]
- 7. Moreno-Reyes R, Suetens C, Mathieu F, Begaux F, Zhu D, et al. (1998) Kashin-Beck osteoarthropathy in rural Tibet in relation to selenium and iodine status. N Engl J Med 339: 1112–1120. [DOI] [PubMed] [Google Scholar]
- 8. Utiger RD (1998) Kashin-Beck disease–expanding the spectrum of iodine-deficiency disorders. N Engl J Med 339: 1156–1158. [DOI] [PubMed] [Google Scholar]
- 9. Peng X, Lingxia Z, Schrauzer GN, Xiong G (2000) Selenium, boron, and germanium deficiency in the etiology of Kashin-Beck disease. Biol Trace Elem Res 77: 193–197. [DOI] [PubMed] [Google Scholar]
- 10. Moreno-Reyes R, Egrise D, Neve J, Pasteels JL, Schoutens A (2001) Selenium deficiency-induced growth retardation is associated with an impaired bone metabolism and osteopenia. J Bone Miner Res 16: 1556–1563. [DOI] [PubMed] [Google Scholar]
- 11. Zhang WH, Neve J, Xu JP, Vanderpas J, Wang ZL (2001) Selenium, iodine and fungal contamination in Yulin District (People's Republic of China) endemic for Kashin-Beck disease. Int Orthop 25: 188–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moreno-Reyes R, Mathieu F, Boelaert M, Begaux F, Suetens C, et al. (2003) Selenium and iodine supplementation of rural Tibetan children affected by Kashin-Beck osteoarthropathy. Am J Clin Nutr 78: 137–144. [DOI] [PubMed] [Google Scholar]
- 13. Zou K, Liu G, Wu T, Du L (2009) Selenium for preventing Kashin-Beck osteoarthropathy in children: a meta-analysis. Osteoarthritis Cartilage 17: 144–151. [DOI] [PubMed] [Google Scholar]
- 14.Fairweather-Tait SJ, Bao Y, Broadley MR, Collings R, Ford D, et al.. (2011) Selenium in Human Health and Disease. Antioxid Redox Signal. [DOI] [PubMed]
- 15. Marasas WF, Van Rensburg SJ (1986) Mycotoxicological investigations on maize and groundnuts from the endemic area of Mseleni joint disease in Kwazulu. S Afr Med J 69: 369–374. [PubMed] [Google Scholar]
- 16. Wright GC Jr, Marasas WF, Sokoloff L (1987) Effect of fusarochromanone and T-2 toxin on articular chondrocytes in monolayer culture. Fundam Appl Toxicol 9: 595–597. [DOI] [PubMed] [Google Scholar]
- 17. Chasseur C, Suetens C, Nolard N, Begaux F, Haubruge E (1997) Fungal contamination in barley and Kashin-Beck disease in Tibet. Lancet 350: 1074. [DOI] [PubMed] [Google Scholar]
- 18. Ying G, Jia Y, Luan R (2007) [Research advance on relation between humic acid and chondrocyte injuries]. Wei Sheng Yan Jiu 36: 238–241. [PubMed] [Google Scholar]
- 19. Li SJ, Yang LS, Wang WY, Hu X, Li YH, et al. (2007) [Determination of trace elements in drinking water of Kashin-Beck disease (KBD) affected and non-affected areas in Tibet by ICP-AES]. Guang Pu Xue Yu Guang Pu Fen Xi 27: 585–588. [PubMed] [Google Scholar]
- 20.Shi Y, Lu F, Liu X, Wang Y, Huang L, et al.. (2011) Genetic variants in HLA-DRB1 gene are associated with Kashin-Beck disease in the Tibetan population. Arthritis Rheum. [DOI] [PubMed]
- 21. Xiong YM, Mo XY, Zou XZ, Song RX, Sun WY, et al. (2010) Association study between polymorphisms in selenoprotein genes and susceptibility to Kashin-Beck disease. Osteoarthritis Cartilage 18: 817–824. [DOI] [PubMed] [Google Scholar]
- 22.Greulich WW PS (1959) Radiographic atlas of skeletal development of the hand and wrist. Stanford, Calif: Stanford University Press.
- 23. Shi Y, Lu F, Liu X, Wang Y, Huang L, et al. (2011) Genetic variants in the HLA-DRB1 gene are associated with Kashin-Beck disease in the Tibetan population. Arthritis Rheum 63: 3408–3416. [DOI] [PubMed] [Google Scholar]
- 24. Ravn-Haren G, Olsen A, Tjonneland A, Dragsted LO, Nexo BA, et al. (2006) Associations between GPX1 Pro198Leu polymorphism, erythrocyte GPX activity, alcohol consumption and breast cancer risk in a prospective cohort study. Carcinogenesis 27: 820–825. [DOI] [PubMed] [Google Scholar]
- 25. Raaschou-Nielsen O, Sorensen M, Hansen RD, Frederiksen K, Tjonneland A, et al. (2007) GPX1 Pro198Leu polymorphism, interactions with smoking and alcohol consumption, and risk for lung cancer. Cancer Lett 247: 293–300. [DOI] [PubMed] [Google Scholar]
- 26. Arsova-Sarafinovska Z, Matevska N, Eken A, Petrovski D, Banev S, et al. (2009) Glutathione peroxidase 1 (GPX1) genetic polymorphism, erythrocyte GPX activity, and prostate cancer risk. Int Urol Nephrol 41: 63–70. [DOI] [PubMed] [Google Scholar]
- 27. Cox DG, Tamimi RM, Hunter DJ (2006) Gene x Gene interaction between MnSOD and GPX-1 and breast cancer risk: a nested case-control study. BMC Cancer 6: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bhatti P, Stewart PA, Hutchinson A, Rothman N, Linet MS, et al. (2009) Lead exposure, polymorphisms in genes related to oxidative stress, and risk of adult brain tumors. Cancer Epidemiol Biomarkers Prev 18: 1841–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steinbrecher A, Meplan C, Hesketh J, Schomburg L, Endermann T, et al. (2010) Effects of selenium status and polymorphisms in selenoprotein genes on prostate cancer risk in a prospective study of European men. Cancer Epidemiol Biomarkers Prev 19: 2958–2968. [DOI] [PubMed] [Google Scholar]
- 30. Kohrle J, Jakob F, Contempre B, Dumont JE (2005) Selenium, the thyroid, and the endocrine system. Endocr Rev 26: 944–984. [DOI] [PubMed] [Google Scholar]
- 31. Patrick L (2008) Iodine: deficiency and therapeutic considerations. Altern Med Rev 13: 116–127. [PubMed] [Google Scholar]
- 32. Kohrle J, Gartner R (2009) Selenium and thyroid. Best Pract Res Clin Endocrinol Metab 23: 815–827. [DOI] [PubMed] [Google Scholar]
- 33. Bock A, Forchhammer K, Heider J, Leinfelder W, Sawers G, et al. (1991) Selenocysteine: the 21st amino acid. Mol Microbiol 5: 515–520. [DOI] [PubMed] [Google Scholar]
- 34. Forchhammer K, Boesmiller K, Bock A (1991) The function of selenocysteine synthase and SELB in the synthesis and incorporation of selenocysteine. Biochimie 73: 1481–1486. [DOI] [PubMed] [Google Scholar]