Abstract
Background
Mutans streptococci are a group of bacteria significantly contributing to tooth decay. Their genetic variability is however still not well understood.
Results
Genomes of 6 clinical S. mutans isolates of different origins, one isolate of S. sobrinus (DSM 20742) and one isolate of S. ratti (DSM 20564) were sequenced and comparatively analyzed. Genome alignment revealed a mosaic-like structure of genome arrangement. Genes related to pathogenicity are found to have high variations among the strains, whereas genes for oxidative stress resistance are well conserved, indicating the importance of this trait in the dental biofilm community. Analysis of genome-scale metabolic networks revealed significant differences in 42 pathways. A striking dissimilarity is the unique presence of two lactate oxidases in S. sobrinus DSM 20742, probably indicating an unusual capability of this strain in producing H2O2 and expanding its ecological niche. In addition, lactate oxidases may form with other enzymes a novel energetic pathway in S. sobrinus DSM 20742 that can remedy its deficiency in citrate utilization pathway.
Using 67 S. mutans genomes currently available including the strains sequenced in this study, we estimates the theoretical core genome size of S. mutans, and performed modeling of S. mutans pan-genome by applying different fitting models. An “open” pan-genome was inferred.
Conclusions
The comparative genome analyses revealed diversities in the mutans streptococci group, especially with respect to the virulence related genes and metabolic pathways. The results are helpful for better understanding the evolution and adaptive mechanisms of these oral pathogen microorganisms and for combating them.
Keywords: Mutans Streptococci, Streptococcus Mutans, Streptococcus Ratti, Streptococcus Sobrinus, Comparative Genomics, Core-genome, Pan-genome, Pathogenicity, Lactate Oxidase, Metabolic Network
Background
Traditionally and supported by 16S rRNA gene and rnpB gene sequence analyses, the genus Streptococcus is divided into several groups, with the mutans group streptococci consisting of the species S. mutans, S. sobrinus, S. ratti, S. criceti, S. downei, S. macacae, and – but controversially discussed – S. ferus (for update refer to http://www.bacterio.net/s/streptococcus.html) [1]. Mutans streptococci are considered significant contributors to the development of dental caries [2]. By attaching to the tooth surfaces and forming biofilms, they can tolerate and adapt to the harsh and rapidly changing physiological conditions of the oral cavity such as extreme acidity, fluctuation of nutrients, reactive oxygen species, and other environmental stresses [3]. They occasionally also cause bacteremia, abscesses, and infective endocarditis [4,5]. Many strains of mutans streptococci are genetically competent, i.e. they can take up DNA fragments from the environment and recombine them into their chromosome, an important mechanism for horizontal gene transfer (HGT). The ability of some bacteria to generate diversity through HGT provides a selective advantage to these microbes in their adaptation to host eco-niches and evasion of immune responses [6,7]. Due to diversities in the genetic contents between different isolates, the genome content of a single isolate does not necessarily represent the genomic potential of a certain species. With the rapid development of DNA sequencing technologies, the steadily increasing genome data enable us to dig the evolutionary and genetic information of a species from a pan-genome perspective. In 2002, the release of the genome sequence of S. mutans UA159, the first genome sequence of mutans group streptococci, has greatly helped in understanding the robustness and complexity of S. mutans as an oral and odontogenic (e.g. infective endocarditis and abscesses) pathogen [8]. Later, after the genome sequence of S. mutans NN2025 became available, a comparative genomic analysis of S. mutans NN2025 and UA159 provides insights into chromosomal shuffling and species-specific contents [9]. Recently, Cornejo et al. have studied the evolutionary and population genomics of S. mutans based on 57 S. mutans draft genomes and revealed a high lateral gene transfer (LGT) rate of S. mutans[10].
In this study, we determined the whole genome sequences of six S. mutans strains (5DC8, KK21, KK23, AC4446, ATCC 25175 and NCTC 11060), one S. ratti strain (DSM 20564) and one S. sobrinus strain (DSM 20742) and performed cross-comparison with the genome contents of S. mutans UA159 and NN2025, focusing on issues that are highly related to pathogenicity. The core- and pan-genome of S. mutans was analyzed by including 67 currently available S. mutans genome sequences. By constructing and comparing the genome-scale metabolic networks, the diversities in sub-networks (pathways) are systematically revealed. The results should be helpful for understanding the evolution and pathogenicity, as well as for prevention and treatment, of these very common opportunistic pathogens.
Results and discussion
Genome sequencing, assembly and annotation of eight mutans streptococci strains
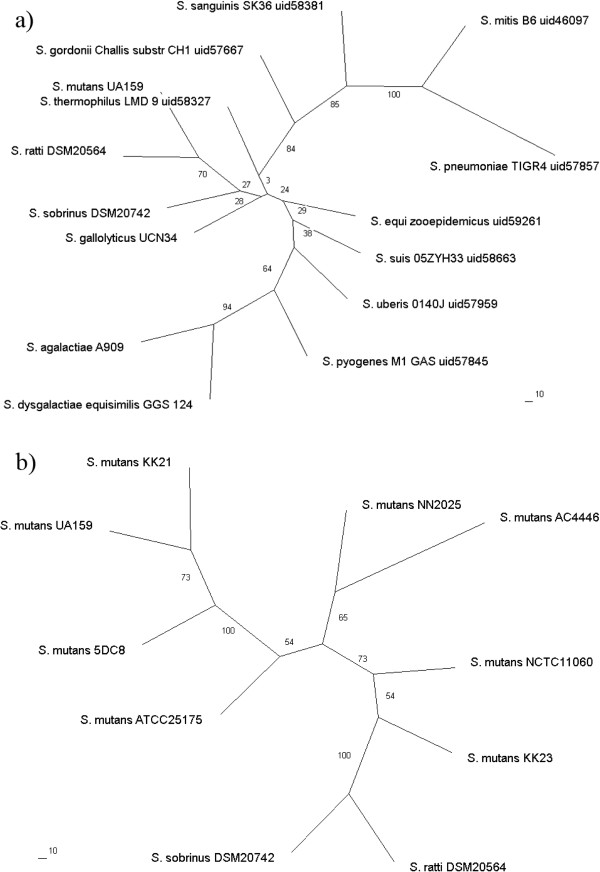
As expected, the overall genomic features of the eight S. mutans strains are more close to each other than to S. ratti and S. sobrinus. This is consistent with the results of the phylogenetic analysis, as visualized by the phylogenetic tree constructed based on 16S rRNA and core genes single-nucleotide polymorphisms (SNPs) information shown in Figure 1. An overview of the genome assemblies and annotations of the 6 S. mutans isolates as well as S. ratti DSM 20564 and S. sobrinus DSM 20742 is summarized in Table 1 in comparison with two previously sequenced S. mutans strains, namely UA159 and S. mutans NN2025. The average GC contents are in the range of low GC organisms [8]. The genome sizes are very close to each other, with the largest one from S. sobrinus DSM 20742 and the smallest one from S. mutans KK23 showing merely 5.7% differences. The total numbers of protein-coding sequences per genome are also similar among all the strains compared.
Figure 1.
Phylogenetic analysis of the 10 mutans streptococci strains compared in this study and their phylogenetic relationship to other Streptococcus species with genomes known before 01/01/2011. a) 16S rRNA phylogenetic tree of Streptococcus species with genomes known before 01/01/2011 (Since the 16S rRNA sequences were almost identical between the different S. mutans strains, only UA159 is shown as representative). b) Phylogenetic tree of mutans streptococci constructed with the core-genome SNPs obtained by PGAP pipeline [11]. All phylogenetic trees were constructed using ClustalX and Phylip by applying the maximum likelihood (ML) method with bootstrap value set to 100. Values beside branches depict ML bootstrap support values. The scale bars in the unit of “substitution/site” are shown below the trees.
Table 1.
Genome assembly and annotation of eight newly sequenced mutans streptococci strains in comparison with previously sequenced S. mutans strains UA159 and S. mutans NN2025
| S. mutans UA159 | S. mutans NN2025 | S. mutans 5DC8 | S. mutans KK21 | S. mutans KK23 | S. mutans AC4446 | S. mutans ATCC 25175 | S. mutans NCTC 11060 | S. ratti DSM 20564 | S. sobrinus DSM 20742 | |
|---|---|---|---|---|---|---|---|---|---|---|
| |
NC_004350.2 |
NC_013928.1 |
AOBX 00000000 |
AOBY 00000000 |
AOBZ 00000000 |
AOCA 00000000 |
AOCB 00000000 |
AOCC 00000000 |
AOCD 00000000 |
AOCE 00000000 |
|
Total Length |
2,030,921 |
2,013,587 |
2,010,935 |
2,034,586 |
1,976,057 |
2,003,537 |
1,999,532 |
2,021,202 |
2,037,184 |
2,096,203 |
|
Contigs |
Complete |
Complete |
9 |
2 |
38 |
42 |
10 |
36 |
182 |
283 |
|
N50 size |
Complete |
Complete |
354,736 |
1,622,660 |
134,323 |
167,413 |
233,425 |
94,580 |
23,860 |
12,417 |
|
N90 size |
Complete |
Complete |
325,634 |
411,935 |
38,851 |
26,425 |
107,076 |
43,987 |
6,098 |
3,659 |
|
G + C content |
36.82% |
36.85% |
36.90% |
36.81% |
36.68% |
36.90% |
36.87% |
36.98% |
40.29% |
43.46% |
|
Total Genes |
2040 |
1975 |
2,004 |
2,031 |
1,933 |
1,999 |
1,982 |
1,982 |
1,995 |
2,057 |
| Protein Coding Genes | 1,960 | 1,895 | 1,924 | 1,949 | 1,907 | 1,919 | 1,903 | 1,915 | 1,965 | 2,032 |
Chromosomal rearrangement of the S. mutans strains
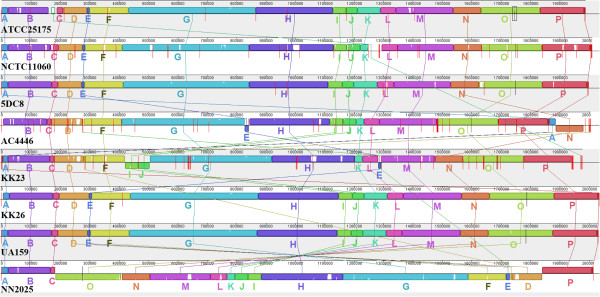
Genome rearrangements have important effects on bacterial phenotypes and the evolution of bacterial genomes [12]. A comparison of the genomes of S. mutans NN2025 and UA159 discovered a large genomic inversion across the replication axis and similar genomic variations were also confirmed among 95 clinical S. mutans isolates using long-PCR analysis [9]. In this study, genome rearrangements among the eight S. mutans strains are determined by genome alignment using the MAUVE software [13]. The results are presented in Figure 2, which shows the locally collinear blocks (LCBs) representing the landmarks, i.e. the homologous/conserved regions shared by all the input sequences in the chromosomes. A LCB is defined as a collinear (consistent) set of exactly matched subsequences (multiple maximal unique matches, namely ‘multi-MUMs’) which are shared by all the chromosomes considered, appear once in each chromosome and are bordered on both sides by mismatched nucleotides. The weight (the sum of the lengths of the included multi-MUMs) of a LCB serves as a measure of confidence that it is a true homologous region rather than a random match.
Figure 2.
Comparison of local collinear blocks (LCBs) of chromosomal sequences of the eight S. mutans strains. In total 16 local LCBs, marked as A to P, were generated and compared by applying the MAUVE software [13,14] with default settings and using strain UA159 as reference. The red vertical bars indicate contig ends. The white areas inside each LCB show regions with low similarities.
As shown in Figure 2, 16 LCBs (marked as A to P) are identified by multi-alignment of the eight S. mutans genome sequences. Compared to UA159 and NN2025, the chromosome segment represented by LCB E is reversely inserted between the LCB G and H in the strain AC4446, and between the LCB L and M in the strain KK23. This segment is related to the genomic island “SMU.100-SMU.116” of S. mutans UA159 which mainly contains sorbitol phosphotransferase system (PTS), transposase and hypothetical proteins [15]. LCB N is found to be reversed and relocated to the position between LCB A and B in the strain AC4446. A cluster of tRNA genes is found to be located downstream of LCB N. In KK23, LCB I and J are moved to position between LCB F and G. A tRNA-Gln and a tRNA-Tyr is found to be adjacent to the left of LCB I. LCB K in NCTC 11060, AC4446, KK23 and NN2025 are very similar to each other but much smaller than those of other strains (with sequence length reduced about two-thirds). The missing sequence corresponds to the genomic island TnSmu2 of S. mutans which harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments [16]. Using the known information about genomic islands in S. mutans UA159, additional genomic islands were found to be present/absent in the mutans streptococci strains of this study [15,17] (see Additional file 1). Furthermore, there are much more diversities as shown by the white areas inside the LCBs which show regions with low similarities. However, it should be noticed that there might be more genome rearrangements among the strains, because draft genome sequences are used in current analysis and all contigs in each genome are sorted according to the reference genome sequence of the strain UA159.
Core- and pan-genome analysis of S. mutans
The genetic variability within species in the domain Bacteria is much larger than that found in other domains of life. The gene content between pairs of isolates can diverge by as much as 30% in species like Streptococcus pneumoniae[18]. This unexpected finding led to the introduction of the pan-genome concept, which describes the sum of genes that can be found in a given bacterial species [19,20]. The genome of any isolate is thus composed of a “core-genome” shared with all strains of this particular species, and a “dispensable genome” that accounts for the phenotypic differences between strains. The pan-genome is usually much larger than the genome of any single isolate, constituting a reservoir that could enhance the ability of many bacteria to survive in stressful environments. The pan-genome concept has important consequences for the way we understand bacterial evolution, adaptation, and population structure, as well as for more applied issues such as vaccine design or the identification of virulence genes [21]. In this study, we performed core-genome and pan-genome analyses of 67 S. mutans strains, including the eight S. mutans strains sequenced in this study and 59 S. mutans strains with genome available in NCBI till April 2013.
Core-genome
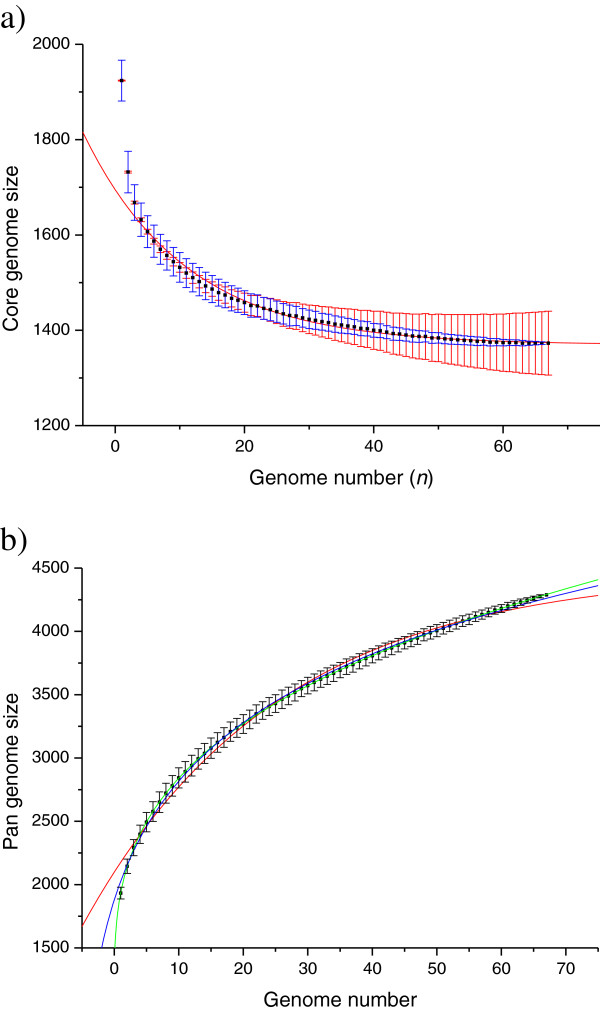
The core-genome size of the 67 S. mutans strains was calculated to be 1,373. Detailed information of the core genes are provided in the Additional file 2. To estimate the theoretical core-genome size achievable with an infinite number of S. mutans genomes, core-genome size medians corresponding to different genome numbers as shown in Figure 3a by the red rectangles were first calculated by random sampling 1,000 genome combinations of n genomes out of the 67 S. mutans genomes. Then, we applied the exponential regression core-genome model proposed previously by Tettelin et al. [19,20] to fit the median data points of the core-genome sizes, where Kc, τc and Ω are parameters, n represents the number of genomes, and Ω stands for the theoretical core-genome size. To take into consideration the different deviations of the core-genome size medians, as clearly indicated by the blue error bars in Figure 3a, we modified the fitting process by introducing the genome number as weight to the corresponding data point. The fitting parameters thus obtained are as follows: r2 = 0.97403, Kc = 325.74718 ± 10.00912,Ω = 1,369.41225 ± 1.986,τc = 15.90248 ± 0.66807 (Detailed information of all core and pan-genome modeling are given in Additional file 3). Using this fitting result to describe the core-genome of S. mutans, the theoretical core-genome size (Ω) was estimated to be around 1,370 genes, which is slightly lower than the calculated core-genome size (1,373) using 67 genomes. Compared with other streptococcus species, the core-genome of S. mutans is at the same level to the core-genome of S. pyogenes (1,400 genes determined using 11 strains), less than that of S. pneumoniae (1,647 genes determined using 47 strains) and S. agalactiae (1,800 genes determined using eight strains) [19,22,23]. However, we should be cautious with such comparison. In a recent study of Cornejo et al. [10], the core genome size of S. mutans was determined as 1,490 by using 57 S. mutans genomes which is obviously different to the core genome size of S. mutans we estimated, although we included the 57 S. mutans genomes used by Cornejo et al. in our study. The difference can be caused by different reasons, such as difference in the correction step for core gene determination and, very likely, different methods and parameter settings used for determining orthologs. Apparently, we have used a more stringent process to determine orthologs which led to smaller core genome size of S. mutans estimated.
Figure 3.
Core and pan-genome model of 67 S. mutans genomes. a) Core-genome model of S. mutans. The core-genome size (number of common genes) is plotted as a function of the number (n) of genomes according to a previously proposed model where Kc, τc and Ω are model parameters. Red rectangles are the medians of the core-genome sizes calculated by random sampling 1000 different genome combinations of n genomes out of 67 genomes. Blue bars are the standard deviation of the medians. The red bars are weights used for model fitting and the red curve is the fitting result. b) Pan genome modeling of S. mutans genomes using three models, y = a + bxc, y = a − bln(x + c) and y = a × e− x/b + c (where a, b and c are parameters), represented by green, blue and red curves respectively. Black rectangles are the medians of the pan-genome sizes calculated by random sampling 1000 different genomes combination of n genomes out of 67 genomes, and black bars are the standard deviations of the medians.
Pan-genome
We applied three models, namely y = a + bxc, y = a − bln(x + c) and y = a × e− x/b + c (where a, b and c are parameters) for modeling the pan-genome of S. mutans, as shown in Figure 3b by green, blue and red curves respectively (all fitting results are detailed in Additional file 3). Both the fitting results of using y = a + bxc and y = a − bln(x + c) indicated an infinite pan-genome, while the fitting result of using y = a × e− x/b + c resulted in a negative value of the parameter a, suggesting a finite pan-genome However, the last fitting shows obvious deviations to many of the data points. Especially, the deviations even become larger with increased genome numbers, indicating that this model is not suitable. The best fitting result obtained with the model y = a + bxc shows fittings to all the data points with very high confidence. Based on this model, the pan-genome of S. mutans is still “open” while 67 genomes were included, and the expected average new gene number with the addition of a new genome is estimated to be 15. The infinite pan-genome was first proposed by Tettelin et al. for S. agalactiae based on the use of 9 S. agalactiae genomes. The three regression models used in this study are all based on the assumption that contingency genes are independently sampled from the pan-genome with equal probability, except in the case of “specific/unique genes”, which are modeled as unique events that appear only once in the entire global population. Hogg et al. [24] proposed a finite supragenome model for pan-genome based on a different supposition that contingency genes are sampled from the pan-genome with unequal probability. By applying this finite supragenome model to 44 S. pneumoniae genomes, the predicted number of new genes drops sharply to zero when the number of genomes exceeds 50. However, in the case of S. mutans we could not observe such sharp decrease of new gene number even after 67 genomes were included. In the study of Cornejo et al. [10], they proposed a finite pan-genome for S. mutans, after they used a special “pseudogene cluster” identification process to exclude about 30% of the rare genes that are considered to be pseudogenes. However, they didn’t provide detailed parameters they obtained from fitting. Our modeling using the 67 S. mutans genomes by applying the model described above without any restrictions pointed to an infinite pan-genome of S. mutans. However, we would like to understand this predicted “infinite” pan-genome as follows: 1) a “pan-genome” should be considered as “dynamic” rather than “static”, which means the pan-genome content is changing during the evolution, it does not matter if its size is infinite or finite; 2) The change of a pan-genome content can be caused by the acquirement of new genes or by the loss of genes; 3) The actual pan-genome size can be more stable than the content of the pan-genome but can also change during evolution coupled with the change of the environment. Thus, without considering the “gene loss events”, it’s quite understandable to have a “growing” or “infinite” pan-genome as gene acquirement occurs no matter how slow it might be. Interestingly, Cornejo et al. found a high rate of LGT in S. mutans, where many genes were acquired from related streptococci and bacterial strains predominantly residing not only in the oral cavity, but also in the respiratory tract, the digestive tract, cattle, genitalia, in insect pathogens and in the environment in general [10]. Such high rate of LGT might also lead to a continuously growing pan-genome.
Gene content-based comparative analysis of 10 mutans streptococci strains
The annotated protein sequences of all the genomes studied were cross-compared based on alleles/ortholog groups established by the program OrthoMCL [25]. In total, 2,211 putative alleles/ortholog groups are established, as documented in Additional file 4. A pair-wise comparison of the protein coding sequences between each two strains is shown in Table 2. It is clear to see that remarkable differences in protein coding sequences exist between the strains, even inside the same species of S. mutans. In the following sections, systems that are highly related to stress resistance and pathogenicity are presented and discussed. As all the following results are based on putative alleles/ortholog groups established by OrthoMCL, if not otherwise specified, the word “putative allele(s)/ortholog(s)” is omitted in the following text.
Table 2.
Unique protein coding sequences (CDSs) revealed by ortholog analysis between the different strains of this study
| Unique CDSs in comparision to | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| |
S. mutans UA159 |
S. mutans NN2025 |
S. mutans 5DC8 |
S. mutans KK21 |
S. mutans KK23 |
S. mutans AC4446 |
S. mutans ATCC 25175 |
S. mutans NCTC 11060 |
S. ratti DSM 20564 |
S. sobrinus DSM 20742 |
All others |
|
S. mutans UA159 |
|
216 |
125 |
63 |
230 |
221 |
166 |
212 |
427 |
566 |
42 |
|
S. mutans NN2025 |
150 |
|
150 |
150 |
133 |
102 |
182 |
167 |
358 |
510 |
24 |
|
S. mutans 5DC8 |
85 |
176 |
|
52 |
164 |
161 |
132 |
153 |
379 |
522 |
31 |
|
S. mutans KK21 |
47 |
200 |
76 |
|
190 |
184 |
127 |
175 |
402 |
544 |
3 |
|
S. mutans KK23 |
183 |
152 |
157 |
159 |
|
146 |
173 |
175 |
387 |
525 |
56 |
|
S. mutans AC4446 |
145 |
92 |
125 |
124 |
117 |
|
159 |
146 |
364 |
502 |
31 |
|
S. mutans ATCC 25175 |
117 |
199 |
123 |
94 |
171 |
186 |
|
146 |
373 |
525 |
33 |
|
S. mutans NCTC 11060 |
126 |
147 |
107 |
105 |
136 |
136 |
109 |
|
334 |
488 |
34 |
|
S. ratti DSM 20564 |
432 |
429 |
424 |
423 |
439 |
445 |
427 |
425 |
|
564 |
289 |
| S. sobrinus DSM 20742 | 616 | 626 | 612 | 610 | 622 | 628 | 624 | 624 | 609 | 492 | |
High diversities of the competence development regulation module
In a previous study we have systematically discussed the two-component signal transduction systems (TCSTSs) in the 10 mutans streptococci strains [26]. ComDE, one of the TCSTS is directly related to competence development. Competence development is a complex process involving sophisticated regulatory networks that trigger the capacity of bacterial cells to take up exogenous DNA from the environment. This phenomenon is frequently encountered in bacteria of the oral cavity, e.g., S. mutans[27]. In S. mutans, ComX, an alternative sigma factor, drives the transcription of the so called ‘late-competence genes’ required for genetic transformation. ComX activity is modulated by the inputs from two types of signal pathways, namely the competence-stimulating peptide (CSP) dependent competence regulation system and CSP-independent competence regulation system. ComX and the ‘late-competence genes’ regulated by ComX as labeled by boldface in Table 3, are highly conserved even between the species, indicating that all mutans streptococci studied here might have the potential ability of transforming to genetic competence state. On the other hand, the upstream signal pathways regulating the activity of ComX show high variety as discussed in details below.
Table 3.
Distribution of competence development-related systems in the 10 mutans streptococci strains
| Name | Function |
S. mutans |
S. mutans |
S. mutans |
S. mutans |
S. mutans |
S. mutans |
S. mutans |
S. mutans |
S. ratti |
S. sobrinus |
|---|---|---|---|---|---|---|---|---|---|---|---|
| UA159 | NN2025 | 5DC8 | KK21 | KK23 | AC4446 | ATCC 25175 | NCTC 11060 | DSM 20564 | DSM 20742 | ||
| ComA/ |
Competence factor & nonlantibiotic mutacin transporting ATP-binding/permease protein |
SMU.286 |
GI|290581206 |
D816_01150 |
D817_01300 |
D818_01134 |
D819_01163 |
D820_01336 |
D821_01208 |
D822_01584 |
D823_05343 |
| SMU.1881c |
GI|290579788 |
D816_08453 |
D817_08643 |
- |
D819_07724 |
|
D821_08449 |
D822_08325- |
D823_01400 |
||
| NlmT | |||||||||||
| ComB/ |
Accessory factor for NlmT |
SMU.287 |
GI|290581205 |
D816_01155 |
D817_01305 |
D818_01139 |
D819_01168 |
D820_01341 |
D821_01213 |
D822_01589 |
D823_05923 |
| NlmE | |||||||||||
| ComC |
S. mutans specific competence stimulating peptide, precursor |
SMU.1915 |
GI|290579762 |
D816_08588 |
D817_08778 |
D818_08368 |
D819_07839 |
D820_08520 |
D821_08549 |
- |
- |
| SepM |
cell surface-associated protease; Cleavage CSP. |
SMU.518 |
GI|290580977 |
D816_02205 |
D817_02448 |
D818_02735 |
D819_02254 |
D820_02420 |
D821_02274 |
D822_04126 |
D823_08607 |
| ComD |
histidine kinase |
SMU.1916 |
GI|290579761 |
D816_08593 |
D817_08783 |
D818_08373 |
D819_07844 |
D820_08525 |
D821_08554 |
|
D823_05333 |
| ComE |
response regulator |
SMU.1917 |
GI|290579760 |
D816_08598 |
D817_08788 |
D818_08378 |
D819_07849 |
D820_08530 |
D821_08559 |
|
D823_05328 |
| D823_7992 | |||||||||||
| HtrA |
serine protease |
SMU.2164 |
GI|290581420 |
D816_09733 |
D817_00015 D817_09913 |
D818_00020 |
D819_09056 |
D820_09650 |
D821_09748 |
D822_05851 |
D823_03191 |
| HdrM |
high density responsive membrane protein |
SMU.1855 |
GI|290579809 |
D816_08353 |
D817_08543 |
D818_08143 |
D819_07614 |
D820_08345 |
D821_08319 |
D822_08240 |
D823_08222 |
| HdrR |
high density responsive regulator |
SMU.1854 |
GI|290579810 |
D816_08348 |
D817_08538 |
D818_08138 |
D819_07609 |
D820_08340 |
D821_08314 |
- |
- |
| BrsM |
|
SMU.2081 |
GI|290581347 |
D816_09358 |
D817_09538 |
D818_09198 |
D819_08671 |
D820_09275 |
D821_09348 |
- |
- |
| BrsR |
|
SMU.2080 |
GI|290581346 |
D816_09353 |
D817_09533 |
D818_09193 |
D819_08666 |
D820_09270 |
D821_09343 |
D822_05085 |
- |
| OppD |
oligopeptide ABC transporter |
SMU.258 |
GI|290581226 |
D816_01025 |
D817_01175 |
D818_01039 |
D819_01063 |
D820_01211 |
D821_01051 |
D822_05611 |
D823_04322 |
| ComS |
comX-inducing peptide (XIP) precursor |
NC_004350.2 |
NC_013928.1 |
D816_00277 |
D817_00297 |
D818_00297 |
D819_00203 |
D820_00247 |
D821_00253 |
D822_01077 |
- |
| (62613 - 62666)a |
(60952- 61005)b |
||||||||||
| ComR |
ComS receptor |
SMU.61 |
GI|290579576 |
D816_00275 |
D817_00295 |
D818_00294 |
D819_00200 |
D820_00245 |
D821_00250 |
D822_01080 |
|
| ComX(SigX) |
competence-specific sigma factor |
SMU.1997 |
GI|290579687 |
D816_08973 |
D817_09163 |
D818_08748 |
D819_08219 |
D820_08900 |
D821_08929 |
D822_07328 |
D823_08887 |
|
ComEA |
competence protein |
SMU.625 |
GI|290580890 |
D816_02675 |
D817_02923 |
D818_03217 |
D819_02694 |
D820_02880 |
D821_02784 |
D822_02674 |
D823_08107 |
|
ComEC |
competence protein; possible integral membrane protein |
SMU.626 |
GI|290580889 |
D816_02680 |
D817_02928 |
D818_03222 |
D819_02699 |
D820_02885 |
D821_02789 |
D822_02679 |
D823_08117 |
|
CoiA |
competence protein CoiA |
SMU.644 |
GI|290580870 |
D816_02775 |
D817_03018 |
D818_03322 |
D819_02786 |
D820_02970 |
D821_02879 |
D822_02739 |
D823_01025 |
|
EndA |
competence associated membrane nuclease (DNA-entry nuclease) |
SMU.1523 |
GI|290580108 |
D816_06842 |
D817_07008 |
D818_06659 |
D819_06647 |
D820_06860 |
D821_06857 |
D822_03254 |
D823_09687 |
|
ComG |
competence protein G |
SMU.1981c |
GI|290579702 |
D816_08898 |
D817_09088 |
D818_08673 |
D819_08144 |
D820_08825 |
D821_08854 |
D822_07418 |
D823_01170 |
|
ComYD |
competence protein ComYD |
SMU.1983 |
GI|290579700 |
D816_08908 |
D817_09098 |
D818_08683 |
D819_08154 |
D820_08835 |
D821_08864 |
D822_07408 |
D823_01160 |
|
ComYC |
competence protein ComYC |
SMU.1984 |
GI|290579699 |
D816_08913 |
D817_09103 |
D818_08688 |
D819_08159 |
D820_08840 |
D821_08869 |
D822_07403 |
D823_01155 |
| |
possible competence-induced protein |
SMU.2075c |
GI|290581342 |
D816_09328 |
D817_09508 |
D818_09168 |
D819_08641 |
D820_09245 |
D821_09318 |
D822_05110 |
D823_03558 |
|
CinA |
competence damage-inducible protein A |
SMU.2086 |
GI|290581351 |
D816_09383 |
D817_09563 |
D818_09218 |
D819_08691 |
D820_09295 |
D821_09368 |
D822_05060 |
D823_03593 |
|
ComYB |
competence protein; general (type II) secretory pathway protein |
SMU.1985 |
GI|290579698 |
D816_08918 |
D817_09108 |
D818_08693 |
D819_08164 |
D820_08845 |
D821_08874 |
D822_07398 |
D823_01150 |
|
ComYA |
late competence protein; type II secretion system protein |
SMU.1987 |
GI|290579697 |
D816_08923 |
D817_09113 |
D818_08698 |
D819_08169 |
D820_08850 |
D821_08879 |
D822_07393 |
D823_01145 |
|
ComFC |
late competence protein required for DNA uptake |
SMU.499 |
GI|290580999 |
D816_02100 |
D817_02348 |
D818_02650 |
D819_02154 |
D820_02290 |
D821_02159 |
D822_06218 |
D823_02981 |
|
ComFA |
late competence protein F |
SMU.498 |
GI|290581000 |
D816_02095 |
D817_02343 |
D818_02645 |
D819_02149 |
D820_02285 |
D821_02154 |
D822_06223 |
D823_02986 |
| CinA | competence damage-inducible protein A; | SMU.2086 | GI|290581351 | D816_09383 | D817_09563 | D818_09218 | D819_08691 | D820_09295 | D821_09368 | D822_05060 | D823_03593 |
In the case that more than one paralogs were found, the most similar ortholog is marked in italic. The rows related to highly conserved ‘late-competence genes’ were shown in boldface. The missing of ComDE in S. ratti DSM 20564 has been published in our previous study [26].
alocation of comS in S. mutans UA159.
blocation of comS in S. mutans NN2025.
CSP-dependent competence regulation system
It has been reported that the ComABCDE system in S. mutans combines the action of the two ortholog systems which are present as ComABCDE and BlpABCRH in S. pneumoniae and involved in competence regulation and bacteriocins regulation, respectively. It should be noticed that, ComAB have been primarily considered to be the transporter of ComC, the precursor of CSP. Later, ComAB have been renamed as NlmTE as they were found to function together as transporter of nonlantibiotic bacteriocins, while another gene pair CslAB was supposed to be the transporter of ComC [28]. However, a recent study confirms that ComAB is indeed a transporter both for nonlantibiotic bacteriocin and the peptide pheromone CSP [29].
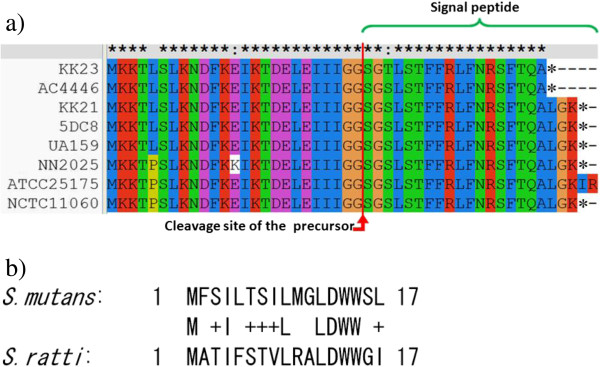
In S. mutans, the comC-encoded prepeptide of CSP has a leader sequence containing a conserved double glycine (GG), at which the leader sequence is cleaved during transporting by ComAB to generate the mature signal peptide (CSP-21) containing 21 amino acid residues [28,30,31]. Recent studies show that an extracellular protease, SepM (SMU.518), is involved in the further processing of CSP-21 by removing the “LGK” residues in the C-terminal to generate a 18-residue peptide (CSP-18), which can work at a concentration much lower than that of CSP-21 [29,32]. SepM is identified in all the 10 strains compared in this study, although putative comC alleles are present only in the eight S. mutans strains, not in the S. sobrinus DSM 20742 and S. ratti DSM 20564. Multi-alignment of the ComC sequences shows clear variations among different S. mutans strains (Figure 4a). Genetic variation of ComC in S. mutans has been reported previously [33]. Interestingly, the C-terminal amino acid sequence “LGK” of ComC is absent in the ComC prepetides of S. mutans KK23 and AC4446, which have also been observed previously in other S. mutans strains by Allan et al. [33]. ATCC 25175 possesses a unique ComC sequence ended with “LGKIR” at its C-terminal. In addition to the variations at the carboxyl end, substitutions of single amino acid residues at different positions are also found.
Figure 4.
Alignment of ComC and ComS amino acid sequences. a) Alignment of ComC amino acid sequences identified in S. mutans species using CLUSTALX. Conserved residues are marked with “*” above the figure. The diversity in the ComC sequences have been verified by PCR experiments (data not shown). b) BlastP alignment of the ComS sequence of S. mutans (identical among the eight S. mutans strains) with that of S. ratti DSM 20564 (No ComS was identified in S. sobrinus). “+” stands for similar amino acid residues.
We have verified all the variants of comC revealed in this study by PCR experiments. Although Allan et al. pointed out that different comC alleles in some clinical strains of S. mutans exist but their products are functionally equivalent and there is no evidence of phenotype specificity [33], considering the complexity of phenotype evaluation, whether and how the variations found in this study may affect the natural genetic competence of these S. mutans strains requires further investigation.
The CSP-initiated activation of the response regulator ComE, through its cognate receptor kinase ComD, leads to the induction of competence through the alternative sigma factor ComX, and at the same time ComE directly induces a set of bacteriocin-related genes [28,30,34-38]. In our previous study focused on the comparison of the two-component signal transduction systems of these mutans streptococci strains, we have reported the complete missing of ComDE in S. ratti DSM 20564 and the low similarities of putative ComDE in S. sobrinus DSM 20742 to the ComDE of S. mutans strains [26]. Accordingly, no comC-like genes could be identified in S. ratti DSM 20564 and S. sobrinus DSM 20742. Thus, it can be inferred that S. ratti DSM 20564 and S. sobrinus DSM 20742 are totally different to the S. mutans strains regarding cellular functions including genetic competence associated with the ComABCDE system.
In S. mutans, no binding motif for ComE is present in the promoter region of ComX, suggesting that ComE is not a direct regulator of ComX, whereas a new peptide regulator system (ComSR) downstream of ComE that directly activates ComX has been identified by Mashburn-Warren et al. ComR activates the expression of the ComS, which is secreted, processed, and internalized through the peptide transporter OppD. The processed peptide, designated XIP (for sigma X-inducing peptide), modulates the activity of ComR, which in turn activates the expression of ComX. Deletion of comR or comS gene completely abolished the competence in S. mutans[39]. In this study, the ComSR regulating system is identified in most of the strains, except for S. sobrinus DSM 20742 which lacks the ComSR-coding genes. This well explains the fact that despite the presence of comX and the ‘late-competence genes’ we were not able to obtain the genetic competence state of S. sobrinus DSM 20742 (see discussion later in the “Variability and specificity in metabolic pathways and network” part). It is also worth to mention that the putative ComS ortholog found in S. ratti DSM 20564 is quite different to those of S. mutans strains, as shown in Figure 4b.
CSP-independent competence regulation system
It has been reported that a basal level of competence remains (referred as CSP-independent competence) after the deletion of comE from S. mutans, suggesting that the CSP-dependent regulation system is one of the several signaling pathways involved in ComX activation [34]. Indeed, under conditions of biofilm growth the HdrMR system, a novel two-gene regulatory system, has been shown to contribute to competence development through the activation of ComX by a yet unknown signal [40]. Moreover, microarray analysis revealed that both regulators, ComE and HdrR, activate a large set of overlapping genes [40,41]. Recently, Xie et al. identified in S. mutans another regulatory system, designated BsrRM, that primarily regulates bacteriocin-related genes but also affects the HdrMR system and thus indirectly contributes to competence development [42]. In this study, HdrR, the response regulator of the HdrMR system, is found neither present in S. ratti DSM 20564 nor in S. sobrinus DSM 20742. Furthermore, the response regulator BrsR of the BsrRM system is also absent in S. ratti DSM 20564, whereas S. sobrinus DSM 20742 lacks the complete BsrRM system. It’s also worth to mention that a competence damage-inducible protein CinA, which is regulated via ComX and has been proven to be related to DNA damage, genetic transformation and cell survival [43], is present in all strains.
Taking together, both the CSP-dependent and CSP-independent competence regulation systems in S. ratti DSM 20564 and especially in S. sobrinus DSM 20742 are very different to those of the S. mutans strains.
Distribution of bacteriocin-related proteins and antibiotic resistance-related proteins
Bacteriocin-related proteins
Bacteriocins are proteinaceous toxins produced by bacteria to kill or inhibit the growth of similar or closely related bacterial strain(s). Bacteriocins produced by mutans streptococci are named “mutacins”. As dental plaque, the dominating niche of mutans streptococci, is a multispecies biofilm community that harbors many microorganism species, mutans group strains have developed a variety of mutacins to inhibit the growth of competitors, such as mitis group streptococci [44-46]. In this study, information about known mutacins as well as mutacin-immunity proteins was collected from the NCBI (http://www.ncbi.nlm.nih.gov) and Oralgen (http://www.oralgen.lanl.gov/) databases, as well as by searching for related publications. The collected protein sequences, as listed in Additional file 5, were used to blast against the proteomes of the 10 strains to see whether or not these known mutacins and mutacin-immunity proteins do exist in the mutans streptococci strains of this study. Distributions of identified mutacins and mutacin-immunity proteins are summarized in Table 4. Using this approach it is, however, not possible to identify any new types of mutacins.
Table 4.
Distribution of mutacins and mutacin immunity proteins in the 10 mutans streptococci strains
| S. mutans UA159 | S. mutans NN2025 | S. mutans 5DC8 | S. mutans KK21 | S. mutans KK23 | S. mutans AC4446 | S. mutans ATCC 25175 | S. mutans NCTC 11060 |
S. ratti |
S. sobrinus DSM 20742 | References | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DSM 20564 | |||||||||||
|
Mutacin-SMB (lantibiotic mutacin) |
- |
- |
- |
- |
- |
- |
- |
- |
D822_07608 D822_07613 |
- |
[47,48] |
|
Mutacin-I (lantibiotic mutacin) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
[49,50] |
|
Mutacin-II (lantibiotic mutacin) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
[51] |
|
Mutacin-III (lantibiotic mutacin) |
- |
- |
- |
- |
- |
- |
- |
- |
- |
- |
[52] |
|
Mutacin-K8 (lantibiotic mutacin) |
- |
GI|290579849 GI|290579848 GI|290579850 |
- |
- |
D818_07928 D818_07933 D818_07938 |
- |
- |
- |
- |
- |
[53] |
|
Mutacin-IV (NlmA) |
SMU.150 |
- |
D816_00655 |
D817_00675 |
D818_00659 |
- |
D820_00642 |
D821_00661 |
- |
- |
[54] |
|
Mutacin-IV (NlmB) |
SMU.151 |
- |
D816_00660 |
D817_00680 |
D818_00664 |
- |
D820_00647 |
D821_00666 |
- |
- |
[54] |
|
Mutacin-IV like (SMU.283) |
SMU.283 |
GI|290581209 |
D816_01135 |
D817_01285 |
D818_01099 |
D819_01148 |
D820_01321 |
D821_01193 |
D822_03404 |
- |
[8] |
|
Immunity protein of Mutacin-IV |
SMU.152 |
GI|290580110 |
D816_06832 |
D817_06998 |
D818_06649 |
D819_06637 |
D820_06850 |
D821_06847 |
D822_03264 |
D823_04636 |
[55,56] |
|
Mutacin-V (CipB) |
SMU.1914c |
GI|290579763 |
D816_08583 |
D817_08773 |
D818_08363 |
D819_07834 |
- |
- |
D822_03354 |
- |
[55,56] |
|
CipI, immunity protein of CipB (Mutacin-V) |
SMU.925 |
|
D816_04020 |
D817_04283 |
D818_04522 |
D819_04119 |
D820_04232 |
D821_04089 |
D822_03349 |
|
[55,56] |
|
Homolog of CipI |
SMU.1913c |
GI|290579764 |
D816_08578 |
D817_08768 |
D818_08358 |
D819_07829 |
|
|
|
D823_03992 |
[56] |
|
SMU.423 possible bacteriocin |
SMU.423 |
GI|290581063 |
D816_01775 |
D817_01930 |
D818_01847 |
D819_01823 |
D820_01975 |
D821_01862 |
|
D823_05348 |
[8] |
|
NlmT/ComA transporting ATPase |
SMU.286 SMU.1881c |
GI|290581206 GI|290579788 |
D816_01150 D816_08453 |
D817_01300 D817_08643 |
D818_01134 - |
D819_01163 D819_07724 |
D820_01336 - |
D821_01208 D821_08449 |
D822_01584 D822_08325 - |
D823_05343 D823_01400 |
[28,29] |
| NlmE/ComB accessory factor for NlmT | SMU.287 | GI|290581205 | D816_01155 | D817_01305 | D818_01139 | D819_01168 | D820_01341 | D821_01213 | D822_01589 | D823_05923 | [28,29] |
In the case that more than one paralogs were found, the most similar ortholog is marked in bold font.
Note: as a multi-function exporter, the entries of NlmTE(ComAB) have been shown in Table 3 and here again.
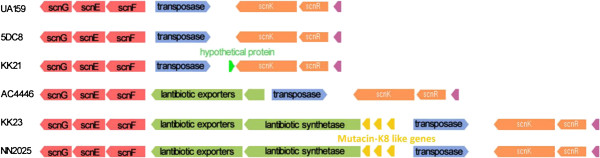
Diversity of Streptococcus bacteriocins has been reported previously [57,58]. The mutacin assortments among the 10 strains in this study also demonstrate certain variations. An interesting result is that in contrast to S. mutans strains and S. ratti DSM 20564, S. sobrinus DSM 20742 does not possess any genes coding for mutacin-like proteins. Mutacin-SMB has been identified in S. mutans and S. ratti previously [47,48]. In our study, mutacin-SMB cluster was only identified in S. ratti DSM 20564 comprising 7 genes, including the mutacin-coding genes smbA and smbB, as well as 5 mutacin-related genes (smbG- > D822_07603, smbT- > D822_07593, smbM- > D822_07578, smbF- > D822_07588, and smbM2- > D822_07598). Lantibiotic mutacins, mutacin-I [49], mutacin-II [51] and mutacin-III [52], are completely absent in the 10 mutans streptococci strains. However, three gene copies possibly encoding the precursor of the lantibiotic mutancin mutacin-K8 are identified in the S. mutans strains KK23 and NN2025. Mutacin-K8 is an ortholog of the bacteriocin Streptococcin A-FF22 identified in group-A streptococci [59], and its production system has previously also been identified in the S. mutans strain K8 [53]. By carefully examining the genes surrounding mutacin-K8 precursor genes the gene cluster coding for a complete mutacin-K8 production system is also revealed in the strains KK23 and NN2025 (Figure 5). A partial ortholog of the mutacin-K8 production system is found in S. mutans UA159, 5DC8 and KK21, with only genes responsible for the immunity (scnFEG) left behind. Orthologous genes coding for a part of the mutacin-K8 production system are also found in S. mutans AC4446, consisting of only scnFEG, scnT (coding a lantibiotic exporter) and a part of scnM (coding the lantibiotic synthetase). Since a gene encoding ISSmu2-type transposase is found to be located upstream of mutacin-K8 precursor genes, we infer that the variety of mutacin-K8 production system in S. mutans strains studied here is highly possible to be caused by transposase actions.
Figure 5.
Cluster structure of the mutacin-K8 production system across six S. mutans strains. The ORFs colored in yellow are the possible mutacin-K8 precursor genes. scnGEF: bacteriocin related ABC element; possible immunity system; scnK: histidine kinase of two component system; scnR: response regulator of two component system (Note: mutacin-K8 production system was failed to be identified in S. mutans NCTC 11060, S. mutans ATCC 25175, S. ratti DSM 20564 and S. sobrinus DSM 20742).
Mutacin-IV, nonlantibiotic bacteriocins coded by nlmA/B (SMU.150/151, Note: hereinafter whenever needed/possible the locus_tag of the reference strain S. mutans UA159 is given for convenience) was discovered first in S. mutans UA140 to be active against the mitis group streptococci [54]. In this study, nlmA/B are found to be present in six of the S. mutans strains, including UA159, 5DC8, KK21, KK23, ATCC 25175 and NCTC 11060, but not in S. mutans NN2025 and AC4446, nor in S. ratti DSM 20564 and S. sobrinus DSM 20742. On the other hand, the immunity protein for mutacin-IV (SMU.152), is identified in all strains, consistent with the fact that no inhibition phenomenon has been observed yet among different mutans streptococci strains. A mutacin-IV like protein found before in the strain UA159 (SMU.283) is identified in all strains except for S. sobrinus DSM 20742.
Mutacin-V, another nonlantibiotic peptide coded by cipB (SMU.1914) is found, in addition to S. sobrinus DSM 20742, also absent in the S. mutans strains ATCC 15175 and NCTC 11060. There are two homologs of mutacin-V immunity protein in S. mutans UA159, namely CipI (SMU.925) and SMU.1913 [8,60]. These two immunity proteins share a sequence identity of 82%. However, it has been reported that though very likely co-transcribed with cipB, SMU.1913 cannot prevent CipB-caused cell lysis in S. mutans UA159, and the key immunity factor of mutacin-V has been supposed to be CipI (SMU.925) rather than SMU.1913 [60]. All the 10 strains including S. sobrinus DSM 20742 possess at least one orthologous gene encoding one of the two mutacin-V immunity proteins. Based on the similarity scores S. mutans NN2025 does not have an ortholog of CipI (SMU.925), but it possesses an ortholog (GI|290579764) of SMU.1913, which is possibly co-transcribed with GI|290579764, the cipB ortholog in S. mutans NN2025. Furthermore, the only putative immunity protein D822_3349 in S. ratti DSM 20564 shows very close similarities to SMU.925 (61%) and SMU.1913 (56%) and is possibly co-transcribed with D822_03354, the CipB ortholog in S. ratti DSM 20564. From these results, we suppose that SMU.1913, which is co-transcribed with cipB (SMU.1914), might be the ancestor gene coding for the mutacin-V immunity factor. The additional copy, like SMU.925 in S. mutans UA159, might be generated by duplication action and evolved as the dominant immunity factor in some of the mutans streptococci strains.
Furthermore, a possible nonlantibiotic bacteriocin peptide (SMU.423) is found to be present in all strains except for S. ratti DSM 20564. Putative ComAB, which has been proved to be the transporter complex of mutacin IV in S. mutans[28], are identified in all strains, supporting the suggestion that ComAB might function as a common transporter for multi-type nonlantibiotic bacteriocins rather than merely for mutacin IV. Moreover, an additional paralog of ComA is present in most of the strains except for S. mutans KK23 and S. mutans ATCC 25175.
To summarize, a differed distribution of mutacin/bacteriocin encoding genes accompanied with a high conservation of genes coding for mutacin-immunity proteins are revealed for the 10 mutans streptococci strains/species. The conservation of mutacin immunity proteins apparently plays an important role for the survival of mutans streptococci strains under a bacteriocin-rich environment.
Antibiotic resistance-related proteins
Bacteria and other microorganisms that cause infections are remarkably resilient and can develop ways to survive drugs meant to kill or weaken them. Antibiotic resistance can be a result of horizontal gene transfer [61], and also of unlinked point mutations in the pathogen genome at a rate of about 1 in 108 per chromosomal replication [62]. The antibiotic action against the pathogen can be seen as an environmental selective pressure and bacteria which have developed mutations allowing them to survive will live on to reproduce. They will then pass this trait to their offsprings, which will result in the evolution of fully resistant colonies. Putative resistance-related genes are identified and listed in Table 5.
Table 5.
Distribution of antibiotic resistance-related proteins
| Name | Putative function | S. mutans UA159 | S. mutans NN2025 | S. mutans 5DC8 | S. mutans KK21 | S. mutans KK23 | S. mutans AC4446 | S. mutans ATCC 25175 | S. mutans NCTC 11060 | S. ratti DSM 20564 | S. sobrinus DSM 20742 |
|---|---|---|---|---|---|---|---|---|---|---|---|
|
UppP |
Putative bacitracin resistant |
SMU.244 |
GI|290581239 |
D816_00960 |
D817_01110 |
D818_00974 |
D819_00998 |
D820_01146 |
D821_00986 |
D822_05517 |
D823_09307 |
|
BceA |
Bacitracin resistant ABC transporter ATP-binding protein |
SMU.1006 |
GI|290580542 |
D816_04484 |
D817_04663 |
D818_04902 |
D819_04489 |
D820_04607 |
D821_04449 |
D822_02154 |
D823_04551 |
|
BceB |
Bacitracin resistant ABC transporter permease protein |
SMU.1007 |
GI|290580541 |
D816_04489 |
D817_04668 |
D818_04907 |
D819_04494 |
D820_04612 |
D821_04454 |
D822_02159 |
D823_04556 |
|
DacF |
Penicillin binding protein; Penicillin sensitive protein |
SMU.75 |
GI|290579588 |
D816_00335 |
D817_00355 |
D818_00354 |
D819_00260 |
D820_00330 |
D821_00310 |
D822_07803 |
D823_05036 |
|
Pbp2X |
Penicillin-binding protein 2X; Penicillin sensitive protein |
SMU.455 |
GI|290581039 |
D816_01905 |
D817_02153 |
D818_01967 |
D819_01954 |
D820_02095 |
D821_01964 |
D822_00802 |
D823_06528 |
| |
metallo-beta-Lactamase superfamily protein; beta-Lactam resistance; |
SMU.368c |
GI|290581108 |
D816_01525 |
D817_01680 |
D818_01583 |
D819_01608 |
D820_01711 |
D821_01583 |
D822_04346 |
D823_00655 |
| |
beta-Lactamase family protein; beta-Lactam resistance; |
SMU.400 |
GI|290581086 |
D816_01660 |
D817_01815 |
D818_01732 |
D819_01708 |
D820_01860 |
D821_01747 |
D822_05706 |
D823_03675 |
|
YqgA |
beta-Lactam resistance; |
SMU.1444c |
GI|290580186 |
D816_06482 |
D817_06653 |
D818_06314 |
D819_06285 |
D820_06483 |
D821_06502 |
D822_08877 |
D823_08387 |
| |
Lactamase_B; |
SMU.1515 |
GI|290580115 |
D816_06807 |
D817_06973 |
D818_06624 |
D819_06612 |
D820_06825 |
D821_06822 |
D822_03289 |
D823_04661 |
|
beta-Lactam resistance; | |||||||||||
|
MurN |
beta-Lactam resistance factor MurN |
SMU.716 |
GI|290580807 |
D816_03100 |
D817_03358 |
D818_03627 |
D819_03104 |
D820_03315 |
D821_03199 |
D822_00265 |
D823_09452 |
|
MurM |
beta-Lactam resistance factor murM; |
SMU.717 |
GI|290580806 |
D816_03105 |
D817_03363 |
D818_03632 |
D819_03109 |
D820_03320 |
D821_03204 |
D822_00260 |
D823_09457 |
| |
Macrolide-efflux protein |
|
GI|290581182 |
|
|
D818_01269 |
D819_01313 |
|
|
|
|
| |
Putative multidrug resistance ABC transporter |
|
GI|290581181 |
|
|
D818_01274 |
D819_01318 |
|
|
|
|
|
VanW |
Vancomycin b-type resistance protein |
|
|
|
|
|
|
|
|
D822_01634 |
|
| |
Putative multidrug resistance protein b |
SMU.745 |
GI|290580783 |
D816_03220 |
D817_03478 |
D818_03732 |
D819_03234 D819_09750 |
D820_03442 |
D821_03314 |
D822_00530 |
D823_08347 |
|
PmrA |
Putative multidrug resistance efflux pump |
SMU.1611c |
GI|290580030 |
D816_07242 |
D817_07403 |
D818_07009 |
D819_07037 |
D820_07260 |
D821_07242 |
D822_07918 |
D823_02317 |
|
YitG |
Putative multidrug resistancepermease |
SMU.1286c |
GI|290580299 |
D816_05764 |
D817_05958 |
D818_02360 |
D819_05785 |
D820_05818 |
D821_05850 |
D822_01559 |
|
| Putative multidrug resistanceABC transport | SMU.905 | GI|290580642 | D816_03940 | D817_04208 | D818_04447 | D819_03949 | D820_04157 | D821_04009 | D822_09885 | D823_08492 |
The S. mutans species is known to be intrinsically resistant to bacitracins produced by Bacillus subtilis. We confirmed this by testing all the 10 strains with a bacitracin-E-test (data not shown). All strains including S. ratti DSM 20564 and S. sobrinus DSM 20742 had a minimum inhibitory concentration between 128 and >256 μg/l. In fact, this antibiotic is used to isolate mutans-streptococci from highly heterogeneous oral microflora. It has been reported that bceABRS (also named as mbrABCD) system, encoding a two component signal transduction system and an ABC-transporter, is required for bacitracin resistance in S. mutans[63,64]. As expected, ortholog of bceABRS system is found to be present in all strains. Furthermore, an ortholog of a putative bacitracin resistance protein UppP (SMU.244, undecaprenyl-diphosphatase) is present in all strains. It has been proved that overexpression of UppP in Escherichia coli and Bacillus subtilis results in bacitracin resistance [65,66]. However, the function of UppP in bacitracin resistance in mutans streptococci has not yet been investigated. Based on its conservation in all strains studied here, we suppose that UppP might play an important role in bacitracin resistance for mutans streptococci species as well.
Two penicillin-binding proteins (SMU.75 and SMU.455) are identified in all strains, indicating that they are potentially all susceptible to penicillin. Phenotypically all strains were tested to be susceptible to penicillin (data not shown). On the other hand, all the strains possess orthologs of SMU.368c, SMU.400, SMU.1444c and SMU.1515, which are homologs to beta-lactamases (EC 3.5.2.6), as well as orthologs of two so called beta-lactam resistance factors (SMU.716, SMU.717). Thus, all the strains are potentially capable of resistance against beta-lactam antibiotics. Orthologs of macrolide-efflux transporter proteins, as coded by GI|290581182 and GI|290581181 in S. mutans NN2025, are found to be also present in S. mutans 5DC8 and S. mutans KK21. A vancomycin b-type resistance-associated protein (D822_01634) is uniquely present in S. ratti DSM 20564, but our phenotypic testing showed as expected that S. ratti DSM 20564 is susceptible to vancomycin. Furthermore, several putative multidrug resistance-associated proteins (SMU.745, SMU.1611c and SMU.905 except for SMU.1286c) are found to be present in all strains.
Oxidative stress defense systems in mutans streptococci
For protection against reactive oxygen species (such as O2-, H2O2, HO·) or adaptation to oxidative stresses aerobes and facultative anaerobes have evolved efficient defense systems, comprising an array of antioxidant enzymes such as catalase, superoxide dismutase (SOD), Dps-like peroxide resistance protein, alkylhydroperoxide reductase (AhpCF), glutathione reductase, and thiol reductase, which have been identified in many bacterial species.
Although the first genome sequence of S. mutans UA159 has already been published in 2002, the oxidative stress defense systems in the group of mutans streptococci have not yet been systematically discussed. By searching for known antioxidant systems in the genomes of the sequenced mutans streptococci strains of this study, we obtained an overview of putative oxidative defense systems in these mutans streptococci strains/species which are composed of superoxide dismutase (SOD), AhpF/AhpC system, Dpr, thioredoxin system and glutaredoxin system, as shown in Table 6.
Table 6.
Distribution of oxidative stress resistance systems
| Class | Name | Function | S. mutans UA159 | S. mutans NN2025 | S. mutans 5DC8 | S. mutans KK21 | S. mutans KK23 | S. mutans AC4446 | S. mutans ATCC 25175 | S. mutans NCTC 11060 | S. ratti DSM 20564 | S. sobrinus DSM 20742 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
SOD |
Sod |
Superoxide dismutase |
SMU.629 |
GI|290580884 |
D816_02695 |
D817_02943 |
D818_03247 |
D819_02714 |
D820_02900 |
D821_02804 |
D822_02694 |
D823_08152 |
| |
|
3′-Phosphoadenosine-5′-phosphate phosphatase |
SMU.1297 |
GI|290580288 |
D816_05819 |
D817_06013 |
D818_02305 |
D819_05840 |
D820_05873 |
D821_05905 |
D822_08440 |
D823_09052 |
|
AhpF/AhpC system |
AhpC |
Alkyl hydroper oxide reductase, subunit C |
SMU.764 |
GI|290580768 |
D816_03290 |
D817_03548 |
D818_03807 |
D819_03314 |
D820_03512 |
D821_03389 |
D822_08028 |
- |
| AhpF (Nox1) |
Alkyl hydroperoxide reductase, subunit F |
SMU.765 |
GI|290580767 |
D816_03295 |
D817_03553 |
D818_03812 |
D819_03319 |
D820_03517 |
D821_03394 |
D822_08023 |
- |
|
|
Dpr |
Dpr |
Peroxide resistance protein / iron binding protein |
SMU.540 |
GI|290580957 |
D816_02305 |
D817_02548 |
D818_02835 |
D819_02354 |
D820_02520 |
D821_02374 |
D822_04226 |
D823_02352 |
|
Thioredoxin system |
TrxB |
Thioredoxin reductase (NADPH) |
SMU.463 |
GI|290581031 |
D816_01940 |
D817_02188 |
D818_02007 |
D819_01989 |
D820_02130 |
D821_01999 |
D822_06878 |
D823_01947 |
| TrxB |
Thioredoxin reductase |
SMU.869 |
GI|290580673 |
D816_03785 |
D817_04038 |
D818_04292 |
D819_03804 |
D820_04002 |
D821_03854 |
D822_03499 |
D823_01550 |
|
| TrxA |
Thioredoxin |
SMU.1869 |
GI|290579800 |
D816_08398 |
D817_08588 |
D818_08193 |
D819_07664 |
D820_08390 |
D821_08394 |
D822_08270 |
D823_06913 |
|
| TrxH |
Thioredoxin family protein |
SMU.1971c |
GI|290579712 |
D816_08848 |
D817_09038 |
D818_08623 |
D819_08094 |
D820_08775 |
D821_08804 |
D822_07458 |
D823_08552 |
|
| |
Thioredoxin family protein |
SMU.1169c |
GI|290580401 |
D816_05229 |
D817_05413 |
D818_05692 |
D819_05219 D819_05259 |
D820_05307 |
D821_05309 |
D822_06958 |
- |
|
| Tpx |
Thiol peroxidase |
SMU.924 |
GI|290580628 |
D816_04015 |
D817_04278 |
D818_04517 |
D819_04114 |
D820_04227 |
D821_04084 |
D822_03359 |
D823_07595 |
|
| Glutaredoxin system | GshAB |
Glutathione biosynthesis bifunctional protein |
SMU.267c |
GI|290581223 |
D816_01065 |
D817_01215 |
D818_01054 |
D819_01078 |
D820_01251 |
D821_01091 |
D822_01287 |
D823_06703 |
| GshR |
Glutathione reductase |
SMU.838 |
GI|290580702 |
D816_03640 |
D817_03893 |
D818_04147 |
D819_03659 |
D820_03857 |
D821_03709 |
D822_01904 |
D823_04976 |
|
| GshR |
Glutathione reductase |
SMU.140 |
- |
D816_00620 |
D817_00640 |
D818_00624 |
- |
D820_00607 |
D821_00626 |
D822_06143 |
- |
|
| NrdH | Glutaredoxin | SMU.669c | GI|290580848 | D816_02885 | D817_03143 | D818_03447 | D819_02894 | D820_03090 | D821_03009 | D822_02899 | D823_05398 |
SOD, which catalyzes the dismutation of superoxide into oxygen and hydrogen peroxide, is an important antioxidant defense in nearly all cells exposed to oxygen [67]. SOD is found in all strains of this study. Catalase, which catalyzes the decomposition of hydrogen peroxide, is not found in any of the mutans streptococci strains of this study. It is known that although most streptococci can grow in the presence of air, they do not possess a catalase, implying that hydrogen peroxide defense mechanism, by which lactic acid bacteria established their growth in air, are very different to those of aerobes. It has been reported that both the bi-component peroxidase system AhpF/AhpC and Dps-like peroxide resistance protein confer tolerance to oxidative stress in S. mutans[68].
The AhpF/AhpC system catalyzes the NADH-dependent reduction of organic hydroperoxides and/or H2O2 to their respective alcohol and/or H2O. Both AhpF and AhpC are present in all S. mutans strains of this study and in S. ratti DSM 20564, but are absent in S. sobrinus DSM 20742. The natural missing of AhpF and AhpC in S. sobrinus indicates that AhpF/AhpC system is not an essential peroxide tolerance system for some mutans streptococci species. While studying a ahpF and ahpC double deletion mutant of S. mutans, Higuchi et al. [69] found that the mutant still showed the same level of peroxide tolerance as did the wild-type strain that led them to the finding of the dpr gene, which encodes a ferritin-like iron-binding protein involved in oxygen tolerance by limiting the nonenzymatic hydroxyl radical synthesis via iron-catalyzed ‘Fenton reaction’ in S. mutans. Their further studies on the biological function of dpr found that dpr gene from S. mutans chromosome was capable of complementing an alkyl hydroperoxide reductase-deficient mutant of E. coli, as well as complementing the defect in peroxidase activity caused by the deletion of ahpF/ahpC in S. mutans, indicating that dpr plays an indispensable role in oxygen tolerance of S. mutans[68,70]. Dpr homologs were found in all strains as expected by the supposed essential function of dpr gene in oxygen tolerance.
Thioredoxins are a class of small redox mediator proteins known to be present in all organisms. They are involved in many important biological processes, including redox signaling. Thioredoxins are kept in the reduced state by the flavor enzyme thioredoxin reductase in a NADPH-dependent reaction [71]. They act as electron donors to many proteins including thiol peroxidases [72]. Thioredoxin, thioredoxin reductase and thiol peroxidase, the components of thioredoxin system, are identified in all the strains of this study. Two putative thioredoxin reductases (SMU.463 and SMU.869) are found in all strains/species. It has been reported that in some species thioredoxin reductases have been evolved to be activated by both NADPH and NADH [73]. We speculate that SMU.463 and SMU.869 might have been evolved to have different preferences to NADPH and NADH (SMU.463 and SMU.869 shares less than 20% similarities). If it holds true, this could be advantageous for these mutans streptococci, as the extra amount of NADH produced from glycolysis/gluconeogenesis pathway under anaerobic conditions could be directly used for oxidative stress resistance. Thioredoxin (SMU.1869) and two thioredoxin family proteins (SMU.1971c and SMU.1169c) are found to be present in nearly all strains, except for S. sobrinus DSM 20742, which lacks any ortholog of SMU1169c. An ortholog of a thiol peroxidase-coding gene (tpx) is identified in all strains.
Glutaredoxins share many functions of thioredoxins but are reduced by glutathione (L-γ-glutamyl-L-cysteinylglycine, GSH) rather than by a specific reductase. This means that glutaredoxins are oxidized by their corresponding substrates, and reduced non-enzymatically by GSH [74]. Oxidized glutathione (GSSG) is then regenerated by glutathione reductase. Together, these components comprise the glutathione system [75]. GSH is a well-characterized antioxidant in eukaryotes and Gram-negative bacteria, where it is synthesized by the sequential action of two enzymes, γ-glutamylcysteine synthetase (γ-GCS) and glutathione synthetase (GS). Among Gram-positive bacteria only a few species contain GSH. It has been reported that streptococci lack the moderate-to-high levels of intracellular glutathione normally found in Gram-negative bacteria [76]. Using Streptococcus agalactiae as a model, it has been discovered that in GSH-containing Gram-positive bacteria GSH synthesis is catalyzed by one bifunctional protein, γ-glutamylcysteine synthetase-glutathione synthetase (γ-GCS-GS), encoded by one gene, gshAB. Homologs of γ-GCS-GS have been identified in the genomes of 19 mostly studied Gram-positive bacteria, including S. mutans[77]. All components of the glutathione system were identified in all the 10 strains of this study. Several S. mutans strains, namely UA159, 5DC8, KK21, KK23, ATCC 25175, and NCTC 11060, as well as S. ratti DSM 20564, possess two glutathione reductase orthologs (SMU.140 and SMU.838). This could possibly convey these strains certain advantages in the re-generation of GSH from GSSG, which in turn would be helpful for oxidative resistance.
In addition, 3′-phosphoadenosine-5′-phosphate phosphatase activity has recently been reported to be required for superoxide stress tolerance in S. mutans[78]. Putative 3′-phosphoadenosine-5′-phosphate phosphatase coding genes were identified in all strains (Table 6).
Variability and specificity in metabolic pathways and network
In order to reveal the metabolic variability of the mutans streptococci systematically, we have reconstructed and analyzed the genome-scale metabolic networks of all the strains sequenced with the method proposed by Ma and Zeng [79] and an updated database [79]. All annotated protein sequences having EC numbers are considered for the network reconstruction. From the functional annotation discussed above, total EC numbers identified in the 10 strains are very close to each other, as shown in Table 7. A summary of the total numbers of the reactions and metabolites in each of the reconstructed metabolic networks is shown in Table 7, and all the constructed metabolic networks are provided in Additional file 6 in *.cys format which can be opened with Cytoscape [80], a software for visualization and analysis of biological networks. The sizes of the constructed metabolic networks of the eight S. mutans strains are very close to each other, with UA159, NN2025, AC4446, 5DC8 and KK21 having almost exactly the same size, and the networks of KK23, ATCC 25175 and NCTC 11060 being merely about 2% larger. While the size of the metabolic network of S. ratti DSM 20564 is comparable to those of the S. mutans strains, the metabolic network of S. sobrinus with 833 reactions and 853 metabolites is the smallest one, which have 62 less reactions and 60 less metabolites compared to the largest one of S. mutans NCTC 11060 (895 reactions and 913 metabolites).
Table 7.
Compositions of the established metabolic networks of the 10 mutans streptococci strains
| Strain | EC numbers | Reactions | Metabolites |
|---|---|---|---|
|
S. mutans UA159 |
454 |
875 |
893 |
|
S. mutans NN2025 |
450 |
874 |
892 |
|
S. mutans 5DC8 |
453 |
875 |
893 |
|
S. mutans KK21 |
453 |
875 |
893 |
|
S. mutans KK23 |
452 |
893 |
911 |
|
S. mutans AC4446 |
449 |
874 |
893 |
|
S. mutans ATCC 25175 |
453 |
891 |
911 |
|
S. mutans NCTC 11060 |
456 |
895 |
913 |
|
S. ratti DSM 20564 |
435 |
888 |
893 |
| S. sobrinus DSM 20742 | 434 | 833 | 853 |
Despite the comparable network sizes, however, all the strains possess or lack certain reactions/metabolites, as revealed by detailed comparative analyses. Using the metabolic network of S. mutans UA159 as reference, the presence and absence of reactions in each of the strains/species compared are discovered and mapped into sub-pathways based on the KEGG pathway classification (http://www.genome.jp/kegg/pathway.html). As a result, among the 416 sub-pathways defined in the KEGG pathway database 46 sub-pathways demonstrated certain variations between the strains/species, as summarized in Additional file 7.
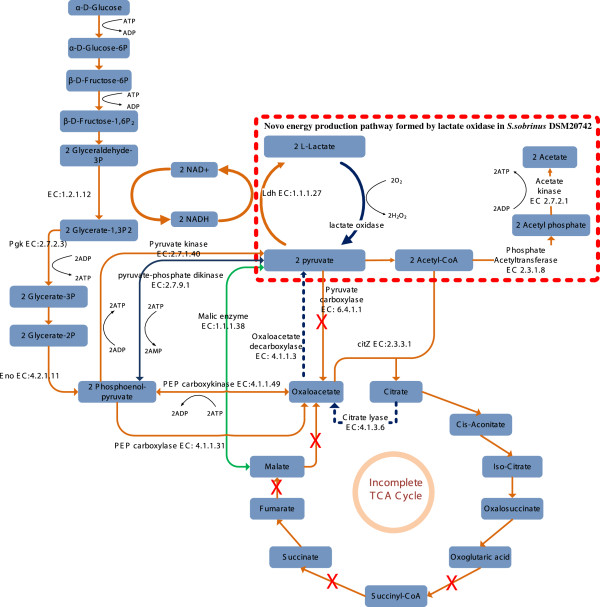
A key feature of the oral environment is that the nutrients available to the oral bacteria are always fluctuating between abundance and famine associated with human diet. Thus, the ability to quickly acquire and metabolize carbohydrates to produce energy and precursors for biosynthesis is essential for the survival of all oral bacteria. Due to their key roles in carbohydrates metabolism and energy production, glycolysis/gluconeogenesis, TCA cycle and pyruvate metabolism pathways are generally considered to be highly conserved among these oral bacteria. Although mutans streptococci strains/species are closely related species as revealed by phylogenetic tree analysis in this study (Figure 1), differences in the central carbon metabolic pathways are found as shown in Figure 6.
Figure 6.
Central metabolism pathways of mutans streptococci. The orange lines represent enzyme reactions conserved across the mutans streptococci strains compared in this study, whereas the blue lines represent enzyme reactions specifically present (solid line) or absent (dashed line) in S. sobrinus DSM 20742. Reaction catalyzed by NAD+-specific malic enzyme (EC: 1.1.1.38) (green line) is absent in S. mutans NN2025 and S. mutans AC4446. Pyruvate-phosphate dikinase, catalyzing the interconversion of PEP and pyruvate (black line), is uniquely present in S. ratti DSM 20564.
Facultative anaerobes such as lactic acid bacteria including Streptococcus lack cytochrome oxidases required for energy-linked oxygen metabolism and energy (in the form of ATP) required for survival and growth are generated by substrate level phosphorylation in the glycolysis pathway [69]. L-lactate oxidase (D823_06598) with a similarity of 73% to YP_003064450.1 (accession number) of Lactobacillus plantarum JDM1 and lactate oxidase (D823_06595) with a similarity of 65% to ZP_09448656.1 (accession number) of Lactobacillus mali KCTC 3596, are found to be uniquely present in S. sobrinus DSM 20742. These two enzymes catalyze the reaction of L-Lactate + O2= > Pyruvate + H2O2 and/or D-Lactate + O2= > Pyruvate + H2O2. It has been reported that in S. pneumoniae concerted action of lactate oxidase and pyruvate oxidase forms a novel energy-generation pathway by converting lactate acid to acetic acid under aerobic growth conditions [81]. Because there is no pyruvate oxidase identified in S. sobrinus DSM 20742, the function of the lactate oxidases in S. sobrinus DSM 20742 should be different to that of S. pneumoniae. By a close examination we hypothesize that lactate oxidase, together with pyruvate dehydrogenase, phosphate acetyl transferase and acetate kinase, could form a novel energy production pathway to convert lactate acid to acetate and simultaneously produce one additional ATP, as depicted Figure 6. By doing so, the lactate oxidases of S. sobrinus DSM 20742 could also play a role in consuming lactate to regulate pH, which would be an advantage for S. sobrinus DSM 20742 in resistance to acid stress. In addition, this pathway could replenish Acetyl-CoA, an important intermediate for the biosynthesis of fatty acids and amino acids. This is for the first time that such an energy production pathway is proposed in Streptococcus species. Furthermore, lactate oxidase and lactate dehydrogenase could form a local NAD+ regeneration system, which would be certainly advantageous to S. sobrinus DSM 20742 under aerobic growth conditions. Moreover, it is known that mutans group streptococci and the mitis group streptococci are competitors, with S. mutans producing mutacins to kill the mitis group streptococci and the mitis group streptococci in turn produce H2O2 to kill mutans group streptococci [16,82]. Favored by possessing the lactate oxidases, S. sobrinus DSM 20742 has the potential ability of producing H2O2 to kill not only competitors (oxygen sensitive S. mutans, oral anaerobes) but also macrophages [83], and defend its ecological niche. The unique presence of lactate oxidases in S. sobrinus DSM 20742 was verified by PCR experiments as shown in Additional file 8. Later, we also found that another S. sobrinus strain AC153 also harbors homologous genes of lactate oxidase, suggesting that lactate oxidase may be conserved and play an important role in S. sobrinus. In the effort to clarify the functionality of lactate oxidase we tried to knock out the two genes encoding the two enzymes by PCR ligation mutagenesis according to the method of Lau PC et al. (2002). We applied different transformation methods (two natural transformation methods and two electroporation methods) but were failed to obtain the desired recombinants. Then, to find out if S. sobrinus DSM 20742 is able to enter genetic competence state at all, we tried to transform S. sobrinus with plasmids replicative in other Streptococcus spp. like pDL278 (Spr, pAT18 Emr, with suicide vector pFW5 Spr in both circular and linearized forms but could not obtain the transformants. Therefore, it is clear that the genetic competence behavior of S. sobrinus DSM 20742 is very different to that of S. mutans, attributing very likely to the lacking of the genes comSR and comC.
In contrast to the unique harboring of lactate oxidases in S. sobrinus DSM 20742, citrate lyase (EC 4.1.3.6), which catalyzes the cleavage of citrate into oxaloacetate and acetate, and oxaloacetate decarboxylase (EC 4.1.1.3), catalyzing the irreversible decarboxylation of oxaloacetate to pyruvate and CO2, are not found in S. sobrinus DSM 20742, as shown in Figure 6 by the blue dotted lines. It has been reported that citrate lyase functions as a key enzyme in initiating the anaerobic utilization of citrate by a number of bacteria, further catabolism of oxaloacetate formed taking place either by decarboxylation or by reduction. In some organisms, oxaloacetate is decarboxylated to pyruvate by oxaloacetate decarboxylase, which is also induced in the presence of citrate. The two enzymatic reactions, which occur sequentially, constitute the ‘citrate fermentation pathway’ [84]. The absence of citrate lyase and oxaloacetate decarboxylase implies that S. sobrinus DSM 20742 might lacks the ability in anaerobic utilization of citrate as a substrate. The disadvantages of S. sobrinus DSM 20742 in citrate utilization could be offset by the novel energy production pathway from lactate to acetate proposed above.
A putative pyruvate-phosphate dikinase (EC 2.7.9.1), which catalyzes the interconversion between PEP and pyruvate, is found to be uniquely present in S. ratti DSM 20564. Pyruvate-phosphate dikinase has been found in propionic acid bacteria [85]. The large difference in the standard free energy of hydrolysis for ATP to AMP and pyrophosphate (−7.6 kcal/mole) and for PEP to pyruvate (−13.6 kcal/mole) at pH 7.0 indicates that the equilibrium for the reaction it catalyzes would strongly favor pyruvate formation. But studies in Acetobacter xylinum clearly indicate that the function of this enzyme under physiological conditions favors the process of gluconeogenesis [86]. Metabolite interconversion at the PEP-pyruvate-oxaloacetate node involves a structurally entangled set of reactions that interconnect the major pathways of carbon metabolism and thus, is responsible for the distribution of the carbon flux among catabolism, anabolism and energy supply of the cell [87]. Under glycolytic conditions oxaloacetate is generated by carboxylation of PEP and/or pyruvate catalyzed by PEP carboxylase (PEPCx) and/or pyruvate carboxylase (PCx). In this study PCx is not found in any of the mutans streptococci strains.
All the 10 strains of this study possess similarly an incomplete TCA cycle and the primary role of the existing TCA enzymes is most likely the synthesis of amino acid precursors as has been reported previously [8,88].
Conclusion
In the present study, the genomes of 8 mutans streptococci strains, including six S. mutans strains, one S. ratti strain and one S. sobrinus strain were sequenced, annotated and compared together with S. mutans UA159 and NN2025. Multiple genome alignment showed extensive genome rearrangement among the eight strains of S. mutans. The core-genome size of S. mutans was determined to be around 1,370 genes by including 67 S. mutans genomes. A possibly open pan-genome of S. mutans was inferred.
Systematic comparative analyses were focused on competence regulation, bacteriocin (mutacin) production, antibiotic resistance, oxidative stress resistance, as well as central carbon metabolism and energy production pathways. Most of these systems show remarkable differences between the strains, except for oxidative stress resistance systems which are found to be well conserved. CSP-dependent and independent competence regulation systems are highly diverse in mutans streptococci: no comC-like genes could be identified in S. ratti and S. sobrinus; putative ComC amino acid sequences of S. mutans show clear variations; ComS and ComR are absent in S. sobrinus which well explains the fact that we were not able obtain genetic competence state of S. sobrinus by experiment, even though the ComX and the downstream competence development genes are well reserved; furthermore, the response regulators of the HdrMR and BsrRM systems, which are known to be involved in competence development, are missing in both S. ratti and S. sobrinus.
Variation in mutacin-encoding genes is accompanied with the conservation of mutacin immunity proteins, which indicates apparently important roles of the mutacin immunity proteins for the survival of these mutans streptococci in a bacteriocin rich environment. The presence of various antibiotic resistance factors, together with the open pan-genome inferred, implies that attention should be paid to the potential of mutans streptococci in the development of antibiotic resistance.
The sizes of the genome-scale metabolic networks of the 10 strains are very close to each other. Comparative analysis of sub-pathways reveals that 46 sub-pathways of all 416 sub-pathways defined in KEGG pathway database show variation using S. mutans UA159 as reference. By identifying lactate oxidases to be uniquely present in S. sobrinus DSM 20742, we proposed for the first time a novel energy production pathway in S. sobrinus. Additional functions of the lactate oxidases in connection with the proposed energy production pathway are also discussed.
In conclusion, the genomes of mutans streptococci display remarkable differences, especially among different species. We believe that the strain-specific information provided in this study should be helpful to understand the evolution and adaptive mechanisms of those oral pathogens.
Methods
Genome sequences and strains
All the newly sequenced strains were described previously [26]. Briefly, serotype c strain S. mutans 5DC8 was isolated from root caries by David Beighton (London, UK); serotype c strain S. mutans AC4446 was isolated from a proven case of infective endocarditis in Dillingen (Germany), serotype c strain S. mutans KK21 was isolated from enamel caries of an adult by Susanne Kneist (Jena, Germany), serotype c strain S. mutans KK23 was isolated from enamel caries of a child by Susanne Kneist (Jena, Germany), Serotype c, type strain S. mutans ATCC 25175 was isolated from carious dentine, serotype f strain S. mutans NCTC 11060 was isolated in Denmark from a patient’s blood, serotype b strain S. ratti DSM 20564(=ATCC 19645) was isolated from caries lesion in rat, and finally, serotype non-d & non-g strain S. sobrinus DSM 20742 (= ATCC 33478) was isolated from human dental plaque. Serotype c is over-represented because 70-80% of all S. mutans isolates are of this serotype. However, non-c serotypes seem to be associated with cardiovascular diseases and this is represented in our study by the serotype f strain. Besides S. mutans, S. sobrinus is considered as a relevant cariogenic species in human. The genome sequences of S. mutans UA159 and S. mutans NN2025 were genome sequenced previously and obtained from NCBI genome database (http://www.ncbi.nlm.nih.gov/genome/, consulted at the beginning of January 2011).
Genome sequencing, assembly and annotation
All of the strains were sequenced using the Solexa sequencing platform at the Helmholtz Center for Infection Research in Braunschweig, Germany. The “high-quality draft” [89] genome sequences of these mutans streptococci strains were assembled by a combined use of the sequence assembly tools SOAPdenovo [90], Maq [91] and Phrap [92]. In brief, we first use SOAPdenovo to obtain the optimal assembly result by using different k-mer from 17 to 41 without scaffolding. Then we map all reads to reference sequence of S. mutans UA159 using Maq and break down the low quality area to obtain a collection of long contigs. Finally, the long contigs were used to close partial gaps of the initial assembly to improve the assembly quality using Phrap. The first version genome annotations were performed using mauve [13,93], tRNAscan-SE 1.21, Glimmer3.02 [94] and Blast2GO [95], and then released through our central genome database (http://biosystem.bt1.tu-harburg.de/) established with PathwayTools [26,96]. This version was used before for the study of TCSTSs of the 10 strains [26]. During this study, all genomes were re-annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (PGAAP, http://www.ncbi.nlm.nih.gov/genomes/static/Pipeline.html) and the whole-genome shotgun sequences have been deposited at DDBJ/EMBL/GenBank under the accessions of AOBX00000000 (S. mutans 5DC8), AOBY00000000 (S. mutans KK21), AOBZ00000000 (S. mutans KK23), AOCA00000000 (S. mutans AC4446), AOCB00000000 (S. mutans ATCC 25175), AOCC00000000 (S. mutans NCTC 11060), AOCD00000000 (S. ratti DSM 20564) and AOCE00000000 (S. sobrinus DSM 20742). The present study used the new version deposited at DDBJ/EMBL/GenBank. As we found out that in the annotated results from PGAAP some coding genes are missing, we did manual curation based on blast searches using known coding nucleotide sequences, the location of the missing coding sequences are given in Additional file 9.
Genome alignment
Multiple genome alignments were computed by using the progressive Mauve algorithm of the Mauve software [13] with default options.
Core-genome and pan-genome analysis
In addition to the 6 S. mutans draft genomes of this study and the previously released complete genomes of S. mutans UA159 and NN2025, 59 newly released S. mutans genomes (2 completed and 57 drafts) available in NCBI till April 2013 were also included in the core- and pan-genome analysis of S. mutans. The accessions of the 59 genomes are as follows:
AGWE00000000, AHRB00000000, AHRC00000000, AHRD00000000, AHRE00000000, AHRF00000000, AHRG00000000, AHRH00000000, AHRI00000000, AHRJ00000000, AHRK00000000, AHRL00000000, AHRM00000000, AHRN00000000, AHRO00000000, AHRP00000000, AHRQ00000000, AHRR00000000, AHRS00000000, AHRT00000000, AHRU00000000, AHRV00000000, AHRW00000000, AHRX00000000, AHRY00000000, AHRZ00000000, AHSA00000000, AHSB00000000, AHSC00000000, AHSD00000000, AHSE00000000, AHSF00000000, AHSG00000000, AHSH00000000, AHSI00000000, AHSJ00000000, AHSK00000000, AHSL00000000, AHSM00000000, AHSN00000000, AHSO00000000, AHSP00000000, AHSQ00000000, AHSR00000000, AHSS00000000, AHST00000000, AHSU00000000, AHSV00000000, AHSW00000000, AHSX00000000, AHSY00000000, AHSZ00000000, AHTA00000000, AHTB00000000, AHTC00000000, AHTD00000000, AHTE00000000, CP003686, AP012336.
Data pre-processing for the core and pan-genome analysis were performed using a self-written perl script (Additional file 10), which is similar as described previously by Tettelin et al. [20]. Briefly, an iterative procedure was carried out to estimate total genes/core genes to be discovered per additional genome sequenced. The number of total genes/core genes provided by each added new genome depends on the selection of previously added genomes. All possible combinations of genomes from 1 to M (the maximal number of available genomes) were calculated. In the case more than 1000 combinations are possible, only 1000 random combinations were used. In order to take into consideration of core genes that are possibly missed during genome sequencing and assembly, for the calculation of core-genome size, an additional correction step was introduced, in which any one gene that is only absent in one of the 63 draft genomes was still regarded as core gene. During the fitting step of the core genome model, the inputted genome numbers were used as fitting weight for corresponding data point.
Gene content-based comparative analysis of 10 mutans streptococci strains
In this work, if not otherwise specified, the uniqueness of genes from “organism A” is defined according to the ortholog groups constructed by using the OrthoMCL program [25]. If the ortholog of a gene from organism A is absent in “organism B”, we define that this gene is unique or specific to organism A in comparison to organism B. This does not imply that there is no homolog (namely paralog) of the gene from organism A in organism B. In some cases, this gene is just an additional copy of another gene whose alleles/orthologs are found in both organisms. This does further not imply that this gene is found in organism A only. For example, the ortholog of this gene may be found in organism C from the relationship table or another strain or species that is not compared in this work.
Genome-scale metabolic networks construction
The bipartite metabolic networks were constructed based on the connection matrix of updated KEGG reactions database according to Stelzer and Zeng [97] with addition of newly identified reaction catalyzed by lactate oxidase (Lactate + O2 = > Pyruvate + H2O2) with provisional R numbers of R10001 (C00186 + C00007 = > C00022 + C00027) and R10002 (C00256 + C00007 = > C00022 + C00027). Compared to the reaction graph or the metabolite graph, wherein either reactions or metabolites (called "nodes") are shown in an interconnected way, the bipartite network is more comprehensible because, similar to the biochemistry textbook, both the reactions and metabolites are visualized at mean time. Seventy-six non-enzymatic automatic reactions were also considered for the network construction. The construction of sub-networks was based on the KEGG pathway classification (http://www.genome.jp/kegg/pathway.html) with slight modification of addition of reaction catalyzed by lactate oxidase into Glycolysis/Gluconeogenesis pathway (MAP00010) and Pyruvate metabolism pathway (MAP00620). The software Cytoscape [80] was used for the visualization and comparative analysis of the genome-scale metabolic networks.
PCR verification
To verify the unique presence of the lactate oxidase (consecutive) coding genes D823_06595 and D823_06598 respectively and to exclude the possibility of contamination with e. g. human DNA during the process of genome sequencing, PCR amplification (using one primer pair covering both genes) with newly isolated DNA from S. sobrinus DSM 20742 as well as a second S. sobrinus strain (AC153) and from strains S. mutans UA159 as well as S. ratti DSM 20564 (as negative controls) was performed. The primers used were: 5′- GAGCAGGATAATTGACAGTC -3′ (forward primer) and 5′- ACTCAGTGACGAATCAGTT -3′ (reverse primer), which were designed by using Primer Premier http://www.premierbiosoft.com/primerdesign/index.html) and Vector NTI 9.0 (InforMax), respectively. Conditions for this conventional PCR were: 94°C, 2 min; followed by 32 cycles of 94°C for 30s; annealing temperature 48°C for 30s; and 72°C for 90s; final extension at 72°C for 5 min; length of amplicon 1,175 bp.
Constructs for lactate oxidase deletion mutants and transformation of S. sobrinus DSM 20742
To clarify the functionality of the two lactate oxidases, namely D823_06598 (Llod) and D823_06595 (lod), PCR ligation mutagenesis according to the method of [98] was used to separately replace the two genes encoding the two enzymes by an erythromycin resistance cassette via double homologous recombination. Primers P1Llod (TTACCGTTATCCGCGAATTAT) and P2Llod (GGCGCGCCAACCACCCAAGGTTGAATC), P1lod (GGCTGGTTTCCTCCATGATA) and P2lod (GGCGCGCCCCAAAACCACCTTGAGGAAT) were used to amplify the 5′flanking regions of both genes, respectively, introducing an AscI restriction site. To amplify the 3′flanking regions of both genes, the primers P3Llod (GGCCGGCCGGGAGCTCAAGGTGTTCAAA) and P4Llod (CAAATTGTTCAAAGCGGGAAC), P3lod (GGCCGGCCGGCAGCAGCCGGTAGTATT) and P4lod (GGGTGCCAACTTATGTCACGA) were used, thereby introducing restriction site for FseI. The erythromycin resistance cassette was amplified from previously constructed gene deletion mutant [99] using primers ErmFor (GGCGCGCCCCGGGCCCAAAATTTGTTTGAT) and ErmRev (GGCCGGCCAGTCGGCAGCGACTCATAGAAT), containing the restriction site for AscI and FseI, respectively. After digestion with the appropriate restriction enzymes, following purification, the three amplicons were ligated together and used for transformation.
For transformation, two natural transformation methods were first used to assay and optimize the natural transformation of the S. sobrinus cells. The first step was the preparation of pre-competent cells of S. sobrinus applying the methods according to [100] and [101]. Afterwards 200 ng of constructs prepared for mutagenesis were used for the transformation. The plasmids like pDL278 (Spr, pAT18 Emr, and suicide vector pFW5 Spr in both circular and linearized form were used as a positive control. Another transformation protocol according to [102] applying pheromone CSP of S. mutans was additionally used to introduce genetic constructs and plasmids into S. sobrinus cells. In this approach two various concentrations of CSP were used: 02 and 1μM, respectively. Transformation of S. mutans was used as a parallel control. All these experiments were carried out at least three times.
Later, electroporation experiment was carried out according to the procedure described by LeBlanc et al. [103]. Various pHs of electroporation mix (EPM) [104] as well as various pulsing conditions were tested. The electroporation was carried out by adding to the chilled electrocompetent cells 200 ng of constructs prepared for mutagenesis or plasmids. Other protocol for electroporation according to [105] was also tested.
Abbreviations
PCR: Polymerase Chain Reaction; PTS: Phosphotransferase system; TCA: Tricarboxylic acid cycle; ATP: adenosine-5′-triphosphate; PEP: Phosphoenolpyruvate; HGT: Horizontal gene transfer; LGT: Lateral gene transfer; SNP: Single-nucleotide polymorphism; LCB: Locally collinear block; multi-MUMs: Multiple maximal-unique-matches; COG: Clusters of orthologous groups of proteins; CSP: Competence stimulating peptide; XIP: Sigma X-inducing peptide; ABC transporter: ATP-binding cassette transporter; SOD: Superoxide dismutase; NAD+: Nicotinamide adenine dinucleotide; NADP+: Nicotinamide adenine dinucleotide phosphate; GSH: L-γ-glutamyl-L-cysteinylglycine; GSSG: Oxidized glutathione; GCS: Glutamylcysteine synthetase; GS: Glutathione synthetase; GCS-GS: Glutamylcysteine synthetase-glutathione synthetase; CoA: Coenzyme A.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
LS carried out the bioinformatics analysis and wrote the draft. WW participated in the conception and coordination of the study and contributed significantly to results analysis and drafting the manuscript. GC provided strains with a verified identity. GC, AR, MR and IWD performed the PCR verification experiments. HS did the experiments on genomic competence of S. sobrinus and the knock-out of the two genes for lactate oxidase. GC and IWD contributed to the microbial aspects and valuable discussions. AZE conceived of and supervised the study and revised the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Presence/absence of genes related to known S. mutans genomic islands of 10 mutans streptococci strains.
Core gene list of S. mutans.
Little square linear fitting details of the core and pan genome models.
Predicted ortholog groups of 10 mutans streptococci strains using OrthoMCL.
Sequences of mutacins used for the identification of putative mutacins in 10 mutans streptococci strains.
Constructed genome wide metabolic networks in Cytoscape (*.cyc) format.
Comparative analysis of the metabolic pathways in the different metabolic networks using S. mutans UA159 as reference. Absent and unique reaction numbers of metabolic networks in strains compared to S. mutans UA159. Absent and unique EC numbers of metabolic networks in strains compared to S. mutans UA159.
PCR verifications of the unique presence of the lactate oxidase genes in S. sobrinus DSM 20742.
The locations of missing genes in NCBI genome annotation results.
The perl script used for core- and pan-genome analysis in this study.
Contributor Information
Lifu Song, Email: lifu.song@tuhh.de.
Wei Wang, Email: wei.wang@tuhh.de.
Georg Conrads, Email: gconrads@ukaachen.de.
Anke Rheinberg, Email: Anke.Rheinberg@rwth-aachen.de.
Helena Sztajer, Email: helena.sztajer@helmholtz-hzi.de.
Michael Reck, Email: Michael.Reck@helmholtz-hzi.de.
Irene Wagner-Döbler, Email: Irene.Wagner-Doebler@helmholtz-hzi.de.
An-Ping Zeng, Email: aze@tuhh.de.
Acknowledgements
This study was done within the project “Development of biofilm inhibitors using a systems biology approach “(0315411) which is financed by the German Federal Ministry of Education and Research (BMBF) in the frame of the Research Program " Medical systems biology - MedSys".
References
- Tapp J, Thollesson M, Herrmann B. Phylogenetic relationships and genotyping of the genus Streptococcus by sequence determination of the RNase P RNA gene, rnpB. Int J Syst Evol Microbiol. 2003;53:1861–1871. doi: 10.1099/ijs.0.02639-0. [DOI] [PubMed] [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology. 2008;154:3247–3255. doi: 10.1099/mic.0.2008/023770-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Nomura R, Matsumoto M, Ooshima T. Roles of oral bacteria in cardiovascular diseases–from molecular mechanisms to clinical cases: Cell-surface structures of novel serotype k Streptococcus mutans strains and their correlation to virulence. J Pharmacol Sci. 2010;113:120–125. doi: 10.1254/jphs.09R24FM. [DOI] [PubMed] [Google Scholar]
- Nomura R, Nakano K, Taniguchi N, Lapirattanakul J, Nemoto H, Gronroos L, Alaluusua S, Ooshima T. Molecular and clinical analyses of the gene encoding the collagen-binding adhesin of Streptococcus mutans. J Med Microbiol. 2009;58:469–475. doi: 10.1099/jmm.0.007559-0. [DOI] [PubMed] [Google Scholar]
- Redfield RJ, Findlay WA, Bosse J, Kroll JS, Cameron AD, Nash JH. Evolution of competence and DNA uptake specificity in the Pasteurellaceae. BMC Evol Biol. 2006;6:82. doi: 10.1186/1471-2148-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich GD, Hu FZ, Shen K, Stoodley P, Post JC. Bacterial plurality as a general mechanism driving persistence in chronic infections. Clin Orthop Relat Res. 2005;437:20–24. doi: 10.1097/00003086-200508000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H. et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama F, Kobata M, Kurokawa K, Nishida K, Sakurai A, Nakano K, Nomura R, Kawabata S, Ooshima T, Nakai K. et al. Comparative genomic analyses of Streptococcus mutans provide insights into chromosomal shuffling and species-specific content. BMC Genomics. 2009;10:358. doi: 10.1186/1471-2164-10-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornejo OE, Lefebure T, Pavinski Bitar PD, Lang P, Richards VP, Eilertson K, Do T, Beighton D, Zeng L, Ahn SJ. et al. Evolutionary and Population Genomics of the Cavity Causing Bacteria Streptococcus mutans. Mol Biol Evol. 2013;30:881–893. doi: 10.1093/molbev/mss278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparon MG, Scott JR. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 1991;204:556–586. doi: 10.1016/0076-6879(91)04028-m. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. PGAP. pan-genomes analysis pipeline. Bioinformatics. 2012;28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Miklos I, Ragan MA. Dynamics of genome rearrangement in bacterial populations. PLoS Genet. 2008;4:e1000128. doi: 10.1371/journal.pgen.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin RE, Ferretti JJ. Electrotransformation of Streptococci. Methods Mol Biol. 1995;47:185–193. doi: 10.1385/0-89603-310-4:185. [DOI] [PubMed] [Google Scholar]
- Darling AC, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling AE, Mau B, Perna NT. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse JC, Swan DC, Russell RR. Comparative genome hybridization of Streptococcus mutans strains. Oral Microbiol Immunol. 2007;22:103–110. doi: 10.1111/j.1399-302X.2007.00330.x. [DOI] [PubMed] [Google Scholar]
- Wu C, Cichewicz R, Li Y, Liu J, Roe B, Ferretti J, Merritt J, Qi F. Genomic island TnSmu2 of Streptococcus mutans harbors a nonribosomal peptide synthetase-polyketide synthase gene cluster responsible for the biosynthesis of pigments involved in oxygen and H2O2 tolerance. Appl Environ Microbiol. 2010;76:5815–5826. doi: 10.1128/AEM.03079-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse JC, Russell RR. Dispensable genes and foreign DNA in Streptococcus mutans. Microbiology. 2006;152:1777–1788. doi: 10.1099/mic.0.28647-0. [DOI] [PubMed] [Google Scholar]
- Muzzi A, Donati C. Population genetics and evolution of the pan-genome of Streptococcus pneumoniae. Int J Med Microbiol. 2011;301:619–622. doi: 10.1016/j.ijmm.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci USA. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Riley D, Cattuto C, Medini D. Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol. 2008;11:472–477. doi: 10.1016/j.mib.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Mira A, Martin-Cuadrado AB, D'Auria G, Rodriguez-Valera F. The bacterial pan-genome:a new paradigm in microbiology. Int Microbiol. 2010;13:45–57. doi: 10.2436/20.1501.01.110. [DOI] [PubMed] [Google Scholar]
- Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, Oggioni M, Dunning Hotopp JC, Hu FZ, Riley DR. et al. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebure T, Stanhope MJ. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 2007;8:R71. doi: 10.1186/gb-2007-8-5-r71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg JS, Hu FZ, Janto B, Boissy R, Hayes J, Keefe R, Post JC, Ehrlich GD. Characterization and modeling of the Haemophilus influenzae core and supragenomes based on the complete genomic sequences of Rd and 12 clinical nontypeable strains. Genome Biol. 2007;8:R103. doi: 10.1186/gb-2007-8-6-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Sudhakar P, Wang W, Conrads G, Brock A, Sun J, Wagner-Dobler I, Zeng AP. A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains. BMC Genomics. 2012;13:128. doi: 10.1186/1471-2164-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzer JM, Livingston J, Thompson AM. The microbiology of primary dental caries in humans. J Dent Educ. 2001;65:1028–1037. [PubMed] [Google Scholar]
- Hale JD, Heng NC, Jack RW, Tagg JR. Identification of nlmTE, the locus encoding the ABC transport system required for export of nonlantibiotic mutacins in Streptococcus mutans. J Bacteriol. 2005;187:5036–5039. doi: 10.1128/JB.187.14.5036-5039.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Biswas I. An extracelluar protease, SepM, generates functional competence-stimulating peptide in Streptococcus mutans UA159. J Bacteriol. 2012;194:5886–5896. doi: 10.1128/JB.01381-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Tang N, Aspiras MB, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J Bacteriol. 2002;184:2699–2708. doi: 10.1128/JB.184.10.2699-2708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FC, Scheie AA. Genetic transformation in Streptococcus mutans requires a peptide secretion-like apparatus. Oral Microbiol Immunol. 2000;15:329–334. doi: 10.1034/j.1399-302x.2000.150511.x. [DOI] [PubMed] [Google Scholar]
- Petersen FC, Fimland G, Scheie AA. Purification and functional studies of a potent modified quorum-sensing peptide and a two-peptide bacteriocin in Streptococcus mutans. Mol Microbiol. 2006;61:1322–1334. doi: 10.1111/j.1365-2958.2006.05312.x. [DOI] [PubMed] [Google Scholar]
- Allan E, Hussain HA, Crawford KR, Miah S, Ascott ZK, Khwaja MH, Hosie AH. Genetic variation in comC, the gene encoding competence-stimulating peptide (CSP) in Streptococcus mutans. FEMS Microbiol Lett. 2007;268:47–51. doi: 10.1111/j.1574-6968.2006.00593.x. [DOI] [PubMed] [Google Scholar]
- Ahn SJ, Wen ZT, Burne RA. Multilevel control of competence development and stress tolerance in Streptococcus mutans UA159. Infect Immun. 2006;74:1631–1642. doi: 10.1128/IAI.74.3.1631-1642.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Hung DC, Merritt J, Perry J, Zhu L, Goodman SD, Cvitkovitch DG, Shi W, Qi F. The response regulator ComE in Streptococcus mutans functions both as a transcription activator of mutacin production and repressor of CSP biosynthesis. Microbiology. 2007;153:1799–1807. doi: 10.1099/mic.0.2007/005975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol Microbiol. 2005;57:392–404. doi: 10.1111/j.1365-2958.2005.04695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Zhu L, Shi W, Qi F. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol Lett. 2006;265:11–17. doi: 10.1111/j.1574-6968.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- van der Ploeg JR. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J Bacteriol. 2005;187:3980–3989. doi: 10.1128/JB.187.12.3980-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaga T, Niu G, Xie Z, Qi F, Merritt J. The hdrRM operon of Streptococcus mutans encodes a novel regulatory system for coordinated competence development and bacteriocin production. J Bacteriol. 2010;192:1844–1852. doi: 10.1128/JB.01667-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okinaga T, Xie Z, Niu G, Qi F, Merritt J. Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol Oral Microbiol. 2010;25:165–177. doi: 10.1111/j.2041-1014.2010.00574.x. [DOI] [PubMed] [Google Scholar]
- Xie Z, Okinaga T, Niu G, Qi F, Merritt J. Identification of a novel bacteriocin regulatory system in Streptococcus mutans. Mol Microbiol. 2010;78:1431–1447. doi: 10.1111/j.1365-2958.2010.07417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair RW, Senadheera DB, Cvitkovitch DG. CinA is regulated via ComX to modulate genetic transformation and cell viability in Streptococcus mutans. FEMS Microbiol Lett. 2012;331:44–52. doi: 10.1111/j.1574-6968.2012.02550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaluusua S, Takei T, Ooshima T, Hamada S. Mutacin activity of strains isolated from children with varying levels of mutants streptococci and caries. Arch Oral Biol. 1991;36:251–255. doi: 10.1016/0003-9969(91)90094-B. [DOI] [PubMed] [Google Scholar]
- Baba T, Schneewind O. Instruments of microbial warfare: bacteriocin synthesis, toxicity and immunity. Trends Microbiol. 1998;6:66–71. doi: 10.1016/S0966-842X(97)01196-7. [DOI] [PubMed] [Google Scholar]
- Hossain MS, Biswas I. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol. 2011;77:2428–2434. doi: 10.1128/AEM.02320-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa H, Kuramitsu HK. Genetic analysis of a unique bacteriocin, Smb, produced by Streptococcus mutans GS5. Antimicrob Agents Chemother. 2005;49:541–548. doi: 10.1128/AAC.49.2.541-548.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyink O, Balakrishnan M, Tagg JR. Streptococcus rattus strain BHT produces both a class I two-component lantibiotic and a class II bacteriocin. FEMS Microbiol Lett. 2005;252:235–241. doi: 10.1016/j.femsle.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Zhang Z, Huang IH, Wu C, Merritt J, Shi W, Qi F. Genes involved in the repression of mutacin I production in Streptococcus mutans. Microbiology. 2009;155:551–556. doi: 10.1099/mic.0.021303-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl Environ Microbiol. 2000;66:3221–3229. doi: 10.1128/AEM.66.8.3221-3229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Qi F, Novak J, Caufield PW. The specific genes for lantibiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl Environ Microbiol. 1999;65:1356–1360. doi: 10.1128/aem.65.3.1356-1360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl Environ Microbiol. 1999;65:3880–3887. doi: 10.1128/aem.65.9.3880-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson CL, Wescombe PA, Klesse NA, Tagg JR. Isolation and partial characterization of the Streptococcus mutans type AII lantibiotic mutacin K8. Microbiology. 2007;153:1631–1641. doi: 10.1099/mic.0.2006/003756-0. [DOI] [PubMed] [Google Scholar]
- Qi F, Chen P, Caufield PW. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl Environ Microbiol. 2001;67:15–21. doi: 10.1128/AEM.67.1.15-21.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JD, Ting YT, Jack RW, Tagg JR, Heng NC. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl Environ Microbiol. 2005;71:7613–7617. doi: 10.1128/AEM.71.11.7613-7617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour D, Cordova M, Cvitkovitch DG, Levesque CM. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J Bacteriol. 2011;193:6552–6559. doi: 10.1128/JB.05968-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nes IF, Diep DB, Holo H. Bacteriocin diversity in Streptococcus and Enterococcus. J Bacteriol. 2007;189:1189–1198. doi: 10.1128/JB.01254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekal-Si Ali S, Hurtubise Y, Lavoie MC, LaPointe G. Diversity of Streptococcus mutans bacteriocins as confirmed by DNA analysis using specific molecular probes. Gene. 2002;283:125–131. doi: 10.1016/S0378-1119(01)00875-7. [DOI] [PubMed] [Google Scholar]
- Johnson DW, Tagg JR, Wannamaker LW. Production of a bacteriocine-like substance by group-A streptococci of M-type 4 and T-pattern 4. J Med Microbiol. 1979;12:413–427. doi: 10.1099/00222615-12-4-413. [DOI] [PubMed] [Google Scholar]
- Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Levesque CM. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol Microbiol. 2009;72:905–917. doi: 10.1111/j.1365-2958.2009.06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee AK, Starr MP. Transfer among Erwinia spp. and other enterobacteria of antibiotic resistance carried on R factors. J Bacteriol. 1972;112:576–584. doi: 10.1128/jb.112.1.576-584.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano H, Kuga A, Okamoto R, Kitasato H, Kobayashi T, Inoue M. Plasmid-encoded metallo-beta-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob Agents Chemother. 2001;45:1343–1348. doi: 10.1128/AAC.45.5.1343-1348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Tian XL, Versey J, Wishart A, Li YH. The BceABRS four-component system regulates the bacitracin-induced cell envelope stress response in Streptococcus mutans. Antimicrob Agents Chemother. 2010;54:3895–3906. doi: 10.1128/AAC.01802-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda H, Yamashita Y, Shibata Y, Nakano Y, Koga T. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob Agents Chemother. 2002;46:3756–3764. doi: 10.1128/AAC.46.12.3756-3764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghachi M, Bouhss A, Blanot D, Mengin-Lecreulx D. The bacA gene of Escherichia coli encodes an undecaprenyl pyrophosphate phosphatase activity. J Biol Chem. 2004;279:30106–30113. doi: 10.1074/jbc.M401701200. [DOI] [PubMed] [Google Scholar]
- Bernard R, El Ghachi M, Mengin-Lecreulx D, Chippaux M, Denizot F. BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J Biol Chem. 2005;280:28852–28857. doi: 10.1074/jbc.M413750200. [DOI] [PubMed] [Google Scholar]
- McCord JM, Fridovich I. Superoxide dismutase: the first twenty years (1968–1988) Free Radic Biol Med. 1988;5:363–369. doi: 10.1016/0891-5849(88)90109-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Higuchi M, Poole LB, Kamio Y. Role of the dpr product in oxygen tolerance in Streptococcus mutans. J Bacteriol. 2000;182:3740–3747. doi: 10.1128/JB.182.13.3740-3747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Yamamoto Y, Kamio Y. Molecular biology of oxygen tolerance in lactic acid bacteria: Functions of NADH oxidases and Dpr in oxidative stress. J Biosci Bioeng. 2000;90:484–493. [PubMed] [Google Scholar]
- Yamamoto Y, Poole LB, Hantgan RR, Kamio Y. An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J Bacteriol. 2002;184:2931–2939. doi: 10.1128/JB.184.11.2931-2939.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustacich D, Powis G. Thioredoxin reductase. Biochem J. 2000;346(Pt 1):1–8. [PMC free article] [PubMed] [Google Scholar]
- Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Seo HJ, Lee YN. Characterization of Deinococcus radiophilus thioredoxin reductase active with both NADH and NADPH. J Microbiol. 2010;48:637–643. doi: 10.1007/s12275-010-0283-7. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- Fernandes AP, Holmgren A. Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid Redox Signal. 2004;6:63–74. doi: 10.1089/152308604771978354. [DOI] [PubMed] [Google Scholar]
- Fahey RC, Brown WC, Adams WB, Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133:1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowiak BE, Griffith OW. Glutathione synthesis in Streptococcus agalactiae. One protein accounts for gamma-glutamylcysteine synthetase and glutathione synthetase activities. J Biol Chem. 2005;280:11829–11839. doi: 10.1074/jbc.M414326200. [DOI] [PubMed] [Google Scholar]
- Zhang J, Biswas I. 3′-Phosphoadenosine-5′-phosphate phosphatase activity is required for superoxide stress tolerance in Streptococcus mutans. J Bacteriol. 2009;191:4330–4340. doi: 10.1128/JB.00184-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zeng AP. Reconstruction of metabolic networks from genome data and analysis of their global structure for various organisms. Bioinformatics. 2003;19:270–277. doi: 10.1093/bioinformatics/19.2.270. [DOI] [PubMed] [Google Scholar]
- Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol Biol. 2011;696:291–303. doi: 10.1007/978-1-60761-987-1_18. [DOI] [PubMed] [Google Scholar]
- Taniai H, Iida K, Seki M, Saito M, Shiota S, Nakayama H, Yoshida S. Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J Bacteriol. 2008;190:3572–3579. doi: 10.1128/JB.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. 2005;187:7193–7203. doi: 10.1128/JB.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okahashi N, Nakata M, Sumitomo T, Terao Y, Kawabata S. Hydrogen peroxide produced by oral streptococci induces macrophage cell death. PLoS One. 2013;8:e62563. doi: 10.1371/journal.pone.0062563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Sivaraman C. Bacterial citrate lyase. J Biosci. 1984;6:379–401. doi: 10.1007/BF02703895. [DOI] [Google Scholar]
- Evans HJ, Wood HG. The mechanism of the pyruvate, phosphate dikinase reaction. Proc Natl Acad Sci USA. 1968;61:1448–1453. doi: 10.1073/pnas.61.4.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benziman M, Eisen N, Palgi A. Properties and physiological role of the pep-synthase of A. xylinum. FEBS Lett. 1969;3:156–159. doi: 10.1016/0014-5793(69)80123-7. [DOI] [PubMed] [Google Scholar]
- Sauer U, Eikmanns BJ. The PEP-pyruvate-oxaloacetate node as the switch point for carbon flux distribution in bacteria. FEMS Microbiol Rev. 2005;29:765–794. doi: 10.1016/j.femsre.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Cvitkovitch DG, Gutierrez JA, Bleiweis AS. Role of the citrate pathway in glutamate biosynthesis by Streptococcus mutans. J Bacteriol. 1997;179:650–655. doi: 10.1128/jb.179.3.650-655.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain PS, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, Ali J, Birren B, Bruce DC, Buhay C. et al. Genomics. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Bastide M, McCombie WR. Assembling genomic DNA sequences with PHRAP. Curr Protoc Bioinformatics. 2007;Chapter 11:Unit11 14. doi: 10.1002/0471250953.bi1104s17. [DOI] [PubMed] [Google Scholar]
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl 1):S225–232. doi: 10.1093/bioinformatics/18.suppl_1.S225. [DOI] [PubMed] [Google Scholar]
- Stelzer M, Sun J, Kamphans T, Fekete SP, Zeng AP. An extended bioreaction database that significantly improves reconstruction and analysis of genome-scale metabolic networks. Integr Biol (Camb) 2011;3:1071–1086. doi: 10.1039/c1ib00008j. [DOI] [PubMed] [Google Scholar]
- Lau PC, Sung CK, Lee JH, Morrison DA, Cvitkovitch DG. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J Microbiol Methods. 2002;49:193–205. doi: 10.1016/S0167-7012(01)00369-4. [DOI] [PubMed] [Google Scholar]
- Reck M, Rutz K, Kunze B, Tomasch J, Surapaneni SK, Schulz S, Wagner-Dobler I. The biofilm inhibitor carolacton disturbs membrane integrity and cell division of Streptococcus mutans through the serine/threonine protein kinase PknB. J Bacteriol. 2011;193:5692–5706. doi: 10.1128/JB.05424-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefrancois J, Samrakandi MM, Sicard AM. Electrotransformation and natural transformation of Streptococcus pneumoniae: requirement of DNA processing for recombination. Microbiology. 1998;144(Pt 11):3061–3068. doi: 10.1099/00221287-144-11-3061. [DOI] [PubMed] [Google Scholar]
- Ween O, Teigen S, Gaustad P, Kilian M, Havarstein LS. Competence without a competence pheromone in a natural isolate of Streptococcus infantis. J Bacteriol. 2002;184:3426–3432. doi: 10.1128/JB.184.13.3426-3432.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Lau PC, Lee JH, Ellen RP, Cvitkovitch DG. Natural genetic transformation of Streptococcus mutans growing in biofilms. J Bacteriol. 2001;183:897–908. doi: 10.1128/JB.183.3.897-908.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc D, Chen Y-Y, Buckley N, Lee L. Genetic transfer methods for Streptococcus sobrinus and other oral streptococci. Methods Cell Sci. 1998;20:85–93. doi: 10.1023/A:1009855312270. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Presence/absence of genes related to known S. mutans genomic islands of 10 mutans streptococci strains.
Core gene list of S. mutans.
Little square linear fitting details of the core and pan genome models.
Predicted ortholog groups of 10 mutans streptococci strains using OrthoMCL.
Sequences of mutacins used for the identification of putative mutacins in 10 mutans streptococci strains.
Constructed genome wide metabolic networks in Cytoscape (*.cyc) format.
Comparative analysis of the metabolic pathways in the different metabolic networks using S. mutans UA159 as reference. Absent and unique reaction numbers of metabolic networks in strains compared to S. mutans UA159. Absent and unique EC numbers of metabolic networks in strains compared to S. mutans UA159.
PCR verifications of the unique presence of the lactate oxidase genes in S. sobrinus DSM 20742.
The locations of missing genes in NCBI genome annotation results.
The perl script used for core- and pan-genome analysis in this study.