Abstract
Citrobacter rodentium is a member of a group of pathogens that colonize the lumen of the host gastrointestinal tract via attaching and effacing (A/E) lesion formation. C. rodentium, which causes transmissible colonic hyperplasia in mice, is used as an in vivo model system for the clinically significant A/E pathogens enterohemorrhagic and enteropathogenic Escherichia coli. These bacteria all contain a pathogenicity island called the locus of enterocyte effacement (LEE), which encodes a type III secretion system that is designed to deliver effector proteins into eukaryotic host cells. These effectors are involved in the subversion of host eukaryotic cell functions to the benefit of the bacterium. In this study we used mutant strains to determine the effects of the C. rodentium LEE-encoded effectors EspF, EspG, EspH, and Map on virulence in the mouse model. In addition, we identified a novel secreted protein, EspI encoded outside the LEE, whose secretion is also dependent on a functional type III secretion system. Mutant strains with each of the effectors investigated were found to be outcompeted by wild-type bacteria in mixed-infection experiments in vivo, although the effects of EspF and EspH were only subtle. In single-infection experiments, we found that EspF, EspG, and EspH are not required for efficient colonization of the mouse colon or for the production of hyperplasia. In contrast, strains producing EspI and Map had significant colonization defects and resulted in dramatically reduced levels of hyperplasia, and they exhibited very different growth dynamics in mice than the wild-type strain exhibited.
The induction of attaching and effacing (A/E) lesions is a key mechanism used by a group of clinically important enteric pathogens, including enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC), to successfully colonize the host gastrointestinal tract. EPEC is a significant cause of infantile diarrhea in developing countries (34), whereas EHEC is more of a concern in the developed world, with around 73,000 cases reported annually in the United States (30). In addition to causing diarrhea, EHEC infection can result in the life-threatening complications of hemorrhagic colitis and hemolytic-uremic syndrome due to the production of verocytotoxins (34). A/E lesions were first described for EPEC strains (32), and similar lesions have also been reported for EHEC (38, 45) and the mouse pathogen Citrobacter rodentium (39). A/E lesions are characterized by localized destruction (effacement) of brush border microvilli and intimate attachment of the bacteria to the host cell plasma membrane through the formation of actin-rich pedestal-like structures beneath the adherent bacteria (reviewed in reference 14).
EHEC and EPEC exhibit narrow host specificity, and since mice are resistant to infection, one difficulty with studying EHEC and EPEC pathogenesis is the lack of a simple small-animal model that simulates an in vivo situation. Consequently, many of the current models and concepts of EHEC and EPEC pathogen-host interactions were developed from studies of infected cultured epithelial cells in vitro. However, a growing body of evidence now suggests that this infection model cannot be extrapolated wholesale to colonization and disease processes in vivo (8). For these reasons, infection of mice with C. rodentium has become a popular surrogate model for in vivo studies of the mechanisms and processes of A/E E. coli pathogenesis (39; reviewed in reference 25). The clinical symptoms associated with infection by C. rodentium include weight loss, soft stools, and enlargement of the descending colon through hyperplasia (2).
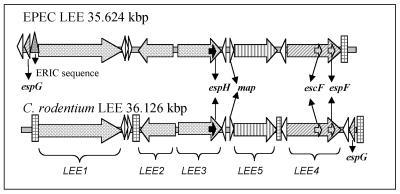
The genes required for A/E lesion formation are encoded on a pathogenicity island termed the locus of enterocyte effacement (LEE) (27). The LEE encodes a type III secretion system (TTSS), a macromolecular complex spanning both bacterial membranes that is used by many gram-negative bacteria to inject virulence factors directly into host cells to subvert host cell functions for the benefit of the pathogen (17). The complete LEE has been sequenced in EPEC O127:H6, EHEC O157:H7, C. rodentium, and two rabbit EPEC (REPEC) strains (7, 13, 36, 43, 47). Analysis of these LEE regions has revealed that all 41 open reading frames (ORFs) are present in C. rodentium, EPEC, and EHEC (orf3 in REPEC strains lacks the ATG start codon, but translation is likely to start with an alternative Val codon [E. Creasey and G. Frankel, unpublished data]). There are some differences among the LEE regions of C. rodentium, EPEC, and EHEC, including size, insertion site in the chromosome, and location of the rorf1 and espG genes (7) (Fig. 1). However, despite this divergence the overall high levels of similarity in the content and organization of these pathogenicity islands suggest that the roles of specific proteins are likely to be conserved in A/E pathogens.
FIG. 1.
Comparison of the LEE regions from EPEC and C. rodentium. The positions and orientations of major operons and genes, including the escF, espF, espH, and map genes, are identical in EPEC and C. rodentium, except for the positions of rorf1 and rorf2/espG. The C. rodentium LEE also differs from that of EPEC by extra insertion sequences that result in an increase in size. ERIC, enterobacterial repetitive intergenic consensus.
The majority of the LEE genes are organized into five polycistronic operons (LEE1, LEE2, LEE3, LEE5, and LEE4) (31) (Fig. 1). Structural components of the TTSS are mainly encoded on the LEE1, LEE2, and LEE3 operons. The LEE5 operon encodes the outer membrane adhesin intimin (18), the translocated intimin receptor (Tir) (19), and CesT (the Tir chaperone) (1, 11). The LEE4 operon encodes the structural needle protein EscF (46), the translocator proteins EspA, EspD, and EspB (10, 22, 24), and the effector protein EspF (28). Other LEE-encoded effector proteins include Tir, EspG, EspH, and Map (12, 19, 21, 44). However, it has been shown that EspF, EspG, EspH, and Map are not essential for A/E lesion formation in vitro, as determined by the fluorescent actin staining test (12, 28, 44).
EspF has been shown to be involved in disruption of tight junctions and in induction of apoptosis (5, 29). EspH is believed to downregulate filopodium formation and enhance pedestal formation (44). Map is thought to have at least two independent functions in the host cell; first, Map has been reported to be targeted to host mitochondria, where it appears to interfere with membrane potential (21), and second, Map has been reported to be involved in cytoskeletal rearrangements, resulting in the transient formation of Cdc42-dependent filopodia (20). The role of EspG has not been determined yet (12), although a rabbit diarrheagenic E. coli strain carrying a mutation in espG had diarrheal attack rates similar to those of the wild type but exhibited diminished intestinal colonization (12).
To date, intimin and EspB have been shown to be essential for pathogenesis of EPEC in human volunteer studies (9, 42) and for virulence of C. rodentium in mice (35, 40). In addition, recent studies have also implicated Tir in C. rodentium infection (8). Moreover, by using signature-tagged mutagenesis, it has been shown that mutations in C. rodentium orf4, espD, and escD result in attenuated virulence following oral challenge (33). In this paper we describe identification of espI, a gene identified by signature-tagged mutagenesis screening, which is homologous to the gene encoding EHEC type III secreted protein P54 (Z6024), which was first identified as an overexpressed and secreted protein in a sepL EHEC mutant strain (23). Indeed, we obtained direct evidence that secretion of EspI is dependent on the C. rodentium LEE-encoded TTSS.
The aim of this study was to determine the contributions of the type III secreted proteins EspF, EspG, EspH, EspI, and Map to virulence in vivo. Mutant strains of C. rodentium were tested in mixed and single infections, and bacterial burden, mucosal association, colonic hyperplasia, and recruitment of immune cells to the mucosa were used as criteria for pathogenicity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are shown in Table 1. Bacteria were grown on Luria-Bertani (LB) medium unless otherwise specified. When appropriate, additional antibiotics were added at the following concentrations: nalidixic acid, 50 μg ml−1; kanamycin, 100 μg ml−1; chloramphenicol, 50 μg ml−1; and ampicillin, 100 μg ml−1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| DBS100 | C. rodentium wild-type strain ATCC 51459 | 39 |
| ICC176 | DBS100 escF mutant; Kanr | This study |
| ICC177 | DBS100 espF mutant; Kanr | This study |
| ICC178 | DBS100 espG mutant; Kanr | This study |
| TX18 | DBS100 espH mutant; Kanr | This study |
| ICC179 | DBS100 espI mutant; Kanr | This study |
| P6C6 | DBS100 Tn::map; Kanr | 33 |
| P10H2 | DBS100 Tn::map; Kanr | This study |
| Plasmids | ||
| pCR2.1 | Ampr Kanr; high-copy-number cloning vector | Invitrogen |
| pGEM-T | Ampr; high-copy-number cloning vector | Promega |
| pUC19 | Ampr; high-copy-number cloning vector | Invitrogen |
| pACYC184 | Chlr Tetr; medium-copy-number cloning vector | New England Biolabs |
| pKD46 | Ampr; expresses λ red recombinase | 6 |
| pUC4K | Kanr | Pharmacia |
| pSA10 | lacI gene in the HindIII site of expression vector pKK177-3 | 41 |
| pICC270 | escF in pUC19 | This study |
| pICC272 | 2.1-kb fragment containing escF and flanking DNA in pACYC184 | This study |
| pICC273 | AphT cassette in BglII site of escF of pICC272 | This study |
| pICC290 | 3.6-kb fragment containing espF in pCR2.1 | This study |
| pICC291 | 2.6-kb EcoRV fragment from pICC290 in pACYC184 | This study |
| pICC292 | AphT cassette in SnaBI site of deleted espF of pICC291 | This study |
| pICC293 | 2.6-kb fragment containing espG plus flanking regions in pGEM-T | This study |
| pICC294 | AphT cassette in SfoI site of espG of pICC293 | This study |
| pICC295 | 2.4-kb BamHI fragment containing flanking regions of espH separated by the kan cassette from pUC4K | This study |
| pICC296 | 1.3-kb BamHI espI ORF in pACYC184 | This study |
| pICC297 | AphT cassette in BglII site of espI of pICC296 | This study |
| pICC298 | pSA10 expressing EspI with C-terminal His tag | This study |
| pICC299 | 0.7-kb XbaI/EcoRI map ORF in pWSK29 | This study |
| pICC300 | 0.7-kb EagI/SphI map ORF in pACYC184 | This study |
Signature-tagged mutagenesis, cloning, and sequencing of espI.
A pool of 48 preselected signature-tagged mini-Tn5Km2 transposons in plasmid pUT was used to generate 48 sets of mutants. For screening of the pool, each of the mutants was grown individually in the well of a microtiter dish in 200 μl of LB medium containing nalidixic acid and kanamycin and incubated for 18 h at 37°C under static conditions. The growth of mutants was assessed by determining the optical density of each well with a microtiter plate reader. One hundred milliliiters of bacterial cells was pooled, centrifuged at 3,400 × g for 10 min, and resuspended in 1 ml of phosphate-buffered saline (PBS); this resulted in a concentration of approximately 5 × 1010 CFU ml−1, which was confirmed by plating dilutions onto LB agar containing nalidixic acid and kanamycin. Three mice received approximately 1 × 1010 CFU consisting of 24 to 28 different mutants by oral gavage. The remaining inoculum was used to harvest DNA for preparation of the input probe. Stool samples were taken from the mice on days 2 and 3 postinoculation, and the mice were killed and the colons were removed on day 5 postinoculation. Stool samples and colons were mashed in PBS and plated onto LB agar containing nalidixic acid and kanamycin. More than 10,000 colonies were used to isolate DNA at each time point. Signature tags were amplified and radiolabeled by using primers P2 and P4, as described previously (16). The input and recovered probes were used in hybridizations with replica dot blots generated by using plasmid DNA from each of the 48 tagged transposons cloned in the high-copy-number vector pCR2.1 TOPO (constructs were supplied by Microscience). Mutants with tags that hybridized with the input probe but not with the recovered probe from day 3 stools and day 5 colon samples were considered to have a colonization defect and were characterized further.
Southern analysis was performed on genomic DNA from colonization-defective mutant P4G1 that was digested with each of six restriction enzymes (BglII, EcoRI, KpnI, PstI, SalI, and SphI). DNA was electrophoresed through 0.8% agarose gels and transferred to nylon membranes (Hybond N+; Amersham) by standard methods. The membranes were probed with the kanamycin resistance cassette from mini-Tn5 labeled with the Megaprime DNA labeling system (Amersham). The SalI enzyme gave a 3.6-kb hybridizing fragment for mutant P4G1 and was used to digest 40 μg of genomic DNA that was size fractionated on a 0.8% agarose gel, and the appropriate fragments were excised and purified (Qiaquick; Qiagen). The fragments were then ligated into pUC18 digested with the same enzyme; the resulting ligation mixture was used to transform E. coli DH5α, and kanamycin-resistant colonies were selected. The DNA adjacent to the tagged mini-Tn5Km2 insert was sequenced by using transposon primers I and O (16). Sequence homology analyses were performed by using GenBank databases and BLAST programs found at the www.ncbi.nih/gov website. To clone the entire ORF into which the transposon had inserted into mutant strain P4G1, a PCR was carried out with primers EspIfor and EspIrev (Table 2), which were designed by using the EHEC sequence of gene Z6024.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| CR1 | 5′-TGATATCGCCAGCCGCCTGCGTGAT-3′ |
| CR2 | 5′-GATATCCTCGCTAAATTTATCCCAC-3′ |
| CR3 | 5′-CGGGATCCATGAATCTGACTCAAAT-3′ |
| CR4 | 5′-GCGTCGACTTAAGACGTACGGTTGG-3′ |
| EspBfor | 5′-CGGGATCCTGAGACAGTTGGCACATTGC-3′ |
| rORF1rev | 5′-CGGGATCCGGATGATTTCCTGTATGCCAC-3′ |
| EspF1 | 5′-TACGTACCGTCCTAGTGTAGAAACGGCCTGAC-3′ |
| EspF2 | 5′-TACGTAAATTTTAATGTGAGTGGAGTTACAACG-3′ |
| EspGfor | 5′-GCTCTAGAGCTGAGATTGCCAAACTTGATCGC-3′ |
| EspGrev | 5′-GGGGTACCCCTGCATGATCCGACCCACCCGGT-3′ |
| Mut18C/1 | 5′-GATCCGTTATGTATTATCAGCAACGAGAC-3′ |
| Mut18C/2 | 5′-ATGGATCCATCCATTCCTATTAATCACATACTACGC-3′ |
| Mut18C/3 | 5′-ATGGATCCATGGTTTAAGAATTATTTAAGGTGTTCAAGG-3′ |
| Mut18C/4 | 5′-ACCACTGGTCTAAGAGTTTGTGTAGC-3′ |
| EspIfor | 5′-GGATCCATGAACATTCAACCGA-3′ |
| EspIrev | 5′-GAATTCTTAGACTCTTGTTTCTTGGATTAT-3′ |
| EspI-1 | 5′-GGAATTCATGAACATTCAACCGAAC-3′ |
| EspI-2 | 5′-GCTGCAGCTTAATGATGATGATGATGATGGACTCTTGTTTCTTGGATTATTTC-3′ |
| Mapfor-1 | 5′-GCTCTAGAATGTTTAATCCAACGGCAATGG-3′ |
| Maprev-1 | 5′-GGAATTCCTACAGCCTGGTATCCTGCA-3′ |
| Mapfor-2 | 5′-CGGCCGTTATTATTTTATTATTCATTTAAT-3′ |
| Maprev-2 | 5′-ACATGCATGCTACAGCCTGGTATCCTGCAC-3′ |
Construction of the nonpolar mutations.
The C. rodentium mutants were all constructed by using a modification of the one-step method developed by Datsenko and Wanner (6). In each case the gene and flanking regions were amplified from C. rodentium wild-type strain DBS100 genomic DNA by PCR by using primers (Table 2) based on the C. rodentium sequence (accession number AF311901) (7). The PCR product was ligated into a vector, and then in order to inactivate the gene and facilitate mutant strain identification, the nonpolar aphT cassette (15) or kan cassette (pUC4K; Pharmacia) conferring kanamycin resistance was inserted into the gene of interest. The insert containing the kanamycin cassette in the correct orientation was then removed from the plasmid by digestion. In order to enhance allelic exchange, plasmid pKD46, an easily curable low-copy-number plasmid that expressed lambda red recombinase and was described by Datsenko and Wanner (6), was transformed into wild-type C. rodentium strain DBS100 by electroporation, generating strain DBS100(pKD46). pKD46 has been shown to aid chromosomal recombination of foreign DNA in E. coli K-12. The kanamycin-resistant fragment was transformed by electroporation into strain DBS100(pKD46). Clones were grown on LB medium containing kanamycin to select for kanamycin resistance. pKD46 was cured by growth at 42°C. PCR, Southern blot analysis, and DNA sequencing were used to verify the mutations.
Construction of the nonpolar mutation in escF.
The escF gene (222 bp) and flanking regions were amplified from C. rodentium wild-type strain DBS100 genomic DNA by PCR by using primers CR1 and CR2 (Table 2). The resulting PCR product was purified and blunt ended with the DNA polymerase I large (Klenow) fragment from New England Biolabs. The blunt-ended PCR product was purified and ligated into EcoRV-digested, chloramphenicol-resistant plasmid pACYC184, generating plasmid pICC272. The aphT cassette was ligated into the BglII site at bp 85 of the escF gene, generating plasmid pICC273. Plasmid pICC273 was used as a template for PCR performed with primers CR1 and CR2; the PCR product was purified and digested with the DpnI enzyme to remove parental DNA, and 100 ng was transformed by electroporation into strain DBS100(pKD46). Transformants were grown on LB agar with kanamycin to select for insertion of the aphT cassette into the chromosome of C. rodentium. Deletion of plasmid pKD46 was achieved by selection at 42°C and was confirmed by a loss of ampicillin resistance and plasmid analysis. The mutation in escF in strain ICC176 was verified by PCR and sequencing.
Construction of the nonpolar deletion in espF.
A 3.6-kb BamHI fragment was amplified from C. rodentium chromosomal DNA by using primers EspBfor and rORF1rev (Table 2) and was cloned into pCR2.1, generating plasmid pICC290. A 2.6-kb EcoRV fragment was excised from plasmid pICC290; this fragment contained the espF ORF plus 1.2 kb upstream of the start codon and 0.5 kb downstream of the stop codon and was cloned into pACYC184, generating plasmid pICC291. Primers were designed for inverse PCR; these primers, EspF1 and EspF2, resulted in deletion of a 138-bp region of the espF gene (from bp 43 to 181) and incorporation of an SnaBI restriction site. The kanamycin-resistant aphT cassette was incorporated into the SnaBI site of pICC291, creating plasmid pICC292. A 3.3-kb EcoRI fragment was excised from plasmid pICC292; this fragment contained the espF gene with an in-frame insertion of the aphT cassette, 1.2 kb of flanking DNA upstream of espF, and 0.5 kb of flanking DNA downstream of espF. This fragment was transformed by electroporation into strain DBS100(pKD46). Clones were grown on LB containing kanamycin to select for kanamycin resistance. pKD46 was cured by growth at 42°C. The mutation in espF in strain ICC177 was verified by PCR and DNA sequencing.
Construction of the nonpolar mutation in espG.
The espG gene and 1-kb flanking regions were amplified by PCR by using primers EspGfor and EspGrev (Table 2) and were ligated into the pGEM-T vector (Promega), creating plasmid pICC293. The aphT cassette was incorporated into the SfoI restriction site of espG, generating plasmid pICC294. The mutated espG and the flanking region were then removed from pICC294 by digestion with XbaI and KpnI (New England Biolabs) and used to transform DBS100(pKD46). Clones were grown on LB medium containing kanamycin to select for kanamycin-resistant mutants, and pKD46 was cured by growth at 42°C. The mutation in espG in strain ICC178 was verified by PCR and Southern blot analysis.
Construction of the nonpolar deletion in espH.
The upstream and downstream sequences of espH were amplified by using two pairs of primers, primers Mut18C/1 and Mut18C/2 and primers Mut18C/3 and Mut18C/4, containing BamHI and EcoRI sites (Table 2). The products (600 bp each) were cloned into pGEM-T (Promega), and the flanking regions of espH were confirmed by sequencing. Plasmid pEspH::kan, containing the two segments separated by a kan cassette (pUC4K; Pharmacia), was constructed by subcloning. A 2.4-kb BamHI fragment containing the espH flanking sequences separated by the kan gene was isolated from this plasmid and used to transform DBS100(pKD46). Clones were grown on LB medium containing kanamycin to select for kanamycin-resistant mutants, and pKD46 was cured by growth at 42°C. The mutation in espH in strain TX18 was verified by PCR.
Construction of the nonpolar mutation in espI.
A 1.3-kb BamHI fragment containing the espI ORF was amplified by PCR by using primers EspIfor and EspIrev (Table 2) and was cloned into pACYC184, generating plasmid pICC296. The kanamycin-resistant aphT cassette was inserted into the BglII site at bp 470 of the C. rodentium espI gene, creating plasmid pICC297. The mutated espI gene and flanking regions were then removed from pACYC184 by digestion with BamHI and transformed into DBS100(pKD46). Clones were grown on LB medium containing kanamycin to select for kanamycin-resistant mutants, and pKD46 was cured by growth at 42°C. The mutation in espI in strain ICC179 was verified by PCR and sequencing.
Cloning of wild-type escF, espI, and map for complementation studies.
The escF gene was amplified from C. rodentium genomic DNA with primers CR3 and CR4 (Table 2), which incorporated BamHI and SalI restriction enzyme sites into the PCR product. The PCR product was purified, treated with the Klenow polymerase to create blunt ends, repurified, ligated into SmaI-digested pUC19, and transformed into E. coli TG1. Plasmids were isolated and screened, and plasmid pICC270 contained the correct insert in the correct orientation. The PCR product described above was also digested with BamHI and SalI, purified, and ligated into BamHI- and SalI-digested plasmid pACYC184. The ligation was transformed into E. coli TG1, plasmids were isolated and screened, and plasmid pICC271 contained the correct escF gene insert. Plasmids pICC270 and pICC271 were transformed into competent ICC176 (ΔescF) C. rodentium by electroporation.
Plasmid pICC296 contained the complete ORF of espI cloned in pACYC184 (see construction of nonpolar mutation in espI).
The map gene was amplified from C. rodentium genomic DNA with primers Map-for1 and Map-rev1 (Table 2), which incorporated XbaI and EcoRI restriction enzyme sites into the PCR product, and was cloned into low-copy-number plasmid pWSK29, creating plasmid pICC299. map was also amplified with primers Mapfor-2 and Maprev-2 (Table 2), which incorporated EagI and SphI restriction sites, and was cloned into plasmid pACYC184, generating plasmid pICC300.
Generation of a His-tagged EspI construct.
The 1,293-bp coding sequence of the C. rodentium espI gene was amplified by PCR by using primers EspI-1 and EspI-2 (Table 2). The primers were designed so that EcoRI and PstI restriction sites were introduced at the 5′ end of EspI-1 and at the 3′ end of EspI-2, respectively, and six histidine codons were introduced into primer EspI-2 near the 3′ end. The PCR amplicon was first cloned into pGEM-T (Promega) and then subcloned into the expression vector pSA10 at the EcoRI-PstI sites, generating plasmid pICC298.
Preparation of C. rodentium secreted proteins.
To isolate C. rodentium secreted proteins, strains were grown in Dulbecco modified Eagle medium at 37°C to an optical density at 600 nm of 1. Bacteria were pelleted by centrifugation at 8,000 × g for 10 min, and the supernatant containing secreted Esp proteins was filtered through a 0.45-μm-pore-size filter syringe and concentrated 10-fold by using centrifugal filter devices (Millipore Corp., Bedford, Mass.). Ten microliters of filtered concentrated supernatant or total cell protein was analyzed by Western blotting and probed with commercially available mouse monoclonal antibodies raised against the histidine tag (Sigma).
Infection of mice.
Male, specific-pathogen-free, C3H/Hej mice that were 4 to 5 weeks old were purchased from Harlan Olac (Bichester, United Kingdom). All mice were housed in individual ventilated cages with free access to food and water. Unanesthetized mice were orally gavaged with 200 μl of a bacterial suspension by using a gavage needle. The viable count of the inoculum was determined by retrospective plating on LB agar containing appropriate antibiotics.
In mixed-infection experiments, mutant bacteria (Nalr Kanr) and wild-type bacteria (Nalr) were grown to the stationary phase in LB broth; equal amounts of bacteria (∼109 CFU of each strain in 100 μl of PBS) were mixed and then orally gavaged into mice. To determine the ratio of wild-type bacteria to mutant bacteria, dilutions of the inoculum were plated onto medium containing only nalidixic acid and onto LB medium containing nalidixic acid and kanamycin. After 9 days the mice were killed by cervical dislocation, and bacteria were recovered by plating dilutions of homogenized colon onto medium containing nalidixic acid and medium containing nalidixic acid and kanamycin. The virulence of each mutant was analyzed by using two or more animals, and the results are expressed below as averages. The competitive index (CI) was calculated by dividing the ratio of mutant bacteria to wild-type bacteria recovered from animals by the ratio of mutant bacteria to wild-type bacteria in the inoculum.
In single-infection experiments, bacteria were grown to the stationary phase in LB broth containing the appropriate antibiotic. The bacteria were pelleted by centrifugation, resuspended in an equal volume of PBS, and gavaged into mice (approximately 1 × 108 CFU per mouse). Each mutant strain was tested in independent experiments at least twice by using groups of at least three mice per strain. Stool samples were recovered aseptically at various times after inoculation, and the number of viable bacteria per gram of stool was determined by plating samples onto LB agar containing the appropriate antibiotics. Mice were killed 13 days postchallenge, and the distal 8 cm of colon was aseptically removed from each mouse and weighed after removal of fecal pellets. The colons were then homogenized mechanically with a Seward 80 stomacher (Seward, London, United Kingdom), and the numbers of viable bacteria per colon were determined by plating onto LB agar containing the appropriate antibiotics.
Histological and immunofluorescence analyses.
Distal colons of uninfected mice and of mice infected with C. rodentium strains were snap frozen and stored at −80°C until they were cut. Cryosections (thickness, 6 μm) were cut, air dried for 1 h, fixed with ice-cold acetone for 10 min, and processed for histological analysis or immunofluorescence analysis. For histological analysis cryosections were stained with hematoxylin and eosin and analyzed with a Nikon Eclipse E600 microscope. For immunofluorescence analysis cryosections were blocked with 7% horse serum; then a rabbit universal intimin serum (4) was added, and the cryosections were incubated for 1 h at room temperature. After extensive washing, the cryosections were incubated for 45 min with goat anti-rabbit immunoglobulin G labeled with fluorescein isothiocyanate (Sigma). Host cell F-actin was stained with tetramethyl rhodamine isothiocyanate-conjugated phalloidin (Sigma). Specimens were mounted in Mowiol (Aldrich) and viewed with an inverted Zeiss LSM 510 Meta confocal microscope.
Nucleotide sequence accession number.
The nucleotide sequence of espI has been deposited in the GenBank database under accession number AY373261.
RESULTS
Identification of EspI, a novel type III secreted protein.
Further screening of signature-tagged C. rodentium mutants by using the murine model in our laboratory and the conditions described previously (33) led to identification of strain P4G1. This strain was present in the input pool that was orally inoculated into three mice but was not recovered from stools taken 3 days postinoculation or from colons removed 5 days postinoculation. Mutant P4G1 was found to contain a transposon insertion 200 bp into an ORF with strong identity to gene Z6024 (part of prophage CP-933P) of EHEC strain EDL933 (37), to ORF Ecs1812 of the EHEC Sakai strain (26), and to ORF 364 of EPEC strain E2348/E69, which is currently being sequenced by the Sanger Centre (http://www.sanger.ac.uk/Projects/Microbes).
The original 1.9-kb SalI fragment flanking the transposon insertion that was cloned from mutant P4G1 contained only the remaining 1.1 kb of an ORF. The complete ORF from C. rodentium was then amplified by using primers designed from gene Z6024 and sequenced. There was 81% identity at the amino acid level between the product of the ORF identified in strain P4G1 and the EHEC protein encoded by Z6024 in EDL933 and by ORF Ecs1812 in the Sakai strain, and there was 78% identity to the EPEC protein encoded by ORF 364. A multiple alignment of these ORFs is shown in Fig. 2.
FIG. 2.
Alignment of the deduced amino acid sequences of EspI and the homologs encoded by Z6024 of EHEC and ORF 364 of EPEC. Identical amino acids are indicated by a grey background. Gaps introduced for alignment are indicated by dashes.
There is previous evidence that the EHEC protein encoded by gene Z6024 is secreted through the LEE-encoded TTSS, as the first 20 amino acids match those of EHEC type III secreted protein P54 identified by Kresse et al. (23). This 54-kDa protein was found to be hypersecreted by an EHEC sepL mutant, and the first 20 N-terminal amino acids were sequenced. In order to determine if the C. rodentium homolog of Z6024 Can be secreted into the supernatant via the TTSS, we constructed a plasmid carrying an inducible His-tagged version of the P4G1 ORF, pICC298. This plasmid was then electroporated into C. rodentium wild-type strain DBS100, the escD transposon mutant P10G3 (33), and the escF mutant strain ICC176. It has previously been shown that EscF is necessary for secretion of the EspA, EspB, and EspD proteins in EPEC (46). A Citrobacter escF mutant strain also proved to be unable to secrete these proteins into the supernatant (data not shown).
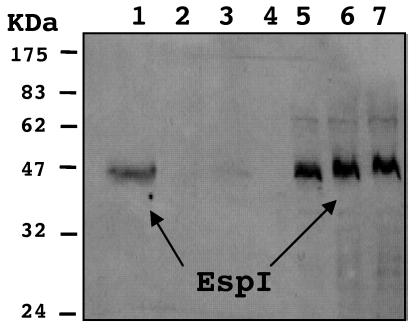
Expression of the approximately 47-kDa His-tagged P4G1 protein in cell pellets was confirmed by Western blotting by using anti-His antiserum (Fig. 3). The His-tagged P4G1 protein was also detected in the culture supernatant of the wild-type strain. However, no His-tagged P4G1 protein was secreted into the culture supernatant by mutant strain P10G3 (Tn::escD). Only a trace amount was secreted by mutant ICC176 (ΔescF) (Fig. 3), although it is not clear if this amount represented genuine type III secretion. This suggests that the C. rodentium ORF identified in strain P4G1 encodes a protein that requires a functional TTSS for secretion into the culture supernatant. The 1,293-bp ORF was therefore designated espI, and the sequence has been deposited in the GenBank database. Importantly, the fluorescent actin staining test revealed that EspI was not required for A/E lesion formation by C. rodentium in vitro (data not shown).
FIG. 3.
Immunoblot analysis of His-tagged EspI in cell pellets and concentrated supernatants. Equivalent amounts of His-tagged EspI were detected in cell pellets of wild-type, Tn::escD (P10G3), and ΔescF (ICC176) strains (lanes 5, 6, and 7). His-tagged EspI was also detected strongly in concentrated supernatant of the wild-type strain (lane 1), but no His-tagged EspI was detected in the equivalent amount of concentrated supernatant of Tn::escD strain P10G3 (lane 2) and only a trace amount was detected in the supernatant of ΔescF strain ICC176 (lane 3). Lane 4 was left empty to prevent overspill between wells. The position of a protein corresponding to His-tagged EspI (47.2 Da) is indicated by arrows.
Construction of nonpolar mutations in C. rodentium genes escF, espF, espG, espH, and espI.
Mutant strains were constructed to determine whether the secreted proteins EspF, EspG, EspH, EspI, and Map are required for colonization and/or colonic hyperplasmia in the C. rodentium mouse model. Nonpolar mutations were constructed in the escF, espG, and espI genes by using the nonpolar kanamycin resistance cassette aphT (15), which generated strains ICC176, ICC178, and ICC179, respectively. Deletions were made in the espF gene (by using aphT), creating strain ICC177, and in the espH gene (by using the kan cassette) (pUC4K; Pharmacia), resulting in strain TX18. The mutations in the strains were confirmed by PCR and Southern blot analyses (data not shown). Strain P6C6 contains a transposon insertion in the predicted promoter region upstream of the start codon of map (33). No nonpolar mutant with a mutation in map was constructed as this gene is known to be monocistronic, suggesting that the transposon insertion in strain P6C6 was unlikely to have any polar effects on neighboring genes. As the context of espI in C. rodentium is not yet known, a nonpolar mutation was constructed in this gene to eliminate any possibility of polar effects on downstream genes.
CI of each mutant strain as determined by mixed infection.
The colonization potential of each of the mutant strains was established by performing mixed-infection experiments with inocula containing approximately 50% wild-type C. rodentium (Nalr) and 50% mutant (Nalr Kanr). The exact ratios of mutant CFU to wild-type CFU were calculated for the inocula, for the stool contents on days 3 and 6, and for the colon contents on day 9 by using the different antibiotic resistance characteristics of the two strains. The CI was defined as the output ratio (mutant CFU/wild-type CFU) divided by the input ratio (mutant CFU/wild-type CFU). A CI of 1 indicated that the test strain was able to compete well with the wild-type strain and colonized the mouse colon at a similar level. A CI of less than 1 indicated that a strain was attenuated and was unable to colonize as effectively as the wild-type strain, and a CI of 0 meant that the test strain was completely unable to colonize.
Previous experiments have shown that C. rodentium strains with mutations in the intimin, tir, or espD gene have a CI of virtually 0 by day 3 (33). The data obtained for strain ICC176 (ΔescF) are similar; a CI of 0 was obtained for all three mice by using stool samples taken on days 3 and 6 and colon samples taken on day 9 (Fig. 4). No kanamycin-resistant ICC176 (ΔescF) mutant colonies were present in these samples, whereas large numbers of wild-type C. rodentium cells were present in stool and colon samples at all time points.
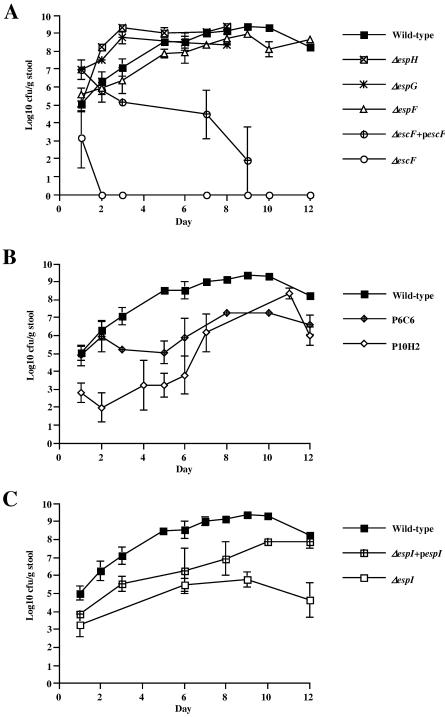
FIG. 4.
Virulence of C. rodentium mutant strains in mixed-infectionexperiments with the wild-type strain. Mice were challenged with inocula containing approximately 50% wild-type C. rodentium (Nalr) and 50% mutant (Nalr Kanr). The CI were calculated for the Nalr and Kanr CFU recovered from stools on day 3 (A) and day 6 (B) and from colon contents on day 9 (C). The ΔescF, ΔespG, ΔespI, and Δmap mutant strains were rapidly outcompeted by the wild-type strain, and the levels of these mutants quickly declined over the course of the infection. In contrast, the ΔespF and ΔespH mutant strains were able to compete with the wild-type strain more effectively and were only slightly outcompeted over time.
The other mutant strains were all also unable to colonize mice as efficiently as the wild-type strain but generally were present in the colon for longer times. Strain ICC179 (ΔespI) was the most attenuated of these mutants, and it was rapidly outcompeted by the wild-type strain. Very few colonies were isolated from stools on days 3 and 6 (CI, 0.022 and 0.00005, respectively), and no colonies were isolated from the colon on day 9 (CI, 0). Strain ICC178 (ΔespG) and map mutant P6C6 exhibited similar patterns, except that a few mutant colonies were still recovered from the colons on day 9 (CI, 0.0006 and 0.035, respectively).
In contrast, strains ICC177 (ΔespF) and TX18 (ΔespH) were only mildly attenuated in mixed-infection experiments. Both of these strains were able to efficiently compete with the wild-type strain on day 3, but over time significantly lower numbers of these strains than of the wild-type strain were recovered from stools and colons, resulting in CI of 0.629 (ΔespF) and 0.522 (ΔespH) for day 9 colons.
Shedding patterns of the mutant strains as determined by single infections.
Single-infection experiments were carried out to determine the level of virulence attenuation of each C. rodentium strain. Mice were challenged orally with ∼1 × 108 CFU of the wild-type strain or a mutant or complemented strain. Stool samples were collected at various times during the first 8 to 12 days postinoculation, and the numbers of CFU per gram of stool were determined by plating samples onto LB medium containing the appropriate antibiotics. The results are shown in Fig. 5. The wild-type strain had a growth curve typical of C. rodentium infection of C3H/Hej mice; the number of CFU per gram of stool slowly increased over the first few days postinoculation, reached a peak at around day 10, and then started to decline.
FIG. 5.
Colonization of mouse colons by different C. rodentium strains, as indicated by shedding of CFU in stools. The levels of colonization were indicated by the viable bacterial counts from stool samples taken at different times for 12 days postchallenge. The error bars indicate standard errors. (A) Strains ICC177 (ΔespF), ICC178 (ΔespG), and TX18 (ΔespH) were all shed in stools at levels very similar to the levels of the wild-type strain, whereas strain ICC176 (ΔescF) was not recovered after 2 days postinoculation, a phenotype which was partially rescued by plasmid pUC19-escF. (B) Mutant map strains P6C6 and P10H2 exhibited very similar patterns. The levels of these strains that were shed were approximately 3 logs lower than the levels of the wild type that were shed over the first 10 days postinoculation and then slowly increased to levels which were more similar to the wild-type levels. (C) ΔespI strain ICC179 exhibited growth dynamics similar to those of the map mutant strains. The levels of this strain that were shed were 3 to 4 logs lower than the levels of the wild-type strain that were shed over 12 days. This phenotype was rescued by pACYC184-espI, which resulted in higher numbers of challenge bacteria shed in stools, although the levels were not as high as the wild-type levels.
In contrast, strain ICC176 (ΔescF) was shed in stools only for the first 24 h following inoculation (Fig. 5A). It therefore appeared that this mutant strain was incapable of colonizing the gastrointestinal tract and instead passed straight through the mice via the stools. Two plasmids were constructed to try to complement the ΔescF mutant strain ICC176, pACYC-escF (pICC271) and pUC19-escF (pICC270). Both of these plasmids were able to restore wild-type levels of secretion of EspB and EspD into the concentrated supernatant of ΔescF mutant ICC176 (unpublished data). However, both plasmids were unstable in vivo in the absence of antibiotic selection (data not shown). Plasmid pUC19-escF was the more stable construct of the two and resulted in partial complementation of a colonization phenotype to the ΔescF mutant for 9 days postinoculation, before all challenge bacteria containing the plasmid were lost from the population.
The numbers of strain ICC178 (ΔespG) and TX18 (ΔespH) CFU recovered from stools were comparable to the numbers of the wild-type strain CFU recovered over the course of the infection (Fig. 5A), suggesting that these strains had no colonization defect. Strain ICC177 (ΔespF) also exhibited a pattern similar to that of the wild-type strain; however, at each time point approximately 1 log less CFU was consistently recovered from stools compared to the number of wild-type CFU. At most time points the difference was not statistically significant, but this finding is worth noting as it was a consistent finding for every mouse infected in the course of two independent experiments.
In contrast to the wild-type strain, the espI and map mutants both had an intermediate colonization phenotype, and the number of CFU shed in the stools at each time point was 3 to 4 logs less than the number of wild-type CFU shed (Fig. 5B and C). Complementation of ICC179 with pACYC-espI resulted in partial complementation, and the number of CFU per gram of stool shed at each time point was slightly higher than the number of CFU of the ΔespI mutant strain shed. This complementation peaked at day 12, when comparable numbers of CFU were shed from mice infected with the wild type and from mice infected with the ΔespI mutant containing pACYC-espI.
The map mutant strain P6C6 also had an intermediate colonization phenotype in mice, and the number of bacteria shed in stools at each time point was approximately 3 logs less than the number of bacteria shed in stools of the mice infected with the wild type (Fig. 5B). Two plasmids were constructed to try to complement this strain, pWSK29-map (pICC299) and pACYC-map (pICC300). Each plasmid was introduced into strain P6C6 and was tested by single infection of mice. Unfortunately, both plasmids were unstable in vivo, and very low numbers were recoverable from stools after 3 days postinoculation (data not shown). For this reason, a second strain with a transposon insertion in map, P10H2, was also tested in mice. Strain P10H2 had a transposon insertion 6 bp from the end of the map ORF (Mundy, Simpson, and Frankel, unpublished data). This strain had a phenotype very similar to that of strain P6C6.
Mucosal association of the mutants following single infection.
Thirteen days postchallenge, the pathogen burdens in the colons of mice infected with wild-type, mutant, and complemented strains were determined, as were the abilities of the strains to induce colonic hyperplasia. Mice infected with the wild-type strain and with mutant strains ICC177 (ΔespF), ICC178 (ΔespG), and TX18 (ΔespH) all had high levels of challenge bacteria in their colons (Fig. 6A), and the strains induced similar levels of colonic hyperplasia, as indicated by increased colon weight (Fig. 6B) and microscopic visualization of crypt length in frozen colon sections (Fig. 7).
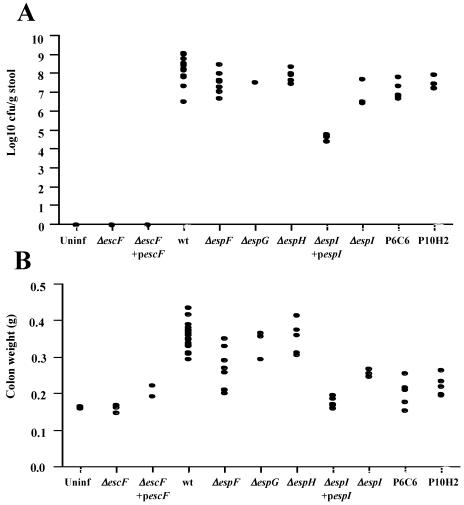
FIG. 6.
Virulence of C. rodentium strains in the mouse colon. (A) Numbers of C. rodentium CFU recovered from colonic tissues of individual mice on day 13 postchallenge. Mice infected with the wild-type strain and with espF, espG, espH, and map mutants all had high pathogen burdens. In contrast, mice infected with the espI mutant strains had lower bacterial loads, although the levels were still higher than the levels in mice infected with the escF mutant and complemented strains, from which no challenge bacteria were recovered. The numbers of CFU obtained from mice infected with the espI mutant strain complemented with pICC296 were significantly higher than the numbers of CFU obtained from mice infected with the espI mutant strain (6.56 ± 042 CFU compared to 4.81 ± 0.19 CFU; P > 0.001), although the levels were not quite the same as the wild-type levels (7.98 ± 0.20 CFU). (B) The distal 8 cm of colon was removed and weighed 13 days postchallenge. Mice infected with ΔespG and ΔespH strains had colon weights that were not significantly different from the colon weights of mice infected with the wild type (P > 0.001). There was no significant difference between the colon weights of mice infected with the avirulent ΔescF strain and the colon weights of uninfected mice (P > 0.001). In contrast, the colon weights of mice infected with map mutants P6C6 and P10H2 and those of mice infected with the complemented ΔescF strain containing pICC270 and the complemented ΔespI strain containing pICC296 were significantly greater than those of uninfected mice (P > 0.05) but still significantly less than the colon weights of mice infected with the wild type (P > 0.001). Uninf, uninfected; wt, wild type.
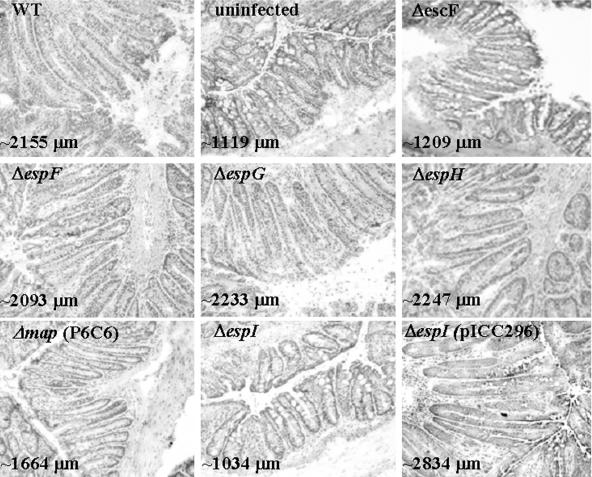
FIG. 7.
Hematoxylin- and eosin-stained frozen colonic sections from mice infected with different C. rodentium strains for 13 days. Mice infected with the wild-type strain or ΔespF, ΔespG, and ΔespH mutant strains all exhibited crypt hyperplasia and inflammation of the distal colon. In contrast, no crypt hyperplasia and inflammation were observed in the distal colons of mice infected with the escF mutant or the espI mutant or in uninfected control mice. Crypt hyperplasia was restored in the ΔespI mutant strain by addition of complementing plasmid pICC296. In contrast, mutant strain P6C6 (Δmap) exhibited an intermediate level of hyperplasia, as measured by crypt length. The average colonic crypt height in each sample is indicated. Magnification, ×2,000. WT, wild type.
In contrast, no challenge bacteria were isolated from the colons of mice infected with the ΔescF mutant strain ICC176 (Fig. 6A). There was also no colonic hyperplasia, as determined by colon weight (Fig. 6B) or by microscopic examination of crypt length (Fig. 7); for both of these parameters the data were similar to the data obtained for the uninfected control mice. No challenge bacteria were isolated from the colons of mice inoculated with the complementing ΔescF strain containing pUC19-escF (Fig. 6A); however, some hyperplasia was observed, as determined by colon weight (Fig. 6B). These results correlate with the shedding of bacteria observed in the stools (Fig. 5A), as few challenge bacteria containing plasmid pUC19-escF were isolated after 9 days postinoculation. However, the presence of reasonably high numbers of challenge bacteria for the first 7 days of infection explains the small but significant level of hyperplasia as determined by colon weight on day 13 postinoculation (212 ± 10 mg for mice infected with the ΔescF strain containing pUC19-escF strain; 160 ± 6 mg for mice infected with the ΔescF strain; 161 ± 3.5 mg for uninfected mice; P < 0.01).
The strains having mutations in espI and map had phenotypes that fell between the extremes of wild-type C. rodentium colonization and disease and the ΔescF mutant strain's complete lack of colonization and pathology. The espI mutant ICC179 was isolated from mouse colons 13 days postinoculation, but the levels were 3 to 4 logs lower than the levels of bacteria isolated from mice infected with the wild-type strain (Fig. 6A). This correlates with the finding that the number of CFU of this strain isolated from stools was 3 to 4 logs less than the number of CFU of the wild type isolated from stools over the first 12 days of infection (Fig. 5C). The mice infected with the espI mutant strains also had significantly lower levels of hyperplasia, as determined both by colon weight (Fig. 6B) (178 ± 6 mg for the ΔespI strain; 354 ± 8 mg for the wild type; P < 0.001) and by measurement of crypt length (Fig. 7). Partial complementation of ΔespI mutant strain ICC179 was achieved by addition of plasmid pACYC-espI (pICC296) carrying a wild-type copy of espI. The numbers of CFU of this complementing strain in the colons of mice were significantly higher than the numbers of CFU of the mutant strain, although the numbers of CFU were still 1 log lower than the numbers of CFU recovered from mice infected with the wild-type strain (6.56 ± 0.42 CFU for the ΔespI strain containing pACYC-espI; 4.81 ± 019 CFU for the ΔespI strain; 7.98 ± 0.20 CFU for the wild type). The level of hyperplasia in colons from mice infected with the complementing strain was also higher than the level of hyperplasia in the colons from mice infected with the mutant strain, as measured by increased colon weight (Fig. 6B), although again the plasmid did not restore the wild-type level of hyperplasia (240 ± 17 mg for the ΔespI strain containing pICC296; 177 ± 6 mg for the ΔespI strain; 354 ± 8 mg for the wild type). However, microscopic examination of the crypt lengths revealed crypts that on average were longer than the crypts from mice infected with the wild-type strain (Fig. 7).
Mice infected with map mutants P6C6 and P10H2 all exhibited levels of bacterial colonization in the colon similar to those in mice infected with the wild-type strain (Fig. 6A) (7.10 ± 0.22 CFU for P6C6; 7.45 ± 0.12 CFU for P10H2; 7.98 ± 0.20 CFU for the wild type). However, they exhibited significantly reduced levels of colonic hyperplasia compared to the levels in the mice infected with the wild type, as measured by colon weight (Fig. 6B) (203 ± 17 mg for P6C6; 222 ± 13 mg for P10H2; 354 ± 8 mg for the wild type; P < 0.001). However, there were signs of hyperplasia when these mice were compared to uninfected mice (161 ± 3.5 mg for uninfected mice; P < 0.5 for the comparison with P6C6 and P < 0.01 for the comparison with P10H2). The low level of hyperplasia could be explained by the fact that significantly lower numbers (approximately 3 logs lower) of the map mutant strains than of the wild type were isolated from mouse stools over the first 12 days of infection. Measurement of crypt length by microscopy also revealed an intermediate level of hyperplasia; crypts from mice infected with map mutant strains were significantly longer than crypts from uninfected mice but were significantly shorter than crypts from mice infected with the wild-type strain (Fig. 7).
Challenge bacteria present in the frozen colon sections were also stained by using anti-intimin antisera and were visualized by using immunofluorescence. Actin staining was used to counterstain the tissue. High numbers of C. rodentium cells were observed on the epithelial surfaces of mice infected with the wild-type strain, ICC177 (ΔespF), ICC178 (ΔespG), and TX18 (ΔespH) (Fig. 8). In contrast, no C. rodentium cells were observed in the colons of uninfected mice or the colons of mice infected with ΔescF strain ICC176. Between these extremes and in agreement with the viable counts, very low levels of challenge bacteria, mainly in the lumen, were observed in mice infected with map or espI mutants (Fig. 8).
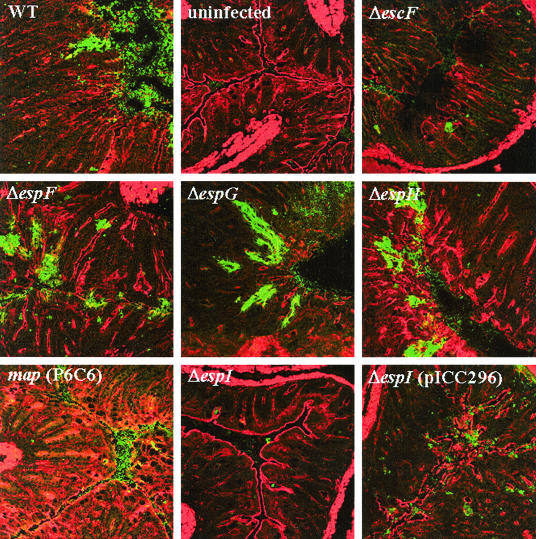
FIG. 8.
Intimin and actin staining of colonic sections from C. rodentium-infected mice. Colonic sections were stained with polyclonal antiserum to the outer membrane adhesin intimin to directly label adherent bacteria (green) and were counterstained with phalloidin to stain cellular actin (red). Large numbers of bacteria were present on the colonic epithelia of mice infected with the wild-type strain, as well as mice infected with ΔespF, ΔespG, and ΔespH mutant strains; bacteria were not present on the colonic epithelia of mice infected with the ΔescF mutant and in the colons of uninfected mice. Low numbers of bacteria were associated with tissue taken from mice infected with the ΔespI mutant and the Δmap mutant. Complementing the ΔespI mutant with plasmid pICC296 resulted in more bacteria associated with the colon than the number in mice infected with the mutant strain alone, but the level did not reach the level associated with infection by the wild-type strain. Magnification, ×2,000. WT, wild type.
espI and map mutant strains have very different growth dynamics in mice compared to the growth dynamics of the wild-type and espF mutant strains.
The ability of the mutant strains with an intermediate colonization phenotype (espI and map) to establish, expand, and then be cleared by mice was investigated by monitoring the viable counts recovered from stools over an extended time period (40 days) until all the mice in each group had cleared the infection (Fig. 9). The wild-type strain produced the classic growth curve previously reported by Barthold et al. (3). The size of the challenge bacterial population steadily increased, reaching a peak of approximately 109 CFU on around day 9 or 10 of infection, and then it steadily decreased until the infection was cleared at day 26 postinoculation. Mutant strain ICC177 (ΔespF) exhibited a very similar pattern (Fig. 9A), except that it slightly lagged behind wild type, with the population size peaking after 12 days at a slightly lower level, perhaps explaining the slightly reduced levels of hyperplasia indicated by lower colon weights (Fig. 6B). It also took several days longer to clear the ΔespF mutant strain, which finally disappeared on day 30.
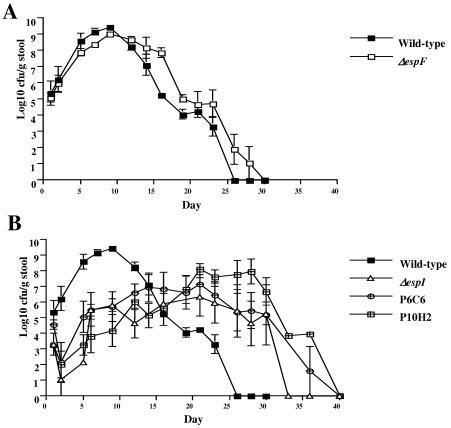
FIG. 9.
Growth dynamics of several C. rodentium strains in the mouse gastrointestinal tract as determined from viable counts in stools. Colonization was indicated by viable counts of C. rodentium from stool samples taken over the course of 40 days postinoculation. The error bars indicate standard errors.
In contrast, the map and espI mutant strains exhibited very different growth dynamics than the wild type exhibited. The P6C6 and P10H2 strains had very similar growth patterns. After an initial decline in the viable counts on days 2 to 4, the number of bacteria began to increase slowly, reaching approximately 105 to 106 CFU/g of stool by day 12; then this level was maintained for an extended period of time (10 to 15 days) (Fig. 9B) before it started to decline, until no CFU were recovered from stools by day 40. The size of the ΔespI mutant strain ICC179 population also never reached more than 105 to 106 CFU/g of stool and remained relatively steady between days 6 and 24, and this organism was not cleared from mice until day 33 (Fig. 9B).
DISCUSSION
In this study, we identified a novel type III secreted protein, EspI, in C. rodentium which is encoded outside the LEE and is present in the sequenced A/E EPEC and EHEC pathogens. Although the function of EspI is not yet known, we show here that it plays an important role in both bacterial colonization of the colonic epithelium of infected mice and induction of hyperplasia in the colonic epithelium of infected mice. In addition, by using targeted mutagenesis of LEE-encoded effectors, we identified three distinct virulence phenotypes in vivo: complete attenuation, intermediate attenuation, and no attenuation.
With complete virulence attenuation, the mutant strains appear to pass straight through the gastrointestinal tract and exit via the stools without colonizing the animal. This phenotype is associated with proteins involved in the formation of the A/E lesions, including intimin (40), Tir (8), TTSS apparatus and needle complex proteins EscD (33) and EscF (this study), and secreted translocon proteins EspB (35) and EspD (33).
At the opposite extreme, fully virulent mutant strains behave exactly like the wild-type strain; i.e., large numbers of these strains colonize the colon, and the organisms are shed in stools over more than 24 days and cause pathology such as colonic hyperplasia and inflammation, resulting in loss of weight by the mice, production of soft or runny stools, and development of rectal bleeding and in some cases rectal prolapse. Strains with mutations in the effector genes espF, espG, and espH have this phenotype. However, it is possible that the espF mutant has a subtle growth defect in mice over the first 12 days of infection, which results in levels of hyperplasia and inflammation that are slightly lower than those seen in mice infected with the wild-type strain.
Between these two extremes, certain mutant strains have partial virulence attenuation; these organisms are able to colonize the gut, and they are shed in the stools but at a significantly lower level (3 to 4 logs lower) than the wild type, especially over the first 10 to 12 days of infection. This is correlated with a much lower level of hyperplasia and milder clinical signs, such as soft stools with no signs of rectal bleeding or prolapse. Strains with mutations in genes encoding the secreted proteins EspI and Map have this phenotype.
Figures 4 to 6 show that the mutant strains have consistent phenotypes in vivo, whether they are tested in mixed infections or single infections. Strains that are completely avirulent (e.g., a ΔescF strain) are absent from stools by day 3 regardless of whether they are inoculated singly or as part of a mixture with the wild type. In contrast, strains that have no attenuation in single infections are generally able to compete well in mixed infections over longer periods of time (e.g., ΔespF and ΔespH strains). Strains having an intermediate colonization defect in single infections (e.g., Δmap and ΔespI strains) have an intermediate ability to compete with the wild type in mixed infections, managing to colonize mice at low levels but gradually being ousted by the wild type so that few cells remain 9 days postinoculation (Fig. 4). The only exception to this pattern is the ΔespG mutant that has an intermediate phenotype in mixed infections but appears to have no colonization defect in single-infection experiments. Interestingly, in the REPEC rabbit model of single infection, approximately 1 log more wild-type REPEC bacteria than espG mutant bacteria was recovered from the rectums, suggesting that in this system EspG might play a role in colonization (12).
Previously, it was shown that the map mutant strain P6C6 colonized the colon at virtually wild-type levels by day 13 postinoculation but that it produced much lower levels of hyperplasia (33). However, this snapshot of what was happening in the mouse colon gave no real indication of whether colonization and hyperplasia are linked. By examining the growth dynamics of C. rodentium strains in mice over time, in this study we showed that the outcome of an infection is determined in the first 4 days postinoculation. Our data suggest that at 24 h postchallenge there are already differences between the virulent and more attenuated strains. For example, the wild-type strain and the fully virulent ΔespF, ΔespG, and ΔespH mutant strains all produced around 105 CFU/g of stool at 24 h postinoculation, whereas the levels of the more attenuated ΔescF, ΔespI, and Δmap mutants were only 103 to 104 CFU/g of stool (Fig. 5). These differences became more marked at days 2 to 4 postinoculation, when the virulent strains started to become established and grow, resulting in 106 to 107 CFU/g of stool. In contrast, either the attenuated strains disappeared completely (ΔescF strain) or the levels of these strains remained static or declined (Δmap and ΔespI strains) and were only 102 to 104 CFU/g of stool at days 2 to 4 postinoculation (Fig. 5). From day 5 to day 10 postinoculation, the virulent strains continued to survive, replicate, and expand in the mouse gut, and the levels reached peaks of 108 to 109 CFU/g of stool at around day 10 and then slowly started to decline. The intermediately attenuated strains also started to become established during this time, and the levels started to increase to approximately 105 to 106 CFU/g of stool.
The growth dynamics over the first 12 days of infection were reflected in the levels of bacteria found in association with the colonic mucosa, which was removed 13 days postinoculation. For the fully virulent strains the levels of challenge bacteria were high (∼107 to 109 CFU/colon) (Fig. 6). The levels of the map mutant strains were also high (∼107 to 108 CFU/colon). The numbers of the ΔespI strains recovered were much lower (∼104 to 105 CFU/colon), while no ΔescF mutant bacteria were recovered. The severity of colonic hyperplasia was directly correlated with the number of bacteria associated with the mucosa. For example, the levels of strains ICC178 (ΔespG) and TX18 (ΔespH) recovered from stools were equivalent to the levels of the wild type, and the levels of hyperplasia in mice infected with these strains were indistinguishable from the levels in mice infected with the wild type. The ΔespF strain produced slightly less hyperplasia, which reflected the approximately 1 log less CFU recovered from stools of mice infected with this strain than from stools of mice infected with the wild type over the first 12 days. The mutant map and espI strains both produced low levels of hyperplasia, and the level observed with the ΔespI strain was almost indistinguishable from the level in uninfected mice.
We attempted to complement all the mutants that exhibited measurable virulence attenuation in vivo. However, various degrees of success were achieved despite the use of plasmids such as pACYC184 and pWSK29, which we and other workers found to be useful in the past (8, 33, 35). The biggest problem encountered was the instability of the complementing plasmids in vivo in the absence of antibiotic selection. Plasmids pACYC-escF (pICC271), pWSK29-map (pICC299), and pACYC-map (pICC300) were all lost from the in vivo challenge bacterial population within 3 to 5 days postinoculation (data not shown). Partial complementation of the ΔescF mutant was achieved by using plasmid pUC19-escF (pICC270), although this plasmid was also lost from the challenge bacterial population 9 days postinoculation. Plasmid pACYC-espI (pICC296) did complement ΔespI mutant strain ICC179, remaining stable for the whole 13 days of infection (except in one of four mice in the group) (data not shown). However, this strain took longer to become established and to expand in the mouse gut than the wild-type strain took (Fig. 5B), and the plasmid did not restore hyperplasia to the wild-type level (Fig. 6B). An inability to restore a mutant completely to the wild-type virulent phenotype has been observed by other workers, including Newman et al. (35), who used pACYC-espB (pJVN111) in an espB mutant and observed that the number of CFU was 2 logs less than the number of CFU of the wild type at each time point. Also, Deng et al. (8) used a pACYC construct of tir in a tir mutant background and observed that the number of CFU was 2 to 3 logs less than the number of CFU obtained from the colons of mice infected with the wild-type strain.
The growth dynamics of the wild-type, ΔespF, Δmap, and ΔespI strains were monitored by examining the shedding of bacteria in stools over a longer time until the infection was completely cleared. This revealed that the levels of wild-type bacteria recovered from stools peaked at around day 9 or 10 and then slowly declined until no bacteria were recovered 26 days postinoculation (Fig. 9). The levels of ΔespF bacteria peaked several days later, at day 12, and then also declined at a rate similar to that of the wild type but several days behind, until the infection was cleared at day 28. In contrast, the level of the ΔespI mutant strain peaked at around 105 to 106 CFU/g of stool, which was several logs lower than the wild-type value, but it also took longer to clear this mutant (34 days). The map mutant strains also exhibited a very different growth pattern than the wild-type strain; the levels remained high (∼106 to 107 CFU/g of stool) between days 10 and day 30, and the strains were not completely cleared until day 40. Considering that colonic hyperplasia is an immunopathological reaction to C. rodentium infection, the fact that the map mutants induced a low level of hyperplasia while efficiently colonizing the colon at late stage of the infection suggests that Map can modulate immunological responses, the nature of which are currently being investigated.
This study was, to our knowledge, the first to attempt to compare and contrast the contribution of type III proteins to infection with A/E pathogens. Clearly, the phenotypes of some single mutants are masked or compensated for by other effectors. For example, it is known that two additional copies of EspF-like proteins are present in EHEC. For this reason, we are now testing double and triple C. rodentium mutants in order to determine the potential roles of combinations of gene products in virulence.
Acknowledgments
This work was supported by the BBSRC, the Wellcome Trust, the European Union Fifth Framework Quality of Life Program (grant QLK2-2000-00600), and a grant from the Israeli Academy of Science and Humanities.
Editor: A. D. O'Brien
REFERENCES
- 1.Abe, A., M. de Grado, R. A. Pfuetzner, C. Sanchez-Sanmartin, R. Devinney, J. L. Puente, N. C. Strynadka, and B. B. Finlay. 1999. Enteropathogenic Escherichia coli translocated intimin receptor, Tir, requires a specific chaperone for stable secretion. Mol. Microbiol. 33:1162-1175. [DOI] [PubMed] [Google Scholar]
- 2.Barthold, S. W., G. L. Coleman, P. N. Bhatt, G. W. Osbaldiston, and A. M. Jonas. 1976. The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 26:889-894. [PubMed] [Google Scholar]
- 3.Barthold, S. W., G. L. Coleman, R. O. Jacoby, E. M. Livestone, and A. M. Jonas. 1978. Transmissible murine colonic hyperplasia. Vet. Pathol. 15:223-236. [DOI] [PubMed] [Google Scholar]
- 4.Batchelor, M., S. Knutton, A. Caprioli, V. Huter, M. Zanial, G. Dougan, and G. Frankel. 1999. Development of a universal intimin antiserum and PCR primers. J. Clin. Microbiol. 37:3822-3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crane, J. K., B. P. McNamara, and M. S. Donnenberg. 2001. Role of EspF in host cell death induced by enteropathogenic Escherichia coli. Cell Microbiol. 3:197-211. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deng, W., Y. Li, B. A. Vallance, and B. B. Finlay. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, W., B. A. Vallance, Y. Li, J. L. Puente, and B. B. Finlay. 2003. Citrobacter rodentium translocated intimin receptor (Tir) is an essential virulence factor needed for actin condensation, intestinal colonization and colonic hyperplasia in mice. Mol. Microbiol. 48:95-115. [DOI] [PubMed] [Google Scholar]
- 9.Donnenberg, M. S., C. O. Tacket, S. P. James, G. Losonsky, J. P. Nataro, S. S. Wasserman, J. B. Kaper, and M. M. Levine. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J. Clin. Investig. 92:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., J. Yu, and J. B. Kaper. 1993. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J. Bacteriol. 175:4670-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, S. J., S. W. Hutcheson, M. S. Dubois, J. L. Mellies, L. A. Wainwright, M. Batchelor, G. Frankel, S. Knutton, and J. B. Kaper. 1999. Identification of CesT, a chaperone for the type III secretion of Tir in enteropathogenic Escherichia coli. Mol. Microbiol. 33:1176-1189. [DOI] [PubMed] [Google Scholar]
- 12.Elliott, S. J., E. O. Krejany, J. L. Mellies, R. M. Robins-Browne, C. Sasakawa, and J. B. Kaper. 2001. EspG, a novel type III system-secreted protein from enteropathogenic Escherichia coli with similarities to VirA of Shigella flexneri. Infect. Immun. 69:4027-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y. K. Deng, L. C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 14.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 15.Galan, J. E., C. Ginocchio, and P. Costeas. 1992. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J. Bacteriol. 174:4338-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 17.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenny, B., R. DeVinney, M. Stein, D. J. Reinscheid, E. A. Frey, and B. B. Finlay. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511-520. [DOI] [PubMed] [Google Scholar]
- 20.Kenny, B., S. Ellis, A. D. Leard, J. Warawa, H. Mellor, and M. A. Jepson. 2002. Co-ordinate regulation of distinct host cell signalling pathways by multifunctional enteropathogenic Escherichia coli effector molecules. Mol. Microbiol. 44:1095-1107. [DOI] [PubMed] [Google Scholar]
- 21.Kenny, B., and M. Jepson. 2000. Targeting of an enteropathogenic Escherichia coli (EPEC) effector protein to host mitochondria. Cell Microbiol. 2:579-590. [DOI] [PubMed] [Google Scholar]
- 22.Kenny, B., L. C. Lai, B. B. Finlay, and M. S. Donnenberg. 1996. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol. Microbiol. 20:313-323. [DOI] [PubMed] [Google Scholar]
- 23.Kresse, A. U., F. Beltrametti, A. Muller, F. Ebel, and C. A. Guzman. 2000. Characterization of SepL of enterohemorrhagic Escherichia coli. J. Bacteriol. 182:6490-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, L. C., L. A. Wainwright, K. D. Stone, and M. S. Donnenberg. 1997. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect. Immun. 65:2211-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luperchio, S. A., and D. B. Schauer. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3:333-340. [DOI] [PubMed] [Google Scholar]
- 26.Makino, K., K. Yokoyama, Y. Kubota, C. H. Yutsudo, S. Kimura, K. Kurokawa, K. Ishii, M. Hattori, I. Tatsuno, H. Abe, T. Iida, K. Yamamoto, M. Onishi, T. Hayashi, T. Yasunaga, T. Honda, C. Sasakawa, and H. Shinagawa. 1999. Complete nucleotide sequence of the prophage VT2-Sakai carrying the verotoxin 2 genes of the enterohemorrhagic Escherichia coli O157:H7 derived from the Sakai outbreak. Genes Genet. Syst. 74:227-239. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNamara, B. P., and M. S. Donnenberg. 1998. A novel proline-rich protein, EspF, is secreted from enteropathogenic Escherichia coli via the type III export pathway. FEMS Microbiol. Lett. 166:71-78. [DOI] [PubMed] [Google Scholar]
- 29.McNamara, B. P., A. Koutsouris, C. B. O'Connell, J. P. Nougayrede, M. S. Donnenberg, and G. Hecht. 2001. Translocated EspF protein from enteropathogenic Escherichia coli disrupts host intestinal barrier function. J. Clin. Investig. 107:621-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 32.Moon, H. W., S. C. Whipp, R. A. Argenzio, M. M. Levine, and R. A. Giannella. 1983. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect. Immun. 41:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mundy, R., D. Pickard, R. K. Wilson, C. P. Simmons, G. Dougan, and G. Frankel. 2003. Identification of a novel type IV pilus gene cluster required for gastrointestinal colonization of Citrobacter rodentium. Mol. Microbiol. 48:795-809. [DOI] [PubMed] [Google Scholar]
- 34.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newman, J. V., B. A. Zabel, S. S. Jha, and D. B. Schauer. 1999. Citrobacter rodentium espB is necessary for signal transduction and for infection of laboratory mice. Infect. Immun. 67:6019-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perna, N. T., G. Plunkett, 3rd, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 38.Phillips, A. D., S. Navabpour, S. Hicks, G. Dougan, T. Wallis, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer's patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schauer, D. B., and S. Falkow. 1993. Attaching and effacing locus of a Citrobacter freundii biotype that causes transmissible murine colonic hyperplasia. Infect. Immun. 61:2486-2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schauer, D. B., and S. Falkow. 1993. The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun. 61:4654-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlosser-Silverman, E., M. Elgrably-Weiss, I. Rosenshine, R. Kohen, and S. Altuvia. 2000. Characterization of Escherichia coli DNA lesions generated within J774 macrophages. J. Bacteriol. 182:5225-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tacket, C. O., M. B. Sztein, G. Losonsky, A. Abe, B. B. Finlay, B. P. McNamara, G. T. Fantry, S. P. James, J. P. Nataro, M. M. Levine, and M. S. Donnenberg. 2000. Role of EspB in experimental human enteropathogenic Escherichia coli infection. Infect. Immun. 68:3689-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tauschek, M., R. A. Strugnell, and R. M. Robins-Browne. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol. Microbiol. 44:1533-1550. [DOI] [PubMed] [Google Scholar]
- 44.Tu, X., I. Nisan, C. Yona, E. Hanski, and I. Rosenshine. 2003. EspH, a new cytoskeleton-modulating effector of enterohaemorrhagic and enteropathogenic Escherichia coli. Mol. Microbiol. 47:595-606. [DOI] [PubMed] [Google Scholar]
- 45.Tzipori, S., F. Gunzer, M. S. Donnenberg, L. de Montigny, J. B. Kaper, and A. Donohue-Rolfe. 1995. The role of the eaeA gene in diarrhea and neurological complications in a gnotobiotic piglet model of enterohemorrhagic Escherichia coli infection. Infect. Immun. 63:3621-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, R. K., R. K. Shaw, S. Daniell, S. Knutton, and G. Frankel. 2001. Role of EscF, a putative needle complex protein, in the type III protein translocation system of enteropathogenic Escherichia coli. Cell Microbiol. 3:753-762. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, C., T. S. Agin, S. J. Elliott, L. A. Johnson, T. E. Thate, J. B. Kaper, and E. C. Boedeker. 2001. Complete nucleotide sequence and analysis of the locus of enterocyte effacement from rabbit diarrheagenic Escherichia coli RDEC-1. Infect. Immun. 69:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]