Abstract
Detection of proteins and nucleic acids is dominantly performed using optical fluorescence based techniques, which are more costly and timely than electrical detection due to the need for expensive and bulky optical equipment and the process of fluorescent tagging. In this paper, we discuss our study of the electrical properties of nucleic acids and proteins at the nanoscale using a nanoelectronic probe we have developed, which we refer to as the Nanoneedle biosensor. The nanoneedle consists of four thin film layers: a conductive layer at the bottom acting as an electrode, an oxide layer on top, and another conductive layer on top of that, with a protective oxide above. The presence of proteins and nucleic acids near the tip results in a decrease in impedance across the sensing electrodes. There are three basic mechanisms behind the electrical response of DNA and protein molecules in solution under an applied alternating electrical field. The first change stems from modulation of the relative permittivity at the interface. The second mechanism is the formation and relaxation of the induced dipole moment. The third mechanism is the tunneling of electrons through the biomolecules. The results presented in this paper can be extended to develop low cost point-of-care diagnostic assays for the clinical setting.
INTRODUCTION
Direct electrical detection of biomolecules without the need for any labeling can help greatly advance point-of-care diagnostics. Applications include the study of virology,1, 2, 3 ligand fishing,4, 5 bacteriology,6, 7, 8 apoptosis,9 cell biology and adhesion,10, 11 epitope mapping,12, 13, 14 signal transduction,15, 16 immune regulation,17 nucleic acid–nucleic acid interactions,18, 19, 20 and nucleic acid–protein protein interactions,21, 22 and study of post-translational modifications.23, 24 Detection of proteins and nucleic acids is often performed using optical fluorescence based techniques, which are more costly and timely than electrical detection due to the need for expensive and bulky optical equipment and the process of fluorescent tagging. Thus, a robust label-free electrical detection technique can provide for a promising solution in lowering both reagent costs and instrumentation costs. Optical detection of nucleic acids in miniaturized systems is also challenging because the signal originates from dye molecules in solution, and thus the strength of the signal scales with sample volume. Therefore, there is a direct conflict between the goals of obtaining a strong optical signal and reducing reagent consumption in a microfluidic system. Furthermore, optical readout requires that PCR product markers such as Sybr Green and Taqman probes be added to the reagents. This process of labeling and adding reagents makes this process of real time PCR unsuitable as a point-of-use diagnostic technique in the clinical setting.
Protein detection is typically performed using the sandwich ELISA technique, which involves several steps of incubating test sample, then a polyclonal antibody, and then finally a secondary antibody tagged with a fluorescent or luminescent label, with several wash steps in between. A label-free technique, which could directly detect the binding of a target protein to the surface antibody would be much more suitable as a point of care diagnostic. Various label-free nanoelectronic25, 26, 27, 28 sensors including nanowires have been demonstrated exhibiting femtomolar detection limits.29
The detection limit or the minimum detectable concentration of target biomarkers in the test sample is dependent on two parameters: the transducer sensitivity (the minimum number of binding events on the sensor surface required to generate sensor response greater than the noise level) and the capture rate of target molecules on the surface of the sensor. Both of these parameters are affected by the flow rate, diffusion time of target molecules, and the sensor geometry. In general, most of the electrical impedance based biosensors suffers from low transducer signal to noise ratio due to various noise processes in the system such as flicker (1/f), Johnson-Nyquist noise, and also the noise resulting from the amplification circuitry. The contribution of these noise sources are relatively higher at frequencies below 100 Hz where electrical impedance measurements are typically made, since the desired signal to be measured results from modulation of the double layer as target biomarkers bind to the sensor surface.
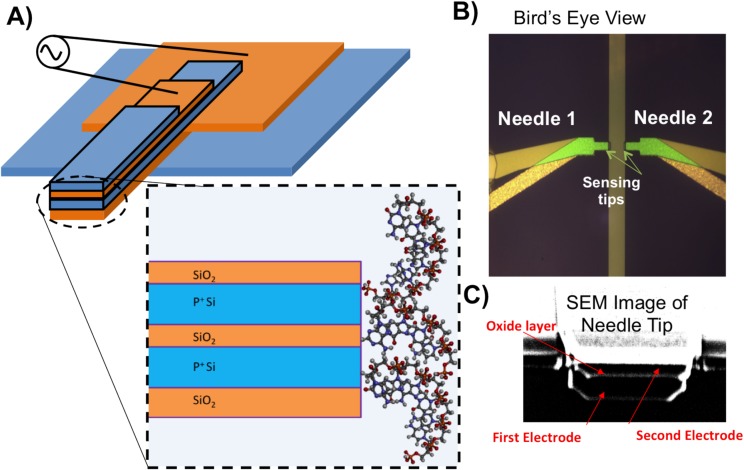
To overcome these various problems mentioned, we propose a novel method of directly measuring the electrical response of the DNA and protein molecules of interest at the nanoscale by using a novel ultra-sensitive, real time, label-free sensing platform, which we refer to as the nanoneedle biosensor. The nanoneedle biosensor structure consists of three thin-film layers, as shown in Fig. 1. There are two conductive layers with an insulator layer in between. The interface of this middle oxide layer with the electrolyte is the active region of the sensor. A protective oxide layer is deposited above the topmost electrode. Underneath the bottom electrode, there is an oxide layer, which is thermally grown insulating the first electrode from the substrate.
Figure 1.
(a) Schematic of nanoneedle biosensor three-dimensional and side view of horizontal nanoneedles (Not to Scale). (b) Optical micrograph of bird's eye view of aluminum-polysiliconhybridnanoneedle biosensor. (c) SEM image of the tip of a nanoneedles biosensor; 1 & 3 are the electrodes; 2 is the oxide in between the electrodes
One of the advantages of this sensor is the ability to directly measure biomolecular binding as a function of time (real-time). The presence of biomolecules in the active sensing region of the needle tip results in modulation of the measured impedance in real time. This can be useful for measuring kinetic constants for various biomolecular species. Also, transducer sensitivity is improved with the nanoneedle biosensor compared to standard micro-electrode label-free impedance sensors due to miniaturization of the nanoneedle tip, resulting in high sensitivity for detection of small numbers of molecules. The sensing area of this sensor is a nano-sized area, which is in the size range of biomolecules of interest. As a result, a small number of binding events in the active sensing area is sufficient to modulate the impedance at the sensor tip to a level greater than the noise resulting in high sensitivity. In addition, the suspended geometry of a nanoneedle in a micro-channel results in diffusion taking place in three dimensions unlike most electrical biosensors which have a planar structure(two dimensional) thus diffusion taking place in only two dimensions. This results in a higher hit rate of target molecules to the probe molecules on the sensor surface and thus a faster detection platform.
Another advantage of the nanoneedles biosensor, due to its rigid yet high aspect ratio solid-state structure, is the ability to measure protein and nucleic acid levels directly in-vivo inside a living cell. A thin functionalized needle can be inserted into a living cell, and impedance measurements can be made directly as proteins bind to the needle surface. This can be used for many different applications such as measuring protein expression for the purposes of drug screening.
In addition to the above-mentioned advantages, the ability to fabricate an array of needles onto a substrate potentially enables high throughput sensing. Fabrication of an array of nanoneedles with the state-of-the-art nanofabrication techniques makes it possible to monitor various binding events simultaneously in over a large area. On chip integration of the sensors with CMOS amplification electronics can further improve the signal to noise ratio of the sensor resulting in a single portable device suitable for point-of-care diagnostics.
DEVICE DESIGN FACTORS
Various thicknesses and geometrical designs have been fabricated and tested. The sensor design used in this study consists of electrodes 100 nm thick and a middle oxide layer 30 nm thick. The top protective oxide layer thickness is 20 nm and the bottom oxide layer thickness is 250 nm. The width of the nanoneedle tip is 5 μm. For specificity, probe molecules (e.g., DNA molecule or protein) can be immobilized on the tip of the nanoneedles. The binding of target molecules to the probe molecules modulates the impedance between the electrodes. In order to maximize the effect of the impedance at the interface of the sensor compared to the total measured impedance, it was necessary to reduce the parasitic impedance resulting from the resistance of the trace and also the body capacitance of the traces leading from the bonding pad to the sensor. To achieve this goal, the sensor has an aluminum/polysilicon hybrid structure, where the sensor electrode is polysilicon, and the trace leading up to it is aluminum, which has a higher conductivity than polysilicon and can minimize the trace resistance leading up to the sensor. And also the pads are separated out from each other to eliminate the body capacitance between the traces. However, the sensing area will still be polysilicon in order to ensure compatibility with surface chemistries optimized for high immobilization capacity of probe molecules.
RESULTS AND DISCUSSIONS
In order to demonstrate the utility of the nanoneedle for label-free sensing while maintaining a high signal-noise ratio we studied the electrical response of the sensor for various types of biological agents. We tested the electrical response of two types of biomolecules: nucleic acids and proteins. In this manuscript, first we present the results of the experimental characterization of the device, then we discuss the theory behind the response and discuss the various physical mechanisms involved, and afterwards we demonstrate the utility of the sensor for affinity based protein sensing.
Nucleic acids
We initially studied the electrical response of nucleic acids using the nanoneedle sensor. As observed in Fig. 2a, the presence of single stranded DNA (20 base pairs long) modulates the measured impedance. Various concentrations of DNA were injected onto our sensor surface sequentially. Between every step that DNA was added we aspirated the measurement well. The DNA is in free solution and unlikely to adsorb to the surface. As the concentration of DNA in the solution decreases, the measured impedance increases getting closer and closer to the baseline value. Fig. 2b shows the measurement performed with error bars. We acknowledge, however, that during the aspiration step it is possible that residues of DNA remain on the substrate resulting in the actual concentration being somewhat higher than the concentration we injected. For example, for the final measured concentration (0.6 fM which equates to 1800 DNA molecules in 5 μl of volume), it is possible that the actual number of molecules is somewhat higher. This observation of increase in conductivity with higher DNA concentration was counter intuitive and the results are contrary to the observations traditionally made with electrical impedance biosensors where the presence of DNA generally results in an increase in impedance as a result of modulation of the double layer.30 We will discuss the mechanisms affecting the modulation of the impedance level in Sec. 4; however, in order to confirm the behavior of the sensor even further, we also performed a next set of experiments with proteins.
Figure 2.
Presence of single stranded DNA modulates the measured impedance. (a) Various concentrations of DNA were injected onto our sensor surface sequentially. Between every step that DNA was added we dried out the measurement well. As the concentration of DNA in the solution decreases, the measured impedance increases getting closer and closer to the baseline value. (b) Impedance change plotted with error bars over three measurements per point.
Proteins
For protein experiments, we measured the electrical response of unconjugated streptavidin in free solution (Fig. 3). Since streptavidin has an isoelectric point of 4, and silicon oxide has an isoelectric point of 3, both will have a negatively charged surface in water, thus minimizing adsorption of molecules to the surface. We measured the impedance across the electrodes as varying concentrations of streptavidin was injected onto the sensor surface. Fig. 3a shows representative results of this experiment, which was performed three times. The behavior of streptavidin is similar to the electrical response of the DNA. Higher concentration of protein resulted in lower levels of impedance between the electrodes. Again, similar to our results with DNA, this goes contrary to the behavior of traditional micro-electrode sensors where the presence of biomolecules at the surface results in an effective decrease in the double layer capacitance, thus resulting in an increase in impedance.30
Figure 3.
Presence of non-adsorbed streptavidin protein modulates the measured impedance. (a) Various concentrations of streptavidin were injected onto our sensor surface sequentially: (1) water, (2) 250 ng/ml streptavidin, (3) 25 ug/ml streptavidin, (4) 25 mg/ml streptavidin. Between every step that protein was added we dried out the measurement well. As the concentration of protein in the solution decreases, the measured impedance increases getting closer and closer to the baseline value. (b) Impedance response of polystyrene beads injected onto sensor resulting in increase in impedance. (c) Impedance change plotted with error bars over three measurements per point.
Polystyrene beads
As a control experiment to verify the physical mechanism resulting in the electrical response of the sensor we injected polystyrene beads. Polystyrene beads have fully insulative electrical properties, and also a lower dielectric constant (∼2.6) compared to water (∼80). As shown in Fig. 3b) an increase in impedance was observed as opposed to the decrease in impedance seen with injection of proteins and DNA. Fig. 3c shows the impedance measurements repeated several times with error bars included. The presence of the beads on the sensor surface results in both an increase in resistance across the electrodes, and also a decrease in the sensor surface capacitances, all of which contribute to an increase in impedance. This behavior is in line with that of traditional micro-scale electrodes. This implies that the charge and the relative dielectric constant of the biomolecules play an important role in the behavior observed for nucleic acids and proteins.
To reconcile this contradiction of our protein/nucleic acid results with the traditional micro-sensors, we theoretically and experimentally characterized the various parasitic components of our circuit model at the sensor-electrolyte interface in our system.
MODELING
To understand the results, we developed a full circuit model to characterize the interface of the nanoneedle sensor with the electrolyte as shown in Fig. 4a. In the full model, our system contains several parasitic impedance components. From here on, we are referring to the parasitic impedances at the electrode-electrolyte interface and the electrolyte itself (rather than the components of the body of the sensor). Above the electrodes and the insulator, we have a double layer resulting from accumulation of ions in the electrolyte at each surface (metal and oxide). The electrical double layer consists of two layers, the stern layer, which is an adsorbed fixed layer, and also a diffuse layer. The stern layer consists of the ions that are adsorbed to the surface and are estimated to be concentrated roughly 1 nm from the sensor surface. The diffuse layer results because of the condition of charge neutrality. That is, another layer of ions accumulates in order to neutralize the charge in the stern layer. The thickness of the stern layer is generally concentration independent, whereas that of the diffuse layer is related inversely to the electrolyte concentration. The stern layer and diffuse layer can be represented as two capacitors Cads and Cdiff
| (1) |
where z is the ion valence number, A is a constant, Ci is the concentration of the ionic solution (mol/liter), e the charge of an electron (1.6 × 10−19), NA is Avogadro's number, εε0 is the absolute dielectric constant of the sample, k is the Boltzmann constant, and T is the absolute temperature. The Cdl is the result of both Cads and Cdiff which are connected in series
| (2) |
At high salt concentrations (>10 mM), the diffuse layer size becomes comparable to that of the stern layer. However, when the salt concentration is low (<0.01 mM), the diffuse layer increases in size to be on the order of 10 nm,31 thus the capacitance becomes smaller than the stern layer capacitance and thus dominates the double layer capacitance. Based on the fact that we are using DI water as our electrolyte, the double layer is estimated to be on the order of 10 nm. This has several implications for our model. The first parasitic impedance is an interface resistance and a capacitance in between the electrodes, which we refer to as the wall capacitance and resistance (Cw and Rw). This is resulting from the fringing field between our electrodes, which extends into our electrolyte on a similar length scale as our double layer. The out of phase response affects Cw and the in-phase response affects Rw. The tunneling of electrons from the conductive electrodes into the biomolecules present at the active sensing region also affects the wall resistance (Rw). In parallel to these two components, we have another loop, which consists of the double layer capacitance (Cdl) at the surface of each metal electrode. In parallel to the double layer capacitance is the faradaic resistance (impedance) or the electron transfer resistance (Rf). This results from tunneling of electrons from the electrodes into the bulk electrolyte. In series with this is the bulk solution resistance (Rs), which results from the resistivity of the electrolyte.
Figure 4.
Circuit model of the nanoneedle sensor-electrolyte interface. (a) Full model where CW represents the fringing capacitance at the sensor interface. Rw represents the resistance across the double layer (on top of the insulator) between the electrodes. Cdl represents the double layer capacitance on each electrode surface. Rf represents the tunneling resistance or the electron transfer resistance from the electrode into the bulk solution. Rb represents the bulk resistance of the electrolyte. Rtr represents the trace resistance of the electrode leading up to the bonding pads. Cb represents the body capacitance between the electrodes along the body of the sensor. (b) Simplified model which is valid at f = 15 kHz.
In the experimental discussion section, we observed that the presence of biomolecules results in a decrease in the impedance across the electrodes. Since the spacing of the sensing electrodes is only 30 nm apart, the electrical response of the protein and DNA molecules measured using our sensor is due to three sources. The first is due to change in the relative permittivity at the sensor interface, which affects the imaginary component of the measured impedance. The presence of the biomolecules results in an increase in relative dielectric permittivity (κεsol) of the solution at the sensor interface, thus resulting in an increase in capacitance and thus a decrease in impedance measured. The second is due to charge transport, which affects the real component of the measured impedance and is due to two different mechanisms. One is the formation and relaxation of the induced dipole moments. The mobile charges in and around the biomolecules allows for a dipole to be induced in the biomolecules when undergoing an AC field. The second mechanism is the tunneling of electrons from the electrodes to and through the biomolecules. The three effects are elaborated in more detail below.
Capacitance modulation
The dominant phenomenon, which affects the out of phase (imaginary) AC response of the impedance, is the change in the wall capacitance at the sensor interface (Cw), which is defined as
| (3) |
where K is a geometrical factor of the electrode structures.32 The κεsol is the relative dielectric permittivity of the solution and is explicitly affected by DNA dipole moment (DNA length and charge) and concentration of the DNA. The changes in the extracted wall capacitance Cw, which represents the DNA capacitance due to the movement of the counterioncharges around the DNA backbone (dielectric relaxation of DNA dipoles), varied with changes in concentration of our DNA solutions. Changes in wall capacitance result in change in the imaginary component of the measured impedance.32 Similar mechanisms are at play for proteins resulting in a decrease in impedance as protein concentration increases at the electrode surface.
AC coupling
The second dominant mechanism affecting change in impedance is the decrease in the resistance at the sensor interface (Rw). This is the dominant mechanism, which results in a change to the real part of the impedance. To understand the electrical response of polyelectrolyte molecules such as DNA, it helps to understand the structure electronically. Polyelectrolyte molecules display a range of complex behaviors due to their connectivity, steric conformation, and strong electrostatic interactions. The backbone of DNA is negatively charged due to the phosphate backbone. The fixed charges on the polyelectrolyte and their electrostatic interactions with counterions are of great significance. The negatively charged surface of the DNA results in accumulation of positively charged counterions into a condensed layer. This accumulation has been previously described using counterion condensation theory33 and also Manning-Oosawa (MO) counterion condensation theory.34 The DNA counterions, free as well as condensed, contribute to the oscillating polarization in the applied electric field and thus determine the dielectric response of the DNA solution.
When an AC field is applied, polarization is induced and a flow of net charge resulting from the DNA counterions35 occurs. Diffusion controls the movement of counterions, thus the dielectric response is dictated by the mean relaxation time,
| (4) |
where Din is the diffusion constant and L is the length scale.36, 37
For pure water DNA solutions, the characteristic length L at low frequencies increases with decreasing DNA. In our view, this result together with the fact that the relaxation at low frequencies happens at the length scale of the average size of the polyelectrolyte chain suggests that low frequency relaxation engages mostly condensed counterions along and in close vicinity of the chain. This means that at least some of the free counterions join the relaxation of the condensed counterions along the segments of the same chain. It is thus impossible to completely separate condensed counterions from free counterions in their contribution to the LF relaxation mode. Dielectric data from previous work35 indicate that at low frequencies the decoupling of MO condensed and free DNA counterions are both responsible for the response, in addition to the counterions along a single DNA chain.
In summary, the electrical response of DNA and protein molecules in solution under an applied alternating electric field stems from the formation and relaxation of the induced dipole moment.38 Since there are mobile charges in and around the biomolecule forming a sheath of ions around the backbone of the molecule, a dipole can be induced in the DNA or protein when electrically probed in solution. As a result by increasing protein or DNA concentration a higher number of counter-ions will be attracted by the backbone charge. The increased number of mobile counter-ions can increase the conductivity of the solution.
Tunneling current modulation
The third mechanism resulting in a change in the real part of the impedance is the increase in tunneling current at the sensor interface (Rf and Rw). Two approaches have been used in investigating delocalization of electrons in polynucleotides for a DNA molecule. One approach involves approximating the backbone of the DNA molecule to a periodic system with nucleotide bases superimposed on top. Another approach developed by Brillouin (1962)39 suggested that nucleotide bases play the role of impurities in the band gap (formed by sugar-phosphate backbone). The assumption of periodicity of the sugar-phosphate backbone allows the DNA molecule to be treated as a semiconductor. Using Brillouin's approach, Suhai (1974)40 showed that electron transport is equally probable both along the sugar-phospate backbone and through the system of bases themselves. Also, comparison of various relative energy bands showed that a charge transfer interaction in DNA could also occur between the poly (bases-pairs) and the backbone. This results in conducting holes in the base pairs and conducting electrons in the DNA backbone.
Transfer of electrons in DNA occurs along the axis of the molecule. There are four basic types of transfer including extra electrons, singlet and triplet π-type molecular excited states, and holes. The delocalized electrons not participating in bonding are in the π-orbitals and are free to move about the carbon nuclei in a molecule. The σ-orbitals are symmetric around the bond axis with localized C-C and C-H bonds. The electrons participating in these bonds are around the line joining the two nuclei and are localized. The nucleotide bases have an electronic π system, exhibiting delocalized electrons. The excited electrons reside in π-type orbitals, which lie low.41, 42, 43, 44, 45 Transfer of electrons can occur from nucleotide to nucleotide given that the nucleotide bases are stacked in proximity. Experimental evidence shows support for both short (less than five bases) and long range tunneling effects. No significant evidence has been found to indicate soliton effects play a role.46 More recently, examples of long range tunneling between intercalated reagents (separated by up to 40 A) have been shown.42, 47, 48, 49, 50 The rate of electron transfer in these studies showed weak dependence on the number of interspersed nucleotides.
Similar effects are seen in proteins. Electrons travel as they tunnel from donor sites to acceptor sites through the various pathways of electronic transport. On the theoretical side, there are two pathways for electronic transport in proteins. The first one is the hydrogen-bonded network
This network runs perpendicular to both α-helical and β-pleated sheet structures and provides an extended electron conjugated pathway. The other pathways for electronic transport in proteins are along the main polypeptide chain:
 These groups of the polypeptide chain can shape the elementary unit cell with different R groups that act as impurity centers. Although this charge transportation in the protein molecules occurs through the hydrogen-bonded crosslinks, it mainly occurs in the main polypeptide chains.51, 52
These groups of the polypeptide chain can shape the elementary unit cell with different R groups that act as impurity centers. Although this charge transportation in the protein molecules occurs through the hydrogen-bonded crosslinks, it mainly occurs in the main polypeptide chains.51, 52
IMPEDANCE CHARACTERIZATION
We are able to empirically determine the contribution of these various parasitic components by measuring the impedance spectrum (Fig. 5) of the sensor and extracting the parasitic impedance components from the impedance curve. In Figure 5, we have shown the spectrum for both the bare sensor in water and also in streptavidin solution. As seen in the figure, the largest difference between the sensor response in water and streptavidin solution occurs between 1 and 100 kHz, thus we chose 15 kHz as our operating frequency (250 mV RMS AC signal). We performed our sensing measurements using a potentiostat (Versa STAT3). For all of our measurements, the sampling rate was one sample per 4 s. Given that the double layer is 10 nm thick, we can estimate the impedance resulting from the double layer capacitance (Cdl) to be ∼1 GΩ at 15 kHz. We can also estimate the faradaic impedance (due to tunneling of electrons from the electrodes to the electrolyte) by looking at the measurement at low frequencies (1 Hz). The total impedance at 1 Hz is 0.5 GΩ, meaning that Rf which is frequency independent can be no more than 0.5 GΩ regardless of the frequency. This means that the equivalent impedance of Cdl and Rf in parallel with each other has to also be greater than 0.5 GΩ. Comparing this to the total impedance of the sensor at 15 kHz which is 3.6 MΩ, we are able to assume that the loop containing Cdl and Rf is essentially an open circuit allowing us to simplify our model significantly as shown in Fig. 4b. Now that we have significantly simplified the model to Cw and Rw in parallel, we can estimate what percentage of the measured real and imaginary impedance comes from which source. The change in the real component of the impedance (data measured but not shown) is influenced by two factors. The major change results from the modulation of the wall resistance (Rw), which is affected by the AC coupling resulting from the induced dipole moment of the molecules. The other effect is the modulation of the tunneling current from the electrodes to the biomolecules, which again affects the wall resistance (Rw). However, this change resulting from tunneling is minor compared to the AC coupling affect as we discussed based on the measured impedance value at f = 1 Hz. Based on impedance measurements, the ratio of the real component of the impedance at 15 KHz to the real component of the impedance at 1 Hz is 0.008 (0.8%). Thus at least 99.2% of the change in the real component of the impedance results from modulation of the AC coupling component of Rw, while the remaining 0.8% is due to modulation of the tunneling current from the electrode to the biomolecules. The change in the imaginary component (data measured but not shown) on the other hand is fully dominated by modulation of the wall capacitance (Cw), which is affected by the increase in relative dielectric permittivity (κεsol) of the solution at the sensor interface due to the presence of the proteins or nucleic acids. As mentioned the relative dielectric permittivity (κεsol) is affected by both the dipole moment and concentration of the biomolecules. Based on the real and imaginary components of real time data (Table TABLE I.), the imaginary component of the measured impedance drops 84.7% compared to the baseline (water) when 1 μmolar DNA is injected, whereas the real component drops 40.8% compared to the baseline (water).
Figure 5.
Plot of the electrical impedance spectrum across the device from f = 1 Hz all the way to f = 1 MHz. The magnitude of the impedance is shown for both water and streptavidin solution.
TABLE I.
Average real and imaginary impedance values for experiments in Figure 2 for steps with DI water and 1 μM Oligo.
| Sample | ZImaginary | ZReal |
|---|---|---|
| DI water | 1.67 × 106 Ω | 137 × 103 Ω |
| 1 μM Oligo (20 base pair) | 0.254 × 106 Ω | 81 × 103 Ω |
| % Drop in impedance (oligo relative to DI water) | 84.7% | 40.8% |
AFFINITY BASED BIOSENSING
After fully characterizing the sensor behavior in the presence of proteins and nucleic acids and having a full understanding of the mechanisms affecting the behavior of the sensor, we proceeded to demonstrate the utility of this sensor in affinity biosensing. Thus, we performed another set of experiments in order to demonstrate the selectivity of this sensor in detecting target proteins. We adsorbed biotinilated BSA on the surface and tested the binding of streptavidin. Biotin-streptavidin binding is one of the strongest non-covalent biochemical interactions. Streptavidin is a tetramer material which has subunits arrayed in D2 symmetry. Each protomer is an 8-stranded beta-barrel, which has a simple up-down topology. Biotin molecules can be bound at one end of any of these barrels. In order to confirm the adsorption of biotinilated BSA on the surface, we injected streptavidin coated beads on the surface and verified the binding optically (Figure 6a). We performed a series of experiments where we physically adsorbed biotinilated BSA (250 mg/ml) on the surface of the nanoneedle sensor, which resulted in a drop in impedance (Fig. 6b). A wash step was then performed where we washed the sensor surface with water, which increased the impedance of the sensor. We then injected streptavidin (5 mg/ml), which resulted in a decrease in impedance, which was followed by a wash step afterwards, which again resulted in an increase in impedance. The final impedance levels after both wash steps (after biotinilated BSA adsorption and after streptavidin binding) are compared with each other to quantify the amount of protein bound to the sensor surface. In Fig. 6c, we show the error bars of our impedance measurements.
Figure 6.
(a) Optical image of streptavidin beads bound to nanoneedle sensor coated with biotinilated BSA. (b) Representative results of real-time measurement of impedance as (1) sensor is covered with water (2) biotinilated BSA (250 mg/ml) is physically adsorbed on the surface of the nanoneedle sensor, which results in a drop in impedance. (3) The sensor surface is washed with water resulting in increase in impedance. (4) Streptavidin (5 mg/ml) was injected in a decrease in impedance, (5) followed by a wash step afterwards, resulting in an increase in impedance. The final impedance levels after both wash steps (after biotinilated BSA adsorption and after streptavidin binding) are compared with each other to quantify the amount of protein bound to the sensor surface. (c) Impedance change plotted with error bars over three measurements per point. ΔZ1 represents the difference in impedance of steps (5) and (3) for the experiments where biotinalated BSA was immobilized on surface. ΔZ2 represents the difference in impedance of steps (5) and (3) for the experiments where unconjugated BSA (control experiment) was immobilized on surface. (d). Control experiments where representative results of real-time measurement of impedance where (1) sensor is covered with water then (2) unconjugated BSA (250 mg/ml) is adsorbed to the surface resulting in a partial drop in impedance. (3) The sensor surface was washed with water to wash out the unbound BSA molecules resulting in an increase in impedance. (4) Streptavidin (5 mg/ml) was injected onto the sensor surface resulting in a decrease in impedance similar to the positive experiment. (5) The sensor surface is washed. The difference in electrical response of the control experiment and positive experiment lies in the final wash step which results in increasing the impedance level all the way back up to the original baseline level where only BSA was immobilized.
Control experiments were performed (Fig. 6d) where instead of biotinilated BSA, unconjugated BSA (250 mg/ml) is adsorbed to the surface resulting in a partial drop in impedance. The sensor surface was washed with water to wash out the unbound BSA molecules resulting in an increase in impedance. Streptavidin (5 mg/ml) was injected onto the sensor surface resulting in a decrease in impedance similar to the positive experiment. The difference in electrical response of the control experiment and positive experiment lies in the final wash step which results in increasing the impedance level all the way back near the original baseline level where only BSA was immobilized. This set of experiments confirmed the ability to use our sensor for affinity based biosensing, giving us hope to use this type of sensor in the future for multiplexed detection of a wide variety of proteins, where instead of biotinilated BSA we would immobilize a probe antibody for the protein of interest.
CONCLUSION
Direct electrical detection of nucleic acids and proteins using nanoneedle sensors has been demonstrated. Instead of resulting in an increase in impedance due to blockage of ionic current passing between the nano-electrodes, the attachment of proteins and nucleic acids results in a decrease in impedance which we attribute to three main mechanisms: the increase in relative dielectric permittivity at the active sensing region, an increase in AC coupling current due to an increase in induced dipole moments, and also the increase of tunneling current passing between the electrodes to the biomolecules present in the active sensing region. The work performed in this study can be extended to various biological applications such as the study of binding affinities of protein-protein interactions, multiplexed protein detection, real-time PCR for point of care diagnostics, and point of care proteomic assays. Further work can be performed to develop new optimized structures to improve signal-to-noise ratio for probing low abundance biomarkers. Also, as mentioned, given the appropriate form factor of this type of sensor, one can envision injecting the needle inside a cell to perform real time in-vivo measurement of protein expression. We believe that our nanoneedle sensor results strongly motivate such effort, given the advantages of this sensor over other labeled and label-free sensing schemes. This work provides a strong starting point for a new class of electronic biosensing devices with the capability of rapid direct large-scale integration.
METHODS
Device fabrication
We performed the following steps in order to fabricate the biosensor devices. Starting out with an undoped silicon wafer, first, 250 nm of silicon oxide was thermally grown on a silicon substrate. This was followed by the deposition of 100 nm of poly-silicon using low-pressure chemical vapor deposition (LPCVD) by using the first mask. It was then doped with phosphorus to achieve a sheet resistance of 210 Ω per square. A 30 nm-thick SiO2 layer was thermally grown on the bottom p+-silicon layer. Then the top electrode was pattern by using the second mask. It was followed by deposition of 100 nm p+-silicon layer using LPCVD following by phosphorus doping to achieve the same conductivity as the bottom electrode. Then, there was a dry etching step followed by a lift off to pattern and makes the aluminum traces by using two new masks. It was followed by the deposition of 200 nm SiO2 layer of the p+-silicon layer and aluminum traces using plasma enhanced chemical vapor deposition (PECVD). We patterned the nanoneedle bundles by etching the 200 nm top-SiO2layer; 100 nm top p+-silicon layer, and 30 nm middle oxide layer,100nmp+-silicon layer down to bottom oxide layer.53, 54, 55, 56, 57 We performed wet etch step to etch out the channel below the bundle of nanoneedles. Afterwards, we performed another etch step to expose the bonding pads to allow wire bonding.
Protein sample preparation
We prepared our protein samples as discussed below. First, we diluted our protein samples in DI water to achieve our desired concentrations. After every dilution step we used a vortex for 30 s to fully mix the contents of the epindorph tube to ensure uniformity. Each sample was prepared approximately 3 ∼ 5 min before than the injection to the well.
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health Grant No. P01HG000205.
References
- Inoue K., Arai T., and Aoyagi M., Biol. Pharm. Bull. 22, 210 (1999). 10.1248/bpb.22.210 [DOI] [PubMed] [Google Scholar]
- B. M.McDermott, Jr., Rux A. H., Eisenberg R. J., Cohen G. H., and Racaniello V. R., J. Biol. Chem. 275, 23089–23096 (2000). 10.1074/jbc.M002146200 [DOI] [PubMed] [Google Scholar]
- Xing L., Tjarnlund K., Lindqvist B., Kaplan G. G., Feigelstock D., Cheng R. H., and Casasnovas J. M., EMBO J. 19, 1207–1216 (2000). 10.1093/emboj/19.6.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catimel B., Weinstock J., Nerrie M., Domagala T., and Nice E., J. Chromatogr. A 869, 261–273 (2000). 10.1016/S0021-9673(99)01098-5 [DOI] [PubMed] [Google Scholar]
- Guermazi S., Regnault V., Gorgi Y., Ayed K., Lecompte T., and Dellagi K., Blood Coagul. Fibrinolysis 11, 491–498 (2000). 10.1097/00001721-200007000-00012 [DOI] [PubMed] [Google Scholar]
- Charalambous B. M. and Feavers I. M., FEMS Microbiol. Lett. 191, 45–50 (2000). 10.1111/j.1574-6968.2000.tb09317.x [DOI] [PubMed] [Google Scholar]
- Chen H., Clayton A., Wang W., and Sawyer W., Eur. J. Biochem. 268, 1659–1669 (2001). 10.1046/j.1432-1327.2001.02039.x [DOI] [PubMed] [Google Scholar]
- Elliott J. L., Mogridge J., and Collier R. J., Biochemistry 39, 6706–6713 (2000). 10.1021/bi000310u [DOI] [PubMed] [Google Scholar]
- Uegaki K., Otomo T., Sakahira H., Shimizu M., Yumoto N., Kyogoku Y., Nagata S., and Yamazaki T., J. Mol. Biol. 297, 1121–1128 (2000). 10.1006/jmbi.2000.3643 [DOI] [PubMed] [Google Scholar]
- Andersen O. M., Christensen L. L., Christensen P. A., Sørensen E. S., Jacobsen C., Moestrup S. K., Etzerodt M., and Thøgersen H. C., J. Biol. Chem. 275, 21017–21024 (2000). 10.1074/jbc.M000507200 [DOI] [PubMed] [Google Scholar]
- Holaska J. M., Black B. E., Love D. C., Hanover J. A., Leszyk J., and Paschal B. M., J. Cell Biol. 152, 127–140 (2001). 10.1083/jcb.152.1.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achen M. G., Roufail S., Domagala T., Catimel B., Nice E. C., Geleick D. M., Murphy R., Scott A. M., Caesar C., and Makinen T., Eur. J. Biochem. 267, 2505–2515 (2000). 10.1046/j.1432-1327.2000.01257.x [DOI] [PubMed] [Google Scholar]
- Jokiranta T. S., Hellwage J., Koistinen V., Zipfel P. F., and Meri S., J. Biol. Chem. 275(36 ), 27657–27662 (2000). 10.1074/jbc.M002903200 [DOI] [PubMed] [Google Scholar]
- Vogel M., Miescher S., Kuhn S., Zürcher A. W., Stadler M. B., Ruf C., Effenberger F., Kricek F., and Stadler B. M., J. Mol. Biol. 298, 729–735 (2000). 10.1006/jmbi.2000.3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellson C. D., Gobert-Gosse S., Anderson K. E., Davidson K., Erdjument-Bromage H., Tempst P., Thuring J. W., Cooper M. A., Lim Z.-Y., and Holmes A. B., Nat. Cell Biol. 3, 679–682 (2001). 10.1038/35083076 [DOI] [PubMed] [Google Scholar]
- Gaullier J.-M., Rønning E., Gillooly D. J., and Stenmark H., J. Biol. Chem. 275, 24595–24600 (2000). 10.1074/jbc.M906554199 [DOI] [PubMed] [Google Scholar]
- Ablamunits V., Henegariu O., Hansen J. B., Opare-Addo L., Preston-Hurlburt P., Santamaria P., Mandrup-Poulsen T., and Herold K. C., Diabetes 61, 145–154 (2012). 10.2337/db11-1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier F. F., Kleinjung F., and Scheller F. W., Sens. Actuators B 38, 78–82 (1997). 10.1016/S0925-4005(97)80174-8 [DOI] [Google Scholar]
- Jensen K. K., Ørum H., Nielsen P. E., and Nordén B., Biochemistry 36, 5072–5077 (1997). 10.1021/bi9627525 [DOI] [PubMed] [Google Scholar]
- Nakatani K., Sando S., and Saito I., Nat. Biotechnol. 19, 51–55 (2001). 10.1038/83505 [DOI] [PubMed] [Google Scholar]
- Blaesing F., Weigel C., Welzeck M., and Messer W., Mol. Microbiol. 36, 557–569 (2002). 10.1046/j.1365-2958.2000.01881.x [DOI] [PubMed] [Google Scholar]
- Hart D. J., Speight R. E., Blackburn J. M., Cooper M. A., and Sutherland J. D., Nucleic Acids Res. 27, 1063–1069 (1999). 10.1093/nar/27.4.1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scire A., Tanfani F., Saccucci F., Bertoli E., and Principato G., Proteins: Struct., Funct., Bioinf. 41, 33–39 (2000). [DOI] [PubMed] [Google Scholar]
- Steinrücke P., Aldinger U., Hill O., Hillisch A., Basch R., and Diekmann S., Anal. Biochem. 286, 26–34 (2000). 10.1006/abio.2000.4780 [DOI] [PubMed] [Google Scholar]
- Sang J., Du H., Wang W., Chu M., Wang Y., Li H., Zhang H. A., Wu W., and Li Z., Biomicrofluidics 7, 024112 (2013). 10.1063/1.4802936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H.-J. and Velev O. D., Biomicrofluidics 7, 031501 (2013). 10.1063/1.4804249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senapati S., Basuray S., Slouka Z., Cheng L.-J., and Chang H.-C., in Microfluidics (Springer, 2011), pp. 153–169. [DOI] [PubMed] [Google Scholar]
- Basuray S., Senapati S., Aijian A., Mahon A. R., and Chang H.-C., ACS Nano 3, 1823–1830 (2009). 10.1021/nn9004632 [DOI] [PubMed] [Google Scholar]
- Chen K.-I., Li B.-R., and Chen Y.-T., Nano Today 6, 131–154 (2011). 10.1016/j.nantod.2011.02.001 [DOI] [Google Scholar]
- Katz E. and Willner I., Electroanalysis 15, 913–947 (2003). 10.1002/elan.200390114 [DOI] [Google Scholar]
- Hong J., Yoon D. S., Park M.-I., Choi J., Kim T. S., Im G., Kim S., Pak Y. E., and No K., Jpn. J. Appl. Phys., Part 1 43, 5639–5645 (2004). 10.1143/JJAP.43.5639 [DOI] [Google Scholar]
- Liu Y.-S., Banada P. P., Bhattacharya S., Bhunia A. K., and Bashir R., Appl. Phys. Lett. 92, 143902 (2008). 10.1063/1.2908203 [DOI] [Google Scholar]
- Hansen P. L., Podgornik R., and Parsegian V. A., Phys. Rev. E 64, 021907 (2001). 10.1103/PhysRevE.64.021907 [DOI] [PubMed] [Google Scholar]
- Schmitz K. S., Macroions in Solution and Colloidal Suspension (VCH, New York, 1993). [Google Scholar]
- Tomić S., Babić S. D., Vuletić T., Krča S., Ivanković D., Griparić L., and Podgornik R., Phys. Rev. E 75, 021905 (2007). 10.1103/PhysRevE.75.021905 [DOI] [PubMed] [Google Scholar]
- Angelini T. E., Golestanian R., Coridan R. H., Butler J. C., Beraud A., Krisch M., Sinn H., Schweizer K. S., and Wong G. C., Proc. Natl. Acad. Sci. U.S.A. 103, 7962–7967 (2006). 10.1073/pnas.0601435103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi F., Cametti C., and Colby R., J. Phys.: Condens. Matter 16, R1423 (2004). 10.1088/0953-8984/16/49/R01 [DOI] [Google Scholar]
- Jungner G., Jungner I., and Allgen L., Nature 164, 1009 (1949). 10.1038/1641009a0 [DOI] [PubMed] [Google Scholar]
- Brillouin L., Kasha M., and Pullman B., in Horizons in Biochemistry (Academic Press, London, 1962). [Google Scholar]
- Suhai S. and Ladik J., Int. J. Quantum Chem. 7, 547–560 (1973). 10.1002/qua.560070310 [DOI] [Google Scholar]
- Dee D. and Baur M., J. Chem. Phys. 60, 541–560 (1974). 10.1063/1.1681073 [DOI] [Google Scholar]
- Murphy C., Arkin M., Jenkins Y., Ghatlia N., Bossmann S., Turro N., and Barton J., Science 262, 1025–1029 (1993). 10.1126/science.7802858 [DOI] [PubMed] [Google Scholar]
- Priyadarshy S., Risser S., and Beratan D., J. Phys. Chem. 100, 17678–17682 (1996). 10.1021/jp961731h [DOI] [Google Scholar]
- Risser S. M., Beratan D. N., and Meade T. J., J. Am. Chem. Soc. 115, 2508–2510 (1993). 10.1021/ja00059a057 [DOI] [Google Scholar]
- Williams R., Mol. Phys. 68, 1–23 (1989). 10.1080/00268978900101931 [DOI] [Google Scholar]
- Baverstock K. and Cundall R., Int. J. Rad Appl. Instrum. C 32(3 ), 553–556 (1988). 10.1016/1359-0197(88)90063-X [DOI] [Google Scholar]
- Arkin M., Stemp E., Holmlin R., Barton J., Hörmann A., Olson E., and Barbara P., Science (New York, NY) 273, 475 (1996). 10.1126/science.273.5274.475 [DOI] [PubMed] [Google Scholar]
- Holmlin R. E., Stemp E. D., and Barton J. K., J. Am. Chem. Soc. 118, 5236–5244 (1996). 10.1021/ja953941y [DOI] [Google Scholar]
- Kelley S. O. and Barton J. K., Chem. Biol. 5, 413–425 (1998). 10.1016/S1074-5521(98)90158-2 [DOI] [PubMed] [Google Scholar]
- Kelley S. O., Holmlin R. E., Stemp E. D., and Barton J. K., J. Am. Chem. Soc. 119, 9861–9870 (1997). 10.1021/ja9714651 [DOI] [Google Scholar]
- Bakhshi A., Prog. Biophys. Mol. Biol. 61, 187 (1994). 10.1016/0079-6107(94)90001-9 [DOI] [PubMed] [Google Scholar]
- Beratan D. N., J. Am. Chem. Soc 108, 4321–4326 (1986). 10.1021/ja00275a014 [DOI] [Google Scholar]
- Esfandyarpour R., Esfandyarpour H., Javanmard M., Harris J. S., and Davis R. W., “ Microneedle biosensor: A method for direct label-free real time protein detection,” Sens. Actuators B: Chem. 177, 848–855 (2012). 10.1016/j.snb.2012.11.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esfandyarpour R., Esfandyarpour H., Javanmard M., Harris J. S., and Davis R. W., Electrical Detection of Protein Biomarkers Using Nanoneedle Biosensors (Cambridge University Press, 2012).
- Esfandyarpour R., Javanmard M., Harris J. S., and Davis R. W., Thin Film Nanoelectronic Probe for Protein Detection (Cambridge Univ Press, 2013). [Google Scholar]
- Esfandyarpour R., Javanmard M., Koochak Z., Harris J. S., and Davis R. W., “ Nanoelectronic impedance detection of target cells,” Biotechnol. Bioeng. (in press). [DOI] [PubMed]
- Esfandyarpour R., Esfandyarpour H., Harris J. S., and Davis R. W., “ Simulation and fabrication of a new novel 3D injectable biosensor for high throughput genomics and proteomics in a lab-on-a-chip device,” Nanotechnology (in press). [DOI] [PMC free article] [PubMed]