Abstract
Williams syndrome (WS) is a condition caused by a contiguous deletion of approximately 26–28 genes from chromosome 7, and is characterized by abnormal social and emotional processing and abnormal structure and function of the amygdala. Prior studies show that the amygdala is relatively enlarged in WS, but very little is known regarding the regional specificity of increased amygdalar volume in this condition. Here we investigated the regional specificity of structural alterations of the amygdala in WS, compared to a typically developing (TD) control group. We acquired high resolution brain MRI data from 79 participants (39 WS, 40 TD) and used a surface‐based analytical modeling approach. The WS group exhibited several areas of increased radial expansion of the amygdalar surface and no areas of decreased radial expansion of the amygdalar surface compared to TD controls. The areas found to exhibit particularly increased radial expansion in WS included the bilateral posterior cortical nucleus, lateral nucleus, and the central nucleus. This greater regional and anatomical specificity of altered amygdala structure in WS contributes to a model relating genetic risk in WS to the development of key brain regions for social and emotional functioning. Hum Brain Mapp 35:866–874, 2014. © 2012 Wiley Periodicals, Inc.
Keywords: Williams syndrome, genetics, amygdala, shape
INTRODUCTION
Williams syndrome (WS) is a neurodevelopmental condition that affects approximately 1 in every 10,000 individuals and is caused by a contiguous deletion of ∼26–28 genes on chromosome 7q11.23. WS is paired with a compelling neuropsychological phenotype characterized by cognitive delays, visuo‐spatial deficits, heightened non‐social anxiety, and an abnormally increased drive toward social interaction [Martens et al., 2008; Meyer‐Lindenberg et al., 2006]. Investigating WS with brain imaging offers a unique opportunity to elucidate the association between genes, the brain, and social‐cognitive function.
Individuals with WS tend to exhibit abnormalities in social behavior and emotion processing [Haas and Reiss, 2012; Jarvinen‐Pasley et al., 2008]. Persons with WS are generally more driven toward social interaction [Doyle et al., 2004], more socially fearless [Gosch and Pankau, 1994], and relatively less inhibited toward strangers [Dodd et al., 2010], compared to age‐ or mentally age‐matched controls. In terms of emotion processing, individuals with WS tend to exhibit more fears and anxieties in response to non‐social stimuli (e.g., spiders) [Leyfer et al., 2006, 2009] but are generally less sensitive to fearful stimuli that are socially related (e.g., fearful facial expressions) [Plesa‐Skwerer et al., 2006, 2009; Santos et al., 2010]. Together, these findings support the hypothesis that WS is associated with abnormal structure and function in brain regions important for social behavior and processing emotions.
One brain region particularly important for social and emotional function is the amygdala [Aggleton, 2000]. Interestingly, WS is associated with both functional and structural aberrations of the amygdala [Haas et al., 2009; Meyer‐Lindenberg et al., 2005; Reiss et al., 2004]. In particular, several studies have shown that individuals with WS exhibit disproportionately enlarged overall amygdalar volumes (in reference to whole brain volume), compared to typically developing (TD) controls [Capitao et al., 2011; Jabbi et al., 2012; Martens et al., 2009; Reiss et al., 2004] while some studies have not [Chiang et al., 2007; Meda et al., 2012]. The inconsistency of findings may in part be due to localized structural abnormalities of the amygdala in this condition.
The structural and functional connections of the amygdala with other brain regions are also abnormal in WS. For example, Jabbi et al. 2012 demonstrated increased fractional anisotropy within the amygdoloid white‐matter pathway, the uncinate fasciculus, and Sarpal et al. 2008 demonstrated reduced functional connectivity between the amygdala and the fusiform face area in WS as compared to TD controls. Combined, these findings support the hypothesis that WS is associated with abnormal structural morphology of the amygdala and related brain regions.
The amygdala is a key brain region for processing emotions, especially fear‐inducing stimuli [LeDoux, 1998, 2003]. For example, the amygdala is involved in assessing the emotional salience of environmental cues [Phan et al., 2004], fear conditioning [Sehlmeyer et al., 2009], social and non‐social anxieties [Shin and Liberzon, 2010], and in processing facial expressions [Fusar‐Poli et al., 2009]. The amygdala is a complex structure composed of a network of interconnected and related nuclei [Aggleton, 2000]. Amygdala nuclei work together to process various types of social and emotional stimuli. For example, the lateral nucleus is the main input region within the amygdala and is important for forming stimulus‐value associations, such as during fear conditioning [LeDoux, 2007; LeDoux et al., 1990]. Conversely, the central nucleus is the main output region within the amygdala and is thought to influence the expression of innate emotional responses and associated physiological responses [LeDoux, 2007]. Lastly, there is some evidence suggesting that the lateral nucleus is more important to processing threatening stimuli (e.g., angry facial expressions) as compared to other amygdala nuclei such as the central nucleus [Hoffman et al., 2007].
Recent advancements in MRI analysis techniques enable us to investigate the regional specificity of structural alterations of subcortical brain regions, such as the amygdala, in vivo in humans [Chung et al., 2010; Tamburo et al., 2009; Thompson et al., 2004a]. Although surface‐based subcortical shape analyses are based on estimations that are most likely around the vicinity of specific nuclei in question, this approach has demonstrated localized structural alterations of the amygdala associated with traits such as psychopathy [Boccardi et al., 2011; Yang et al., 2009] and diseases such as Alzheimer's [Cavedo et al., 2011].
In this study, we used surface‐based modeling [Thompson et al., 2004b] to investigate the regional specificity of structural alterations of the amygdala in a group of participants with WS, compared to a TD control group. Based on prior evidence [Capitao et al., 2011; Jabbi et al., 2012; Martens et al., 2009; Reiss et al., 2004], we expected WS to be associated with greater overall amygdalar volume, relative to TD controls. Based on previous evidence of abnormal emotion processing in WS [Munoz et al., 2010; Plesa‐Skwerer et al., 2006, 2009; Santos, et al., 2010], we further predicted that WS would be associated with regionally specific alterations in specific amygdalar nuclei important for the processing and expression of emotions (lateral and central nuclei).
METHODS
Participants
Demographic data for participants in this study are presented in Table 1. A total of 79 individuals, 39 WS and 40 TD, participated in this study. WS participants (39 total; 24 females; mean age = 26.1, SD = 11.0) were recruited via advertisements through national agencies, physicians within local clinics, and the Stanford University Medical School. All participants were diagnosed based on exhibiting the WS genetic deletion and on exhibiting the medical and clinical features of the WS phenotype, including cognitive, behavioral, and physical profiles [Martens et al., 2008; Meyer‐Lindenberg et al., 2006]. The genetic diagnosis of WS was confirmed with fluorescent in situ hybridization (FISH) in all participants using probes for the elastin gene. These studies were performed as a component of the present study (n = 31) or confirmed via medical records provided by each participant's legal guardian (n = 8). Participants in this study partially overlap with participants reported in previous studies from our laboratory [Haas et al., 2009, 2010; Reiss et al., 2004; Thompson et al., 2005].
Table 1.
Demographic data for each participant group
| n | M/F | IQ | Age | |
|---|---|---|---|---|
| WS | 39 | 15/24 | 63.28 ± 11.71 | 26.06 ± 11.03 |
| TD | 40 | 17/23 | 114.48 ± 11.31 | 21.34 ± 11.41 |
WS: Williams syndrome, TD: typically developing, M: males, F: females, IQ: intellectual quotient.
TD subjects were recruited locally (Palo Alto, CA) and were financially compensated for their participation (40 total; 23 females; mean age = 21.3, SD = 11.4). TD subjects were screened for a history of psychiatric or neurologic problems. There were no subjects that exhibited any history of psychiatric or neurologic problems.
Each participant's full scale intellectual functioning was assessed using either the Wechsler Intelligence Scale for Children—Fourth Edition (WISC‐IV) [Wechsler, 2003] for participants 16 years of age or younger or the Wechsler Abbreviated Scale of Intelligence (WASI) [Wechsler, 1999] for participants 17 years of age or older. As expected, the TD group (mean IQ = 114.5, SD = 11.3) exhibited higher IQ scores as compared to the WS group (mean IQ = 63.28, SD = 11.7) (t = 17.79, P < 0.01). There were no significant differences in IQ scores derived from the WISC‐IV vs. WASI within the WS (t = 1.63, P > 0.10) or TD (t = 0.86, P > 0.10) groups. There were no significant between‐group differences in age t(1, 77) = 1.86, P > 0.05, or proportion of males to females, (X2 = 0.13, P = .72), or handedness (X2 = 1.52, P = 0.29). None of the participants had a contraindication for MRI. Written informed consent and assent were obtained according to the Declaration of Helsinki. This study was approved by the Stanford University Administrative Panel on Human Subjects in Medical Research.
Image Acquisition
Images were collected on a 3T GE Signa scanner (Lucas Center of Radiology, Stanford University) using a custom build birdcage head coil. Coronally oriented T1‐weighted MR images were acquired using fast spoiled gradient recall (FSPGR) parameters: repetition time (TR) = 6.4–6.6 ms; inversion time (TI) = 1.6 ms; echo time (TE) = 300 ms, flip angle = 15°; field of view (FOV) = 220 × 176 mm2; matrix size = 256 × 256; pixel size = 0.8594 × 0.8594 mm2; and slice thickness = 1.5–1.7 mm (adjusted for brain size in anterior/posterior direction).
Image Analysis
All whole‐brain MR images underwent bias correction with a regularization parameter set to 0.00001 and a full‐width half‐maximum (FWHM) of 30 mm in SPM8 (http://www.fil.ion.ucl.ac.uk/spm/). T1‐weighted anatomical FSPGR images were bias corrected in SPM8 in order to account for spatially varying artifacts that arise due to the pulse sequence and MR scanner environment. Based on previous testing with segmentation of FSPGR images in Freesurfer, we used the bias regularization and FWHM modules within SPM8 “New Segment” and specified a regularization of “extremely light (0.00001)” and a FWHM of 30 mm cutoff. Freesurfer version 5.1 was then used to segment and measure amygdalar structure volumes (http://surfer.nmr.mgh.harvard.edu/) [Fischl et al., 2002]. Freesurfer is an automated MRI analysis tool designed to segment cortical and subcortical brain structures (http://surfer.nmr.mgh.harvard.edu/fswiki). The use of automated approaches such as Freesurfer reduces error related variance such as inter‐ or intra‐rater bias. Segmentation of the amygdala using Freesurfer is reliable across various pulse sequences and voxel geometries [Wonderlick et al., 2009]. Freesurfer has been shown to be effective for measuring condition‐specific alterations in amygdalar volume [Dewey et al., 2010; Lehmann et al., 2010].
Based on our a priori hypothesis of altered shape of the amygdala in WS, we designated the amygdala as the region of interest (ROI) in this study (Fig. 1). Following segmentation by Freesurfer, each amygdalar ROI was visually inspected for validity using Freesurfer's built‐in FreeView utility and manually edited when necessary. Manual editing of the amygdala was performed on 13 of the 79 images (15%) by a trained rater (KS), blind to subject demographics. Edits were performed in accordance with a standardized procedure used to manually delineate the amygdala [Karchemskiy et al., 2011; Reiss et al., 2004]. Brain and amygdalar volume measures were then extracted in Freesurfer for between‐group analyses. Brain size was calculated by summing total gray and white matter volumes. Brain size did not include cerebellar, brain stem, and ventricular volumes. To perform surface modeling, the segmented brains and their corresponding amygdalar ROIs were oriented and normalized to a standard brain template (the ICBM‐152) using a 9‐parameter transformation with FMRIB's Linear Image Registration Tool (FLIRT) [Jenkinson et al., 2002]. FLIRT is a robust and accurate automated linear (affine) registration tool based on a multi‐start, multi‐resolution global optimization method (FLIRT: http://www.fmrib.ox.ac.uk/analysis/research/flirt/).
Figure 1.
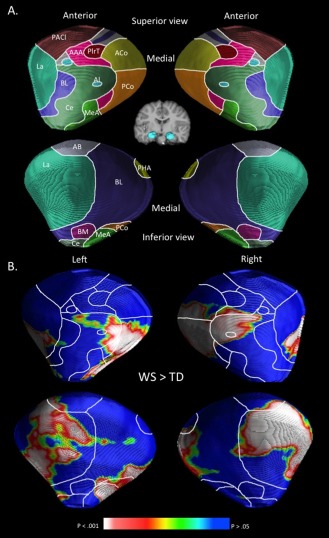
Schematic representation of amygdalar nuclei and localized volume increases in WS compared to TD controls rendered onto a standard template of the amygdala. A: Delineation of the amygdalar nuclei were interpolated from a histological study [Mai et al., 1997], and from previous surface‐based modeling analysis of the amygdala [Boccardi et al., 2011; Cavedo et al., 2011; Yang et al., 2009]. AAA indicates anterior amygdaloid area; AB, accessory basal nucleus; ACo, anterior cortical nucleus; AI, amygdaloid island; BL, basolateral nucleus; BM, basomedial nucleus; Ce, central nucleus; La, lateral nucleus; MeA, medial amygdaloid nucleus; PACl, preamygdalar claustrum; PCo, posterior cortical nucleus; PHA, parahippocampal‐amygdaloid transition area; PirT, piriform cortex. B: Areas corresponding to significantly greater radial distances in WS relative to TD controls are rendered on a standard amygdalar template. No areas corresponding to significantly greater radial distances in TD controls relative to WS were observed.
Statistical Analysis
Overall adjusted amygdala volume was compared between groups. Amygdalar volumes were adjusted for total brain volume by calculating the ratio between the average amygdala (non‐normalized, left and right combined) and total brain volume for each subject [Capitao et al., 2011]. Two‐sample t‐tests were then used to compare adjusted amygdalar volumes between groups using a statistical threshold of P = 0.05. To account for any effect of demographic factors on adjusted amygdalar volumes, subsequent between‐group t‐test analyses were conducted, while sex and age were entered as covariates. We also investigated laterality differences in volume between groups. Specifically, we performed a repeated measures analysis, with hemisphere (left vs. right adjusted amygdala volume) entered as the within subjects factor and group (WS vs. TD) as the between subjects factor. Lastly, post hoc correlation analyses between IQ scores and amygdalar volume within each group (WS and TD) were conducted.
Statistical surface modeling was applied to the transformed, normalized amygdalar ROIs [Thompson et al., 2004b]. In this type of analysis, the surface maps of the amygdala are analyzed by computing the radial distance of each surface point on the amygdala from a centroid (medial curve) threading down the center of each ROI. The medial curve was defined as the 3‐dimensional curve traced out by the centroid of the amygdalar boundary in each coronal image slice. ROIs were initially oriented to standard space based on each subject's whole brain transformation (9‐parameter) to the ICBM‐152 brain template using FMRIB's Linear Image Registration Tool (FLIRT). The radial distance surface points (across subjects) were then aligned by overlaying a standardized frequency of radial distance points (i.e., the distance between a 3‐dimensional curve traced out by the centroid and the surface on each coronal image slice) over the surface of each ROI. This created a mesh surface, on each ROI, with a homologous spatial frequency of each radial distance point across subjects (i.e., point‐by‐point correspondence). Thus, statistical comparisons were performed on radial distances that were in homologous locations across subjects.
Student's t‐tests were used to compare the radial distance for each point on the amygdala between the WS and TD groups. To assess the overall significance of any group differences in the maps, parametric surface point P‐values were corrected for multiple comparisons using permutation tests of group assignment (400,000 total: [Thompson et al., 2004b]). P‐values were then overlaid onto a 3‐D surface rendering of amygdalar subregions, which were interpolated based on a standardized histological atlas [Mai et al., 1997] as in prior surface‐based amygdalar studies [Boccardi et al., 2011; Cavedo et al., 2011; Yang et al., 2009] (Fig. 1A). Specifically, in 3‐dimensional space, the average mesh model of the left and right amygdalae, from our sample, was oriented along an anterior–posterior axis. Next, the contour of each subregion was outlined on a digital 3‐dimensional template and their boundaries were projected onto the surface of the 3‐dimensional mesh model [Cavedo et al., 2011].
RESULTS
Overall Volume of the Amygdala in WS and TD
The adjusted amygdalar volume was compared between the WS and the TD group by using a two sample t‐test. The WS group exhibited a larger adjusted volume of the amygdala, compared to the TD group, t(1, 77) = 3.66, P < 0.001. The overall adjusted amygdala volume (left and right combined) was 10.2% larger in WS (M = 16.11 × 10−4, SD = 0.19 × 10−3) as compared to controls (M = 14.62 × 10−4, SD = 0.17 × 10−3). The left amygdala was 8.7% larger in WS (t = 2.96, P < 0.005) and the right amygdala was 11.6% larger in WS (t = 3.65, P < 0.001) as compared to controls. The difference in adjusted amygdalar volume between the WS and TD group remained significant when age, sex, and handedness were entered as covariates (F = 6.57, P < 0.05). There was no significant interaction observed between hemisphere (left vs. right adjusted amygdala volume) and participant group F(1,77) = 1.41, P = 0.24. There were no significant positive or negative correlations between IQ and adjusted amygdalar volume within the WS or TD groups (all P's > 0.05).
Localized Structural Alterations of the Amygdala in WS
Analyses comparing radial distances between groups (WS and TD) were conducted. Overall, mean radial distances were larger in the WS group (M = 5.49 mm, SD = 0.36) as compared to the TD group (M = 5.16, SD = .30) (t = 4.44, P < 0.001). Figure 1B shows a surface rendering of areas exhibiting significantly (corrected for multiple comparisons) greater radial distances of the amygdala in WS as compared to the TD group. No areas of the left or right amygdala were found to exhibit significantly greater radial distances in the TD, compared to the WS group.
An inspection of the surface rending of regions containing significantly greater radial distances in WS as compared to the TD group (Fig. 1) indicates that structural alterations were localized to several amygdalar nuclei. Within the left amygdala, WS was associated with greater radial distance of the posterior cortical nucleus, central nucleus, medial amygdaloid nucleus, lateral nucleus, and basolateral nucleus. Within the right amygdala, WS was associated with increased radial distance of the posterior cortical nucleus, central nucleus, lateral nucleus, amygdaloid island, anterior cortical nucleus, and basolateral nucleus. A qualitative view of which nuclei show the greatest between‐group differences (in terms spatial extent) indicates that large areas of the bilateral posterior cortical nucleus, moderate areas of the bilateral lateral nucleus, and small areas of the bilateral central nucleus are affected in WS.
DISCUSSION
In this study, we demonstrate that WS is associated with localized alterations in amygdalar volume and/or morphology. Compared to controls, the WS group exhibited localized greater radial distance of the amygdala (i.e., hypertrophy) and no localized decreases (i.e., atrophy). This finding is consistent with previous MRI studies demonstrating greater overall amygdalar volume in WS compared to controls [Capitao et al., 2011; Jabbi et al., 2012; Martens et al., 2009; Reiss et al., 2004]. These results provide an improved understanding of the regional specificity of structural alterations of the amygdala in WS.
By using surface‐based modeling, we obtained evidence of regionally specific increased size of the amygdala in WS. The resolution of MRI, even at 3 T, is not optimal for distinguishing individual nuclei within the amygdala. We therefore used an interpolation approach based on histological and neuroimaging studies to estimate the nuclear localization of morphological alterations observed in this study [Boccardi et al., 2011; Cavedo et al., 2011; Yang et al., 2009]. We found regionally specific increased radial distances in areas corresponding to several amygdalar nuclei. WS was associated with moderate to greatly increased radial distances in areas corresponding to the bilateral posterior cortical nuclei and lateral nuclei along with a small area of increased radial distances in regions corresponding to the bilateral central nucleus.
The finding of increased radial distances associated with the posterior cortical nucleus in WS was unexpected. Along with the medial amygdaloid nucleus, the posterior cortical nucleus maintains several connections with brain regions involved in olfaction [Swanson and Petrovich, 1998]. There is some evidence that WS is associated with abnormalities within the olfactory system. For example, studies of mouse models of WS find that some genes affected in WS (LimK and Gtf2ird1), influence the development of the olfactory system in mice [Ang et al., 2006; Palmer, et al., 2007]. Clearly, more research is necessary to associate structural abnormalities of the amygdala with sensory processing in WS.
A moderate area of the lateral nucleus also exhibited increased radial distances in WS, compared to controls. The lateral nucleus is the main input region within the amygdala and maintains connections with the temporal lobes (including the primary auditory cortex), fusiform gyrus, and the lateral orbitofrontal cortex [LeDoux, 2007; Saygin et al., 2011]. Functionally, the lateral nucleus is involved in learning stimulus–affect associations [Johansen et al., 2010; LeDoux et al., 1990] and fear conditioning [LeDoux, 1998, 2007; Maren, 2003]. Persons with WS tend to exhibit abnormalities in fear processing. More specifically, individuals with WS are generally less sensitive to socially related fear signals [Plesa‐Skwerer et al., 2006; Santos et al., 2010], but exhibit more fears and anxieties as related to non‐socially related fearful information [Green et al., in press; Leyfer et al., 2006, 2009; Stinton et al., 2010] as compared to controls. Additionally, those with WS exhibit reduced amygdala response to fearful facial expressions [Haas et al., 2009; Meyer‐Lindenberg et al., 2005] and greater amygdala response to non‐socially related fearful stimuli (e.g., spiders) [Munoz et al., 2010] as compared to controls. Interestingly, WS is also often associated with hyperacusis (i.e., heightened sensitivity to noise) [Elsabbagh et al., 2011; Klein et al., 1990]. Taken together, these findings suggest that functional abnormalities of the lateral nucleus of the amygdala (and/or amygdalar connections with the auditory cortex) may contribute to aberrant fear processing and hyperacusis in WS.
Lastly, a small area of the central nucleus exhibited increased radial distances in WS as compared to controls. The central nucleus is the main output region of the amygdala and maintains connections with the hypothalamus, basal forebrain, and the brainstem [Saygin et al., 2011]. Functionally, the central nucleus is thought to be involved in the expression of innate emotional responses and associated physiological responses [LeDoux, 2007]. For example, during fear‐conditioning paradigms in animals, the central nucleus controls response behaviors such as freezing and fear potentiated startle [Pare et al., 2004]. There is also evidence that people with WS exhibit abnormalities in autonomic responses to emotional stimuli. For example, Plesa‐Skwerer et al. 2009 showed that individuals with WS exhibit reduced autonomic responses (heart rate and skin conductance) to dynamically presented face stimuli as compared to age‐ and IQ‐matched controls. Combined, abnormal structure of the central nucleus may be a neural substrate associated with abnormalities in emotional reactivity in WS.
In this study, the location of amygdala subregions were estimated based on a histological atlas [Mai et al., 1997] and neuroimaging studies using MRI data acquired at 3 Tesla [Boccardi et al., 2011; Cavedo et al., 2011; Yang et al., 2009]. Recently, several neuroimaging studies have estimated the location of amygdala subregions using complementary methods. For example, Solano‐Castiella et al. 2011 used ultra high field, 7 Tesla, MRI, Entis et al., 2012 used a geometrically based segmentation protocol, and Saygin et al. 2011 used a tractography‐based segmentation approach of diffusion tensor imaging data. Future studies of localized amygdala structural alterations in psychiatric conditions will benefit by considering the efficacy of novel segmentation techniques.
As in several prior MRI studies on the shape of subcortical structures using surface‐based analyses techniques [Apostolova et al., 2010; Lepore et al., 2009, 2010; Morra et al., 2009a, 2009b, 2010; Nicolson et al., 2006], we spatially normalized each participant's brain data into standard stereotaxic space using a 9‐parameter transformation. This procedure allowed for statistical analyses to be focused on localized morphological alterations of the amygdala rather than large‐scale brain volume abnormalities typically associated with WS. Indeed, in native space, brain volumes of WS participants in this study were 17.1% smaller than in TD controls, while amygdala volumes were 9.0% smaller than in TD controls (data not shown). Although our findings support the association between localized amygdala alterations and the WS phenotype, the association between reduced brain volume and the WS phenotype remains poorly understood.
The findings of this study advance a model linking genetic risk in WS to aberrations of neural structure ultimately influencing social and emotional functioning. However, this study is also limited in several important ways. We compared WS to a TD control group characterized by normal IQ. WS is most often associated with cognitive delays and IQ scores that fall 3–5 standard deviations below norms [Martens et al., 2008]. Based on evidence associating the neuroanatomy of the amygdala with social and emotional processing in WS [Martens et al., 2009], we predicted that regionally specific alterations of amygdala volume would not be associated with IQ, but may be associated with the atypical social phenotype in this condition. To investigate the association between IQ and structural alterations of the amygdala in this study, we performed a correlation analysis between IQ and amygdala volume within each group (WS and TD). As expected, this analysis did not provide evidence in support of IQ being associated with structural alterations of the amygdala. Currently, this study does not provide any data specifically associating social and affective functioning to structural alterations of the amygdala in WS. The inclusion of behavioral data representative of social cognition, emotion processing, or anxiety would have allowed for greater neurofunctional specificity in this study. However, we were not able to collect behavioral measures from a sufficient number of participants in this regard, and thus, such statistical analyses were not justified. Future studies on the neural substrates of social and emotional disturbances in WS are warranted to address the specificity of behavioral, cognitive, and emotional factors associated with amygdalar structure in WS. Additionally, future studies that incorporate fear conditioning with neuroimaging techniques will be well suited to test hypotheses that associate altered amygdala volume with specific social and emotional processes. Furthermore, this study only included participants with a typical (i.e., classical) genetic deletion associated with WS. Several recent studies have described distinctive patterns of social and neurocognitive processing in individuals with WS having partial deletions [Antonell et al., 2010; Dai et al., 2009; Karmiloff‐Smith et al., 2012]. Thus, future studies designed to investigate how structural and functional abnormalities of the amygdala differ in partial deletion conditions as compared to typical (or classical) deletion conditions of WS are warranted. Lastly, the atlas that was used to interpolate the amygdalar nuclei in this study was based on a single subject [Mai et al., 1997]. Future studies may benefit by utilizing other techniques, such as probabilistic tractography [Saygin et al., 2011], to localize amygdalar nuclei across larger samples of subjects and patient groups.
In conclusion, this study provides evidence of localized increased size of the amygdala in WS. Alterations of amgydalar structure may be a neural construct associated with abnormal fear processing and reactivity to emotions in this condition. These findings build on a model that associates the WS genetic deletion to abnormal development of brain regions important for social and emotional processing.
REFERENCES
- Aggleton JP (2000): The Amygdala: A Functional Analysis. New York:Oxford University Press. [Google Scholar]
- Ang LH, Chen W, Yao Y, Ozawa R, Tao E, Yonekura J, Uemura T, Keshishian H, Hing H (2006): Lim kinase regulates the development of olfactory and neuromuscular synapses. Dev Biol 293:178–190. [DOI] [PubMed] [Google Scholar]
- Antonell A, Del Campo M, Magano LF, Kaufmann L, de la Iglesia JM, Gallastegui F, Flores R, Schweigmann U, Fauth C, Kotzot D, Perez‐Jurado LA (2010): Partial 7q11.23 deletions further implicate GTF2I and GTF2IRD1 as the main genes responsible for the Williams‐Beuren syndrome neurocognitive profile. J Med Genet 47:312–320. [DOI] [PubMed] [Google Scholar]
- Apostolova LG, Thompson PM, Green AE, Hwang KS, Zoumalan C, Jack CR Jr, Harvey DJ, Petersen RC, Thal LJ, Aisen PS, Toga AW, Cummings JL, Decarli CS (2010): 3D comparison of low, intermediate, and advanced hippocampal atrophy in MCI. Hum Brain Mapp 31:786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccardi M, Frisoni GB, Hare RD, Cavedo E, Najt P, Pievani M, Rasser PE, Laakso MP, Aronen HJ, Repo‐Tiihonen E, Vaurio O, Thompson PM, Tiihonen J (2011): Cortex and amygdala morphology in psychopathy. Psychiatry Res 193:85–92. [DOI] [PubMed] [Google Scholar]
- Capitao L, Sampaio A, Sampaio C, Vasconcelos C, Fernandez M, Garayzabal E, Shenton ME, Goncalves OF (2011): MRI amygdala volume in Williams Syndrome. Res Dev Disabil 32:2767–2772. [DOI] [PubMed] [Google Scholar]
- Cavedo E, Boccardi M, Ganzola R, Canu E, Beltramello A, Caltagirone C, Thompson PM, Frisoni GB (2011): Local amygdala structural differences with 3T MRI in patients with Alzheimer disease. Neurology 76:727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, Reiss AL, Lee AD, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Thompson PM (2007): 3D pattern of brain abnormalities in Williams syndrome visualized using tensor‐based morphometry. Neuroimage 36:1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Nacewicz BM, Dalton KM, Davidson RJ (2010): General multivariate linear modeling of surface shapes using SurfStat. Neuroimage 53:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Bellugi U, Chen XN, Pulst‐Korenberg AM, Jarvinen‐Pasley A, Tirosh‐Wagner T, Eis PS, Graham J, Mills D, Searcy Y, Korenberg JR (2009): Is it Williams syndrome? GTF2IRD1 implicated in visual‐spatial construction and GTF2I in sociability revealed by high resolution arrays. Am J Med Genet A 149:302–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey J, Hana G, Russell T, Price J, McCaffrey D, Harezlak J, Sem E, Anyanwu JC, Guttmann CR, Navia B, Cohen R, Tate DF (2010): Reliability and validity of MRI‐based automated volumetry software relative to auto‐assisted manual measurement of subcortical structures in HIV‐infected patients from a multisite study. Neuroimage 51:1334–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd HF, Porter MA, Peters GL, Rapee RM (2010): Social approach in pre‐school children with Williams syndrome: The role of the face. J Intellect Disabil Res 54:194–203. [DOI] [PubMed] [Google Scholar]
- Doyle TF, Bellugi U, Korenberg JR, Graham J (2004): “Everybody in the world is my friend” hypersociability in young children with Williams syndrome. Am J Med Genet A 124:263–273. [DOI] [PubMed] [Google Scholar]
- Elsabbagh M, Cohen H, Cohen M, Rosen S, Karmiloff‐Smith A (2011): Severity of hyperacusis predicts individual differences in speech perception in Williams Syndrome. J Intellect Disabil Res 55:563–571. [DOI] [PubMed] [Google Scholar]
- Entis JJ, Doerga P, Barrett LF, Dickerson BC (2012): A reliable protocol for the manual segmentation of the human amygdala and its subregions using ultra‐high resolution MRI. Neuroimage 60:1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM (2002): Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355. [DOI] [PubMed] [Google Scholar]
- Fusar‐Poli P, Placentino A, Carletti F, Landi P, Allen P, Surguladze S, Benedetti F, Abbamonte M, Gasparotti R, Barale F, Perez J, McGuire P, Politi P (2009): Functional atlas of emotional faces processing: A voxel‐based meta‐analysis of 105 functional magnetic resonance imaging studies. J Psychiatry Neurosci 34:418–432. [PMC free article] [PubMed] [Google Scholar]
- Gosch A, Pankau R (1994): Social‐emotional and behavioral adjustment in children with Williams‐Beuren syndrome. Am J Med Genet 53:335–339. [DOI] [PubMed] [Google Scholar]
- Green T, Avda S, Dotan I, Zarchi O, Basel‐Vanagaite L, Zalsman G, Weizman A, Gothelf D: Phenotypic psychiatric characterization of children with Williams syndrome and response of those with ADHD to methylphenidate treatment. Am J Med Genet B Neuropsychiatr Genet 159B:13–20. [DOI] [PubMed] [Google Scholar]
- Haas BW, Reiss AL (2012): Social brain development in Williams syndrome: The current status and directions for future research. Front Psychol 3:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BW, Mills D, Yam A, Hoeft F, Bellugi U, Reiss A (2009): Genetic influences on sociability: Heightened amygdala reactivity and event‐related responses to positive social stimuli in Williams syndrome. J Neurosci 29:1132–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BW, Hoeft F, Searcy YM, Mills D, Bellugi U, Reiss A (2010).Individual differences in social behavior predict amygdala response to fearful facial expressions in Williams syndrome. Neuropsychologia 48:1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman KL, Gothard KM, Schmid MC, Logothetis NK (2007).Facial‐expression and gaze‐selective responses in the monkey amygdala. Curr Biol 17:766–772. [DOI] [PubMed] [Google Scholar]
- Jabbi M, Kippenhan JS, Kohn P, Marenco S, Mervis CB, Morris CA, Meyer‐Lindenberg A, Berman KF (2012): The Williams syndrome chromosome 7q11.23 hemideletion confers hypersocial, anxious personality coupled with altered insula structure and function. Proc Natl Acad Sci USA 109:E860–E866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvinen‐Pasley A, Bellugi U, Reilly J, Mills DL, Galaburda A, Reiss AL, Korenberg JR (2008): Defining the social phenotype in Williams syndrome: A model for linking gene, the brain, and behavior. Dev Psychopathol 20:1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S (2002): Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17:825–841. [DOI] [PubMed] [Google Scholar]
- Johansen JP, Hamanaka H, Monfils MH, Behnia R, Deisseroth K, Blair HT, LeDoux JE (2010): Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc Natl Acad Sci USA 107:12692–12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchemskiy A, Garrett A, Howe M, Adleman N, Simeonova DI, Alegria D, Reiss A, Chang K (2011): Amygdalar, hippocampal, and thalamic volumes in youth at high risk for development of bipolar disorder. Psychiatry Res 194:319–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmiloff‐Smith A, Broadbent H, Farran EK, Longhi E, D'Souza D, Metcalfe K, Tassabehji M, Wu R, Senju A, Happe F, Turnpenny P, Sansbury F (2012): Social cognition in Williams syndrome: genotype/phenotype insights from partial deletion patients. Front Psychol 3:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein AJ, Armstrong BL, Greer MK, Brown FR III (1990): Hyperacusis and otitis media in individuals with Williams syndrome. J Speech Hear Disord 55:339–344. [DOI] [PubMed] [Google Scholar]
- LeDoux J (1998): Fear and the brain: where have we been, and where are we going? Biol Psychiatry 44:1229–1238. [DOI] [PubMed] [Google Scholar]
- LeDoux J (2003): The emotional brain, fear, and the amygdala. Cell Mol Neurobiol 23:727–738. [DOI] [PubMed] [Google Scholar]
- LeDoux J (2007): The amygdala. Curr Biol 17:R868–R874. [DOI] [PubMed] [Google Scholar]
- LeDoux JE, Cicchetti P, Xagoraris A, Romanski LM (1990): The lateral amygdaloid nucleus: Sensory interface of the amygdala in fear conditioning. J Neurosci 10:1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Douiri A, Kim LG, Modat M, Chan D, Ourselin S, Barnes J, Fox NC (2010): Atrophy patterns in Alzheimer's disease and semantic dementia: A comparison of FreeSurfer and manual volumetric measurements. Neuroimage 49:2264–2274. [DOI] [PubMed] [Google Scholar]
- Lepore N, Shi Y, Lepore F, Fortin M, Voss P, Chou YY, Lord C, Lassonde M, Dinov ID, Toga AW, Thompson PM (2009): Pattern of hippocampal shape and volume differences in blind subjects. Neuroimage 46:949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore N, Vachon P, Lepore F, Chou YY, Voss P, Brun CC, Lee AD, Toga AW, Thompson PM (2010): 3D mapping of brain differences in native signing congenitally and prelingually deaf subjects. Hum Brain Mapp 31:970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyfer OT, Woodruff‐Borden J, Klein‐Tasman BP, Fricke JS, Mervis CB (2006): Prevalence of psychiatric disorders in 4 to 16‐year‐olds with Williams syndrome. Am J Med Genet B Neuropsychiatr Genet 141:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyfer O, Woodruff‐Borden J, Mervis CB (2009): Anxiety disorders in children with williams syndrome, their mothers, and their siblings: Implications for the etiology of anxiety disorders. J Neurodev Disord 1:4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai JK, Assheuer J, Paxinos G (1997): Atlas of the Human Brain. San Diego, CA:Academic Press. [Google Scholar]
- Maren S (2003): The amygdala, synaptic plasticity, and fear memory. Ann NY Acad Sci 985:106–113. [DOI] [PubMed] [Google Scholar]
- Martens MA, Wilson SJ, Reutens DC (2008): Research review: Williams syndrome: A critical review of the cognitive, behavioral, and neuroanatomical phenotype. J Child Psychol Psychiatry 49:576–608. [DOI] [PubMed] [Google Scholar]
- Martens MA, Wilson SJ, Dudgeon P, Reutens DC (2009): Approachability and the amygdala: Insights from Williams syndrome. Neuropsychologia 47:2446–2453. [DOI] [PubMed] [Google Scholar]
- Meda SA, Pryweller JR, Thornton‐Wells TA (2012): Regional brain differences in cortical thickness, surface area and subcortical volume in individuals with williams syndrome. PLoS One 7:e31913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer‐Lindenberg A, Hariri AR, Munoz KE, Mervis CB, Mattay VS, Morris CA, Berman KF (2005): Neural correlates of genetically abnormal social cognition in Williams syndrome. Nat Neurosci 8:991–993. [DOI] [PubMed] [Google Scholar]
- Meyer‐Lindenberg A, Mervis CB, Berman KF (2006): Neural mechanisms in Williams syndrome: A unique window to genetic influences on cognition and behaviour. Nat Rev Neurosci 7:380–393. [DOI] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Hua X, Toga AW, Jack CR Jr, Schuff N, Weiner MW, Thompson PM (2009a): Automated 3D mapping of hippocampal atrophy and its clinical correlates in 400 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Hum Brain Mapp 30:2766–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Avedissian C, Madsen SK, Parikshak N, Toga AW, Jack CR Jr, Schuff N, Weiner MW, Thompson PM (2009b): Automated mapping of hippocampal atrophy in 1‐year repeat MRI data from 490 subjects with Alzheimer's disease, mild cognitive impairment, and elderly controls. Neuroimage 45:S3–S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morra JH, Tu Z, Apostolova LG, Green AE, Toga AW, Thompson PM (2010): Comparison of AdaBoost and support vector machines for detecting Alzheimer's disease through automated hippocampal segmentation. IEEE Trans Med Imaging 29:30–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz KE, Meyer‐Lindenberg A, Hariri AR, Mervis CB, Mattay VS, Morris CA, Berman KF (2010): Abnormalities in neural processing of emotional stimuli in Williams syndrome vary according to social vs. non‐social content. Neuroimage 50:340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolson R, DeVito TJ, Vidal CN, Sui Y, Hayashi KM, Drost DJ, Williamson PC, Rajakumar N, Toga AW, Thompson PM (2006): Detection and mapping of hippocampal abnormalities in autism. Psychiatry Res 148:11–21. [DOI] [PubMed] [Google Scholar]
- Palmer SJ, Tay ES, Santucci N, Cuc Bach TT, Hook J, Lemckert FA, Jamieson RV, Gunnning PW, Hardeman EC (2007): Expression of Gtf2ird1, the Williams syndrome‐associated gene, during mouse development. Gene Expr Patterns 7:396–404. [DOI] [PubMed] [Google Scholar]
- Pare D, Quirk GJ, Ledoux JE (2004): New vistas on amygdala networks in conditioned fear. J Neurophysiol 92:1–9. [DOI] [PubMed] [Google Scholar]
- Phan KL, Taylor SF, Welsh RC, Ho SH, Britton JC, Liberzon I (2004): Neural correlates of individual ratings of emotional salience: A trial‐related fMRI study. Neuroimage 21:768–780. [DOI] [PubMed] [Google Scholar]
- Plesa‐Skwerer D, Faja S, Schofield C, Verbalis A, Tager‐Flusberg H (2006): Perceiving facial and vocal expressions of emotion in individuals with Williams syndrome. Am J Ment Retard 111:15–26. [DOI] [PubMed] [Google Scholar]
- Plesa‐Skwerer D, Borum L, Verbalis A, Schofield C, Crawford N, Ciciolla L, Tager‐Flusberg H (2009): Autonomic responses to dynamic displays of facial expressions in adolescents and adults with Williams syndrome. Soc Cogn Affect Neurosci 4:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AL, Eckert MA, Rose FE, Karchemskiy A, Kesler S, Chang M, Reynolds MF, Kwon H, Galaburda A (2004): An experiment of nature: Brain anatomy parallels cognition and behavior in Williams syndrome. J Neurosci 24:5009–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A, Silva C, Rosset D, Deruelle C (2010): Just another face in the crowd: Evidence for decreased detection of angry faces in children with Williams syndrome. Neuropsychologia 48:1071–1078. [DOI] [PubMed] [Google Scholar]
- Sarpal D, Buchsbaum BR, Kohn PD, Kippenhan JS, Mervis CB, Morris CA, Meyer‐Lindenberg A, Berman KF (2008): A genetic model for understanding higher order visual processing: Functional interactions of the ventral visual stream in Williams syndrome. Cereb Cortex 18:2402–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saygin ZM, Osher DE, Augustinack J, Fischl B, Gabrieli JD (2011): Connectivity‐based segmentation of human amygdala nuclei using probabilistic tractography. Neuroimage 56:1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehlmeyer C, Schoning S, Zwitserlood P, Pfleiderer B, Kircher T, Arolt V, Konrad C (2009): Human fear conditioning and extinction in neuroimaging: A systematic review. PLoS One 4:e5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin LM, Liberzon I (2010): The neurocircuitry of fear, stress, and anxiety disorders. Neuropsychopharmacology 35:169–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano‐Castiella E, Schafer A, Reimer E, Turke E, Proger T, Lohmann G, Trampel R, Turner R (2011): Parcellation of human amygdala in vivo using ultra high field structural MRI. Neuroimage 58:741–748. [DOI] [PubMed] [Google Scholar]
- Stinton C, Elison S, Howlin P (2010): Mental health problems in adults with Williams syndrome. Am J Intellect Dev Disabil 115:3–18. [DOI] [PubMed] [Google Scholar]
- Swanson LW, Petrovich GD (1998): What is the amygdala? Trends Neurosci 21:323–331. [DOI] [PubMed] [Google Scholar]
- Tamburo RJ, Siegle GJ, Stetten GD, Cois CA, Butters MA, Reynolds CF III, Aizenstein HJ (2009): Amygdalae morphometry in late‐life depression. Int J Geriatr Psychiatry 24:837–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, De Zubicaray GI, Janke AL, Rose SE, Semple J, Hong MS, Herman DH, Gravano D, Doddrell DM, Toga AW (2004a): Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage 22:1754–1766. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Sowell ER, Gogtay N, Giedd JN, Rapoport JL, de Zubicaray GI, Janke AL, Rose SE, Semple J, Doddrell DM, Wang Y, van Erp TG, Cannon TD, Toga AW (2004b): Mapping cortical change in Alzheimer's disease, brain development, and schizophrenia. Neuroimage 23 ( Suppl 1):S2–S18. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Lee AD, Dutton RA, Geaga JA, Hayashi KM, Eckert MA, Bellugi U, Galaburda AM, Korenberg JR, Mills DL, Toga AW, Reiss AL (2005): Abnormal cortical complexity and thickness profiles mapped in Williams syndrome. J Neurosci 25:4146–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler D (1999): Wechlser Abbreviated Scale of Intelligence. San Antonio:Harcourt Assessment. [Google Scholar]
- Wechsler D (2003): Wechsler Intelligence Scale for Children, 4th ed (WISC‐IV) (4th ed) San Antonio:The Psychological Corporation. [Google Scholar]
- Wonderlick JS, Ziegler DA, Hosseini‐Varnamkhasti P, Locascio JJ, Bakkour A, van der Kouwe A, Triantafyllou C, Corkin S, Dickerson BC (2009): Reliability of MRI‐derived cortical and subcortical morphometric measures: effects of pulse sequence, voxel geometry, and parallel imaging. Neuroimage 44:1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Raine A, Narr KL, Colletti P, Toga AW (2009): Localization of deformations within the amygdala in individuals with psychopathy. Arch Gen Psychiatry 66:986–994. [DOI] [PMC free article] [PubMed] [Google Scholar]


