Abstract
We have developed a rigorous computational screening protocol to identify novel fragment-like inhibitors of N5-CAIR mutase (PurE), a key enzyme involved in de novo purine synthesis that represents a novel target for the design of antibacterial agents. This computational screening protocol utilizes molecular docking, graphics processing unit (GPU)-accelerated molecular dynamics and Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) free energy estimations to investigate the binding modes and energies of fragments in the active sites of PurE. PurE is a functional octamer comprised of identical subunits. The octameric structure, with its eight active sites, provided a distinct advantage in these studies because, for a given simulation length, we were able to place eight separate fragment compounds in the active sites to increase the throughput of the MM/PBSA analysis. To validate this protocol, we have screened an in-house fragment library consisting of 352 compounds. The theoretical results were then compared with the results of two experimental fragment screens, Nuclear Magnetic Resonance (NMR) and Surface Plasmon Resonance (SPR) binding analyses. In these validation studies, the protocol was able to effectively identify the competitive binders that had been independently identified by experimental testing, suggesting the potential utility of this method for the identification of novel fragments for future development as PurE inhibitors.
Keywords: Fragment-based drug discovery, Molecular dynamics, MM/PBSA, N5-CAIR mutase, PurE
INTRODUCTION
The rise of antibiotic-resistant bacteria has become so pronounced that it now threatens global public health on an unprecedented scale. Multidrug-resistant (MDR) bacterial strains, emerging bacterial pathogens and bioterrorism threat organisms have all emerged as new public health concerns.1 Furthermore, the economic and societal impact of bacterial resistance is pronounced, due to the greater expense of treatment, longer treatment durations, and decreased prognosis in resistant infections. Alarmingly, recent studies on deaths caused by just a portion of MDR organisms revealed that each year these infections result in an estimated 12,000 deaths in the U.S. and 29,000 deaths in European countries.2 Unfortunately, despite the increasing prevalence of drug resistance in bacterial pathogens, the rate of new antibacterial drugs entering the market is extremely low and most large pharmaceutical companies and many biotechnology companies have abandoned this research area altogether.3 Amongst the solutions to these critical issues are the identification and validation of new antibacterial drug targets and the design and application of novel screening methods for hit and lead identification.
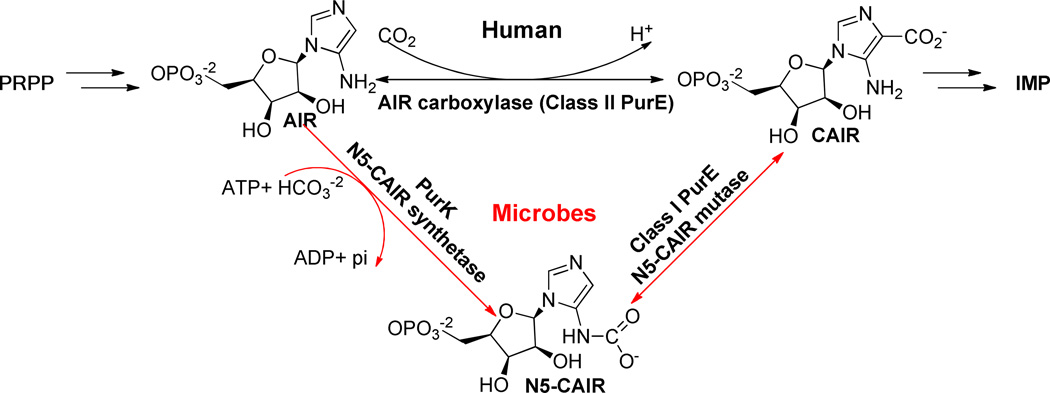
One promising, but largely unexplored pathway for antibacterial drug design is the de novo purine biosynthesis pathway. Purine biosynthesis is essential to all life forms except protozoa.4 Decreases in the virulence of purine auxotrophs have been reported in Salmonella5, Escherichia coli6, Bacillus anthracis6, 7, Yersinia pestis8, Staphylococcus aureus9 and Streptococcus pneumoniae10, demonstrating this pathway’s essentiality in key bacterial organisms. Recent studies have shown that there are key differences in the purine biosynthesis pathways between microbes and humans that can potentially be exploited for antibacterial drug discovery (Figure 1).11, 12 These differences are centered on the synthesis of a key intermediate, 4-carboxyaminoimidazole ribonucleotide (CAIR). In bacteria, two enzymes are required to synthesize this intermediate. First, carboxyaminoimidazole ribonucleotide (AIR) is first converted to N5-CAIR in an ATP-dependent manner by N5-CAIR synthetase (PurK). N5-CAIR is subsequently converted to CAIR in a second step by N5-CAIR mutase (Class I PurE). In humans, AIR is directly carboxylated by AIR carboxylase (Class II PurE). Biochemical studies have shown that the Class I and Class II PurE enzymes are different and highly specific for substrate selection; Class II PurE cannot use N5-CAIR as a substrate and Class I PurE cannot directly use AIR and CO2.13 Genetic studies have shown that microorganisms deficient in the purE gene are unable to grow in minimal media, human serum or mouse models.6, 9, 10, 14 Furthermore, PurE has been identified as a virulence factor and mutations that affect enzymatic activity significantly attenuated virulence.6, 10 Despite the demonstrated essentiality and novelty of this target, very few inhibitors, typically substrate mimics, for PurE have been reported, confirming the need to search for PurE inhibitors.15
Figure 1. Differences in de novo purine biosynthesis between microbes and humans.
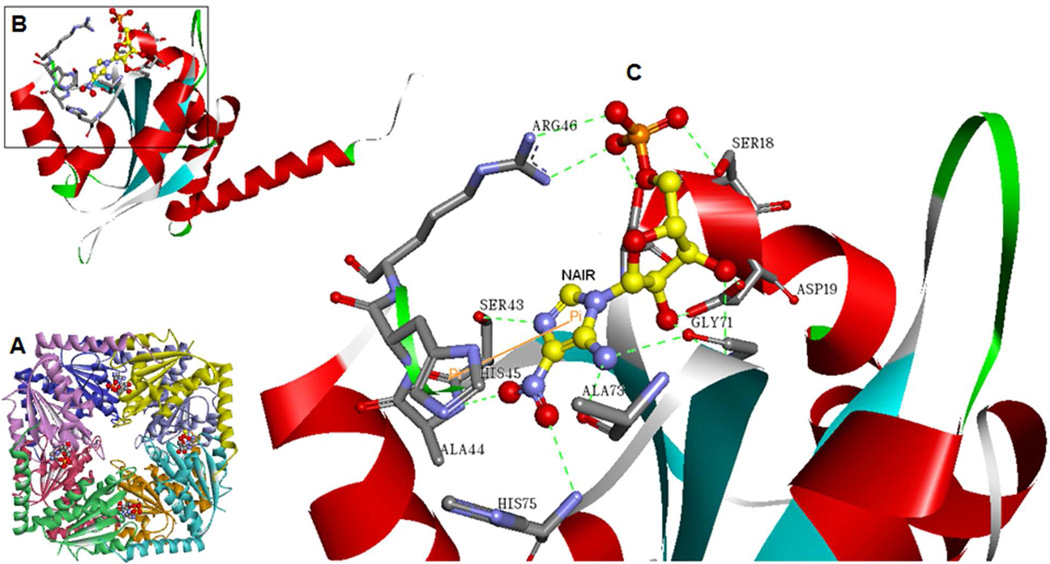
PurE is a functional octamer comprised of eight identical subunits with eight active sites at the interface of three monomers each (Figure 2).16 The crystal structure of E. coli PurE in complex with CAIR has revealed that the active site is solvent exposed, yet still contains a small, hydrophobic pocket. The phosphate-binding site is the most solvent exposed, and here the substrate forms hydrogen bonds with serine and arginine residues. The substrate’s ribose hydroxyl groups form hydrogen bonds with an aspartate residue and a glycine amide group. The substrate’s imidazole ring is buried in a small, hydrophobic pocket and forms hydrogen bonds with backbone groups. A conserved histidine residue is believed to serve as the acid and base for this reaction.13 A channel runs through the center of the octamer along the fourfold-symmetry axis and we propose that this could be a relatively promiscuous binding site for small molecules (see Discussion below). The octameric structure, with its eight active sites, provided a distinct advantage in our molecular dynamics (MD) based fragment studies. This is because, for a given simulation length (assuming independence of the active sites, see Discussion below), we were able to improve the throughput of our MD-based virtual screens against this drug target by simultaneously incorporating eight different ligands in one simulation run.
Figure 2. Structure of Class I PurE.
A. Ribbon diagram of the E. coli PurE octamer (PDB: 2ATE) with the eight monomers shown in different colors. 4-Nitroaminoimidazole ribonucleotide (NAIR), an analog of the product CAIR, is shown in ball-and-stick representation. B. Ribbon diagram of the PurE monomer, color-coded by secondary structure and with NAIR shown in ball-andstick representation. C. Close-up view of the active site residues associated with NAIR. Dashed lines represent hydrogen bonds. Images were prepared using Discovery Studio Visualizer 3.0, Accelrys, 2011.
Because the active site of PurE is relatively small, our laboratory has a keen interest in screening fragment libraries (molecular weight <300 Da) against this target. Fragment-based screening is now established as an emerging paradigm for drug discovery due to its efficient sampling of chemical space and high hit rates.17 Several recently described fragment-based lead discovery campaigns provide illustrative examples of the development of low-affinity fragments into high-affinity inhibitors against various targets.18–22 Because the binding affinities of fragments are much weaker (usually in 0.1–10 mM range) compared to drug-like compounds, sensitive biophysical screening methods are typically used to detect binding.23 There are two strategies in the practical application biophysical fragment screening; the first is to detect fragment binding only, without further information related to the binding site; while the second is able to provide detailed binding site information, albeit at a higher cost in terms of through-put and method complexity. Initial fragment screens are typically conducted by using the first strategy, using techniques such as Thermal Shift (TS), Surface Plasmon Resonance (SPR), Mass Spectrometry (MS), and non-competition NMR spectrometry. Secondary screening using techniques such as competition NMR spectrometry and/or X-ray crystallography is able to provide additional information regarding hit fragments binding specificity and even the exact binding site and conformation.
Experimental fragment-based screening has proven to be useful in drug discovery but is limited by low-throughput and high expense. Computational fragment-based drug design approaches can bypass these issues but suffer from limited representations of protein flexibility and solvation effects, which can lead to difficulties with the prediction of binding poses and energies for fragments. To overcome these limitations, we have combined molecular docking with molecular dynamics and Molecular Mechanics/Poisson-Boltzmann Surface Area (MM/PBSA) free energy estimations for the identification of novel, fragment-like compounds with PurE inhibitory activity. In order to validate this method and optimize the protocol, we have screened a small, in-house fragment library which consists of 352 compounds using MD-MM/PBSA and compared the results with experimental screens of the same library, including both initial screens using NMR and SPR, and a s secondary screen to confirm active site specificity by competition NMR spectrometry. AU-ROC and enrichment metrics have been used to compare the computational and experimental results from both the primary and secondary experimental screens and the results are compared with standard docking approaches.
MATERIALS AND METHODS
Protein and fragment library preparation
The X-ray crystal structure of E. coli PurE in complex with CAIR (PDB code 1D7A) was prepared using the Protein Preparation Wizard available in the Schrödinger Suite 2011. Histidine 45 was protonated due to its essential role in catalysis.13 Hydrogens and charges were added using the OPLS2005 force field and hydrogens were refined using restrained minimization. The fragments were prepared using Schrödinger’s LigPrep software.24 Ligands were desalted and tautomers generated. The OPLS2005 force field was used for geometric optimization. Generation of multiple ligand ionization states and tautomeric forms at pH 7.5 ± 0.5 was completed using Schrödinger’s EPIK software.25 A maximum of two stereoisomers were retained in prepared libraries in situations where fragment compounds contained chiral centers. Single low energy ring conformations were generated for fragment compounds containing non-aromatic ring systems. Additional settings for both protein and ligand preparation utilized default parameters.
Molecular docking and molecular dynamics simulations
Molecular docking was performed with GOLD v5.0.1.26 The binding site sphere was defined with a 10 Å radius around the AIR ligand present in the crystal structure, and 100 docking runs were performed for each fragment ligand. GOLD score was used as the scoring function and “Generate Diverse Solutions” was set to TRUE (Cluster Size = 2, rmsd = 1). Other fitness and search options were set to default settings unless otherwise stated below. Search efficiency genetic algorithm (GA) settings were set to 200% and default values were used for other GA settings. Two solutions of each compound (the best scoring and the most diverse/greatest RMSD from the best scoring conformation) were selected for the molecular dynamics studies discussed below.
All MD calculations in this study were performed using the Amber 11 suite of programs. The protein parameters used the ff99SB force field, and ligands were parameterized using antechamber with the GAFF force field and AM1-BCC charges.27–29 Each protein-ligand complex was solvated in an octahedral box of TIP3P water molecules extending 10 Å outside the protein on all sides, resulting in more than 19,000 waters added per complex.30 The electrostatics were treated using the particle-mesh Ewald method.31 The simulations used a residue-based long-range electrostatic cutoff of 8 Å and 2 femtosecond time steps. Bonds involving hydrogen atoms were constrained using the SHAKE algorithm.32 The solvated complexes were minimized using 10000 steps of conjugate gradient minimization, which was followed by MD equilibration for 600 picoseconds. The equilibration included heating to 300 K over 50 picoseconds, 50 picoseconds of density equilibration using 2 kcal mol−1 Å−2 restraints on the complex, and 500 picoseconds of constant pressure equilibration with 0.5 kcal mol−1 Å−2 restraints on the complex at 300K. After equilibration, 8 nanosecond production runs were performed to assess free energy convergence and coordinates were extracted every 2 picoseconds. Simulation stability was assessed by root mean squared deviation (RMSD) analyses, which were performed using the ptraj module of Amber 11 (see Supporting Information).
MM/PBSA, normal mode and ligand efficiency calculations
MM/PBSA was used to calculate fragment compound binding free energies.33 The binding free energy, ΔGbind, is calculated using a simple thermodynamic cycle from the energy difference between the complex and the unbound forms. The values for the free energy of binding of each fragment compound were calculated according to the following equation:
| (1) |
Where Gcomplex is the calculated Gibb’s free energy of the fragment-enzyme complex, Greceptor is the Gibb’s free energy for the apo-enzyme receptor, and Gligand is the Gibb’s free energy calculated for the ligand. The free energy values for each of these terms were estimated as the sum of the four terms shown here:
| (2) |
Where EMM is the molecular mechanics energy of the molecule expressed as the sum of the internal energy of the molecule plus the electrostatics and van der Waals interactions, Gpsolv is the polar contribution to the solvation energy of the molecule, Gnpsolv is the nonpolar solvation energy, T is the absolute temperature, and S is the entropy of the molecule estimated by normal mode analysis.
The python script, MMPBSA.py, included in AMBER11 was used to perform the MM/PBSA calculations.34 Snapshots for the calculations were taken every 20 picoseconds from the last 2 nanoseconds of the MD production runs, resulting in a total of one hundred snapshots per run. The three monomers forming one complete active site were defined as “receptor” for the ligand located in that particular active site for MM/PBSA calculations. The ionic strength in molarity was set to 0.1 and the internal dielectric constant was set as 2.0 (see Discussion below). Other options were set to default settings unless otherwise stated below. Both normal-mode analysis (NMA) and quasi-harmonic analysis (QHA) were employed to compute the vibrational, rotational, and translational entropy for the complex, receptor and ligand using a single trajectory protocol wherein the snapshots used in the individual calculations were extracted from a single MD trajectory of the complex system (see Discussion below). The results were averaged across all snapshots to obtain the estimated binding entropy. Ionic strength used in nmode calculations (in molarity) was set as 0.1. QHA was calculated using the ptraj program in AmberTools with default settings. Because of the high computational cost and considering that the deviation of entropy is relatively small for different conformations,35 we selected 12 regularly spaced snapshots along the last 2 ns production trajectory for the entropy calculations when using normalmode analysis.
Ligand efficiency (LE) values were calculated using the method of Hopkins, et al. by dividing calculated free energy (ΔG) by the number of heavy (non-hydrogen) atoms (NHA) for ranking compounds and a cutoff of 0.3 was chosen for virtual hits selection.36, 37
| (3) |
Enrichment Metrics
The virtual screening performance of each method was quantified by AU-ROC (the Area Under the Receiver-Operating Characteristic), enrichment factor at 5% and 10%, and TPR (true positive rate) at 5% and 10% FPR (false positive rate). One-sided p-values were calculated to test whether the virtual screening methods performed better than random, and two sided p-values were used to compare the results of the different methods. Detailed procedures have been described in a previous study,38 and the recommended statistical analyses were employed here as shown in the equations below:
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
where erfc(x) is the complementary error function, ΔAUROC is the difference in AUC values, SEΔ is the standard error in ΔAUROC, VarΔ,d is the variance associated with the mean of TPR, and VarΔ,a is the variance associated with the mean of FPR.
Protein expression and purification
The Bacillus anthracis PurE enzyme was used in all experimental binding studies. B. anthracis PurE was cloned, expressed, and purified as previously described.39 Briefly, the purE gene was PCR amplified from B. anthracis genomic DNA using primers designed in-house and cloned into the pET15b plasmid vector, which adds an N-terminal His6 tag for purifications and contains an ampicillin resistance cassette. After sequence confirmation, the plasmid vector was transformed into E. coli BL21-Gold (DE3) cells for expression. Cells were grown at 37 °C in TB (Terrific Broth) medium containing 100 µg/mL ampicillin until the optical density (OD) at 600 nm reached approximately 0.6 (approximately four hours) and induced with 1 mM IPTG for four additional hours at 37 °C, at which time the cells were harvested by centrifugation and lysed by sonication. Recombinant PurE was purified by affinity chromatography on a HisTrap HP column (GE Healthcare); enzyme eluting fractions were confirmed by SDS-PAGE, pooled and concentrated to 5 mg/mL. The His6 tag was cleaved by thrombin from PurE for 1 hour at 37 °C for enzyme kinetics measurements and SPR experiments. After thrombin digestion, samples were reloaded on to HisTrap HP column again to remove His6 tags and uncleaved His6-PurE. Flow through was collected and concentrated. The uncleaved protein was used in NMR experiments described below.
Cooperativity analysis and enzymatic assay
The Hill coefficient was calculated by fitting the activity data to the three Parameter Hill equations by OriginPro 8.5 (OriginLab, Inc.) using the following equation:
| (9) |
Where v is enzyme activity (velocity), [S] is the substrate concentration, K’ is a constant consisting of interacting terms for each binding site and the intrinsic dissociation constant, Ks, Vmax is the maximal activity from three independent assays, and n is the Hill coefficient, which describes the cooperativity of the reaction. The activity of the B. anthracis PurE enzyme was measured using a spectrophometric assay with the CAIR product, by following a decrease in the absorbance at 260 nm as CAIR is converted to the N5-CAIR substrate in the reverse reaction. The assay was performed in an assay buffer containing 50 mM Tris, pH 7.5 and 75 mM NaCl. A series of substrate concentrations (0 to 200 µM final concentration) were prepared and 100 µL each was distributed in 96-well UV microplates (Corning Inc.). The enzyme reaction was initiated by adding 100 µL PurE (10 nM final concentration). The plate was shaken for 30 seconds, and UV absorbance at 260 nm was monitored continuously for 10 minutes with an Envision 2104 Multilabel plate reader (Perkin Elmer). The same series of substrate concentrations without any enzyme was also measured as a control.
NMR and SPR experimental screening and competition binding assays
The in-house fragment library of 352 compounds (Zenobia Therapeutics) was tested at 200 µM in 8-fragment cocktail mixtures, using both Saturation Transfer Difference (STD) and WaterLOGSY NMR experiments. The samples were prepared in 50 mM phosphate buffer (pH 7.5) with 75 mM NaCl and 10% D2O. The PurE enzyme concentration was 20 µM. Fragment cocktail deconvolution was performed and individual fragment compounds tested when activity was noted. A small subset of fragments was further characterized for the specificity of binding by STD competition experiments (see Discussion below). In these experiments the same buffer and PurE concentration was used. The samples were prepared as 5-fragment cocktail mixtures, using 400 µM of each compound. After the first STD spectrum was collected, 500 µM CAIR was added to the sample and an additional STD spectrum was collected. All spectra were acquired at 10 °C with a Brucker 900 MHz NMR spectrometer with water and DMSO double solvent suppression STD pulse sequence. All NMR spectra were processed with the TopSpin3.1 (Bruker Biospin Ltd.) software. Competitive binding compounds were defined as those with greater than a 10% decrease in binding response after CAIR was added.
For surface plasmon resonance (SPR) experiments, the PurE enzyme was prepared in PBS buffer (10 mM phosphate, pH 7.4, 2.7 mM KCl, 137 mM NaCl) and immobilized on a CM5 sensor chip using standard amine-coupling at 20 °C with running buffer HBS-P (10 mM HEPES, 150 mM NaCl, 0.05% surfactant P-20, pH7.4) from GE healthcare. Flow channels 1 and 3 were activated by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/ N-hydroxy succinimide (NHS) mixture, and the activated surfaces were blocked using ethanolamine (pH 8.5) as controls. The PurE enzyme was diluted in 10 mM sodium acetate (pH 4.5) and immobilized to flow channel 2 and 4 after sensor surface activation with EDC/NHS with a 7 minutes injection followed by ethanolamine blocking on nonimmobilized surface area. PurE immobilization level of flow channel 2 and 4 were ~10,500 response units (RU) and ~11,500 RU, respectively. All fragment compounds were prepared as 10 mM stock in 100% DMSO and were diluted in the assay buffer (10 mM phosphate, pH 7.4, 2.7 mM KCl, 137 mM NaCl, and 2% DMSO) to 150 µM final concentration in 384-well plates. Each compound was injected for 30 seconds of association and 30 second of dissociation time at flow rates of 50 µL/min into all four channels. All data were referenced for blank injections on flow channel 1 and 3. All assays were done in duplicate. Compounds with a cut-off of 4 RU or greater were selected as hits.
RESULTS AND DISCUSSIONS
With over 400,000 fragment compounds available commercially,40 the attractiveness of an accurate fragment virtual screening method with reasonable throughput that can be utilized as a pre-filter for more traditional low-throughput methods, such as NMR or crystallography for hit confirmation, is readily apparent. With respect to the PurE target, the nature of the active site makes a fragment-based drug discovery approach particularly appealing. The site is shallow, solvent-exposed with a low volume of approximately 800 Å3 and maximum depth of 12.5 Å (Figure 2). However, difficulties with docking and scoring of fragment compounds have been well documented.41 Most scoring functions have been parameterized using drug-like compounds that are larger and more complex in terms of protein interactions than fragment compounds. These same scoring functions may also underestimate the entropy penalty to fragment compound binding, which have more translational and rotational freedom compared with drug-like compounds.
Strategies which utilize more complex energy functions, such as QM/MM, MM/PBSA, and MM/GBSA have increasingly been employed to improve computational predictions of fragment binding.42 Quantum mechanical approaches such as QM/MM, while accurate, are still too computationally intensive for virtual screening of large libraries, while the MM/GBSA and MM/PBSA approaches have generally been shown to lead to improved enrichments when used to rescore docked fragments, and are significantly less computationally expensive.43–46 Other approaches to binding energy predictions such as free energy perturbation (FEP), thermodynamic integration (TI), or linear interaction energy (LIE) calculations are less applicable to hit identification by virtual screening than they are to lead optimization due to their requirement for known active compounds for comparisons (FEP/TI) or training (LIE). For screening against the PurE target, the MM/PBSA method that was employed here can give the improvement in energy predictions that we sought, while still allowing for a reasonable screening throughput, as discussed below.
It should be noted that the crystal structure of the E. coli PurE was used in all computational studies, while the B. anthracis PurE enzyme was used in experimental studies. Although there is a B. anthracis structure available, it is an apo structure, while the E. coli structure contained the bound ligand (AIR) that facilitated the preparation and completion of the computational studies. We note the high overall identity and similarity between the two enzymes, 59% and 89%, respectively, and the 100% identity of the active site residues within five angstroms of the bound AIR ligand.
Enzyme Cooperativity Analysis
A key factor influencing the utility of the multiple binding site method described here is the effect of cooperativity on binding. The method we have described assumes that there is no positive or negative cooperativity influencing the fragment binding, whereby fragments that bind to one active site of the octamer influence the binding of successive fragments. In order to confirm this lack of cooperativity we performed detailed enzyme kinetics studies to determine the Hill coefficient of binding. The Hill coefficient is used to determine the stoichiometry of substrate-enzyme interactions and is modeled using Equation 9 as described above. In cases where there is no significant positive or negative cooperativity, the Hill coefficient will be near unity and Equation 9 reduces to the standard Michaelis-Menten equation. Hill coefficients significantly greater than 1 indicate positive cooperativity while those significantly less than 1 are indicative of negative cooperativity.47
The Hill coefficient for binding of CAIR to PurE was close to one (n =1.38 ± 0.04), which indicates a homogenous population of non-cooperative binding sites, meaning that ligand binding to one site did not significantly alter ligand affinity at other binding sites. Additionally, the crystal structures of apo E. coli PurE (PDB code 1QCZ) and its complexes with AIR (PDB code 1D7A) and NAIR (PDB code 2ATE) show nearly identical active sites, which suggests that there is no significant local conformational change upon ligand binding that causes conformational changes at nearby binding sites and thereby affecting their ligand affinity. Furthermore, the weak molecular interactions of fragment compounds with PurE are not expected to produce any significant induced fit effects on local or neighboring binding sites. Taken together, we feel that this molecular and structural data justifies our placement of different fragment compounds in each of the eight active sites of the PurE octamer for MD-MM/PBSA screening to increase the throughput of the protocol. Additionally, our use of fragment compounds and the negligible induced fit seen with the PurE system justifies our use of a single trajectory approach for MM/PBSA calculations, as discussed in Methods.48
Comparison of virtual methods with experimental results
384 fragment compounds (352 unique compounds and 32 tautomers/stereoisomers) were prepared and docked into eight separate active sites of PurE as discussed above (see Methods). In order to account for possible inaccuracies in fragment docking pose selection as well as a likely dependence of the final MD results on fragment starting position, we selected two conformations for each fragment compound to take into the MD-MM/PBSA simulations. The highest scoring pose for each fragment as well as the pose with the largest RMSD from the highest scoring pose were selected. In total, 96 MD simulation runs were performed with eight different ligands in each run. The lowest delta G estimated by MM/PBSA from the multiple docked conformations and tautomers/stereoisomers for each compound was selected for ligand efficiency calculation. Virtual ‘hits’ were defined as compounds with a calculated ligand efficiency of 0.3 or greater (see Methods).
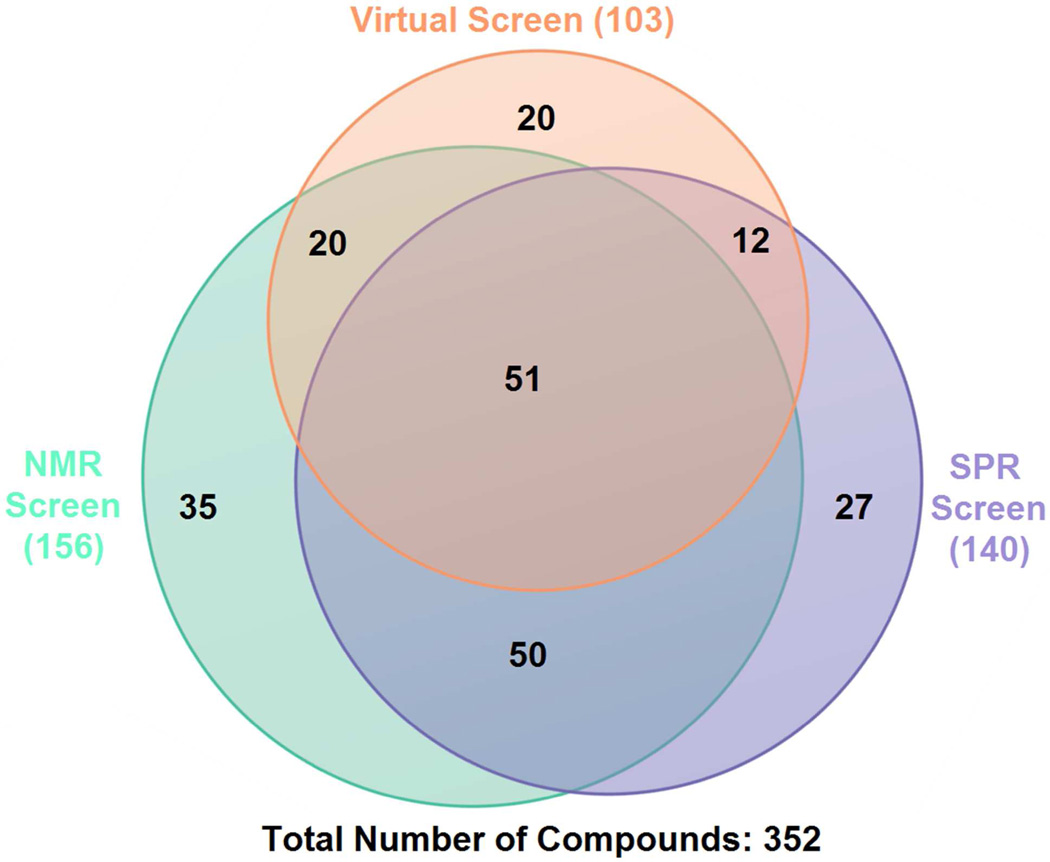
The full results of the virtual and experimental screens are included in Table S1 of the Supporting Information. Virtual screening using this MD-MM/PBSA protocol against our in-house fragment library (Zenobia Therapeutics) yielded 103 virtual hits (~30% virtual hit rate), 83 of which agreed with NMR and/or SPR studies (Figure 3). To evaluate the predictive power of the MM/PBSA protocol, receiver operator characteristic (ROC) curves were generated and compared along with calculated enrichment factors at 5%, 10% and true positive rate (TPR) at 5%, 10% false positive rate (FPR).49 ROC analysis is a powerful method for the evaluation and comparison of the predictive abilities of virtual screening methods.50 A comparison of the areas under ROC curves (AU-ROCs) for the different methods being tested allows for the direct comparison of the overall sensitivity and specificity of a given method. To quantify the significance of any observed difference in the AUC values of virtual screens compared to random selection and with each other, p-values were calculated as described above (see Methods). At a significance level (α) of 0.05, p-values less than 0.05 indicate significant improvement over random selection when one-sided p-values are calculated, and significant differences (significantly better or significantly worse) when two-sided p-values are calculated for other methods under comparison, while p-values greater than 0.05 indicate no significant improvement over random selection or significant difference from other methods under comparison. Enrichment factors were calculated to allow for the comparison of the early enrichment of the methods. Values of 5% and 10% were selected in this case for enrichment factors due to the smaller size of the test library. TPR at 5% and 10% FPR were further calculated for the comparison of the early enrichment ability between the different datasets as enrichment factor values are influenced by composition differences.51
Figure 3. Venn diagrams showing overlaps of hits from different screenings.
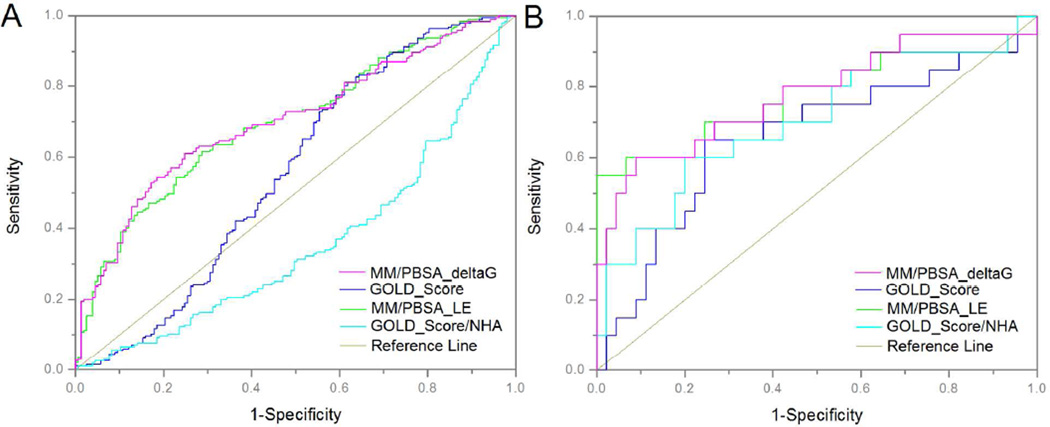
Table 1 summarizes the results of the protocol validation. When comparing standard docking with the MM/PBSA protocol against the full fragment library (with actives defined as those fragments determined to bind using the experimental methods) a significant improvement in enrichment factors, TPR at fixed FPR, and AU-ROC was seen with the MM/PBSA protocol. The MM/PBSA protocol showed almost two-fold enrichment over random selection, while the docking protocol performed under random selection (EF = 1) at both EF 5% and EF 10%. The difference in AU-ROC values (Figure 4A, Table 1) is statistically significant for the pairs under comparison except one pair (MM/PBSA with or without NHA weighting). The AU-ROC values (Figure 4A, Table 1) for the MM/PBSA protocol were significantly improved over the standard docking, which performed at or under the levels expected from random compound selection (AU-ROC = 0.5). For the MM/PBSA and standard docking protocols against the complete dataset, we further tested whether a heavy atom (NHA) weighted method improved the performance. There was no significant improvement associated with heavy atom (NHA) weighting compared to the original, non-weighted, results. We note that the enrichment factor and AU-ROC values, although improved over standard fragment docking performed here, are still lower than what is typically seen with validated docking protocols using drug-like compounds.52–54 We believe the reasons for this are two-fold: the first being the inherent difficulty of scoring fragments bound to a protein receptor, even using a more rigorous method such as MM/PBSA; the second being the possibility that the ‘actives’ used here, which were identified as binders using the experimental methods, may be nonspecific, or allosteric, binders. We discuss this second possibility in further detail below.
Table 1.
Performance of different VS methods against the complete data set and a 65 compound subset that has undergone CAIR competition studies.a
| Data Set |
VS Methods | Enrichment Factor 5% |
Enrichment Factor 10% |
TPR at 5% FPR |
TPR at 10% FPR |
AUROC | p-value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Random | MM/PBSA _ΔG |
Gold_Score /NHA |
Gold_Score | |||||||
| Complete set | MM/PBSA_LE | 1.60 | 1.55 | 0.25 | 0.39 | 0.70 | <0.01* | 0.76 | <0.01* | <0.01* |
| MM/PBSA_ΔG | 1.60 | 1.70 | 0.24 | 0.36 | 0.70 | <0.01* | <0.01* | <0.01* | ||
| GOLD_Score/NHA | 0.60 | 0.72 | 0.03 | 0.06 | 0.35 | 1.00 | <0.01* | |||
| GOLD_Score | 0.60 | 0.72 | 0.02 | 0.06 | 0.55 | 0.10 | ||||
| 65 compound subset | MM/PBSA_LE | 3.25 | 3.25 | 0.6 | 0.6 | 0.78 | <0.01* | 0.24 | 0.03* | <0.01* |
| MM/PBSA_ΔG | 3.25 | 2.79 | 0.50 | 0.60 | 0.79 | <0.01* | 0.03* | <0.01* | ||
| GOLD_Score/NHA | 2.17 | 2.79 | 0.30 | 0.30 | 0.70 | <0.01* | 0.01* | |||
| GOLD_Score | 2.17 | 1.39 | 0.15 | 0.2 | 0.66 | 0.01* | ||||
A one-side p-value is shown for comparison of AU-ROCs with random, a two-sided p-value is shown for each pair, and those below 0.05 are marked with an asterisk to emphasize significance at the 95% level.
Figure 4. ROC Comparison of MM/PBSA vs. GOLD Docking.
A. The complete data set is shown; actives are defined as compounds determined to bind PurE by experimental methods. B. A 65 compound subset that has undergone CAIR competition studies is shown; actives are defined as compounds competitive for the CAIR product in the PurE active site.
Comparison of virtual methods with NMR competition binding assay
Although the NMR and SPR methods described above (see Methods) are able to identify fragment compounds that bind to the PurE enzyme target, they are not able to directly distinguish between specific (active-site) binders and non-specific binders. As discussed in the introduction, the PurE octameric structure contains a large, central channel which may serve as a promiscuous binding site for fragment compounds (Figure 2). For this reason, some of the compounds identified as ‘actives’ in the validation study discussed above may be false positives that would not be identified as a ‘hit’ using the computational protocols, which may explain in part the low EF and AU-ROC values for both the docking and MM/PBSA protocols. In order to confirm active site binding of the fragments identified experimentally, we have tested a subset of the fragment ‘binders’ using a CAIR competition binding assay that employed Saturation Transfer Difference (STD) NMR (see Methods). A limited amount of the CAIR substrate (synthesized in-house) prevented the testing of all experimental binders. Fragment compounds were selected for testing based upon their high signal-to-noise ratio (greater than five) which typically indicates stronger binding; and compounds were classified as active-site, or competitive binders, if they showed a greater than 10% decrease in binding response after the CAIR ligand was added.
Of the 156 unique fragment compounds that were identified as binding by NMR methods, we selected 65 compounds for testing in the CAIR competition assay. Of the 65 compounds selected for testing, 36 had also been identified as hits by the MM/PBSA protocol, as discussed above. Twenty fragment compounds, roughly one-third of the tested subset, were confirmed to bind to the PurE active site by the CAIR competition assay, which confirmed our suspicions that a large number of ‘binders’ were non-specific. Encouragingly, 16 of the 20 active site binders had also been identified as hits by our MM/PBSA protocol. A representative, competitive fragment binder identified and characterized by SPR, competition NMR and computational predictions is shown in Figure 5. The ability of our protocol and standard docking to discriminate the active-site binders (confirmed competition with CAIR) from noncompetitive binders was further examined by ROC analysis, enrichment factors and TPR at fixed FPR; similar to the full library studies discussed above. The results of these studies are listed in Table 1 and shown in Figure 4B.
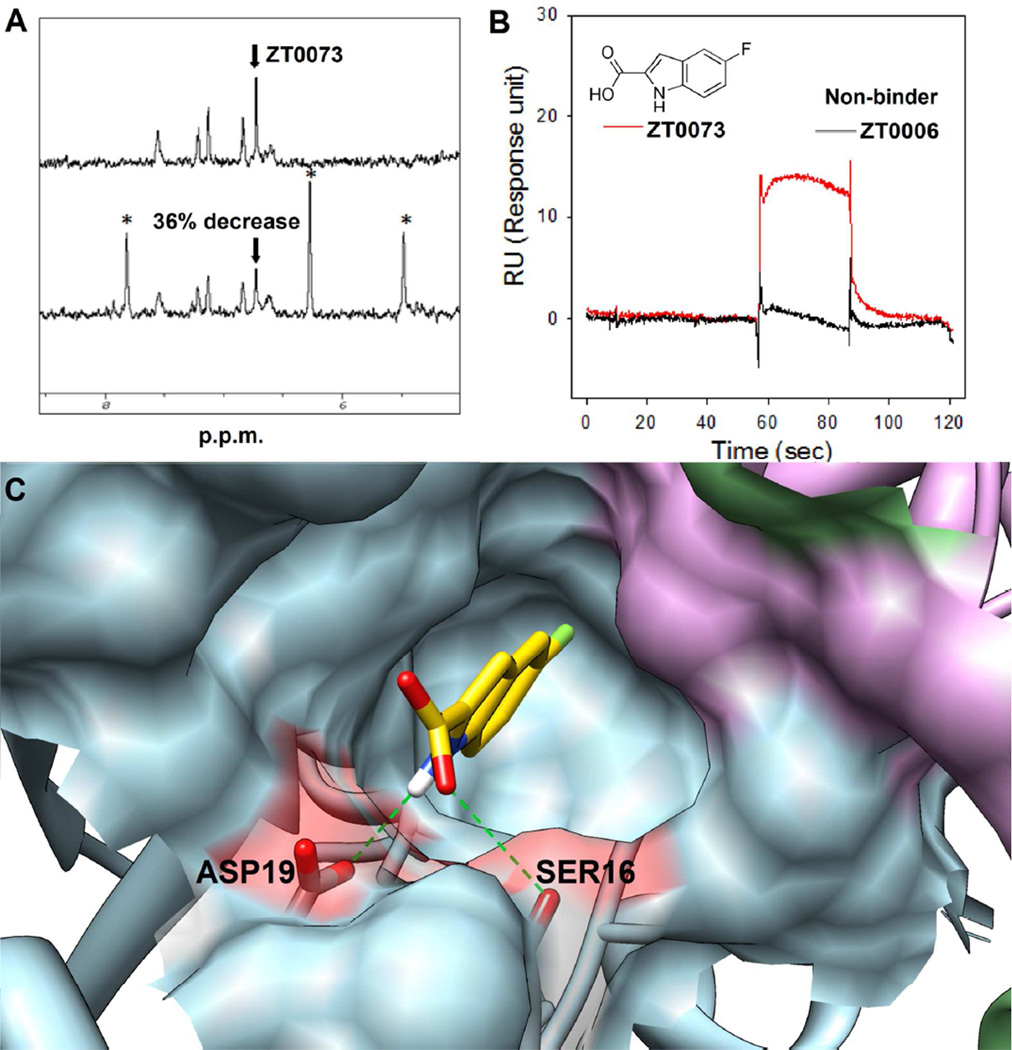
Figure 5. Identification and characterization of a novel fragment binder, ZT0073.
A. The NMR spectrum of compound ZT0073 (top) shows a decrease in integration following addition of CAIR (bottom, asterisks). B. SPR sensor grams of ZT0073 colored in red and a non-binder (ZT0006) colored in black as reference. C. Computational binding prediction by MD-MM/PBSA protocol: three monomers forming the complete active site are colored as blue, plum and green; ZT0073 is colored in yellow; hydrogen bond is shown in green dash lines. The surface of two residues which forming hydrogen bonds with the ligand is highlighted in red. Images were prepared using Chimera v1.6.1, UCSF, 2012.64
Significant improvements were seen in the early enrichment metrics, EF at 5%, 10% and TPR at 5%, 10% FPR, for both MM/PBSA and traditional docking, with MM/PBSA outperforming traditional docking for both heavy atom weighted and non-weighted metrics. The greater than 3-fold enrichment seen with the MM/PBSA protocol is more closely aligned with the enrichments seen with traditional docking of drug-like compounds, which indicates that non-specific binders, or false-positives, had affected the results, at least in part, described above. The difference in AU-ROC values (Figure 4B, Table 1) is statistically significant for the pairs under comparison except one pair (MM/PBSA with or without NHA weighting). The AU-ROC values (Figure 4B, Table 1) for the MM/PBSA protocol were also significantly improved over the standard docking. Significant improvement was also observed with heavy atom (NHA) weighted GOLD scores compared to non-weighted scores. For the MM/PBSA and standard docking protocols, additional performance testing was completed for comparison of the 65- compound subset to the complete data set, partly due to this composition difference noted above. Noticeable improvement was seen in the early enrichment ability as significant differences were observed for TPR at 5% and 10% FPR for both the MM/PBSA protocol (~ 2 fold differences) and the traditional docking method (~ 8 fold differences). The AU-ROC values were also improved, modestly for the MM/PBSA protocol, and significantly for the traditional docking method. This improvement in the performance of docking for the 65-compound subset over the full dataset can be explained in part by the binding affinity differences between the two datasets. Because the compounds in the subset were selected by their higher signal-to-noise, their binding affinities are expected to generally be higher than the full fragment set. Docking and scoring of compounds with high ligand efficiencies (LE) is believed to be more accurate than that for low LE compounds, as suggested by the studies conducted by Verdonk and coworkers at Astex Pharmaceuticals.55
Comparison of MM/PBSA with standard docking
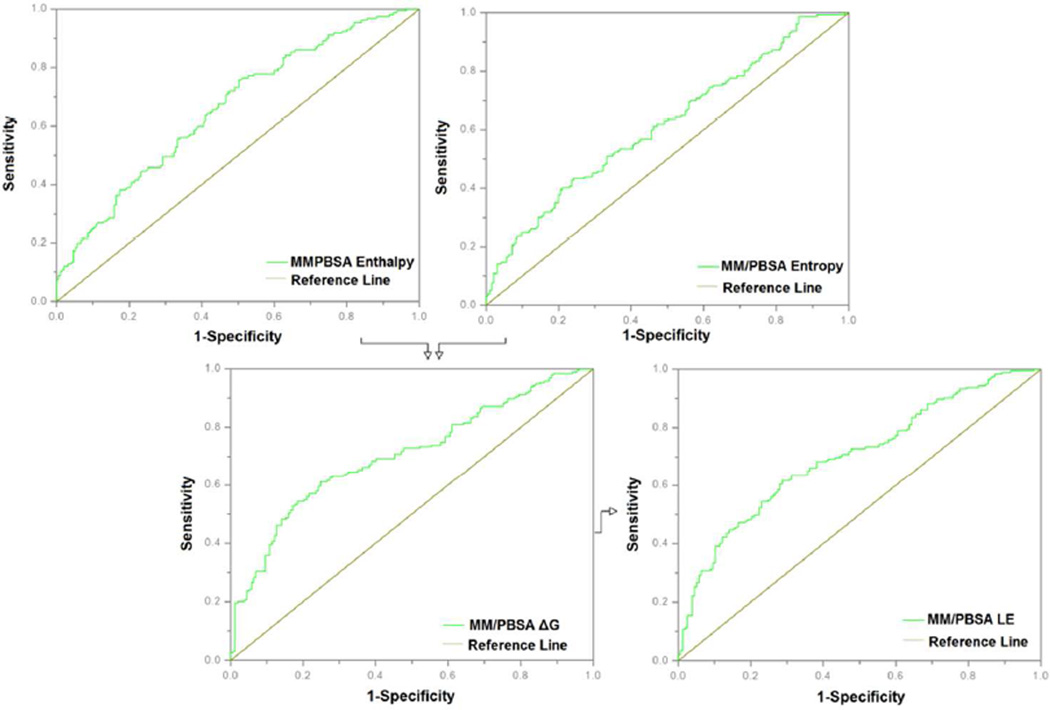
The improvement in scoring efficiency discussed above for the MD-MM/PBSA protocol over traditional docking can be attributed to several factors, including the conformational refinement of docking poses by molecular dynamics, an unbiased energy function, and the more detailed treatment of the entropic contribution to binding in the energy function. Entropy is commonly estimated by one of two methods: normal-mode analysis (NMA) and quasi-harmonic analysis (QHA). It has been shown that QHA can provide more accurate and efficient entropy estimations than NMA.56, 57 In these studies, we have applied both methods of entropy calculation (See Methods) and performed a cross-comparison of the results with respect to the enrichment values seen with the 65 compound subset (See Supporting Information, Figure S1). We noted that in our system QHA yielded much larger entropy values which appeared to be unrealistic leading to positive free energies of binding in many cases. This overestimation of entropy has been reported in other studies.58, 59 Due to this observation, all subsequent studies utilized NMA as the method for entropy calculations. Figure 6 shows an ROC analysis of the contributions of the enthalpic and entropic MM/PBSA components of the overall binding energy to the fragment screening enrichment for the complete fragment set. The AU-ROC for the enthalpy component was 0.66 and the AU-ROC for the entropy component was 0.61, while the overall AU-ROC was 0.7. The overall improvement in enrichment upon the combination of the two components was modest; however a significant improvement in early enrichment is readily apparent. Similar results were seen when the same analysis was applied to the competing fragment subset: enthalpy AU-ROC 0.76, entropy AU-ROC 0.59, overall 0.79, and a noticeable improvement in early enrichment (see Supporting Information, Figure S2). We observed a marked difference between the shapes of the enthalpy ROC, entropy ROC and overall ROC between the complete set and the competitive compounds subset. We attribute this observed phenomenon to the difference in compound composition and binding affinities between the two compound sets.
Figure 6. Effect of MM/PBSA components on enrichment.
The contribution of enthalpy and entropy to overall binding energy and fragment screening enrichment are shown here using ROC plots.
MM/PBSA and MM/GBSA methods combined with MD have recently emerged as relatively fast and accurate methods to estimate binding free energy. MD simulations using programs such as AMBER29 are still relatively computationally expensive and high-throughput screening using MD-MM/PBSA protocol is admittedly more time consuming compared to traditional docking methods. In our studies, two strategies were employed in order to reduce the running time and achieve higher throughput: GPU acceleration of AMBER MD simulations and simultaneously incorporating different ligands in one simulation run. Early GPU vs. CPU benchmarking studies performed in-house (data not shown) indicated that for the PurE system we could achieve approximately 3x acceleration using GPU’s over CPU’s for the MD and MM/PBSA calculations (10 ns per day vs. 3 ns per day at maximum efficiency). The time for one run (8 compounds in the octamer) is approximately 500–600 CPU/GPU hours (roughly 48 hours on a 2-GPU/12-CPU node). We believe this is in a practical time range for screening the small to medium size libraries typically seen with fragment sets, if one is utilizing parallel processing resources such as the National Science Foundation’s Extreme Science and Engineering Discovery Environment (XSEDE). By way of comparison, in our studies the docking (384 compounds) was performed on a 4-processor (CPU) in-house workstation in approximately six hours, while the MD-MM/PBSA (768 compounds) was accomplished in just under 30 days on the TACC Lonestar cluster which contains eight GPU nodes, each consisting of two NVIDIA M2070 GPU’s (448 cores each) and two Intel Xeon hexa-core CPUs. With the availability of cluster resources such as this and others, as well as the relatively low-cost and accessibility of GPU workstations, the screening of larger fragment libraries using MD-MM/PBSA or MM/GBSA-based methods is becoming more realistic.
Effect of solute dielectric constant, simulation time, and multiple ligand conformations
A dielectric constant (εin) of unity (1.0) is typically used for the solute in simulations such as these. However, for a highly charged binding interface, higher εin values have been recommended to consider the electronic polarization effect.60, 61 In order to optimize εin, we compared the experimental binding free energies of CAIR (Km = 22.2 ± 4.6 µM) and AIR (Ki = 236 ± 15 µM)62 for PurE to values computed theoretically using three different solute dielectric constants (1.0, 2.0, and 4.0). The results are listed in Table 2. The results shown indicate that the calculations are sensitive to the value of solute dielectric constant. The calculated experimental ΔG values for CAIR and AIR show that the CAIR ligand has a higher binding affinity for the PurE enzyme than the AIR ligand (a decarboxylation product of CAIR as well as a substrate for PurK, the precursor enzyme to PurE in this pathway). By using a value of εin = 2 over εin = 1 or εin = 4 we were better able to differentiate the two compounds by relative binding free energy, although the absolute binding free energies of these two ligands was more closely calculated using εin = 1. In addition, we have compared the effects of different solute dielectric constant values on the overall MM/PBSA virtual screening performance. The difference in AU-ROC values (Figure S1, Table 2) is statistically significant for the pairs under comparison. The AU-ROC value (Figure S1, Table 2) for εin = 2 was significantly improved over εin = 1 or εin = 4. The active site of PurE is solvent exposed and there are two charged residues in the active site of PurE, Arg46 and Asp19. Because these charged residues are likely to form ionic interactions with charged groups of the fragment sets, we felt that the larger value for the solute dielectric constant was justified.
Table 2.
The binding energy of CAIR and AIR along with the areas under ROC curves (AU-ROCs) against the 65 compounds subset computed using different solute dielectric constants and simulation lengths in the MM/PBSA calculations.
| ΔGexp1 (kcal/ mol) |
εin = 1 | εin = 2 | εin = 4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔGcal (kcal/mol) | AUROC2 | ΔGcal (kcal/mol) | AUROC | p-value3 | ΔGcal (kcal/mol) | AUROC | |||||||||
| εin=1 | εin=4 | ||||||||||||||
| 2ns | 4ns | 8ns | 8ns | 2ns | 4ns | 8ns | 8ns | 2ns | 4ns | 8ns | 8ns | ||||
| CAIR | −6.2 | −15.3 ± 5.2 | −16.1 ± 5.1 | −17.3 ± 4.3 | 0.72 <0.01* |
−32.5 ± 8.2 | −32.8 ± 7.6 | −33.0 ± 6.2 | 0.78 <0.01* |
0.01* | <0.01* | −24.1 ± 6.7 | −24.3 ± 7.2 | −25.7 ± 5.3 | 0.64 0.04* |
| AIR | −4.9 | −16.0 ± 6.2 | −15.8 ± 5.1 | −16.2 ± 3.9 | −24.1 ± 7.3 | −24.0 ± 7.1 | −25.3 ± 5.9 | −21.3 ± 6.1 | −21.1 ± 5.6 | −22.6 ± 5.3 | |||||
Calculated from reported Km value for CAIR and Ki value for AIR, reverse reaction assumes rapid equilibrium kinetics (Km = Ks)
The areas under ROC curves (AU-ROCs) for the different solute dielectric constants were shown for the direct comparison of the overall sensitivity and specificity against the 65 compounds subset that has undergone CAIR competition studies; a one-side p-value is shown for comparison with random and those below 0.05 are marked with an asterisk to emphasize significance at the 95% level.
A two-sided p-value is shown for comparison of εin = 2 with εin = 1 and εin = 4, and those below 0.05 are marked with an asterisk to emphasize significance at the 95% level.
In order to investigate the effect of the simulation length on predictions, we conducted a comparative study of free energy calculations by using three different MD simulation production lengths: 2 ns (100 snapshots), 4 ns (200 snapshots), and 8 ns (400 snapshots). The results are listed in Table 2. The length of MD simulations had only slight impact on the ΔG predictions. In order to demonstrate system equilibration, we calculated the enzyme backbone and ligand RMSD values for the complexes of CAIR and AIR with PurE (shown in Figure S3, Supporting Information). The CAIR/PurE system achieved equilibration after approximately 2 ns, and AIR/PurE system achieved equilibration after approximately 4 ns. To further demonstrate system stability, the fluctuations of enthalpies for CAIR/PurE and AIR/PurE were also calculated over the different time courses (shown in Figure S4, Supporting Information). The enthalpies were variable, but the mean values became stable after short MD simulation lengths of approximately 2 ns. The estimated binding energies between ligands and receptors are not extremely sensitive to fine conformational adjustments, which can occasionally be reflected in the appearance of energy convergence (fast energy convergence), while the system’s conformation appears to have yet converged (slow conformational convergence).53 We observed this phenomenon in our studies and we believe that it can be partly explained by the fragment-like nature of the compounds in our screening sets. Although the data suggested that shorter MD simulations would be justified (particularly for analysis of larger libraries), we chose 8 ns as the length of MD simulations in these studies because the initial complexes of fragments with the PurE enzyme were computationally derived and the test fragment library was relatively small.
The effects of using multiple ligand poses from the docking results versus a single, high-scoring docking pose in the MD simulations were also compared. ROC analysis for both scenarios was performed (shown in Figure S5, Supporting Information). As expected, using multiple conformations improved the performance of the MM/PBSA protocol over the use of a single pose. This was particularly evident when the analysis was applied to the competitive fragments subset, which showed significant improvement with the multiple poses over the single pose. An analysis of the poses that resulted in the best ligand efficiencies for each compound revealed that 49% from the complete set and 43% from the subset resulted from the RMSD poses rather than the highest scoring pose. Docking pose selection of fragments can be more difficult compared to drug-like compounds because the energy differences between possible binding modes are small and difficult to detect using standard scoring functions. While a typical drug-like molecule will only fit into an active site in limited ways, a fragment will often have many more binding modes due to its smaller size and weaker interactions. For these reasons, it has been recommended that multiple, diverse conformations of ligands be used to improve the performance of fragment-based virtual screening protocols.63 Considering the relatively small active site of PurE and the high computational cost of using multiple poses, we only selected two diverse docking poses for the MD-MM/PBSA studies described here.
CONCLUSIONS
Fragment-based approaches and MD-based virtual screening methods are being increasingly utilized in drug discovery. The combination of these two techniques can provide new avenues to efficiently sample chemical space in inhibitor design. Herein, we have described the development of a novel fragment-based, MD-MM/PBSA virtual screening protocol to identify potential inhibitors of antibacterial target, PurE. The protocol was able to effectively identify the weak binders that had been confirmed by experimental testing. By simultaneously incorporating GPU acceleration and the use of multiple, distinct fragment compounds in one simulation run, we were able to improve the throughput of our MD-based virtual screens to reach a time scale that is realistic for the screening of medium- to large-size fragment libraries, depending on the resources available. The virtual screening protocol described here is currently being employed to screen larger fragment libraries to prioritize compounds for purchase and experimental testing against this and other targets, significantly reducing the time and expense of experimental testing.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Defense Threat Reduction Agency; contract HDTRA1-11-C-0011. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number OCI-1053575. KEH was supported during a portion of this work by NIDCR 5T32-DE018381, UIC College of Dentistry, MOST program. The authors wish to acknowledge the work of Ms. Loredana Huma, who contributed to the synthesis of the CAIR ligand used in competition experiments. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081). We thank ChemAxon, Ltd. for an academic research license of their cheminformatics suite including JChem and JChem for Excel for data analysis.
Abbreviations
- MM/PBSA
Molecular Mechanics/Poisson-Boltzmann Surface Area
- NMR
Nuclear Magnetic Resonance
- SPR
Surface Plasmon Resonance
- MD
Molecular Dynamics
- VS
Virtual Screening
- CAIR
4-Carboxyaminoimidazole Ribonucleotide
- AIR
Carboxyaminoimidazole Ribonucleotide
- NAIR
4-Nitroaminoimidazole Ribonucleotide
- N5-CAIR
N5-Carboxyaminoimidazole Ribonucleotide
- RMSD
Root Mean Square Deviation
- STD
Saturation Transfer Difference
- WaterLOGSY
water–ligand observed via gradient spectroscopy
- GPU
Graphics Processing Rnit
- RU
Response Units
- MDR
Multidrug-Resistant
- NMA
Normal Mode Analysis
- QHA
Quasi-Harmonic Analysis
- AU-ROC
Area Under the Receiver-Operating Characteristic
- TPR
True Positive Rate
- FPR
False Positive Rate
Footnotes
SUPPORTING INFORMATION
The full results of the virtual screen and experimental (NMR, SPR) screens, RMSD plots demonstrating MD equilibration, and ROC analyses of the effect of multiple ligand docking poses, different entropy calculation methods and solute dielectric constants are included in Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discovery. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 2.Zell BL, Goldmann DA. Healthcare-associated infection and antimicrobial resistance: moving beyond description to prevention. Infect. Control Hosp. Epidemiol. 2007;28:261–264. doi: 10.1086/513722. [DOI] [PubMed] [Google Scholar]
- 3.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 4.Downie MJ, Kirk K, Mamoun CB. Purine salvage pathways in the intraerythrocytic malaria parasite Plasmodium falciparum. Eukaryot. Cell. 2008;7:1231–1237. doi: 10.1128/EC.00159-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McFarland WC, Stocker BA. Effect of different purine auxotrophic mutations on mouse-virulence of a Vi-positive strain of Salmonella dublin and of two strains of Salmonella typhimurium. Microb. Pathog. 1987;3:129–141. doi: 10.1016/0882-4010(87)90071-4. [DOI] [PubMed] [Google Scholar]
- 6.Samant S, Lee H, Ghassemi M, Chen J, Cook JL, Mankin AS, Neyfakh AA. Nucleotide biosynthesis is critical for growth of bacteria in human blood. PLoS Pathog. 2008;4:e37. doi: 10.1371/journal.ppat.0040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanovics G, Marjai E, Dobozy A. The growth of purine mutants of Bacillus anthracis in the body of the mouse. J. Gen. Microbiol. 1968;53:147–162. doi: 10.1099/00221287-53-2-147. [DOI] [PubMed] [Google Scholar]
- 8.Straley SC, Harmon PA. Growth in mouse peritoneal macrophages of Yersinia pestis lacking established virulence determinants. Infect. Immun. 1984;45:649–654. doi: 10.1128/iai.45.3.649-654.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mei JM, Nourbakhsh F, Ford CW, Holden DW. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 10.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He B, Shiau A, Choi KY, Zalkin H, Smith JM. Genes of the Escherichia coli pur regulon are negatively controlled by a repressor-operator interaction. J. Bacteriol. 1990;172:4555–4562. doi: 10.1128/jb.172.8.4555-4562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni L, Guan K, Zalkin H, Dixon JE. De novo purine nucleotide biosynthesis: cloning, sequencing and expression of a chicken PurH cDNA encoding 5-aminoimidazole-4-carboxamide-ribonucleotide transformylase-IMP cyclohydrolase. Gene. 1991;106:197–205. doi: 10.1016/0378-1119(91)90199-l. [DOI] [PubMed] [Google Scholar]
- 13.Hoskins AA, Morar M, Kappock TJ, Mathews II, Zaugg JB, Barder TE, Peng P, Okamoto A, Ealick SE, Stubbe J. N5-CAIR mutase: role of a CO2 binding site and substrate movement in catalysis. Biochemistry. 2007;46:2842–2855. doi: 10.1021/bi602436g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Mushegian A, Lory S, Jin S. Large-scale isolation of candidate virulence genes of Pseudomonas aeruginosa by in vivo selection. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10434–10439. doi: 10.1073/pnas.93.19.10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Firestine SM, Poon SW, Mueller EJ, Stubbe J, Davisson VJ. Reactions catalyzed by 5-aminoimidazole ribonucleotide carboxylases from Escherichia coli and Gallus gallus: a case for divergent catalytic mechanisms. Biochemistry. 1994;33:11927–11934. doi: 10.1021/bi00205a031. [DOI] [PubMed] [Google Scholar]
- 16.Mathews II, Kappock TJ, Stubbe J, Ealick SE. Crystal structure of Escherichia coli PurE, an unusual mutase in the purine biosynthetic pathway. Structure. 1999;7:1395–1406. doi: 10.1016/s0969-2126(00)80029-5. [DOI] [PubMed] [Google Scholar]
- 17.Haider MK, Bertrand HO, Hubbard RE. Predicting Fragment Binding Poses Using a Combined MCSS MM-GBSA Approach. J. Chem. Inf. Model. 2011;51:1092–1105. doi: 10.1021/ci100469n. [DOI] [PubMed] [Google Scholar]
- 18.Efremov IV, Vajdos FF, Borzilleri KA, Capetta S, Chen H, Dorff PH, Dutra JK, Goldstein SW, Mansour M, McColl A, Noell S, Oborski CE, O'Connell TN, O'Sullivan TJ, Pandit J, Wang H, Wei B, Withka JM. Discovery and Optimization of a Novel Spiropyrrolidine Inhibitor of beta-Secretase (BACE1) through Fragment-Based Drug Design. J. Med. Chem. 2012;55:9069–9088. doi: 10.1021/jm201715d. [DOI] [PubMed] [Google Scholar]
- 19.Bamborough P, Diallo H, Goodacre JD, Gordon L, Lewis A, Seal JT, Wilson DM, Woodrow MD, Chung CW. Fragment-based discovery of bromodomain inhibitors part 2: optimization of phenylisoxazole sulfonamides. J. Med. Chem. 2012;55:587–596. doi: 10.1021/jm201283q. [DOI] [PubMed] [Google Scholar]
- 20.Cheng Y, Judd TC, Bartberger MD, Brown J, Chen K, Fremeau RT, Hickman D, Hitchcock SA, Jordan B, Li V, Lopez P, Louie SW, Luo Y, Michelsen K, Nixey T, Powers TS, Rattan C, Sickmier EA, St Jean DJ, Wahl RC, Wen PH, Wood S. From Fragment Screening to In Vivo Efficacy: Optimization of a Series of 2-Aminoquinolines as Potent Inhibitors of Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) J. Med. Chem. 2011;54:5836–5857. doi: 10.1021/jm200544q. [DOI] [PubMed] [Google Scholar]
- 21.Murray CW, Carr MG, Callaghan O, Chessari G, Congreve M, Cowan S, Coyle JE, Downham R, Figueroa E, Frederickson M, Graham B, McMenamin R, O'Brien MA, Patel S, Phillips TR, Williams G, Woodhead AJ, Woolford AJ. Fragment-based drug discovery applied to Hsp90. Discovery of two lead series with high ligand efficiency. J. Med. Chem. 2010;53:5942–5955. doi: 10.1021/jm100059d. [DOI] [PubMed] [Google Scholar]
- 22.Englert L, Silber K, Steuber H, Brass S, Over B, Gerber HD, Heine A, Diederich WE, Klebe G. Fragment-based lead discovery: screening and optimizing fragments for thermolysin inhibition. ChemMedChem. 2010;5:930–940. doi: 10.1002/cmdc.201000084. [DOI] [PubMed] [Google Scholar]
- 23.Scott DE, Coyne AG, Hudson SA, Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry. 2012;51:4990–5003. doi: 10.1021/bi3005126. [DOI] [PubMed] [Google Scholar]
- 24.Suite 2012: LigPrep, version 2.5, version 2.5. New York, NY: Schrödinger, LLC; 2012. [Google Scholar]
- 25.Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J. Comput.-Aided Mol. Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 26.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 27.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts BP, Wang B, Hayik S, Roitberg A, Seabra G, Kolossvary I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh M-J, Cui G, Roe DR, Mathews DH, Seetin MG, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA. AMBER. Vol. 11. San Francisco: University of California; 2010. [Google Scholar]
- 28.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 29.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 31.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 32.Ryckaert JP, Ciccotti G, Berendsen HJC. Numerical-Integration of Cartesian Equations of Motion of a System with Constraints - Molecular-Dynamics of N-Alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 33.Wong S, Amaro RE, McCammon JA. MM-PBSA Captures Key Role of Intercalating Water Molecules at a Protein-Protein Interface. J. Chem. Theory Comput. 2009;5:422–429. doi: 10.1021/ct8003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller BR, McGee DT, Swails JM, Homeyer N, Gohlke H, Roitberg AE. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012;8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 35.Wang J, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 36.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discovery Today. 2004;9:430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka D, Tsuda Y, Shiyama T, Nishimura T, Chiyo N, Tominaga Y, Sawada N, Mimoto T, Kusunose N. A practical use of ligand efficiency indices out of the fragment-based approach: ligand efficiency-guided lead identification of soluble epoxide hydrolase inhibitors. J. Med. Chem. 2011;54:851–857. doi: 10.1021/jm101273e. [DOI] [PubMed] [Google Scholar]
- 38.Craig IR, Essex JW, Spiegel K. Ensemble docking into multiple crystallographically derived protein structures: an evaluation based on the statistical analysis of enrichments. J. Chem. Inf. Model. 2010;50:511–524. doi: 10.1021/ci900407c. [DOI] [PubMed] [Google Scholar]
- 39.Boyle MP, Kalliomaa AK, Levdikov V, Blagova E, Fogg MJ, Brannigan JA, Wilson KS, Wilkinson AJ. Crystal structure of PurE (BA0288) from Bacillus anthracis at 1.8 A resolution. Proteins. 2005;61:674–676. doi: 10.1002/prot.20599. [DOI] [PubMed] [Google Scholar]
- 40.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. ZINC: A Free Tool to Discover Chemistry for Biology. J. Chem. Inf. Model. 2012;52:1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandor M, Kiss R, Keseru GM. Virtual fragment docking by Glide: a validation study on 190 protein-fragment complexes. J. Chem. Inf. Model. 2010;50:1165–1172. doi: 10.1021/ci1000407. [DOI] [PubMed] [Google Scholar]
- 42.Gleeson MP, Gleeson D. QM/MM as a tool in fragment based drug discovery. A cross-docking, rescoring study of kinase inhibitors. J. Chem. Inf. Model. 2009;49:1437–1448. doi: 10.1021/ci900022h. [DOI] [PubMed] [Google Scholar]
- 43.Thompson DC, Humblet C, Joseph-McCarthy D. Investigation of MM-PBSA rescoring of docking poses. J. Chem. Inf. Model. 2008;48:1081–1091. doi: 10.1021/ci700470c. [DOI] [PubMed] [Google Scholar]
- 44.Kawatkar S, Wang H, Czerminski R, Joseph-McCarthy D. Virtual fragment screening: an exploration of various docking and scoring protocols for fragments using Glide. J. Comput.-Aided Mol. Des. 2009;23:527–539. doi: 10.1007/s10822-009-9281-4. [DOI] [PubMed] [Google Scholar]
- 45.Lyne PD, Lamb ML, Saeh JC. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J. Med. Chem. 2006;49:4805–4808. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
- 46.Graves AP, Shivakumar DM, Boyce SE, Jacobson MP, Case DA, Shoichet BK. Rescoring docking hit lists for model cavity sites: predictions and experimental testing. J. Mol. Biol. 2008;377:914–934. doi: 10.1016/j.jmb.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perutz MF. Mechanisms of cooperativity and allosteric regulation in proteins. Q. Rev. Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- 48.Sponer J, Spackova N. Molecular dynamics simulations and their application to four-stranded DNA. Methods. 2007;43:278–290. doi: 10.1016/j.ymeth.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linden A. Measuring diagnostic and predictive accuracy in disease management: an introduction to receiver operating characteristic (ROC) analysis. J. Eval. Clin. Pract. 2006;12:132–139. doi: 10.1111/j.1365-2753.2005.00598.x. [DOI] [PubMed] [Google Scholar]
- 50.Triballeau N, Acher F, Brabet I, Pin JP, Bertrand HO. Virtual screening workflow development guided by the "receiver operating characteristic" curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J. Med. Chem. 2005;48:2534–2547. doi: 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
- 51.Jain AN. Bias, reporting, and sharing: computational evaluations of docking methods. J. Comput.-Aided Mol. Des. 2008;22:201–212. doi: 10.1007/s10822-007-9151-x. [DOI] [PubMed] [Google Scholar]
- 52.Cummings MD, DesJarlais RL, Gibbs AC, Mohan V, Jaeger EP. Comparison of automated docking programs as virtual screening tools. J. Med. Chem. 2005;48:962–976. doi: 10.1021/jm049798d. [DOI] [PubMed] [Google Scholar]
- 53.Hevener KE, Zhao W, Ball DM, Babaoglu K, Qi J, White SW, Lee RE. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009;49:444–460. doi: 10.1021/ci800293n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006;49:5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 55.Verdonk ML, Giangreco I, Hall RJ, Korb O, Mortenson PN, Murray CW. Docking performance of fragments and druglike compounds. J. Med. Chem. 2011;54:5422–5431. doi: 10.1021/jm200558u. [DOI] [PubMed] [Google Scholar]
- 56.Swanson JM, Henchman RH, McCammon JA. Revisiting free energy calculations: a theoretical connection to MM/PBSA and direct calculation of the association free energy. Biophys. J. 2004;86:67–74. doi: 10.1016/S0006-3495(04)74084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thorpe IF, Brooks CL., 3rd Molecular evolution of affinity and flexibility in the immune system. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8821–8826. doi: 10.1073/pnas.0610064104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hensen U, Lange OF, Grubmuller H. Estimating absolute configurational entropies of macromolecules: the minimally coupled subspace approach. PLoS One. 2010;5:e9179. doi: 10.1371/journal.pone.0009179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gandhi NS, Mancera RL. Free energy calculations of glycosaminoglycan-protein interactions. Glycobiology. 2009;19:1103–1115. doi: 10.1093/glycob/cwp101. [DOI] [PubMed] [Google Scholar]
- 60.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011;51:69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ravindranathan K, Tirado-Rives J, Jorgensen WL, Guimaraes CR. Improving MM-GB/SA Scoring through the Application of the Variable Dielectric Model. J. Chem. Theory Comput. 2011;7:3859–3865. doi: 10.1021/ct200565u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meyer E. Ph.D. Thesis. Cambridge, MA: Massachusetts Institute of Technology; 1996. Two new activities and a new intermediate in the purine pathway. [Google Scholar]
- 63.Davies TG, Hyvönen M. Topics in current chemistry. Heidelberg; New York: Springer; 2012. p. 1. online resource (xi, 225 p.). [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.