Abstract
Although hantaviruses have been previously considered as rodent-borne pathogens, recent studies demonstrate genetically distinct hantaviruses in evolutionarily distant non-rodent reservoirs, including shrews, moles and bats. The immunological responses to these newfound hantaviruses in humans are unknown. We compared the innate immune responses to Imjin virus (MJNV) and Thottapalayam virus (TPMV), two shrew-borne hantaviruses, with that toward two rodent-borne hantaviruses, pathogenic Hantann virus (HTNV) and nonpathogenic Prospect Hill virus (PHV). Infection of human macrophages and endothelial cells with either HTNV or MJNV triggered productive viral replication and up-regulation of anti-viral responsive gene expression from day 1 to day 3 postinfection, compared with PHV and TPMV. Furthermore, HTNV, MJNV and TPMV infection led to prolonged increased production of pro-inflammatory cytokines from days 3 to 7 postinfection. By contrast, PHV infection failed to induce pro-inflammatory responses. Distinct patterns of innate immune activation caused by MJNV suggest that it might be pathogenic to humans.
Keywords: hantavirus, Imjin virus, shrew, innate immune response, anti-viral response
Introduction
Approximately half of the more than 50 hantavirus species identified in multiple rodent species worldwide are pathogenic to humans (Klein and Calisher, 2007), causing distinct clinical syndromes known as hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS). Widespread throughout Asia and Europe, HFRS is characterized by increased vascular permeability and acute renal insufficiency or failure, whereas HCPS occurs in North and South America with patients exhibiting mild to severe hypoxemia and respiratory failure with cardiogenic shock. Hantaan virus (HTNV), Seoul virus (SEOV) and Puumala virus (PUUV) are the principal HFRS-causing hantaviruses, whereas Sin Nombre virus (SNV) and Andes virus (ANDV) are prototypic hantaviruses that cause HCPS. Best known among the so-called nonpathogenic hantaviruses are Prospect Hill virus (PHV) and Tula virus (TULV) (Easterbrook and Klein, 2008; Klein and Calisher, 2007; Markotic, 2003).
Previously, all hantavirus species were believed to be harbored by rodents. Recent studies, however, indicate that genetically distinct hantaviruses are also hosted by non-rodent reservoirs, including shrews (Arai et al., 2008a; Arai et al., 2012; Arai et al., 2007; Kang et al., 2011a; Kang et al., 2011b; Song et al., 2007b; Song et al., 2009; Song et al., 2007c), moles (Arai et al., 2008b; Kang et al., 2009a; Kang et al., 2011a; Kang et al., 2009b) and bats (Sumibcay et al., 2012; Weiss et al., 2012). Thottapalayam virus (TPMV), initially an unclassified virus isolated from the Asian house shrew (Suncus murinus) (Carey et al., 1971), is now known to be among the most genetically and phylogenetically divergent of all hantaviruses (Song et al., 2007a). Similarly, phylogenetic analysis of Imjin virus (MJNV), recently isolated from the Ussuri white-toothed shrew (Crocidura lasiura) (Song et al., 2009), indicates an early evolutionary divergence from rodent-borne hantaviruses. Although limited data are available about the pathogenic potential of these newfound hantaviruses in humans, the detection of anti-TPMV IgG in a patient with high fever of unknown etiology in Thailand suggests the possibility that TPMV and other shrew-borne hantaviruses might cause a wide spectrum of clinical disease (Okumura et al., 2007; Song et al., 2007a). However, the importance of these and other as yet uncharacterized hantaviruses as human pathogens requires further exploration.
Modulation of innate immune responses by rodent-borne hantaviruses has been previously described (Easterbrook and Klein, 2008; Rang, 2010). Both pathogenic and nonpathogenic rodent-borne hantaviruses were shown to differentially modulate mRNA expression of genes of both anti-viral and pro-inflammatory innate immune responses in human endothelial cells (Alff et al., 2006; Geimonen et al., 2002; Handke et al., 2009; Kraus et al., 2004; Spiropoulou et al., 2007). Interestingly, a different interferon response was observed at one day postinfection (p.i.) with either pathogenic (HTNV or NYV) or putative nonpathogenic (PHV) hantaviruses. PHV infection caused a more rapid and profound expression of interferon (IFN)-related gene transcripts, compared with pathogenic hantaviruses. The inhibition of PHV replication in human endothelial cells suggests that this early up-regulation of IFN-related genes induced by PHV infection would have a detrimental effect on viral replication. Thus, the data suggest that hantavirus pathogenesis and innate immune responses may in part be determined by viral regulation of cellular IFN responses (Alff et al., 2006; Geimonen et al., 2002; Handke et al., 2009; Kraus et al., 2004; Spiropoulou et al., 2007). Presently, it is unknown whether IFN responses limit MJNV or TPMV infection of human endothelial cells.
In this study, we explored the innate immune responses to shrew-borne hantaviruses in human umbilical vein endothelial cells (HUVEC) and a human macrophage cell line (THP-1). Both MJNV and TPMV replicated and infected HUVEC similar to rodent-borne pathogenic or nonpathogenic hantaviruses, but not as efficiently as in THP-1 cells. In contrast to PHV, HTNV and MJNV were shown to differentially up-regulate anti-viral responsive genes, such as myxovirus resistance protein gene (MxA) and interferon-β (IFN-β), from day 1 to day 3 p.i. in THP-1 cells. Similar to the anti-viral responses, HTNV, MJNV and TPMV induced pro-inflammatory cytokine and chemokine production. Our findings provide new insights into innate immune activation by shrew-borne hantaviruses but leave unanswered the pathogenicity of these newfound, still-orphan viruses.
Results
Replication of HTNV, MJNV, TPMV and PHV in HUVEC and THP-1 cells
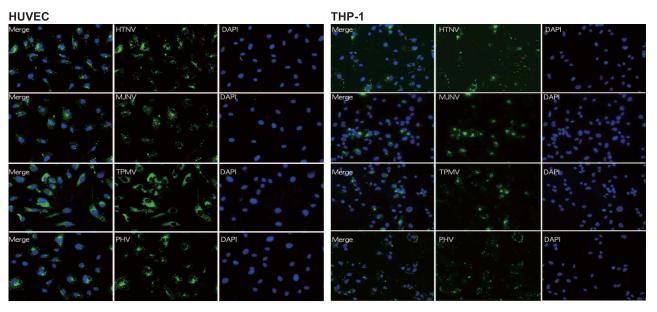
Both endothelial cells and macrophages are considered to be major target cells of hantavirus infection (Easterbrook and Klein, 2008). Accordingly, to study the innate immune responses caused by shrew-borne hantavirus infection in vitro, HUVEC and THP-1, differentiated into macrophages with the addition of PMA, were used. HUVEC and THP-1 cells were infected at a multiplicity of infection (MOI) of 1 with HTNV, MJNV, TPMV and PHV. HTNV, MJNV, TPMV and PHV antigens could be detected in 90-100% of HUVEC (day 1 p.i.) and in 40-50% (day 3 p.i.) of differentiated THP-1 cells by immunofluorescence staining (Fig. 1A and 1B). Microscopic examination of the morphology and viability of HUVEC and THP-1 cells did not show cytopathic effects at any time point during the course of infection (data not shown).
Fig. 1. Hantavirus infection in HUVEC and THP-1 cells.
HUVEC (A) and THP-1 cells (B) were infected with HTNV, MJNV, TPMV or PHV at an MOI of 1.0 and incubated for 24hr (HUVECs) and 72 hr (THP-1). Each virus was detected by IFA staining with incubation of each virus-specific antibody followed by an FITC-conjugated anti-mouse or rat IgG (green) while the corresponding cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI, blue). Original magnification X40
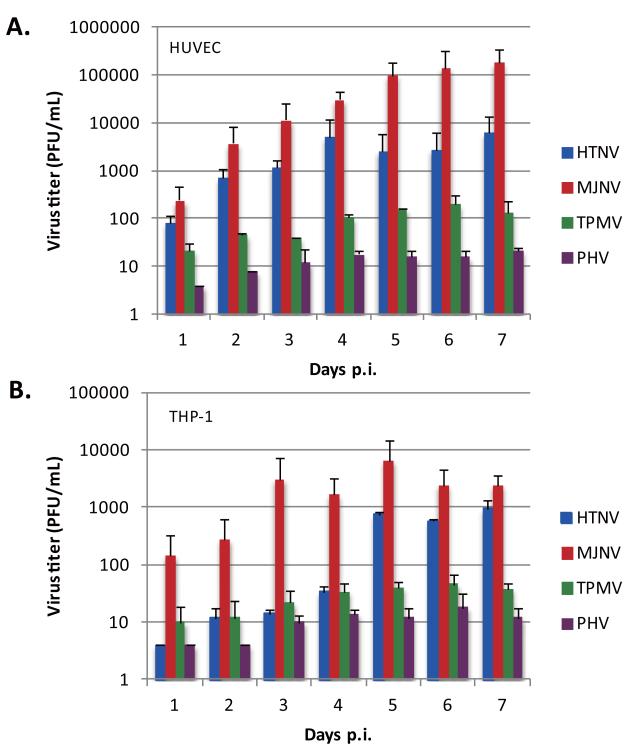
Next, the ability of HTNV, MJNV, TPMV and PHV to replicate in HUVEC and THP-1 cells was determined, using plaque assay. Cells were infected at an MOI of 0.5 and virus infectivity in cell supernatants was titered from days 1 to 7 p.i. At 3 to 4 days p.i., the titers of HTNV and MJNV reached 103 to 105 PFU/mL in HUVEC cells, whereas the titers of TPMV remained at 101 to 102 PFU/mL. There was little PHV replication in HUVEC from days 1 to 7 p.i., with maximal titers of 101 PFU/mL (Fig. 2A). In comparison with HUVEC, HTNV and MJNV replication in THP-1 cells showed lower efficacy and slower kinetics, yielding maximal infectivity titers of ~103 to 104 PFU/mL after 5 days p.i. (Fig. 2B). TPMV and PHV remained at ~101 to 102 PFU/mL in THP-1 cells from days 1 to 7 p.i. These findings suggest that MJNV infection led to the highest replication efficiency in both human endothelial cells and macrophages, which was similar to that of pathogenic HTNV.
Fig. 2. Hantavirus replication in HUVEC and THP-1 cells.
HUVEC (A) and THP-1 (B) cells were infected with HTNV, MJNV, TPMV or PHV at an MOI of 1.0. Viral titers in the supernatant of infected cells were analyzed 1 to 7 days postinfection by a plaque assay. Days postinfection are indicated on the x axis, and titers are represented as PFU/mL. Error bars represent SD.
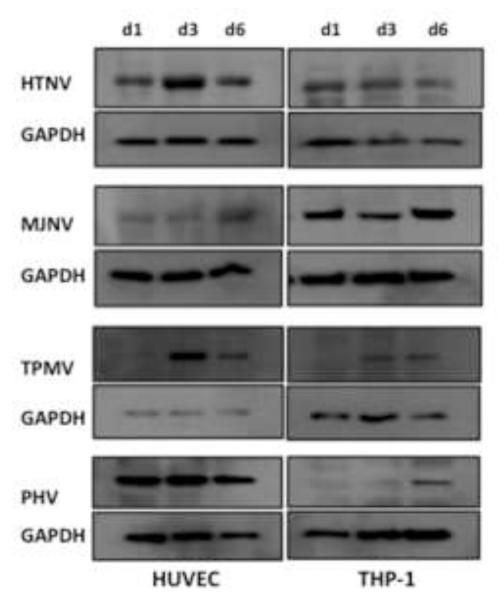
We also determined the level of new protein synthesis, following infection in HUVEC and THP-1. Nucleocapsid protein expression in HTNV-, MJNV-, TPMV- or PHV-infected HUVEC or THP-1 cells was measured by Western blot. Whereas HTNV and MJNV both increased nucleocapsid protein levels since day 1 p.i. and persisted until day 6 p.i., nucleocapsid protein levels of TPMV and PHV increased only after day 3 p.i. in THP-1 cells (Fig. 3). Therefore, these data suggest that protein synthesis of TPMV and PHV occurs at later time points than that of HTNV or MJNV in human macrophages.
Fig 3. Western blot analysis of N protein expression.
HUVEC or THP-1 cells were infected with HTNV, MJNV, TPMV or PHV (MOI of 1.0). Cells were lysed at 1, 3, 6 days postinfection as indicated. Total protein levels were determined, and an equivalent amount of whole-cell lysate was separated by 10% SDS-polyacrylamide gel electrophoresis. Proteins were detected by Western blotting using anti-nucleocapsid monoclonal mouse or polyclonal rat antibody, followed by HRP-conjugated species-specific secondary antibodies. Anti-GAPDH monoclonal antibody was used as a loading control.
Anti-viral responses in HUVEC and THP-1 cells infected with hantaviruses
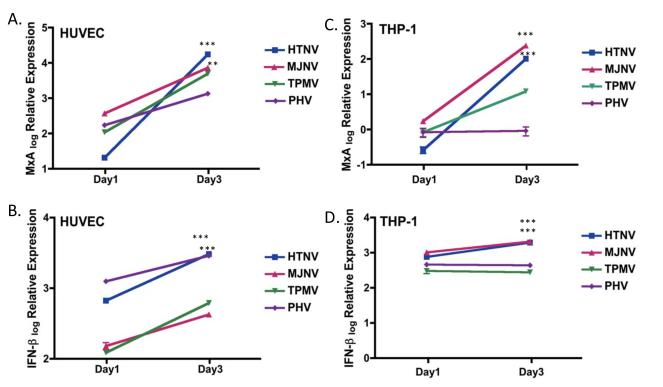
To analyze the anti-viral responses induced by infection with various hantaviruses, transcripts for several anti-viral genes (MxA and IFN-β) were measured using quantitative real-time PCR (RT-PCR). Either HUVEC or THP-1 cells were infected with HTNV, MJNV, TPMV or PHV at an MOI of 0.5 and total RNA was isolated at 1 and 3 days p.i. At day 1 p.i., PHV infection of HUVEC directed a more rapid increase in MxA gene expression than HTNV (Fig. 4A). However, MxA mRNA in HTNV-infected cells increased by 880.9-fold from days 1 to 3, whereas there was only a 7.9-fold increase in MxA mRNA expression in PHV-infected cells. MJNV and TPMV infection caused 19.8-fold and 45.8-fold increase in MxA gene expression from days 1 to 3, respectively. IFN-β mRNA expression was also increased from days 1 to 3 p.i. (4.5-fold by HTNV, 2.7-fold by MJNV, 5.0-fold by TPMV, and 0.4-fold by PHV) (Fig. 4B).
Fig 4. Kinetics of MxA and IFN-β mRNA induction during hantavirus infection.
HUVEC or THP-1 cells were infected with HTNV, MJNV, TPMV or PHV (MOI of 0.5) or mock infected. RNA was collected at days 1 and 3 postinfection. The mRNA expression of MxA (A) and IFN-β (B) in HUVEC or MxA (C) and IFN-β(D) in THP-1 cells treated with media was arbitrarily set to 1 and relative expression (log) is shown in the graph. Expression of target genes was normalized to that of β-actin. The qRT-PCR experiments were performed in duplicate and the average of all experiments is shown. Two-way ANOVA with repeated measures, followed by Bonferroni post-test, (*p <0.05, **p <0.01, ***p <0.001, 1 days p.i. vs 3 days p.i.). Error bars indicate SD. MxA HUVEC: HTNV p<0.001, MJNV p<0.01, TPMV p>0.05, PHV p>0.05; IFN-β HUVEC: HTNV p<0.001, MJNV p<0.05, TPMV p>0.05, PHV p<0.05; MxA THP-1: HTNV p<0.001, MJNV p<0.001, TPMV p>0.05, PHV p>0.05; IFN-β THP-1: HTNV p<0.001, MJNV p<0.001, TPMV p>0.05, PHV p>0.05.
Hantavirus-induced anti-viral responses in THP-1 cells were also analyzed. The mRNA expression of MxA gene in HTNV, MJNV or TPMV-infected cells increased sharply from days 1 to 3 p.i. However, PHV did not induce MxA gene expression in THP-1 cells (Fig. 4C). IFN-β gene expression in cells infected by HTNV or MJNV increased from days 1 to 3 p.i., while no significant change was found from days 1 to 3 p.i. in TPMV- and PHV-infected cells (Fig. 4D). Compared to TPMV or PHV infection, HTNV or MJNV infection resulted in significant increase of IFN-β and MxA gene expression from days 1 to 3 p.i. in THP-1 cells, and HTNV and MJNV may similarly regulate early IFN responses during infection.
Hantaviruses direct the production of pro-inflammatory cytokines and chemokines
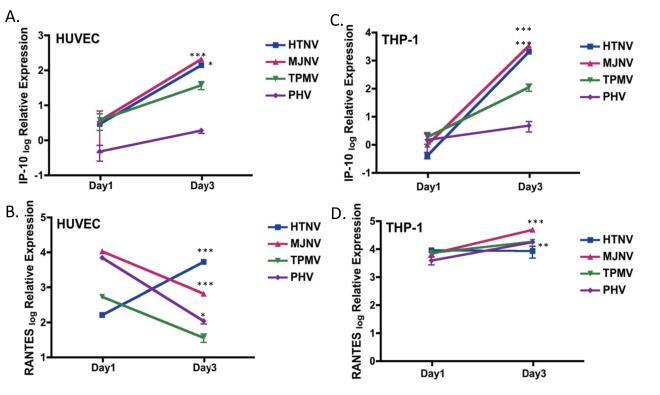
To investigate the inflammatory responses induced by shrew-borne hantaviruses, Regulated upon Activation, Normal T-cell Expressed, and Secreted (RANTES) and interferon gamma-induced protein-10 (IP-10) mRNA triggered in response to hantavirus infection was determined by quantitative RT-PCR. As previously reported, the expression of RANTES was differentially induced in response to HTNV and PHV infection in HUVEC. Although at day 1 p.i., RANTES expression was higher in PHV-infected cells than in HTNV-infected cells, at day 3 p.i., HTNV infection resulted in higher RANTES mRNA expression compared with PHV infection (Fig. 5B). IP-10 mRNA was not detected with any of the four hantaviruses at 1 day p.i. However, IP-10 mRNA was increased at 3 day p.i. by ~47.9- to 58.3-fold in samples from either HTNV- or MJNV-infected cells, compared with 1 day p.i. (Fig. 5A). As demonstrated in Fig. 5C, no detectable levels of IP-10 mRNA was seen with any of the hantaviruses at day 1 p.i. in THP-1 cells, however, HTNV and MJNV infection induced significant increase in IP-10 mRNA expression by day 3 p.i. However, at 3 days p.i., IP-10 mRNA was 103 -log fold higher in HTNV- or MJNV-infected cells and only 4-fold higher in PHV-infected cells, compared with uninfected cells. The maximal expression of RANTES from days 1 to 3 was induced by MJNV infection (Fig. 5D). These data demonstrate that HTNV and MJNV infection similarly induced significantly higher increases in IP-10 chemokine expression from days 1 to 3, differentially from PHV.
Fig. 5. Kinetics of chemokine (IP-10 and RANTES) mRNA expression during hantavirus infection.
HUVEC or THP-1 cells were infected with HTNV, MJNV, TPMV or PHV (MOI of 0.5) or mock infected. RNA was collected at days 1 and 3 postinfection. mRNA expression of IP-10 (A) and RANTES (B) in HUVEC or IP-10 (C) and RANTES (D) in THP-1 cells treated with media was arbitrarily set to 1 and relative expression (log) is shown in the graph. Expression of target genes was normalized to that of β-actin. The qRT-PCR experiments were performed in duplicate and the average of all experiments is shown.
Two-way ANOVA with repeated measures, followed by Bonferroni post-test (*p <05, **p <0.01, ***p <0.001, 1 days p.i. vs 3 days p.i.). Error bars indicate SD. IP-10 HUVEC: HTNV p<0.05, MJNV p<0.001, TPMV p>0.05, PHV p>0.05; RANTES HUVEC: HTNV p<0.001, MJNV p<0.001, TPMV p>0.05, PHV p<0.05; IP-10 THP-1: HTNV p<0.001, MJNV p<0.001, TPMV p>0.05, PHV p>0.05; RANTES THP-1: HTNV p>0.05, MJNV p<0.001, TPMV p<0.05, PHV p<0.01.
To determine the pro-inflammatory cytokine response caused by shrew-borne hantavirus infection, we also collected supernatant from cells on days 1 to 7 p.i. and measured levels of tumor necrosis factor-α (TNF-α), interleukin-1β (IL-1β) and interleukin-8 (IL-8) by ELISA. At days 1 to 2 p.i., hantavirus-directed responses were similar between pathogenic and nonpathogenic viruses. However, at days 3 to 4 p.i., each hantavirus began to induce unique cellular responses. HTNV infection gradually increased the protein secretion levels of TNF-α, IL-8, and IL-1β, peaking at days 3 to 4 p.i. (Fig. 6). Following a similar pattern, TNF-α and IL-1β production gradually increased in MJNV-infected samples until day 4 p.i. (compared with earlier time points) but IL-8 secretion was down-regulated from days 5 to 7. Notably, TPMV infection caused up-regulation of TNF-α and IL-1β at earlier time points than HTNV or MJNV, but induction of IL-8 was not seen. In contrast to these viruses, PHV infection caused relatively static or down-regulation of all three cytokines, as shown in Fig. 6. Thus, these data suggest that the induction patterns of pro-inflammatory cytokines/chemokines differ among various hantaviruses.
Fig 6. Pro-inflammatory cytokine response during hantavirus infection.
THP-1 cells were infected with HTNV, MJNV, TPMV or PHV (MOI of 0.5) or mock infected (M). Supernatant was collected from days 1 to 7 postinfection. Concentrations of TNF-α (A), IL-8 (B) and IL-1β (C) were measured by ELISA. The graph shows the average of all experiments. Error bars represent SDs.
Discussion
Genetically distinct hantaviruses have been found in multiple small mammal hosts and the identification of novel hantaviruses with host specificity suggests the need to study the pathogenesis of these viruses in humans. The pathogenicity and host immune responses to shrew-borne hantaviruses are unknown. In our study, we used two cell types (HUVEC and THP-1 cells) to characterize viral replication and host innate immune responses (anti-viral and inflammatory) caused by pathogenic (HTNV) and nonpathogenic (PHV) rodent-borne hantaviruses compared with shrew-borne hantaviruses (MJNV and TPMV). Of the four hantaviruses tested, MJNV provoked the highest induction of viral replication in both HUVEC and THP-1 cells. Similar to HTNV, MJNV induced increased expression of anti-viral responsive genes in a time-dependent manner. In contrast to PHV, MJNV caused pronounced up-regulation of pro-inflammatory cytokines and chemokines in human macrophages. Collectively, our findings indicate that MJNV and possibly other shrew-borne hantaviruses elicit host responses via mechanisms that are similar to pathogenic hantaviruses, but markedly differ from nonpathogenic hantaviruses.
HTNV and MJNV replicated in HUVEC with similar kinetics and efficiency, yielding maximal titers of 104 to 106 PFU/mL within 7 days p.i. Compared with HUVEC, all of the studied hantaviruses replicated less productively in THP-1 cells, since viral titers ranged from 101 to 103 PFU/mL for HTNV and 102 to 104 PFU/mL for MJNV within the examined timeframe. Both TPMV and PHV failed to replicate as efficiently as HTNV or MJNV, reaching titers of only 101 to 102 PFU/mL. TPMV and PHV replicated less efficiently than HTNV and MJNV in both HUVEC and THP-1 cells. The ability of MJNV to replicate efficiently in these cells, compared to PHV and TPMV, suggests that MJNV might be a human pathogen. On the other hand, the absence of replication by TPMV in these cells also suggests that it may be nonpathogenic like PHV, which similarly is unable to productively replicate in HUVEC and THP-1. Furthermore, this differential viral replication pattern may explain the differential innate immune responses triggered by these viruses.
Our data indicate that MJNV triggered distinct activation of innate immune responses, resembling that of the HTNV-specific induction pattern, in human macrophages. Using primary human endothelial cells, several groups have shown that infection with pathogenic and nonpathogenic hantaviruses differentially activate the innate IFN system (Alff et al., 2006; Geimonen et al., 2002; Handke et al., 2009; Kraus et al., 2004; Spiropoulou et al., 2007). For THP-1 cells, more pronounced increase in expression of MxA was observed with HTNV, MJNV and TPMV infection from days 1 to 3 p.i., compared with PHV infection (Fig. 4C). This observation suggests that high-level replication achieved by MJNV at earlier time frames might have possibly suppressed early detection of viruses by unidentified receptor(s), required for the activation of innate IFN responses.
Immediately after viral infection, innate antiviral and inflammatory responses can be elicited. Our data reveal that beyond anti-viral genes, shrew-borne hantaviruses differentially induce inflammatory responses in THP-1 cells, as indicated by the expression pattern of IP-10 and RANTES, which are known to mobilize and attract mononuclear cells, including lymphocytes and cytokines/chemokines, TNF-α, IL-8 and IL-1β. Compared with PHV, MJNV induced increased expression of both IP-10 and RANTES with higher kinetics at 3 day p.i. (Fig. 5C and 5D). A similar induction pattern of increased TNF-α, IL-8 and IL-1β production was also observed with MJNV infection whereas PHV infection failed to cause an increase in cytokine production (Fig. 6). This suggests that differential virus replication might drive differential induction of cellular antiviral and proinflammatory responses. Our data indicate that hantaviruses, which replicate to higher levels in endothelial cells and macrophages (HTNV and MJNV), also cause more increased induction of antiviral and proinflammatory responses.
Although MJNV infection has not been reported in humans, it may be premature to conclude that MJNV is nonpathogenic. In a recently developed hamster model of MJNV infection, an apparent age-dependent susceptibility and fatal disease were noted, akin to an HCPS-like disease caused by ANDV (S.H. Gu et al., manuscript submitted). Thus, in line with our in vitro data, the hamster model of MJNV infection raises the possibility that MJNV may cause an immunopathogenesis-driven disease in humans.
Recent discovery of shrew-, mole- and bat-borne hantaviruses raise important questions about their phylogenetic origin and potential pathogenicity (Carey et al., 1971; Jung and Kim, 1995; Song et al., 2007a; Song et al., 2009; Song et al., 2007c; Sumibcay et al., 2012; Weiss et al., 2012). Cross-reactivity between routinely used rodent-borne hantavirus antigens has been a limiting factor to identifying patients suffering from clinical diseases possibly caused by novel hantaviruses. The development of sensitive and specific serological assays for newfound hantaviruses will facilitate future studies aimed at identifying clinical diseases or syndromes associated with these viruses.
In conclusion, our study constitutes the first attempt to characterize the innate immune responses induced by shrew-borne hantaviruses. Although cellular receptors for MJNV and TPMV have not been identified, either or both might use αvβ3 integrin receptors, previously reported as receptors for pathogenic HTNV (Gavrilovskaya et al., 1998; Mackow and Gavrilovskaya, 2001). Taken together, our data suggest that shrew-borne hantaviruses differentially induce anti-viral and pro-inflammatory innate immune responses, compared with nonpathogenic PHV. Further elucidation of the molecular mechanisms and viral determinants of pronounced immune responses by shrew-borne hantaviruses would lead to a better understanding of hantavirus pathogenesis in humans.
Materials and methods
Cells and reagents
Vero E6 (African green monkey epithelial kidney cell, ATCC) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM; Lonza, Walkersville, MD) with 5% heat-inactivated fetal bovine serum (FBS) (Lonza), 2 mM L-glutamine and antibiotics (penicillin-streptomycin) at 37°C at 5% CO2. Induced pluripotent stem cell (iPS) HUVEC were purchased from Inopharmascreen (Asan, Korea) and maintained in M199 (Gibco), containing penicillin-streptomycin, 25mM HEPES, 10 unit/mL heparin, 2.2 g/L sodium bicarbonate, 20% FBS / bFGF 20 ng/mL and passaged up to seven times. The human monocytic cell line THP-1, obtained from the Korean Cell Line Bank (KCLB) (Seoul, Korea), was grown in RPMI-1640 (Lonza) supplemented with 5% FBS, 10 mM HEPES buffer, 2 mM l-glutamine, and 50 ng/mL gentamicin. To induce monocyte-to-macrophage differentiation, THP-1 cells were cultured for 24 h in standard culture medium supplemented with 100 nM phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich).
Viruses
HTNV (strain 76-118), TPMV (strain VRC 66412) and PHV (strain PHV-1) were propagated in Vero E6 cells (Shim et al., 2011). MJNV strains 04-55 and 05-11 were isolated directly from shrew lung tissues in Vero E6 cells (Song et al., 2009). Using PCR analysis, all virus stocks and cells were determined to be free of mycoplasma contamination.
Virus infection and plaque assay
Various cells were infected with each virus at MOI of 0.5 (or 1.0 for immunofluorescence staining, plaque assay and Western blotting) for 90 min at 37°C. After incubation, the viruses were washed in phosphate buffered saline (PBS), and the cells were maintained in complete medium. Virus infectivity titers were measured on each postinfection day, using the plaque assay (Shim et al., 2011).
Western blot analysis
Western blot analysis was performed as previously described (Alff et al., 2006). Briefly, hantavirus-infected HUVEC or THP-1 cells were lysed as previously described at various times p.i. Cellular lysates were prepared by lysis buffer (0.05 M Tris pH 7.4, 0.15 M NaCl, 0.5 mM PMSF, 50 μg/mL aprotinin, 10 μg/mL leupeptin, 50 μg/mL pepstatin, 0.4 mM sodium orothovanadate, 10 mM sodium fluoride and 10 mM sodium pyrophosphate) and then separated by SDS-PAGE on 10% acrylamide gels and transferred to a polyvinyldifluoride (PVDF) membrane. The membrane was incubated in blocking buffer (5% milk in 0.2 M Tris base, 1.36 M NaCl with 0.1% Tween 20) (TBS/T) for 1 h at room temperature and washed three times for 5 min each with 15 mL of TBS/T. Nucleocapsid proteins were detected using mouse or rat anti-nucleocapsid sera (1:2,000) followed by anti-mouse or rat HRP-conjugated antibody (1:5,000) (Amersham). Membranes were incubated with the primary and secondary antibodies, respectively, each for 1 h each at 25°C. Anti-GAPDH antibody, which served as an internal control, was purchased from Santa Cruz Biotechnology. After washing three times with TBS/T, the membranes were incubated with HRP-conjugated anti-rabbit IgG for 1 h at 25°C. After washing three times with TBS/T, the membrane was incubated with LumiGLO substrate (Cell Signaling) and exposed to the film.
ELISA
TNF-α, IL-1β and IL-8 were each analyzed in culture media samples using commercial enzyme-linked immunosorbent assays (ELISA), according to the manufacturer’s protocols (R&D Systems).
Immunofluorescence microscopy
Various cells were plated in a 24-well plate containing coverslips. The following day, cells were infected with each hantavirus at an MOI of 1.0 for 90 min at 37°C. Thereafter, fresh cell culture medium was added and incubated for 24-72 h. Virus-infected cells were fixed and permeabilized in methanol for 30 min. For HTNV and MJNV, infected cells were then stained with mouse monoclonal antibody to HTNV and MJNV, respectively (1:1,000 dilution), and were stained with FITC-conjugated anti-mouse IgG (H + L) (Vector Laboratories, Burlingame, CA) that served as a secondary antibody (1:200 dilution). For TPMV and PHV, infected cells were stained with rat polyclonal antibody to TPMV and PHV, respectively (1:1,000 dilution), and were stained with FITC-conjugated anti-rat IgG (H + L) (Vector Laboratories) that was used as a secondary antibody (1:200 dilution). All antibodies were diluted in PBS containing 0.05% Tween-20 and 5% skim milk. The coverslips were mounted onto glass slides using mounting media containing 4,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories) and were examined with fluorescence microscopy using a Zeiss Axioplan 2 microscope. Images were captured with a digital CCD camera (Hamamatsu).
Quantitative real time-PCR (Q-rt-pcr)
RNA was isolated with Trizol reagent (Invitrogen). First-strand synthesis of cDNA from total RNA was performed using ImProm-II (Promega) according to the manufacturer’s instructions. Quantification of cDNA was performed by qRT-PCR using Sybrgreen PCR mix (Applied Biosystems). Cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 30 s and 60°C for 1 min. Primers were as follows: MxA 5′-CGTGGTGATTTAGCAGGAAG-3′ and 5′-TGCAAGGTGGAGCGATTCTG-3′; IFN-β1 5′-AAACTCATGAGCAG TCTGCA-3′ and 5′-AGGAGATCTTCAGTTTCGGAGG-3′; RANTES 5′-GCTGTCATCCTCATTGCTAC-3′ and 5′-TCCATCCTAGCTCATCT CCA-3′; IP-10 5′-CGATTCTGATTTGCTGCCTT-3′ and 5′-CATTTCCTTGCTAACTGCTTTC-3′; β-actin 5′-CCACACCTTCTACAATGAGCTGCG-3′ and 5′CGGAGTCCATCACGATCCA-3′. The specificity of each reaction was checked by melt curve analysis and by agarose gel electrophoresis of PCR products. Expression of target genes was referenced to expression of β-actin. Calculations of expression were normalized by using the ΔCt method where the amount of target, normalized to an endogenous reference and relative to a calibrator, is given by 2ΔΔCt, where Ct is the cycle number of the detection threshold.
Statistical Analysis
qRT-PCR data of all experiments were analyzed using ANOVA for repeated measures, followed by Bonferroni post-test. GraphPad Prism software was used for statistical analysis of the data. All data were expressed as mean±SD, and significance was set at p<0.05.
Acknowledgements
This work was supported in part by U.S. Public Health Service grant R01AI075057 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, and by the 65th Medical Brigade/USAMEDDAC-Korea.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alff PJ, Gavrilovskaya IN, Gorbunova E, Endriss K, Chong Y, Geimonen E, Sen N, Reich NC, Mackow ER. The pathogenic NY-1 hantavirus G1 cytoplasmic tail inhibits RIG-I- and TBK-1-directed interferon responses. J Virol. 2006;80:9676–9686. doi: 10.1128/JVI.00508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Bennett SN, Sumibcay L, Cook JA, Song JW, Hope A, Parmenter C, Nerurkar VR, Yates TL, Yanagihara R. Phylogenetically distinct hantaviruses in the masked shrew (Sorex cinereus) and dusky shrew (Sorex monticolus) in the United States. Am J Trop Med Hyg. 2008a;78:348–351. [PMC free article] [PubMed] [Google Scholar]
- Arai S, Gu SH, Baek LJ, Tabara K, Bennett SN, Oh HS, Takada N, Kang HJ, Tanaka-Taya K, Morikawa S, Okabe N, Yanagihara R, Song JW. Divergent ancestral lineages of newfound hantaviruses harbored by phylogenetically related crocidurine shrew species in Korea. Virology. 2012;424:99–105. doi: 10.1016/j.virol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Ohdachi SD, Asakawa M, Kang HJ, Mocz G, Arikawa J, Okabe N, Yanagihara R. Molecular phylogeny of a newfound hantavirus in the Japanese shrew mole (Urotrichus talpoides) Proc Natl Acad Sci U S A. 2008b;105:16296–16301. doi: 10.1073/pnas.0808942105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Song JW, Sumibcay L, Bennett SN, Nerurkar VR, Parmenter C, Cook JA, Yates TL, Yanagihara R. Hantavirus in northern short-tailed shrew, United States. Emerg Infect Dis. 2007;13:1420–1423. doi: 10.3201/eid1309.070484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey DE, Reuben R, Panicker KN, Shope RE, Myers RM. Thottapalayam virus: a presumptive arbovirus isolated from a shrew in India. The Indian journal of medical research. 1971;59:1758–1760. [PubMed] [Google Scholar]
- Easterbrook JD, Klein SL. Immunological mechanisms mediating hantavirus persistence in rodent reservoirs. PLoS Pathog. 2008;4:e1000172. doi: 10.1371/journal.ppat.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. beta3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc Natl Acad Sci U S A. 1998;95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimonen E, Neff S, Raymond T, Kocer SS, Gavrilovskaya IN, Mackow ER. Pathogenic and nonpathogenic hantaviruses differentially regulate endothelial cell responses. Proc Natl Acad Sci U S A. 2002;99:13837–13842. doi: 10.1073/pnas.192298899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handke W, Oelschlegel R, Franke R, Kruger DH, Rang A. Hantaan virus triggers TLR3-dependent innate immune responses. J Immunol. 2009;182:2849–2858. doi: 10.4049/jimmunol.0802893. [DOI] [PubMed] [Google Scholar]
- Jung YT, Kim GR. Genomic characterization of M and S RNA segments of hantaviruses isolated from bats. Acta virologica. 1995;39:231–233. [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Dizney L, Sumibcay L, Arai S, Ruedas LA, Song JW, Yanagihara R. Host switch during evolution of a genetically distinct hantavirus in the American shrew mole (Neurotrichus gibbsii) Virology. 2009a;388:8–14. doi: 10.1016/j.virol.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Hope AG, Cook JA, Yanagihara R. Shared ancestry between a newfound mole-borne hantavirus and hantaviruses harbored by cricetid rodents. J Virol. 2011a;85:7496–7503. doi: 10.1128/JVI.02450-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Bennett SN, Sumibcay L, Arai S, Hope AG, Mocz G, Song JW, Cook JA, Yanagihara R. Evolutionary insights from a genetically divergent hantavirus harbored by the European common mole (Talpa europaea) PLoS One. 2009b;4:e6149. doi: 10.1371/journal.pone.0006149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HJ, Kadjo B, Dubey S, Jacquet F, Yanagihara R. Molecular evolution of Azagny virus, a newfound hantavirus harbored by the West African pygmy shrew (Crocidura obscurior) in Cote d’Ivoire. Virol J. 2011b;8:373. doi: 10.1186/1743-422X-8-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein SL, Calisher CH. Emergence and persistence of hantaviruses. Curr Top Microbiol Immunol. 2007;315:217–252. doi: 10.1007/978-3-540-70962-6_10. [DOI] [PubMed] [Google Scholar]
- Kraus AA, Raftery MJ, Giese T, Ulrich R, Zawatzky R, Hippenstiel S, Suttorp N, Kruger DH, Schonrich G. Differential antiviral response of endothelial cells after infection with pathogenic and nonpathogenic hantaviruses. J Virol. 2004;78:6143–6150. doi: 10.1128/JVI.78.12.6143-6150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackow ER, Gavrilovskaya IN. Cellular receptors and hantavirus pathogenesis. Curr Top Microbiol Immunol. 2001;256:91–115. doi: 10.1007/978-3-642-56753-7_6. [DOI] [PubMed] [Google Scholar]
- Markotic A. [Immunopathogenesis of hemorrhagic fever with renal syndrome and hantavirus pulmonary syndrome] Acta medica Croatica : casopis Hravatske akademije medicinskih znanosti. 2003;57:407–414. [PubMed] [Google Scholar]
- Okumura M, Yoshimatsu K, Kumperasart S, Nakamura I, Ogino M, Taruishi M, Sungdee A, Pattamadilok S, Ibrahim IN, Erlina S, Agui T, Yanagihara R, Arikawa J. Development of serological assays for Thottapalayam virus, an insectivore-borne Hantavirus. Clin Vaccine Immunol. 2007;14:173–181. doi: 10.1128/CVI.00347-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rang A. Modulation of innate immune responses by hantaviruses. Crit Rev Immunol. 2010;30:515–527. doi: 10.1615/critrevimmunol.v30.i6.20. [DOI] [PubMed] [Google Scholar]
- Shim SH, Park MS, Moon S, Park KS, Song JW, Song KJ, Baek LJ. Comparison of innate immune responses to pathogenic and putative non-pathogenic hantaviruses in vitro. Virus Res. 2011;160:367–373. doi: 10.1016/j.virusres.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Song JW, Baek LJ, Schmaljohn CS, Yanagihara R. Thottapalayam virus, a prototype shrewborne hantavirus. Emerg Infect Dis. 2007a;13:980–985. doi: 10.3201/eid1307.070031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Gu SH, Bennett SN, Arai S, Puorger M, Hilbe M, Yanagihara R. Seewis virus, a genetically distinct hantavirus in the Eurasian common shrew (Sorex araneus) Virol J. 2007b;4:114. doi: 10.1186/1743-422X-4-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Kang HJ, Gu SH, Moon SS, Bennett SN, Song KJ, Baek LJ, Kim HC, O’Guinn ML, Chong ST, Klein TA, Yanagihara R. Characterization of Imjin virus, a newly isolated hantavirus from the Ussuri white-toothed shrew (Crocidura lasiura) J Virol. 2009;83:6184–6191. doi: 10.1128/JVI.00371-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JW, Kang HJ, Song KJ, Truong TT, Bennett SN, Arai S, Truong NU, Yanagihara R. Newfound hantavirus in Chinese mole shrew, Vietnam. Emerg Infect Dis. 2007c;13:1784–1787. doi: 10.3201/eid1311.070492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiropoulou CF, Albarino CG, Ksiazek TG, Rollin PE. Andes and Prospect Hill hantaviruses differ in early induction of interferon although both can downregulate interferon signaling. J Virol. 2007;81:2769–2776. doi: 10.1128/JVI.02402-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumibcay L, Kadjo B, Gu SH, Kang HJ, Lim BK, Cook JA, Song JW, Yanagihara R. Divergent lineage of a novel hantavirus in the banana pipistrelle (Neoromicia nanus) in Cote d’Ivoire. Virol J. 2012;9:34. doi: 10.1186/1743-422X-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Witkowski PT, Auste B, Nowak K, Weber N, Fahr J, Mombouli JV, Wolfe ND, Drexler JF, Drosten C, Klempa B, Leendertz FH, Kruger DH. Hantavirus in bat, Sierra Leone. Emerg Infect Dis. 2012;18:159–161. doi: 10.3201/eid1801.111026. [DOI] [PMC free article] [PubMed] [Google Scholar]