Abstract
Candida albicans and Saccharomyces cerevisiae expressing the adhesins Als5p or Als1p adhere to immobilized peptides and proteins that possess appropriate sequences of amino acids in addition to a sterically accessible peptide backbone. In an attempt to further define the nature of these targets, we surveyed the ability of yeast cells to adhere to 90-μm-diameter polyethylene glycol beads coated with a 7-mer peptide from a library of 197 unique peptide-beads. C. albicans bound to ca. 10% of beads from the library, whereas S. cerevisiae expressing Als5p or Als1p bound to ca. 0.1 to 1% of randomly selected peptide-beads. S. cerevisiae expressing Als1p had a distinctly different adherence phenotype than did cells expressing Als5p. The former adhered in groups or clumps of cells, whereas the latter adhered initially as single cells, an event which was followed by the build up of cell-cell aggregates. Beads with adherent cells were removed, and the peptide attached to the bead was determined by amino acid sequencing. All adhesive beads carried a three-amino-acid sequence motif (τϕ+) that possessed a vast combinatorial potential. Adherence was sequence specific and was inhibited when soluble peptide identical to the immobilized peptide was added. The Als5p adhesin recognized some peptides that went unrecognized by Als1p. The sequence motif of adhesive peptides identified by this method is common in proteins and offers so many possible sequence combinations that target recognition by the Als proteins is clearly degenerate. A degenerate recognition system provides the fungi with the potential of adhering to a multitude of proteins and peptides, an advantage for any microorganism attempting to establish a commensal or pathogenic relationship with a host.
The adherence of Candida albicans to human tissue is an important characteristic of this fungus for its life as both a commensal and an opportunistic pathogen (1, 17). Adherence may be mediated by a variety of proteins expressed on the surface of the microorganism, including up to eight members of the Agglutinin-Like Sequence (ALS) gene family (20). Two of the proteins encoded by these genes, Als5p and Als1p, can mediate adherence of the nonpathogenic yeast Saccharomyces cerevisiae to human cells and extracellular matrix components (4, 6). (In previous publications we referred to ALS5 as ALA1 or ALA1/ALS5 [5, 6, 7].) Als5p mediates the initial adhesion of yeast cells to extracellular matrix protein-coated substrates and human buccal epithelial cells and also cell-cell aggregation, a feature that follows adherence of a single yeast cell to a fixed target (5). Fu et al. refer to the cell-cell association mediated by Als1p as flocculation (3). In a model of oral pharyngeal candidiasis C. albicans cells bearing knockouts of ALS1 adhered less than wild-type cells to the mouse tongue, supporting the concept that an Als protein is important to the adherence process both in vivo and in vitro (10).
Given these findings, how does one go about determining the molecular mechanisms of adherence of C. albicans? One approach is to perform knockouts of selected genes, as has been done with ALS1 (3, 10). Because of the large number of ALS genes in C. albicans we have chosen a different approach, i.e., studying C. albicans alongside S. cerevisiae expressing different Candida ALS genes. The adhesive properties of S. cerevisiae cells expressing either Als5p or Als1p mimic the properties of C. albicans clinical isolates (5). The C. albicans adhesin proteins Als5p and Als1p are sufficient to mediate fungal cell adhesion to extracellular matrix, human cells, and cell-cell aggregation to form microcolonial structures (4-6). This finding has allowed us to delineate the behavior of these proteins when present as the sole active cell surface adhesin when expressed in Saccharomyces spp. Furthermore, this avoids the possibility of unintended and undetected changes introduced by knockout procedures. This same method of heterologous expression has been used to characterize an adhesin of C. glabrata (2).
The binding specificity of Als5p has been partially characterized. Adherence mediated by Als5p occurs within minutes, is strong enough to resist vortexing and is reversibly inhibited by denaturing agents such as formamide, high pH, or urea (5). Adherence with Als5p occurs to a number of amino acid sequences that are characterized, in part, by their steric configuration. For example, homopolymers of serine, threonine, or alanine (6-mers or larger) make ready targets for adherence when immobilized on bead surfaces (7). When these same peptides are formed into a closed loop with the same number of residues, the patches are not “recognized,” and adherence does not ensue (7). Adherence occurs with naturally occurring sequences as well, e.g., that of the serine/threonine-rich fibronectin peptide, FTTTSTSTPV (7). Thus, both sequence and conformation are important ligand recognition characteristics.
Despite the similar amino acid sequence and adherence properties of Als5p and Als1p, there are differences in ligand recognition. S. cerevisiae expressing Als5p or Als1p adhered to magnetic beads coated with peptides in very different numbers (7; V. L. Chan, S. A. Klotz, N. K. Gaur, and P. B. Byrd-Williams, Abstr. Sixth Annu. Candida Candidiasis Meet., abstr. 121, p. 74, 2002). For example, S. cerevisiae expressing Als5p adhered in small numbers, whereas C. albicans and S. cerevisiae expressing Als1p adhered and aggregated in great numbers to beads bearing FTTTSTSTPV. Indeed, Als1p-expressing cells can flocculate in the absence of an added ligand, whereas cells bearing Als5p do not (3, 5). Furthermore, cells expressing Als5p or Als1p recognize and adhere to peptides that do not contain sequences rich in serine, threonine, or alanine. Such an adherent peptide was APRLRFYSL (7). These observations suggest that (i) Als5p and Als1p recognize and bind to many different amino acid sequences and (ii) different Als adhesins may recognize and bind to different amino acid sequences with different affinities.
Therefore, we studied the adherence of C. albicans and S. cerevisiae yeast cells expressing either Als protein to specific peptides identified from a random, polyethylene glycol (PEG)-bead based peptide library. The results show that the two adhesins recognize a broad array of target ligands. The ability of the yeast cells to adhere to a multitude of peptide ligands indicates that the “adherence recognition system” of these adhesins is degenerate, i.e., neither highly specific nor highly organized as to their protein or peptide targets. Such a recognition system provides the microorganism the ability to adhere to a large repertoire of targets and is entirely consistent with previous in vitro measurements of C. albicans adherence to proteins and peptides.
MATERIALS AND METHODS
Bead-peptide library.
7-mer peptides were synthesized onto PEG beads (∼90-μm diameter) according to the method of Lam et al. (16). The complete library consists of 197 unique beads and peptides (cysteine is not used in synthesis in order to avoid cross-linking of peptides). The library was washed thoroughly with Tris-EDTA (TE) buffer and stored at 4°C until use.
Microorganisms.
C. albicans strain CA1 was grown in liquid yeast extract-peptone-dextrose. S. cerevisiae YPH499 or S. cerevisiae YPH499 expressing Als5p or Als1p were grown in liquid yeast extract-peptone-raffinose-galactose medium at 28°C prior to adherence assays as described previously (5, 7). The vectors of Als5p and Als1p are low-copy vectors, and both genes are expressed from GAL1 promoters. Cells were thoroughly washed with TE buffer by centrifugation and suspended at 109/ml prior to use. We have used numerous biological buffers in this adherence assay; all are equivalent (6), and the adherence of the fungal cells is not affected by the addition of sugars or Tween 20 to the incubation mixture. Hence, the binding described in not significantly affected by hydrophobic or lectin interactions. However, yeast cell adherence to the beads is readily reversible by the addition of agents or conditions that break hydrogen bonds, e.g., formamide, high pH, and urea (5, 7). TE buffer is used in order to reduce the spontaneous cell-cell aggregation of S. cerevisiae expressing Als1p. S. cerevisiae YPH499 always served as a negative control.
Assay.
Six-well culture plates were used for the adherence assay. A total of 20 μl of beads (ca. 1,000 beads) and 1 μl of yeast (∼106 cells) was added to each well containing 1.5 ml of TE buffer. The culture plate was then agitated for 30 min on a rotary shaker at 150 rpm at room temperature. For the bovine serum albumin (BSA) experiments, 107 yeast cells were mixed with 100 μg of BSA/ml either in native conformation or denatured by boiling for 10 min in TE buffer. Beads were observed by inverted microscopy for adherent fungi and removed with a plastic pipette.
Scoring of adherence was done with the following notations: −, no adherence; +, adherence of 10 to 25 yeast cells; ++, bead almost covered by 80 to 150 yeast cells; and +++, adherence with aggregation of yeast cells. Four different observers were tested, and each reported similar results in the scoring the assay.
Identification of peptides.
Beads with adherent yeast cells were treated with 8 M guanidine-HCl for 30 min, washed thoroughly with phosphate-buffered saline, and submitted to the Laboratory for Protein Sequencing and Analyses, Department of Chemistry, University of Arizona, for sequence analysis of the attached peptide. Confirmation that the peptide sequences were responsible for adherence was obtained by specific synthesis of PEG beads bearing the sequence to be tested. Adherent sequences resulted in yeast cells adhering in large numbers to >95% of the beads, thus confirming the sequence as adherent.
(These findings were presented in part at the 42nd Interscience Conference on Antimicrobial Agents and Chemotherapy, San Diego, Calif., September 2002.)
RESULTS
Detecting multiple adherent peptide ligands.
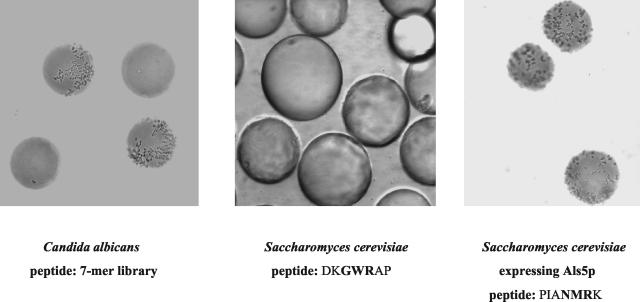
The assay consisted of mixing C. albicans yeast cells with ∼1,000 beads from the 7-mer bead-peptide library in 1.5 ml of TE buffer, all placed in the well of a six-well tissue culture tray. More than 150 wells were observed in this manner with the library beads and bead selection attempted. In each well with its random sample of 1,000 library beads, usually 5 to 100 beads were coated with various amounts of C. albicans. However, the vast majority of beads had no adherent C. albicans whatsoever (Fig. 1, left panel). S. cerevisiae expressing Als5p or Als1p adhered to one to three beads per 1,000 beads from the 7-mer library. S. cerevisiae not expressing an Als protein did not adhere to any peptide-beads (Fig. 1, middle panel). In addition, S. cerevisiae expressing the vector without Candida DNA did not adhere to protein or peptide coated beads (6),; and no yeast cell (Candida nor Saccharomyces) adhered to beads lacking a peptide. As another negative control we sequenced the peptide on a bead that had not bound yeast cells. A population of beads bearing the sequence EHAHTPR did not bind C. albicans or S. cerevisiae expressing either adhesin. If a confirmed sequence was used, >95% of the beads were covered by yeast cells (for example, beads bearing PIANMRK, Fig. 1, right panel [boldfacing is defined below]).
FIG. 1.
Left panel: Candida albicans yeast cells adhering to the surface of 2 PEG beads from the 7-mer library. Two of the library beads are not recognized by C. albicans. Middle panel: Saccharomyces cerevisiae mixed with beads coated with DKGWRAP. The yeast is expressing no Candida DNA. None of the beads have yeast cells adherent to them. Right panel: S. cerevisiae expressing Als5p mixed with beads bearing the peptide, PIANMRK. All beads are coated with yeast cells, some of which are aggregating.
Since a number of library beads were binding yeast cells to their surface, it was apparent that this bead-fungus interaction had fairly broad specificity. We randomly chose five beads that bound C. albicans from a screening of 50,000 beads, and the peptides on these beads were sequenced and confirmed (see Materials and Methods) as adhesive ligands (Table 1). Similar screening and random selection of beads with adherent S. cerevisiae expressing Als5p or Als1p was performed and identified six and three peptide ligands, respectively (Table 1).
TABLE 1.
Peptide sequences to which fungi adhereda
| Peptide-bead | Peptide sequence(s)b |
|---|---|
| Peptide-beads bound by C. albicansc | KTKFLVD, VTHTHRR, DKGWRAP, KLRIPSV, and AYKSLMT |
| Peptide-beads bound by S. cerevisiae expressing ALS5c | FGYPIRR, GHKNATR, PIANMRK, RHMAHKL, STGMKKM, and EFKSWRY |
| Peptide-beads bound by S. cerevisiae expressing ALS1 | HLYASWR, VYYPFKQ, and DLKLVRP |
Fungi were mixed with ≈103 beads from the 7-mer library.
The sequence motif is indicated in boldface.
These peptide sequences were confirmed.
All peptides identified as ligands in this search had a sequence motif “τϕ+,” with “τ” representing a residue with high turn propensity (A, D, G, K, N, P, or S), “ϕ” representing a bulky hydrophobic or aromatic residue (F, H, I, L, M, T, V, W, and Y), and “+” representing R or K. In Table 1 and throughout the remainder of the text this sequence motif is denoted in boldface type.
Yeast cells were reversibly bound to the beads. This was demonstrated by treatment of beads decorated with yeast cells with 50% formamide in water. This treatment dissociates the adherent yeast cells. Upon removal of the formamide and replacing it with buffer, the yeast cells rapidly reattached to the beads (data not shown).
Specificity of ligand sequences.
We have reported previously on the use of tosyl- and carboxylate-activated magnetic beads in peptide-fungal adherence assays (7). The tosyl-activated beads are 5 μm in diameter, the carboxylate-activated beads are 0.7 μm in diameter, and in each case the peptides are covalently bound at the N terminus of the peptides. The adherence of yeast cells to the tosyl- and carboxylate-activated beads was inhibited with a soluble peptide of the same sequence as the bead-bound peptide. The PEG beads used in this assay are ∼90 μm in diameter, and the peptides are covalently bound at the C terminus. Therefore, KLRIPSV, a PEG-bead-bound peptide recognized by C. albicans, was coupled to the carboxylate-modified magnetic beads at the N terminus and tested for adherence. C. albicans recognized and adhered to the peptide attached in this configuration as well (data not shown). Thus, neither the type of bead nor the mode of coupling qualitatively affected recognition of an adhesive peptide by C. albicans.
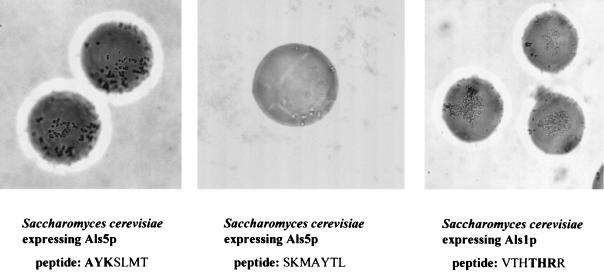
Because the adhesive peptides had similar amino acid compositions (see Discussion), we determined whether adhesion was mediated by composition alone. Accordingly, we synthesized a set of beads bearing the sequence SKMAYTL, a scrambled version of the sequence AYKSLMT that was determined to be adhesive for all three adhesin-expressing cell types (Fig. 2, left panel). The scrambled sequence was poorly adhesive for either adhesin when expressed in S. cerevisiae (Fig. 2, middle panel) and showed slightly reduced binding for C. albicans (Table 2). Sequence specificity was also demonstrated by peptide competition studies. The soluble heptamer VTHTHRR (5.5 × 10−4 M) effectively inhibited adherence of Als5p-bearing S. cerevisiae cells to homologous beads (Table 3). The high concentration of the peptide required for specific blocking in this instance demonstrates the weak interaction of the adhesin with the target peptide when in the nonimmobilized state. C. albicans adhesion was not inhibited, but recall that C. albicans possesses multiple adhesins and the likelihood that a single 7-mer would inhibit all available adhesins is small. A 23-mer, GRGDSPASSKGGGGSRLLLLLLR (S. A. Klotz, N. K. Gaur, J. M. Rauceo, and P. N. Lipke, Abstr., 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. M-375, p. 436, 2003), entirely inhibits the adherence of both C. albicans and S. cerevisiae expressing Als5p or Als1p to beads bearing any of the identified heptamers. However, the use of the 7-mer peptide library has allowed us to detect differences in ligand binding by two adhesins, Als5- and Als1p, a feature we would probably not have been able to detect if stronger binding peptides (such as the 23-mer) were used. Furthermore, the exogenous ligand need not be identical to the immobilized ligand for inhibition of adherence to occur. For example, AYKSLMT and DKGWRAP, but not EHAHTPR (which does not have a consensus sequence), inhibited the adherence of S. cerevisiae expressing Als5p to beads bearing KLRIPSV (data not shown). The lack of requirement of an homologous peptide free in solution and on the bead underscores the degeneracy of the recognition system of these adhesins.
FIG. 2.
(Left panel) S. cerevisiae expressing Als5p mixed with beads bearing AYKSLMT. All beads are covered by aggregating yeast cells. (Middle panel) S. cerevisiae expressing Als5p mixed with beads bearing SKMAYTL, the scrambled peptide of AYKSLMT. (Right panel) S. cerevisiae expressing Als1p adhering and aggregating on the surface of beads bearing the peptide, VTHTHRR. Note that the microcolonial phenotype is different from that of Als5p, since large patches of cells are adherent and aggregating on the surface of the bead.
TABLE 2.
Specificity of the adherence process demonstrated for the peptide sequence, AYKSLMTa
| Peptide-bead | Specificity score
|
|||
|---|---|---|---|---|
| C. albicans | S. cerevisiae expressing ALS5 | S. cerevisiae expressing ALS1 | S. cerevisiae | |
| AYKSLMT | +++ | +++ | +++ | − |
| SKMAYTL | +++ | − | − | − |
The same amino acids were then randomly assembled to prepare a “scrambled peptide sequence”, SKMAYTL.
TABLE 3.
Inhibition of Als5p with free ligand
| Microorganism and adhesin | Inhibition score
|
|
|---|---|---|
| Peptide-bead with VTHTHRR | Peptide-bead plus 500 μg of free VTHTHRR/ml | |
| C. albicans | +++ | +++ |
| S. cerevisiae | − | − |
| S. cerevisiae expressing Als5p | +++ | − |
| S. cerevisiae expressing Als1p | ++ | ++ |
Specificity of Als5p and Als1p.
To determine whether the two adhesins bound to the same ligands, the peptides originally identified as ligands for C. albicans were tested for their ability to be recognized by S. cerevisiae expressing either adhesin (Table 4). Cells expressing Als5p adhered to each of the ligand sequences. In contrast, S. cerevisiae cells expressing Als1p bound less well than Als5p to AYKSLMT and in even smaller numbers to VTHTHRR and not at all to three other adhesive peptides (KLRIPSV, KTKFLVD, and DKGWRAP). These results demonstrate that there is a clear difference in the ability of two Als proteins to recognize and adhere to various peptides. Furthermore, the adherence phenotype of these two adhesins is distinctly different. Als1p-bearing cells adhered initially in large clumps or aggregates of cells, whereas Als5p-bearing cells adhered initially as single cells (compare Fig. 2, left panel, with the right panel).
TABLE 4.
Adherence of fungi expressing different ALS genes to 7-mer peptides
| Peptide-bead | Adherence score
|
|||
|---|---|---|---|---|
| C. albicans | S. cerevisiae expressing ALS5 | S. cerevisiae expressing ALS1 | S. cerevisiae | |
| KTKFLVD | + | ++ | − | − |
| VTHTHRR | ++ | +++ | + | − |
| DKGWRAP | ++ | +++ | − | − |
| KLRIPSV | +++ | +++ | − | − |
| AYKSLMT | ++ | +++ | ++ | − |
| EHAHTPR | − | − | − | − |
The binding sequence motif and its relationship to proteins.
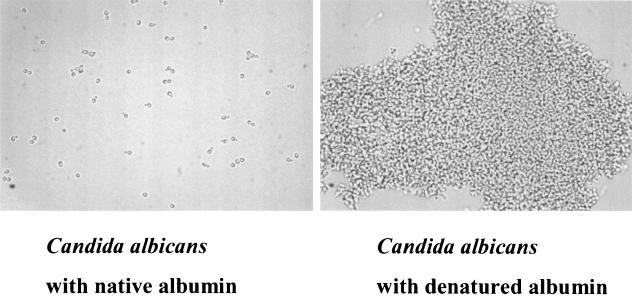
The broad specificity of Als5p binding to such sequences as τϕ+ and threonine, serine, and alanine patches (7) may explain previously observed characteristics of C. albicans binding to numerous proteins (11). C. albicans (or S. cerevisiae expressing an Als adhesin) binds better to many proteins after they are denatured such as disrupted basement membrane and denatured type I collagen known as gelatin (12, 14). BSA is another such example. Inspection of the sequence of BSA reveals 16 τϕ+ sequence motifs. At least 13 of these are buried in the native structure but would become exposed after denaturation. As a proof of this concept, BSA was denatured by boiling and added to C. albicans yeast cells, and this combination was compared to a similar number of yeast cells exposed to native BSA (Fig. 3). As indicated in the figure, there is adherence and aggregation around denatured BSA, whereas these are not detectable when native BSA is added. This same phenomenon occurs when BSA is immobilized on magnetic beads (data not shown), i.e., denatured BSA elicits adherence and aggregation, whereas native BSA does not. Similar results are obtained with S. cerevisiae expressing Als1 or Als5p (data not shown). Thus, denaturation exposes adhesive ligands such as τϕ+ and perhaps others, which elicit adherence followed by aggregation.
FIG. 3.
Photomicrographs of C. albicans yeast cells mixed with BSA in native conformation (left panel) or denatured by boiling (right panel). Note that the cells on the left are present in singlets and doublets, demonstrating little or no interaction with BSA, whereas those on the right are aggregated into a large mass.
DISCUSSION
This study is a logical outgrowth of previous work demonstrating the adherence of C. albicans and S. cerevisiae expressing ALS5 or ALS1 to immobilized peptides when they are presented in the appropriate steric configuration and composition (7). The use of a random peptide library as described herein clearly demonstrates that the recognition of peptide ligands by the Als proteins is degenerate, i.e., many peptides can be recognized and adhered to. This finding was suggested by our previous work (7) but could not easily be tested with the other peptide-beads since each peptide had to be individually synthesized and coupled to the beads-an extremely slow and inflexible method. The screening method used herein allows one to screen large numbers of beads but is not amenable to high throughput screening since the recognition system is degenerate, i.e., so many beads are bound by yeast cells.
Adhesive peptides in the current study (Table 1) had several common properties. Basic residues were overrepresented relative to expectation from random composition (25 occurrences versus 11 expected by chance), and sequential basic residues were more common than expected (5 instances in 15 peptides versus 2 expected). There was a deficit in acidic residues (3 occurrences versus 11 expected), and they occurred only at the N terminus. Each peptide in Table 1, as well as the peptide APRLRFYSL mentioned previously, possesses the sequence motif. Note that the nonadherent control peptide EHAHTPR has no sequence motif. The scrambled peptide SKMAYTL, which is not adhesive, also has no sequence motif. A single naturally occurring adherent peptide without this sequence motif is known: FTTTSTSPV (7). Therefore, there must be additional binding specificity manifested by Als proteins in addition to the motif found in this work. Binding of τϕ+ peptides could be competed and was specific. SKMAYTL, a “scrambled version of the adherent peptide,” AYKSLMT, was inactive with Als5p- and Als1p-bearing S. cerevisiae cells. The specificity of the interaction was further demonstrated by the fact that free peptide ligand inhibited the ability of S. cerevisiae expressing Als5p to adhere to the identical sequence on the surface of a bead (Table 3).
It should be borne in mind that C. albicans has many types of adhesins on the cell surface at any one time, including hydrophobic proteins (18), cell wall proteins that can be linked to human cells by host transglutaminase (19), and numerous Als proteins (9). This could explain why a greater fraction of library beads (>10-fold more) were coated by C. albicans than by S. cerevisiae expressing Als5p or Als1p. It also could explain why C. albicans adheres to sequences chosen by Als5p and Als1p and even “scrambled” sequences (Table 2). Furthermore, because C. albicans has potentially more adhesins present on the cell surface, it would explain why a soluble peptide, VTHTHRR (Table 3), inhibited S. cerevisiae expressing Als5p from adhering to beads bearing the homologous peptide, whereas C. albicans could still adhere by using other Als adhesins. Only a larger peptide, a 23-mer, among peptides we have worked with inhibits C. albicans adherence to target proteins and peptides (15; Klotz et al., unpublished). Lastly, it must be appreciated that Als1p on the surface of C. albicans is not at its greatest magnitude because the cells were harvested in the stationary phase of growth, and this will affect the adherence of this fungus (3, 7a).
As mentioned above, yeast cell adherence to 7-mer peptide-coated beads is reversed upon the addition of formamide. Similarly, yeast cells blocked from adhering to beads by the addition of free peptides (such as VTHTHRR) when treated with formamide release the peptide-yeast cell interaction, and yeast cells can once again adhere to the peptide-coated beads (data not shown). This likely is occurring because the formamide disrupts the hydrogen bonds formed with the exposed peptide backbone of the target (7; Gaur and Klotz, unpublished). In this assay, neither nonionic detergents nor saccharides had an effect on adherence; therefore, a hydrophobic or lectin effect is not the sole mode of adhesive interaction in Als binding to peptides (5-7).
Hawser and Islam (8) measured the adherence of C. albicans to single amino acids immobilized in plastic wells and showed specific adherence to several amino acids and not to others. These authors also demonstrated the ability of BSA, when premixed with the yeast cells, to block the subsequent adherence to immobilized BSA but not to the amino acids. Our study is in concurrence with the findings with BSA but not that of the amino acids. This is very likely due to the nature of the assays used, since physical trapping, hydrophobic interactions, and probably other forces contribute to assays in wells, whereas the use of beads has no confounding background interactions that are measured as adherence. Furthermore, the bead-yeast interaction is not affected to any measurable extent by the type of buffer used or by the addition of sugars or Tween 20.
Differential adherence of the Als adhesins has been noted previously (7; Chan et al., Abstr. Sixth Annu. Candida Candidiasis Meet.; N. V. Lucindo, D. C. Sheppard, S. G. Filler, J. E. Edwards, and M. Zhang, Abstr. 102nd Gen. Meet. Am. Soc. Microbiol. 2002, abstr. F-21, p. 204, 2002; D. C. Sheppard, M. Zhang, A. S. Ibrahim, Y. Fu, S. G. Filler, and J. E. Edwards, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. 385, p. 438, 2003). The data are consistent with the concept that each Als protein has a broad repertoire of preferred amino acid sequences, and these repertoires differ among the different Als proteins. Furthermore, as seen in Fig. 2 (compare Fig. 2, left panel, with Fig. 2, right panel), the adherence phenotypes of these two Als proteins are distinctly different. The phenotype for Als1p can be explained by flocculation (3) or aggregation of the Als proteins (5).
The major finding in the present study relates to the wide range of peptide targets to which C. albicans can adhere. A degenerate “recognition system” among adhesins guarantees a plethora of target proteins for adherence. Such appears to be the case with C. albicans, which can bind to many proteins and peptides (11, 13, 14). Our previous study established that cells bearing Als5p adhere to every protein we studied provided the peptide backbone was accessible for adherence (Gaur and Klotz, unpublished). Furthermore, denaturing proteins provides an adherence target for Als proteins by breaking down the secondary and tertiary structure of the protein and exposing previously buried τϕ+ sequences and likely other sequences for recognition by Als proteins (Gaur and Klotz, unpublished).
Acknowledgments
We thank Wallace Clark of the Department of Chemistry, University of Arizona, for the sequence analysis of the beads and Min Hahn for peptide synthesis on the beads. The ALS1 plasmid was a gift from Scott Filler.
Editor: T. R. Kozel
REFERENCES
- 1.Calderone, R. A., and P. C. Braun. 1991. Adherence and receptor relationships of Candida albicans. Microbiol. Rev. 55:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frieman, M. B., J. M. McCaffery, and B. P. Cormack. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a β1,6-glucan-cross-linked cell wall protein. Mol. Microbiol. 46:479-492. [DOI] [PubMed] [Google Scholar]
- 3.Fu, Y., A. S. Ibrahim, D. C. Sheppard, Y.-C. Chen, S. W. French, J. E. Cutler, S. G. Filler, and J. E. Edwards. 2002. Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol. Microbiol. 44:61-72. [DOI] [PubMed] [Google Scholar]
- 4.Fu, Y., G. Rieg, W. A. Fonzi, P. H. Belanger, J. E. Edwards, and S. G. Filler. 1998. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect. Immun. 66:1783-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaur, N., S. Klotz, and R. Henderson. 1999. Overexpression of the Candida albicans ALA1 gene in Saccharomyces cerevisiae results in aggregation following attachment of yeast cells to extracellular matrix proteins, adherence properties similar to those of Candida albicans. Infect. Immun. 67:6040-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaur, N. K., and S. A. Klotz. 1997. Expression, cloning, and characterization of a Candida albicans gene, ALA1, that confers adherence properties upon Saccharomyces cerevisiae for extracellular matrix proteins. Infect. Immun. 65:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaur, N. K., R. L. Smith, and S. A. Klotz. 2002. Candida albicans and Saccharomyces cerevisiae expressing ALA1/ALS5 adhere to accessible threonine, serine, or alanine patches. Cell Commun. Adhesion 9:45-57. [DOI] [PubMed] [Google Scholar]
- 7a.Gaur, N. K., and S. A. Klotz. 2004. Accessibility of the peptide backbone of protein ligands is a key specificity determinant in Candida albicans SRS adherence. Microbiology 150:277-284. [DOI] [PubMed] [Google Scholar]
- 8.Hawser, S., and K. Islam. 1998. Binding of Candida albicans to immobilized amino acids and bovine serum albumin. Infect. Immun. 66:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoyer, L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176-180. [DOI] [PubMed] [Google Scholar]
- 10.Kamai, Y., M. Kubota, Y. Kamai, T. Hosokawa, T. Fukuoka, and S. G. Filler. 2002. Contribution of Candida albicans ALS1 to the pathogenesis of experimental oropharyngeal candidiasis. Infect. Immun. 70:5256-5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klotz, S. A. 1994. Plasma and extracellular matrix proteins mediate in the fate of Candida albicans in the human host. Med. Hypoth. 42:328-334. [DOI] [PubMed] [Google Scholar]
- 12.Klotz, S. A., and R. D. Maca. 1988. Endothelial cell contraction increases Candida adherence to exposed extracellular matrix. Infect. Immun. 56:2495-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klotz, S. A., and R. L. Smith. 1991. A fibronectin receptor on Candida albicans mediates adherence of the fungus to extracellular matrix. J. Infect. Dis. 163:604-610. [DOI] [PubMed] [Google Scholar]
- 14.Klotz, S. A., and R. L. Smith. 1995. Gelatin fragments block adherence of Candida albicans to extracellular matrix proteins. Microbiology 141:2681-2684. [DOI] [PubMed] [Google Scholar]
- 15.Klotz, S. A., R. L. Smith, and B. W. Stewart. 1992. Effect of an arginine-glycine-aspartic acid-containing peptide on hematogenous candidal infections in rabbits. Antimicrob. Agents Chemother. 36:132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam, K. S., S. E. Salmon, E. M. Hersh, V. J. Hruby, W. M. Kazmierski, and R. J. Knapp. 1991. A new type of synthetic peptide library for identifying ligand-binding activity. Nature 354:82-84. [DOI] [PubMed] [Google Scholar]
- 17.Pendrak, M. L., and S. A. Klotz. 1995. Adherence of Candida albicans to host cells. FEMS Microbiol. Lett. 129:103-114. [DOI] [PubMed] [Google Scholar]
- 18.Singleton, D. R., J. Masuoka, and K. C. Hazen. 2001. Cloning and analysis of a Candida albicans gene that affects cell surface hydrophobicity. J. Bacteriol. 183:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stabb, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1537. [DOI] [PubMed] [Google Scholar]
- 20.Zhao, X., C. Pujol, D. Soll, and L. Hoyer. 2003. Allelic variation in the contiguous loci encoding Candida albicans ALS5, ALS1, and ALS9. Microbiology 149:2947-2960. [DOI] [PubMed] [Google Scholar]