Abstract
DARPP-32 (dopamine and adenosine 3′, 5′-cyclic monophosphate cAMP-regulated phosphoprotein, 32 kDa) is a striatal-enriched protein that mediates signaling by dopamine and other first messengers in the medium spiny neurons. The transcriptional mechanisms that regulate striatal DARPP-32 expression remain enigmatic and are a subject of much interest in the efforts to induce a striatal phenotype in stem cells. We report the identification and characterization of a conserved region, also known as H10, in intron IV of the gene that codes for DARPP-32 (Ppp1r1b). This DNA sequence forms multiunit complexes with nuclear proteins from adult and embryonic striata of mice and rats. Purification of proteins from these complexes identified early growth response-1 (Egr-1). The interaction between Egr-1 and H10 was confirmed in vitro and in vivo by super-shift and chromatin immunoprecipitation assays, respectively. Importantly, brain-derived neurotrophic factor (BDNF), a known inducer of DARPP-32 and Egr-1 expression, enhanced Egr-1 binding to H10 in vitro. Moreover, overexpression of Egr-1 in primary striatal neurons induced the expression of DARPP-32, whereas a dominant-negative Egr-1 blocked DARPP-32 induction by BDNF. Together, this study identifies Egr-1 as a transcriptional activator of the Ppp1r1b gene and provides insight into the molecular mechanisms that regulate medium spiny neuron maturation.
Introduction
Gene expression in striatal medium spiny neurons (MSNs) is altered in a number of neuropsychiatric diseases, e.g., Huntington's disease (HD). There is dysregulation in the expression of ubiquitous and striatal-enriched transcription factors (TFs) in the caudate nucleus in multiple HD mouse models and human HD subjects, which may contribute to the downregulation in the expression of striatal-enriched proteins (Luthi-Carter et al., 2000, 2002; Zucker et al., 2005; Hodges et al., 2006; Thomas et al., 2011). However, these TFs and their targets, i.e., markers of terminally differentiated MSNs, have not been linked.
MSNs are GABAergic and contribute to regulation of movement, organization of motor behavior, and cognition (Gerfen, 1992; Jain et al., 2001; Graybiel, 2005). They project to the globus pallidus or substantia nigra (Gerfen and Young, 1988), and they receive glutamatergic synaptic inputs from the cerebral cortex and dopaminergic afferents from the substantia nigra and ventral tegmental area (Parent and Hazrati, 1995). DARPP-32 (dopamine and adenosine 3′, 5′-cAMP-regulated phosphoprotein, 32 kDa), a central mediator of signal transduction pathways in the dopaminoceptive MSNs, is expressed in >90% of striatal MSNs and is the most commonly used marker of this mature neuronal subtype (Gustafson et al., 1992; Ouimet et al., 1998; Greengard et al., 1999; Svenningsson et al., 2004). Identification of the molecular mechanisms that regulate DARPP-32 expression will aid in a rational design of therapeutic strategies to modulate expression of DARPP-32 and likely other striatal-enriched genes, either in neuropathological conditions of the striatum or for the induction of a striatal phenotype in stem cells.
A number of TFs that regulate neurogenesis, migration, and differentiation of MSNs have been identified, including Dlx1, Dlx2, Dlx5, Dlx6, Nolz-1, Isl-1, Mash1, bcl11b/CTIP2, and Gsh1/2. They are present during embryonic development and are essentially undetectable after the first postnatal week, with the exception of CTIP2 (Jain et al., 2001; Chang et al., 2004; Arlotta et al., 2008). None of these has been demonstrated to bind to Ppp1r1b sequences or to be sufficient for induction of DARPP-32 during development or in the adult.
We demonstrated that the promoter and intragenic regions of DARPP-32, collectively termed D9 (genomic elements from DARPP-32 encompassing 9 kb), selectively direct transgene expression to MSNs (Bogush et al., 2005; Brown et al., 2008), whereas the 2.1 kb of 5′ UTR sequence alone does not direct expression to any region of the CNS (Blau et al., 1995). In this study, we sought to characterize the function of a region in intron IV of the Ppp1r1b gene, also known as H10. Our interest in this particular sequence arose from its high conservation among species and its location in a region of the DARPP-32 gene that is included in D9 (Bogush et al., 2005). After the identification of multiunit complexes between H10 and striatal nuclear proteins, the protein-binding site was determined. We found that the TF early growth response-1 (Egr-1) binds to H10 in vitro and in vivo. Importantly, we report that Egr-1 is sufficient to induce DARPP-32 expression in vitro and is required for the induction of DARPP-32 by brain-derived neurotrophic factor (BDNF).
Materials and Methods
Phylogenetic analysis of Ppp1r1b intronic sequences.
The mouse and human genomic sequences of Ppp1r1b were retrieved from the National Center for Biotechnology Information (NCBI) GenBank database. These gene sequences were aligned pairwise (mouse vs human) using BLAST (basic local alignment search tool; bl2seq) at NCBI website with the following parameters: word size = 11; reward for match = 1; penalty for mismatch = −2; gap open penalty = 5; gap extension penalty = 2; gap ×_dropoff = 50; and low-complexity filter checked. Only those alignments in the intronic regions (mouse) with >70% sequence homology were considered for additional analysis. The mVISTA program (Frazer et al., 2004) was used for comparative sequence analysis of the Ppp1r1b gene across species (mouse, rat, human, orangutan, dog, and horse). F10A (75%) and H10 (91%), two of the most highly conserved sequences, were chosen to conduct this study. “H” is for homology and is the species homology; “F” includes the search of other striatal-enriched genes and stands for “family.” Sequences are as follows: F10A, 5′-TTTTATTGCCTGGGTAGCTTACCCACAGGCAGC-3′; and H10, 5′-AGCCGCCCACACTGTTCCTTTCC-3′.
Preparations of nuclear and cytoplasmic extracts.
For preparation of extracts from brain tissue, mice of either sex were killed by CO2 asphyxiation, and brain regions were rapidly dissected. Nuclear and cytoplasmic extracts were prepared as described previously (Schreiber et al., 1989).
Electrophoretic mobility shift assay and super-shift assays.
Double-stranded oligonucleotides (Integrated DNA Technologies) were prepared in annealing buffer (in mm: 20 Tris, 10 MgCl2, 50 NaCl, and 1 DTT) to a final concentration of 5 μm. The double-stranded DNA fragments were labeled with [γ-32P]ATP and T4 polynucleotide kinase and purified with Illustra Microspin G-25 columns (GE Healthcare). Each electrophoretic mobility shift assay (EMSA) reaction (25–30 μl) contained 5–10 μg of nuclear proteins, 2 μg poly(dI-dC), and 32P-labeled DNA probe in 1× binding buffer (10 mm HEPES, pH 7.9, 30 mm KCl, 1.2% glycerol, 0.5 mm DTT, 1 mm MgCl2, 0.2 mm PMSF, and 0.1 mm EDTA, pH 8). For competition experiments, 100× excess unlabeled oligonucleotides were added before the addition of the labeled probe and incubated for 30 min at 4°C. The protein–DNA complexes were separated on native 6% polyacrylamide gels in 0.5× Tris borate/EDTA running buffer at 200 V for 2 h. The gels were dried and exposed on phosphor-screens (GE Healthcare) using the Typhoon-Trio (GE Healthcare). For super-shift assays, 5 μg of rabbit α-Egr-1 antibody, clone 588 (sc-110X; Santa Cruz Biotechnology), rabbit polyclonal α-SRF antibody, clone G-20 (sc-335; Santa Cruz Biotechnology), mouse α-Sp1 antibody, clone 1C6 (sc-420; Santa Cruz Biotechnology), or rabbit polyclonal α-Egr-2 antibody (PRB-236P; Covance) were added to each reaction and incubated for 30 min at 4°C before the addition of the 32P-labeled H10 probe.
The following sequences were used: putative Sp1 binding site upstream (in bold) of H10 (Ppp1r1b), 5′-GTC CTT GTT CCC TCC CCG CCT TGC GTC TTT GGG AAG CCG CCC ACA CTG TTC CTT TCC-3′ (in which the Sp1 and Egr-1 binding sites are underlined); Drd3, 5′-CTGTGTTCGCCCACAGTCATAT-3′; Baiap2, 5′-CCCGCACCGCCCACACCCCGCG-3′; Ppp1r16b, 5′-CCCACTCCGCCCACAGCTCTTG-3′; Klf16, 5′-TGGGGTGTGTGGGCGTGTCGCG-3′; Penk1, 5′-GTTGGGTTGTGGGCGGGGCTCA-3′; Ppp1r1b (5′ UTR), 5′-CGCACCAAGCCCACACATGTGA-3; Cpne2, 5′-CGCTTCCCGCCCACATCCTCCC-3′; and Stra13, 5′-CTCACAGACACCCGCTCTGAGG-3′ (the putative Egr-1 binding sites are underlined).
Southwestern blot analysis.
Nuclear proteins (200 μg) from either the brainstem or striatum were separated by SDS-PAGE (4–12% Bis Tris) in MES running buffer (Invitrogen) for 1 h at 200 V and transferred to nitrocellulose membranes for 3 h at 30 V. Southwestern blot analysis was performed as per Handen and Rosenberg (1997), with some modifications. The blots were incubated with 20 ml of 1× renaturation buffer (10 mm Tris-Cl, pH 7.5, 0.1 mm EDTA, pH 7.5, 50 mm NaCl, 1 mm DTT, and 5% milk) for 2 h at room temperature and then prehybridized with 1× hybridization buffer (10 mm HEPES and 5% nonfat dry milk) for 30 min at room temperature. Double-stranded H10 oligonucleotides (20 pmol) were end labeled with 32P to specific activity of 2 × 105 cpm/ng. The membranes were hybridized with 1× binding buffer (10 mm HEPES, pH 7.9, 50 mm NaCl, 1 mm EDTA, 1 mm DTT, and 0.25% nonfat dry milk) containing 30 μg of poly(dI-dC) and 6 ng of radiolabeled probe (2 × 105 cpm/ng) for 1 h at room temperature. The blot was washed three times with 1× binding buffer for 20 min each, air dried, and exposed for autoradiography.
Affinity chromatography.
Double-stranded, biotin-tagged (5′ end of forward sequence) DNA oligonucleotides corresponding to the H10 or F10A sequences were prepared to a final concentration of 20 μm (Integrated DNA Technologies). Nuclear extract from rat striatum or brainstem was used, and reactions were performed with 5 mg of nuclear proteins, after the determination that mouse and rat extracts yielded identical patterns on EMSA. Affinity chromatography was performed as described previously (Gadgil et al., 2001; Jiang et al., 2009; Minig et al., 2009) with modifications.
Nuclear extracts were precleared by incubation with 250 μl of streptavidin agarose resin (Pierce) for 1 h at 4°C. To purify the proteins of interest from the precleared extract, we combined it with 1 mg of poly(dI-dC) (Sigma) as carrier DNA and 10 nmol double-stranded biotin-tagged H10 or F10A oligonucleotide as a control in 1× binding buffer (10 mm HEPES, pH 7.9, 30 mm KCl, 1.2% glycerol, 0.5 mm DTT, 5 mm MgCl2, 0.2 mm PMSF, and 0.1 mm EDTA, pH 8) to allow the formation of the protein–DNA complexes in solution. As controls, we combined biotinylated F10A oligonucleotide with striatal tissue and H10 oligonucleotide with brainstem tissue. The reactions were incubated with gentle mixing for 15 min at 4°C, 15 min at room temperature, and then applied to the capture columns, which contained 250 μl of streptavidin agarose resin (capacity of 197 pmol biotin/1 μl resin). The column was washed five times with 1× binding buffer to remove unbound DNA and protein and was then eluted with high-salt buffer at room temperature. The optimal elution condition for the H10-binding proteins as determined by EMSA was 2 m NaCl buffer (12% glycerol, 20 mm Tris, pH 6.8, 2 m NaCl, 5 mm MgCl2, 1 mm EDTA, and 1 mm EGTA) at room temperature. Eluted proteins were analyzed by mass spectrometry.
Mass spectrometry.
The pH of all three protein samples was adjusted to 8.5 with 100 mm NH4HCO3. Proteins were reduced with 5 mm [tris(2-carboxyethyl)phosphine]hydrochloride at 37°C for 20 min and alkylated with 10 mm iodoacetamide for 30 min in the dark at room temperature. A Lys-C digestion (50 ng/sample) was performed for 4 h, followed by overnight trypsin digestion at 37°C (1:50 enzyme:substrate ratio). The digestion was quenched by adding trifluoroacetic acid (5%) to achieve a pH of 2–4. Samples were desalted with ZipTip C18 (Millipore). The eluate was dried down, and peptides were reconstituted with 0.1% formic acid in 2:98 acetonitrile (ACN) in H2O for liquid chromatography/tandem mass spectrometry (LC-MS/MS) analysis. A Waters NanoAcquity UPLC system was interfaced to a Thermo LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). Reversed-phase LC was performed on a Waters BEH130 C18 column (100 μm × 100 mm, 1.7 μm particle size). Samples were trapped and washed in a Waters Symmetry C18 trap column (180 μm × 100 mm, 5 μm particle size) before separation in the nanocolumn. Gradient elution was performed with 0.1% formic acid in water as solvent A and in ACN as solvent B, with solvent B raised from 1 to 50% in 90 min, then 50 to 85% in the next 10 min at a flow rate of 0.6 μl/min. The mass spectrometer was operated in positive mode with spray voltage at 2.5 kV, ion transfer tube voltage at 45 V, and ion transfer tube temperature at 170°C. A normalized collision energy of 35% and activation time of 30 ms were applied in MS/MS acquisitions. The top eight most intense ions were selected for fragmentation in the LTQ-Orbitrap mass spectrometer. The following dynamic exclusion settings were applied to precursor ions chosen for MS/MS analysis: repeat count, 1; repeat duration, 60 s; and exclusion duration, 240 s. MS/MS spectra were searched against the UniProtKB rat database (downloaded February 2011) using Sorcerer-SEQUEST (version 27, revision 11; Sage-N Research), Mascot (version 2.3.02; Matrix Science), X! Tandem (version 2007.01.01.1; The Global Proteome Machine Organization), and Scaffold (version 3.0.8; Proteome Software) algorithms (Perkins et al., 1999; Searle, 2010). Searches were performed with full tryptic specificity (two missed cleavages): carbamidomethylated cysteine residues as static modification, deamidated asparagine and glutamine (+0.9840 Da), and oxidized methionine, histidine, and tryptophan (+15.9949 Da) as differential modifications. A precursor mass error tolerance of 10 ppm and default product ion mass error tolerance of above searching algorithms were used.
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (Thomas et al., 2008), with some modifications. Mouse striata equal to 100 mg wet weight were combined for each ChIP reaction. The tissue was fixed with 1% formaldehyde in PBS for 10 min at room temperature, and 2 m glycine was added at a final concentration of 0.125 m. After washing with PBS, the tissue was homogenized with 500 μl of cell lysis buffer [10 mm HEPES, pH 8, 85 mm KCl, 0.5% NP40, and 1× protease inhibitors cocktail (Roche)] on ice. Samples were centrifuged at 2700 × g for 10 min, and the nuclear pellet was resuspended in 300 μl of nuclei lysis buffer [1% SDS, 10 mm EDTA, pH 8, 50 mm Tris-8.0, and 1× protease inhibitors cocktail (Roche)]. Chromatin was sheared into 200 bp–1 kb DNA fragments with a Branson Sonifier 450 (VWR scientific) 12–15 times at setting 3, duty cycle 50%, for 10 s per pulse, while placing the cells on an ice for 1 min between pulses. The samples were then centrifuged at 20,000 × g at 4°C for 10–15 min, and 10% of each sample was saved as input. For immunoprecipitation (IP) of Egr-1–DNA complexes, each sample was diluted 10 times with 1× dilution buffer (1% Triton X-100, 150 mm NaCl, 2 mm EDTA, pH 8, 20 mm Tris-8.0, and 1× protease inhibitors cocktail), followed by the addition of 6 μg of rabbit Egr-1 antibody, clone 588 (sc-110X; Santa Cruz Biotechnology) or 6 μg of rabbit IgG as a control and incubated overnight at 4°C. Seventy to 100 μl of sheep anti-rabbit IgG beads (Dyna Beads; Invitrogen) were added, and the reactions were incubated overnight at 4°C. The beads were washed three times with low-salt buffer (1% Triton X-100, 0.1% SDS, 150 mm NaCl, 2 mm EDTA, pH 8, and 20 mm Tris-8.0, 1×), two times with high-salt buffer (1% Triton X-100, 0.1% SDS, 500 mm NaCl, 2 mm EDTA, pH 8, and 20 mm Tris-8.0, 1×), and one time with TE buffer (10 mm Tris, pH 8.1, 50 mm NaCl, and 1 mm EDTA) and eluted with 300 μl of elution buffer (1% SDS and 0.1 m NaHCO3) at 65°C for 10 min. To reverse the crosslinks, 5 m NaCl was added to a final concentration of 0.2 m, and 100 μg of proteinase K was added to each sample and incubated at 65°C overnight, after which they were incubated with 1 μg of RNase for 30 min at 37°C and then boiled for 15 min. DNA was purified for quantitative PCR analysis by QIAquick PCR purification kit (Qiagen).
Quantitative reverse transcription-PCR analysis.
Primers were designed with PrimerExpress software (forward, 5′-CCCAGCCTTAACCCAGTACTGTTC-3′; and reverse, >5′-TGGGCAAGTGGACTGTTCAGAT-3′) and analyzed with BLAT (basic local alignment tool) of the University of California, Santa Cruz genome browser. Quantitative reverse transcription-PCR was performed by SYBR Green-based real-time PCR using RT2 SYBR Green qPCR Master Mixes (catalog #PA-012; SABiosciences) according to the instructions of the manufacturer. PCR products were detected by ethidium bromide on a Fujifilm LAS-3000 developer after electrophoresis on 1% Tris borate–EDTA agarose gel for 1 h at 60 V. For data analysis, the threshold cycles (Ct) of all replicates were averaged after dropping reactions with PCR inhibitors or a varying slope as determined from the melt and amplification curves. Each ChIP reaction Ct value was then normalized to the input DNA Ct value as follows: ΔCt (normalized ChIP) = [Ct (ChIP) − Ct (input) − Log2 (input dilution factor)]. The percentage of DNA recovered from each IP reaction relative to the starting DNA was calculated as percentage input as follows: % input = 2 [−ΔCt (normalized ChIP)].
Cell culture and transient transfection.
Striatal primary culture cells were prepared as described previously (Ivkovic and Ehrlich, 1997). Transfection was performed using electroporation (Amaxa Biosystems). Neurons were transfected with Egr-1–pIRES2–EGFP vector (Beck et al., 2008) immediately after dissociation, and electroporation was conducted using 4.0 × 106 cells, 3 μg of plasmid DNA, and 100 μl of mouse neuron nucleofector solution.
Infection assays/viral transduction.
Dominant-negative Egr-1 (adnEgr-1) constructs were in adenovirus (Kundumani-Sridharan et al., 2010). Adenoviral infections were performed after cells had attached for 1–2 h. Virus was added in fresh medium, and cells were harvested after 96 h. BDNF (10 ng/ml) was added concurrently with virus on the day of plating. Multiplicity of infection ranged from 100 to 250.
Immunocytochemistry and Western blot analysis.
Both protocols were performed as described previously (Keilani and Sugaya, 2008). Primary and secondary antibodies included the following: rabbit α-DARPP-32, clone 19A3 (2306; Cell Signaling Technology); mouse α-green fluorescent protein (GFP), clones 7.1 and 13.1 (11814460001; Roche); mouse α-WT-1, clone F-6 (sc-7385; Santa Cruz Biotechnology); mouse α-GAPDH, clone 6C5 (sc-32233; Santa Cruz Biotechnology); rabbit α-Egr-1, clone 588 (sc-110X; Santa Cruz Biotechnology); rabbit α-histone H3 antibody (09-838; Millipore); horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit IgG secondary antibodies at concentration of 1:10,000 (Vector Laboratories); and the secondary antibodies green fluorescent Alexa Fluor 488 and red fluorescent Alexa Fluor 594 (Invitrogen). Signals were detected by chemiluminescence using the enhanced chemiluminescence ECL system (Pierce) on a Fujifilm LAS-3000 developer. The densitometric value of each specific band was obtained using MultiGauge (Fujifilm) software and normalized to GAPDH.
Statistical analysis.
Each data point represents triplicate experiments presented as mean ± SEM. One-way ANOVA with Bonferroni's post hoc test was performed using GraphPad Prism version 5.00 for Windows (GraphPad Software). Significance are reported with a p < 0.05.
Results
Intron IV of the Ppp1r1b gene contains seven conserved sequences
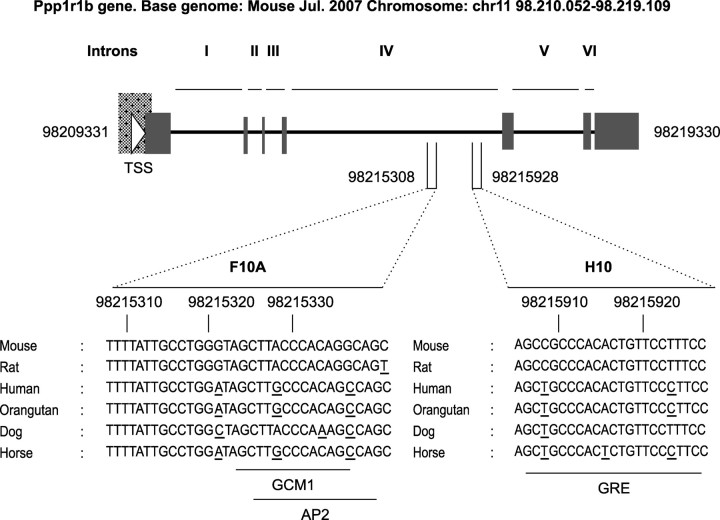
To begin to identify the cis-regulatory elements in the Ppp1r1b genomic elements (D9) that direct gene expression to MSNs (Bogush et al., 2005), we performed an in silico analysis of D9. We identified 30 sequences longer than 11 nt that were >70% conserved across mouse, rat, human, orangutan, dog, and horse (data not shown). Seven of these non-overlapping conserved regions were located in intron IV. In Figure 1, we show the location of two of the most highly conserved sequences, also known as F10A (75%) and H10 (91%).
Figure 1.
H10 and F10A sequences are highly conserved across species (mouse, rat, human, orangutan, dog, and horse). Nucleotides that are not conserved, compared with mouse, are underlined. The numbers depict the location on mouse chromosome 11. TSS, Transcription start site; I–VI, Ppp1r1b introns.
H10 and F10A form complexes with striatal nuclear proteins in vitro
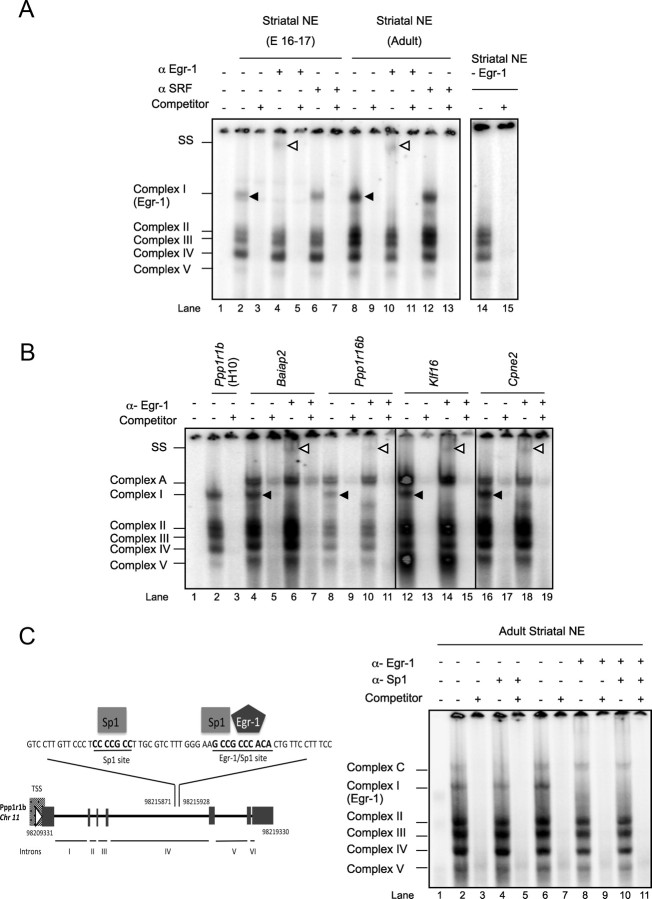
To determine whether H10 and F10A participate in the regulation of the Ppp1r1b gene, we searched each sequence for TF binding sites using MatInspector from Genomatix. Putative sites in F10A matched those for GCM1 (glial cells missing homolog 1) and AP2 (activator protein 2), whereas putative sites in H10 matched those for GREs (glucocorticoid responsive and related elements) (Fig. 1). On EMSA, the H10 oligonucleotide formed DNA–protein complexes in equal levels with both embryonic and adult striatal nuclear extract (Fig. 2A, lanes 11, 13). These complexes did not form with the embryonic (Fig. 2A, lane 9) or adult (Fig. 2B, lane 2) cytoplasmic extract and were depleted on competition with 100-fold molar excess of unlabeled H10 oligonucleotide (Fig. 2A, lanes 10, 12, 14). Conversely, F10A formed specific, high-intensity protein complexes with embryonic striatal nuclear proteins but at very low intensity with adult striatal nuclear proteins (Fig. 2A, lanes 4, 6). Because we are interested in understanding the regulation of DARPP-32 and MSN-specific expression in both embryonic and adult striatum, we focused this study on H10, with F10A as a control in some assays.
Figure 2.
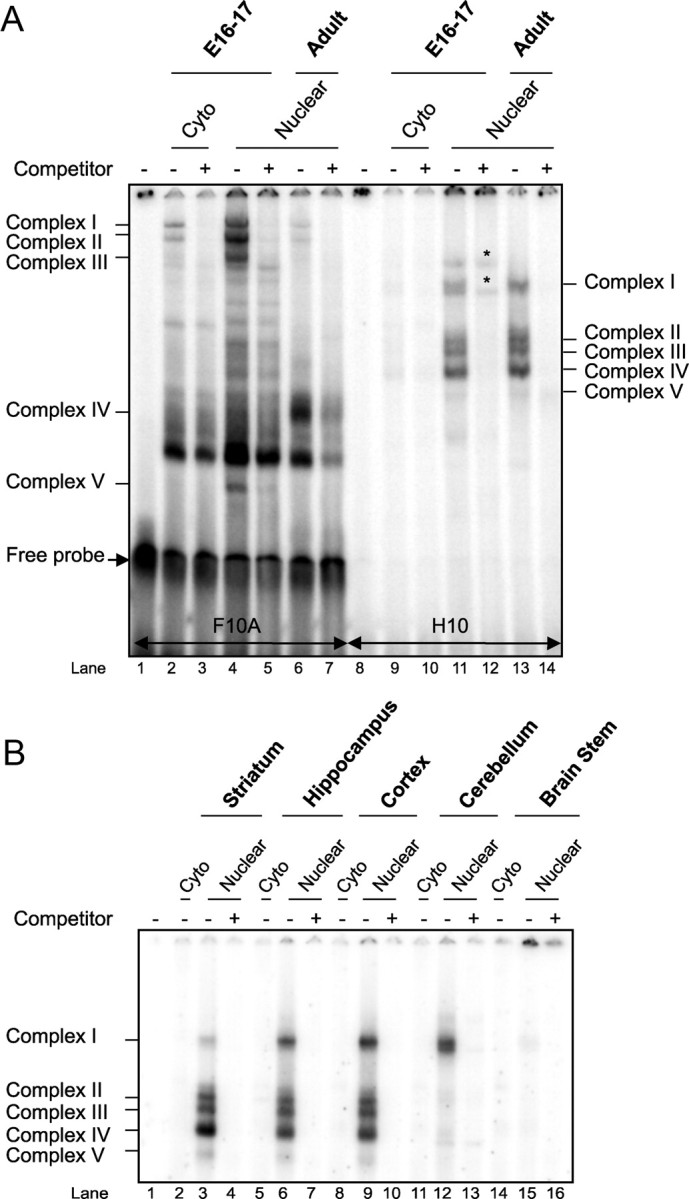
H10 and F10A oligonucleotides interact in vitro with embryonic and adult striatal nuclear proteins. A, EMSA shows that 32P-labeled F10A and H10 oligonucleotides bind with proteins in embryonic (E16–17) and adult striatal nuclear extracts. The specificity of the complexes was determined by the addition of 100-fold excess of unlabeled F10A or H10 oligonucleotide as a competitor. * Refers to nonspecific complexes, i.e., not quenched by competitor. B, EMSA reveals the formation of complexes between H10 and nuclear extracts from adult striatum, hippocampus, cortex, cerebellum, and brainstem.
To determine whether the H10–protein complexes are striatal specific, we repeated the EMSA with nuclear extracts from adult striatum, hippocampus, cortex, cerebellum, and brainstem (Fig. 2B). Complexes I–IV were of markedly greater intensity with nuclear proteins from forebrain regions relative to brainstem. Unlike in most H10 gel shifts with striatal extract, complex I was of lower intensity relative to other forebrain regions. It was the only complex visible with cerebellar extract (Fig. 2B, lane 12). We concluded that the TF(s) complexing with H10 are widely expressed throughout the forebrain. At least one of them is present in cerebellum, and, if they are expressed in brainstem, they are undetectable by EMSA.
H10 contains a novel GC-rich protein-binding site
To determine the precise protein-binding nucleotides in H10, we used EMSA to perform a mutation analysis. H10 variants were created by sequentially mutating 2 nt, resulting in a total of 11 novel sequences (Fig. 3A). Mutant 2 showed lower affinity for the H10-binding proteins relative to the endogenous sequence (Fig. 3B, lane 6), whereas mutants 3–5 failed to complex with striatal nuclear extract (Fig. 3B, lanes 7–9). The new complex with mutant 3 likely resulted from the generation of a non-physiologic binding site and was not further investigated (Fig. 3B, lane 7). Conversely, mutant 2 partially competed with 32P-labeled native H10 oligonucleotide (Fig. 3C, lane 5), whereas mutants 3–5 were totally unable to compete (Fig. 3C, lanes 6–8). We concluded that the sequence 5′-CGCCCACA-3′ or 5′-TGTGGGCG-3′ is the protein-binding site in H10. The presence of multiple complexes derived from TF binding to such a short, single sequence within H10 suggests that multiple TFs likely compete for this binding site.
Figure 3.
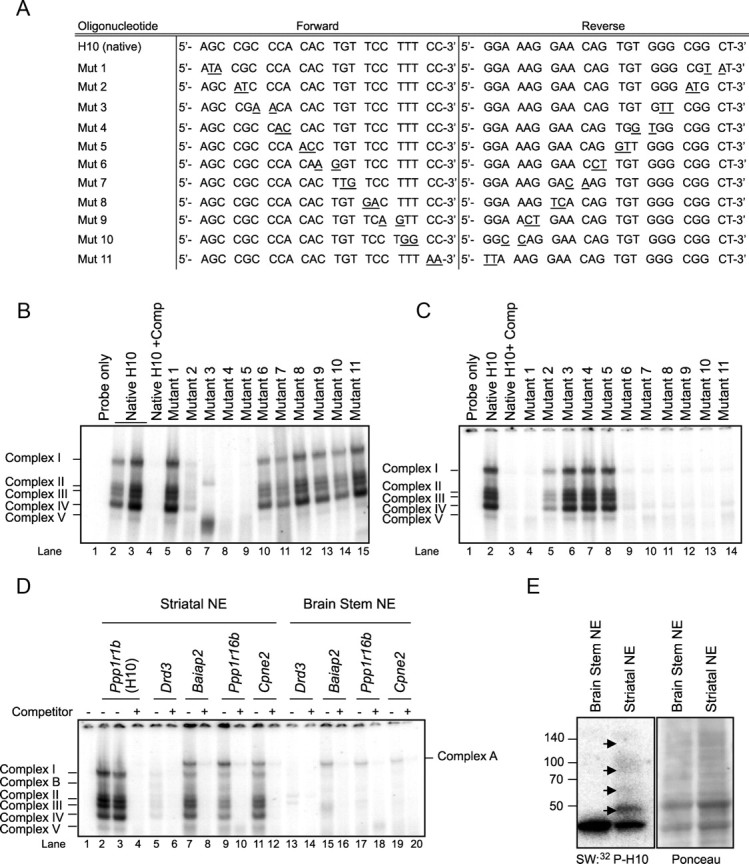
Mutation analysis of H10 identifies the sequence 5′-CGCCCACA-3′, or its complement, as the TF binding site. A, Sequences of 11 mutant (Mut) oligonucleotides generated from the H10 oligonucleotide by sequentially mutating 2 nt (underlined). B, EMSA using native H10 and 11 mutated H10 oligonucleotides with striatal nuclear proteins shows that mutated nucleotides 2–5 are required for TF binding. C, Unlabeled mutant oligonucleotides 2–5 are unable to compete for protein binding with the native radioactive H10 oligonucleotide. Each “cold” mutant was reacted with striatal nuclear proteins before the addition of the 32P-labeled native H10. D, Oligonucleotides containing the sequence 5′-CGCCCACA-3′ with the unique flanking sequences from each gene in the 5′ UTR of Drd3, Baiap2, Ppp1r16b, and Cpne2 genes interact in vitro with striatal nuclear extract (NE) but not with brainstem. The band pattern is similar to that seen with H10 except for an additional, higher-molecular-weight complex (complex A). Complex B forms between complexes I and II with the Drd3, Baiap2, Ppp1r16b, and Cpne2 sequences and also intermittently with H10. E, Southwestern blotting (SW) determines that H10 striatal nuclear binding proteins range from 45 to 130 kDa.
Based on the knowledge that each striatal TF participates in the regulation of multiple genes, we searched for the sequence 5′-CGCCCACA-3′ in the 1 kb 5′ UTR upstream from the transcription start site of 49 striatal-enriched genes and 49 non-striatal genes (Desplats et al., 2008). We found that this sequence is found significantly more frequently in the promoters of striatal-enriched genes (8 of 49) relative to genes not enriched in the striatum (2 of 49) (Fisher's test, one-tailed, p = 0.045). Although we did not search in the introns, we hypothesize that we would have found similar results. We generated oligonucleotides that included 7 nt upstream and 7 nt downstream of the core sequence (sequences are detailed in Materials and Methods) and again used EMSA to determine whether these sequences from other genes bind striatal nuclear proteins in vitro. We tested sequences from five striatal-enriched genes (Drd3, Baiap2, Ppp1r16b, Klf16, and Penk1) in addition to Ppp1r1b and two non-striatal genes (Cpne2 and Stra13). All the sequences tested except for Stra13 formed complexes similar to those seen with H10 with the nuclear extract from the adult striatum, albeit at different intensities. Complexes were not formed with brainstem extract. Examples with sequences derived from Drd3, Baiap2, Ppp1r16b, and Cpne2 are pictured in Figure 3D (lanes 5, 7, 9, 11). The sequences derived from genes other than Ppp1r1b formed an additional complex (complex A) with nuclear extracts from both striatum and brainstem. Also, several of the sequences, including H10 from Ppp1r1b, formed a complex between I and II (complex B).
Egr-1 binds to H10 in vitro and in vivo
We used Southwestern blotting to determine the approximate molecular weights of the H10-binding proteins. Brainstem nuclear extracts were used as a negative control. Relative to brainstem, at least four striatal-enriched proteins were found to bind H10 (Fig. 3E), with estimated molecular weights at 130, 80, 60, and 45 kDa.
The H10-binding proteins were purified by affinity chromatography. The identities of the purified proteins were determined by MS (Table 1). The sequences included two TFs (Egr-1 and Sarnp), four transcription cofactors (Dhx9, Trim28, Nucleolin, and Rbm39), and two transcriptional activators (Psip1 and CAND1). Notably, the glucocorticoid receptor was not identified. We considered Egr-1 to be a strong candidate as a factor that binds to H10 in vitro based on its consensus binding site (5′-GCGC/GGGGCG-3′) and its molecular weight of 82 kDa. We confirmed binding of Egr-1 to H10 by the super-shift of complex I with the addition of α-Egr-1 to nuclear extracts from both embryonic and adult striata (Fig. 4A, lanes 4, 10). Immunodepletion of Egr-1 from striatal nuclear extract abolished formation of complex I but did not disrupt complexes II–IV, indicating that, although they bind the same site, they form independently of Egr-1 (Fig. 4A, lane 14). Finally, super-shift assay with addition of α-Egr-1 to the EMSA striatal mixes with sequences from Baiap2, Ppp1r16b, Klf16, and CpneII also resulted in a super-shift of complex I (Fig. 4B), indicating that Egr-1 binds to the 5′ UTR of these genes in vitro.
Table 1.
List of H10-specific binding proteins identified in affinity chromatography eluate by MS
| Protein name | Molecular weight (kDa) | UniPortKB accession number | Number of unique peptides |
||
|---|---|---|---|---|---|
| Stem–H10 | Striatum–F10A | Striatum–H10 | |||
| Uncharacterized protein (Tpr) | 267 | D3ZNM2 | 0 | 0 | 2 |
| Myosin-9 (Myh9) | 226 | Q62812 | 0 | 0 | 2 |
| Uncharacterized protein (Rock2) | 167 | D3ZZU3 (+1) | 0 | 0 | 2 |
| α-Actinin-4 (Actn4) | 105 | Q9QXQ0 | 0 | 0 | 2 |
| Clathrin coat assembly protein AP180 (Snap91) | 94 | Q05140 (+1) | 0 | 0 | 2 |
| Uncharacterized protein (Ranbp2) | 341 | D4A054 | 0 | 0 | 1 |
| Filamin α (Flna) | 280 | C0JPT7 (+2) | 0 | 0 | 1 |
| Uncharacterized protein (Map2) | 199 | D3ZW41 (+3) | 0 | 0 | 1 |
| Uncharacterized protein (Dhx9)* | 150 | D3Z9Q9 (+1) | 0 | 0 | 1 |
| Cullin-associated NEDD8-dissociated protein 1 (Cand1)* | 136 | P97536 | 0 | 0 | 1 |
| Uncharacterized protein (Atp1a3) | 112 | D3ZE88 (+3) | 0 | 0 | 1 |
| Uncharacterized protein (Mcm2) | 102 | D3ZP96 | 0 | 0 | 1 |
| Sf3b3 protein (Fragment) (Sf3b3) | 101 | B5DF12 | 0 | 0 | 1 |
| Transcription intermediary factor 1-β (Trim28)* | 89 | O08629 | 0 | 0 | 1 |
| Uncharacterized protein (App) | 83 | D3ZP79 (+3) | 0 | 0 | 1 |
| Nucleolin (Ncl)* | 77 | P13383 (+1) | 0 | 0 | 1 |
| ATP-binding cassette, subfamily E (OABP), member 1 (Abce1) | 67 | D3ZD23 | 0 | 0 | 1 |
| Uncharacterized protein (Ythdf1) | 64 | D3ZKJ3 (+5) | 0 | 0 | 1 |
| ATP synthase subunit α, mitochondrial (Atp5a1) | 60 | P15999 | 0 | 0 | 1 |
| Acin1 protein (Fragment) (Acin1) | 60 | Q5BJU9 | 0 | 0 | 1 |
| NADPH:adrenodoxin oxidoreductase, mitochondrial (Fdxr) | 54 | P56522 | 0 | 0 | 1 |
| Early growth response-1 (Egr-1)* | 54 | P08154 | 0 | 0 | 1 |
| Uncharacterized protein (Rbm39)* | 50 | D4A5G0 (+1) | 0 | 0 | 1 |
| Ferrochelatase (Predicted) (Fech) | 48 | D3ZBM3 | 0 | 0 | 1 |
| Uncharacterized protein (RGD1560464) | 45 | D4A063 (+1) | 0 | 0 | 1 |
| Uncharacterized protein (Psip1)* | 37 | D4A7U3 (+3) | 0 | 0 | 1 |
| β-Soluble NSF attachment protein (Napb) | 33 | P85969 | 0 | 0 | 1 |
| Myristoylated alanine-rich C-kinase substrate (Marcks) | 30 | P30009 | 0 | 0 | 1 |
| 14-3-3 protein epsilon (Ywhae) | 29 | P62260 | 0 | 0 | 1 |
| SAP domain-containing ribonucleoprotein (Sarnp)* | 24 | Q498U4 | 0 | 0 | 1 |
| Uncharacterized protein (Ppp1r1b) | 22 | D4A448 (+1) | 0 | 0 | 1 |
| ADP-ribosylation factor 3 (Arf3) | 21 | P61206 (+2) | 0 | 0 | 1 |
| Calcineurin subunit B type 1 (Ppp3r1) | 19 | P63100 (+1) | 0 | 0 | 1 |
| Uncharacterized protein | 14 | D3ZKL0 | 0 | 0 | 1 |
| Uncharacterized protein | 12 | D4ADA8 | 0 | 0 | 1 |
| RCG37200 (Timm8a2) | 11 | D3ZV76 (+1) | 0 | 0 | 1 |
Crosswise comparison of the proteins identified from the Striatum–H10 reaction to the ones identified from the two control columns, Striatum–F10A and Stem–H10, to eliminate the common proteins. This refinement reduced the number of the proteins of interest down to 36.
* Indicates proteins with a reported function in gene transcription.
Figure 4.
Super-shift assay demonstrates that Egr-1 binds to H10 and to oligonucleotides derived from the 5′ UTR of Baiap2, Ppp1r16b, Klf16, and Cpne2 and containing the H10 binding site 5′-CGCCCACA-3′. A, The addition of α-Egr-1, but not α-SRF, super-shifts H10 complex I. SS, Super-shift, indicated by white arrowheads. Complexes II–IV are unaltered after immunodepletion of Egr-1 from striatal nuclear extracts (NE). B, α-Egr-1 super-shifts complex I from oligonucleotide derived from the 5′ UTR of striatal-enriched expressed genes, Baiap2, Ppp1r16b, and Klf16, and of Cpne2, a ubiquitously expressed gene. SS, Super-shift, indicated by white arrowheads. C, Two Sp1 binding sites were identified by Genomatix, the first of which is 15 nt upstream of the Egr-1 binding site and the second overlapping with that for Egr-1. Addition of α-Sp1 did not super-shift any of the H10 complexes. TSS, Transcription start site; I–VI, Ppp1r1b introns.
To identify potential Egr-1 interacting proteins, we searched 30 nt upstream and downstream of H10. Worth noting, the addition of only 1 nt upstream of H10 identified an Egr binding site that falls on the binding site we identified by the mutational analysis. However, this nucleotide is clearly not required for binding of Egr-1, at least in vitro. In addition, we located an Sp1 binding site that overlaps with that for Egr, and another that was 15 nt upstream of the Egr binding site (Fig. 4C). Sp1 and Egr-1 can act synergistically when located on neighboring elements (Lin and Leonard, 1997; Kaiser et al., 2000; Tai and Wong, 2003) or can antagonize each other by competing for the same site (Huang et al., 1997; Papanikolaou and Sabban, 2000; Davis et al., 2003; Tan et al., 2003). To test both possibilities, we performed a super-shift assay on an oligonucleotide that contained both Sp1 elements. Although a complex appeared that migrates slower than the Egr-1 complex (complex C), the addition of α-Sp1, either alone or together with α-Egr-1, did not super-shift any of the complexes (Fig. 4C). We concluded that Sp1 does not bind upstream of Egr-1 nor does it compete with Egr-1 at the complex I site.
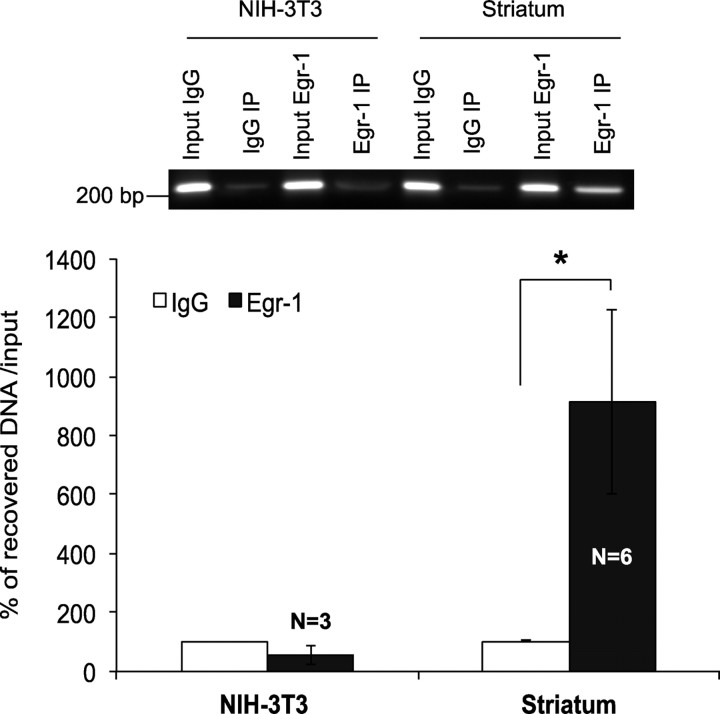
To determine whether Egr-1 binds to H10 in vivo, we performed ChIP followed by quantitative real-time PCR analysis with mouse striatal chromatin. Chromatin from NIH-3T3 cells, which do not express DARPP-32, was used as a negative control. The primers were designed to amplify a 252 bp product that contains the identified Egr-1 binding site (sequences are in Materials and Methods). The percentage of the DNA recovered from the Egr-1 IP reaction was higher than with the IgG IP reaction from striatal chromatin (*p < 0.05) or the Egr-1 IP reaction from DARPP-32-negative NIH-3T3 cells (Fig. 5).
Figure 5.
Egr-1 binds to the H10 sequence in intron IV of the Ppp1r1b gene in vivo. ChIP assays were performed with adult mouse striatal and NIH-3T3 chromatin. Quantitative PCR analysis was performed to measure the amount of DNA recovered with each reaction. Egr-1 enrichment is displayed as percentage of the recovered DNA to the input DNA (*p < 0.05). Error bars indicate SEM.
Egr-1 is sufficient to induce DARPP-32 expression in primary striatal neurons and is necessary for induction of DARPP-32 by BDNF
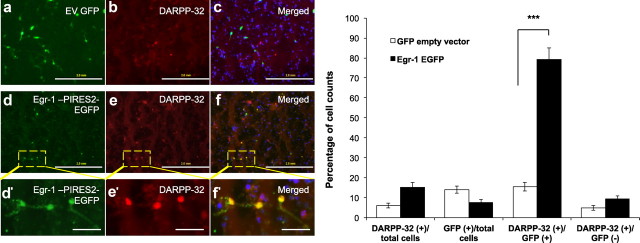
We next sought to determine whether Egr-1 is necessary and/or sufficient for expression of DARPP-32 in embryonic striatal neurons in vitro. Egr-1–IRES–EGFP plasmid (Beck et al., 2008) was transfected into primary striatal neurons via nucleofection and increased the percentage of DARPP-32-immunopositive cells from 6% in neurons transfected with empty vector to 15% in neurons transfected with the Egr-1–IRES–EGFP plasmid. Although only 8% of the neurons were transfected with Egr-1–IRES–EGFP, 79% (***p < 0.001) of the neurons expressing EGFP were also DARPP-32 immunopositive. Of the neurons that were GFP negative in the same wells, only 10% were DARPP-32 immunopositive. In comparison, in empty vector GFP-transfected neurons, only 15% of the EGFP-positive cells expressed DARPP-32, and 5% of the EGFP-negative neurons were DARPP-32 immunopositive (Fig. 6).
Figure 6.
Egr-1 is sufficient to induce DARPP-32 expression in striatal primary neurons in vitro. Primary striatal neurons derived from E15–E17 embryos were transfected with empty vector (EV) GFP (a–c) or Egr-1–EGFP plasmid (d, e). After 24 h, cells were fixed with 4% paraformaldehyde and immunostained with α-GFP (green) and α-DARPP-32 (red). Single- and double-labeled cells were counted. Numbers were derived from 20 fields from two separate platings and transfections (***p < 0.001). Scale bars: a–f, 2.0 mm; d′–f′, 1.2 mm. Error bars indicate SEM.
BDNF induces expression of both DARPP-32 (Ivkovic et al., 1997, 1999) and Egr-1 (Minichiello et al., 2002; Alder et al., 2003; Calella et al., 2007), and addition of BDNF to primary striatal neurons led to an increase in Egr-1 protein after 90 min and an increase in DARPP-32 protein after 24 h (Fig. 7A). We hypothesized that BDNF induces DARPP-32 expression via inducing Egr-1 expression, thereby increasing its binding to intron IV of Ppp1r1b gene, as shown in Figure 7B.
Figure 7.
BDNF increases binding of Egr-1 to H10 and requires Egr-1 to induce DARPP-32 expression in vitro. A, Egr-1 level increases 1.5 h after addition of BDNF to primary striatal neurons. Western blot was performed on total cellular lysates. As reported previously, DARPP-32 level is increased 24 h after treatment. B, The schematic shows how BDNF leads to the induction of DARPP-32 expression after upregulation and binding of Egr-1 to intron IV of the Ppp1r1b gene. C, Egr-1 function is inhibited in BDNF-treated primary striatal neurons via transduction with adnEgr-1 adenovirus (Kundumani-Sridharan et al., 2010), leading to a decrease in induction of DARPP-32 by BDNF. WT1 antibody was used to confirm the transduction of the fusion protein (*p < 0.05, **p < 0.01). Error bars indicate SEM. D, EMSA with equal amounts of nuclear extract from treated and untreated neurons shows that BDNF enhances Egr-1 and Egr-2 binding to H10 3 h after treatment. * Indicates nonspecific complexes, i.e., not quenched by competitor. NE, Nuclear extract; SS, super-shift; TSS, transcription start site.
To determine whether Egr-1 is essential for induction of DARPP-32 by BDNF, we transduced embryonic striatal primary neuronal cultures with either an empty GFP adenovirus (AdGFP) or adnEgr-1. The adnEgr-1 construct contained the transcriptional repressor domain of the Wilms tumor protein WT1 (amino acids 1–307) at the N terminus of the Egr-1 DNA binding domain (amino acids 327–427) (Kundumani-Sridharan et al., 2010), and Western blotting of WT1 confirmed the expression of adnEgr-1 (Fig. 7C). Transduction of primary striatal neurons with adnEgr-1 slightly reduced DARPP-32 protein level relative to control (AdGFP). However, transduction of neurons with adnEgr-1 markedly reduced induction of DARPP-32 by BDNF (*p < 0.05) (Fig. 7C).
To confirm that BDNF increases Egr-1 binding to Ppp1r1b gene, we tested the formation of the H10 complexes before and after treating primary striatal cultures with BDNF for 3 h. The intensity of all H10 complexes, including the Egr-1 complex, increased as early as 3 h after addition of BDNF (Fig. 7D, lanes 5, 6). In addition, a slower migrating complex became apparent, which appeared similar to complex A in Figure 4B. Complex B also appeared between I and II and became much more prominent during BDNF treatment. Egr-1 is a member of a family that includes Egr-2, Egr-3, and Egr-4, all of which are expressed in the striatum. Of these, Egr-2 is the likely candidate to be redundant with Egr-1, because it has a similar DNA binding site and is modulated by BDNF (Calella et al., 2007). As shown in Figure 7D (lane 12), the addition of α-Egr-2 shifted complexes A and B. These results indicate that exposure to BDNF increases the binding of Egr-1 and Egr-2 to Ppp1r1b.
Discussion
This report is part of our continued effort to identify mechanisms of MSN-specific gene expression. We have previously identified a combination of Ppp1r1b genomic regulatory regions, including introns, that directs transgene expression to MSNs (Bogush et al., 2005) and, conversely, does not permit expression in other regions that express endogenous DARPP-32. The next major goal is to identify the required TFs and the sequences to which they bind. In this study, we identified a conserved sequence, also known as H10, in the Ppp1r1b intron IV and demonstrated that the TF Egr-1 binds to this region in vitro and in vivo. Functionally, we showed that Egr-1 is sufficient to induce DARPP-32 expression in striatal-derived embryonic neurons and is required for induction of DARPP-32 by BDNF. It is important to note that BDNF induces expression of β-galactosidase (β-gal) in neurons prepared from mice transgenic for D9–IRES–β-gal, verifying that D9 contains BDNF-responsive sequences. D9–IRES–β-gal was derived by replacing the Cre recombinase cDNA with the β-galactosidase cDNA (Bogush et al., 2005; A. Bogush, B. Tang, K. Tang, E. A. Thomas, M. E. Ehrlich, unpublished observations).
We and others have previously reported that BDNF induces DARPP-32 transcription in MSNs (Ivkovic and Ehrlich, 1999) via phosphatidylinositol 3-kinase/Akt and Cdk5 pathways and is independent of ERK (Stroppolo et al., 2001; Bogush et al., 2007). The Egr-1 promoter contains both an SRE (serum response element) and a CRE (cAMP response element) (Knapska and Kaczmarek, 2004). In the hippocampus, BDNF induces the expression of Egr-1 via the activities of TrkB, MEK, CaMK, PLCγ, and CREB (Minichiello et al., 2002; Alder et al., 2003). In the striatum, BDNF induces the phosphorylation of Elk-1, which targets the SRE site on Egr-1 (Gokce et al., 2009). Of note, however, the latter pathway is ERK dependent. BDNF also transiently increases phosphorylation of CREB in MSNs (Stroppolo et al., 2001), and it is possible that DARPP-32 induction by BDNF is in part dependent on Egr-1 induction by phosphorylated CREB. This process, however, must be context specific because other inducers of CREB phosphorylation, most notably dopamine and its agonists, do not alter expression of DARPP-32 in MSNs (Ehrlich et al., 1990; Grebb et al., 1990).
The striatum develops from the lateral ganglionic eminence, and the first MSNs to mature are located in patches and the lateral streak, whereas later-born neurons populate the matrix (van der Kooy and Fishell, 1987). The mature, terminal phenotype of the MSN is characterized by DARPP-32 and other markers (Desplats et al., 2006; Lobo et al., 2006). Many of these, including DARPP-32, are enriched in patch neurons during the embryonic period and become more diffusely expressed as matrix neurons mature. Others, of course, remain confined to the patch compartment. Relative to other embryonic brain regions, Egr-1 is enriched in the striatum. In the adult, it is expressed in multiple brainstem nuclei and in the striatum (Wells et al., 2011), and it is obviously induced in multiple regions after neuronal activation. Within the embryonic striatum, Egr-1 is further enriched in patches, perhaps suggesting a developmental, regulatory role (Snyder-Keller et al., 2002). Beckwith et al. (1966) hypothesizes that a mature neuronal subtype is determined by a group of TFs that induce a specific set of genes, which together comprise a “regulon,” a term previously restricted to prokaryotes. Furthermore, cis-motifs may be bound by multiple TFs, creating a system for coordinated, master, regulation of phenotype (Flames and Hobert, 2011). Thus, we show that the H10 binding sequence is more frequently found in striatal-enriched genes and that Egr-1 binds to these sequences in vitro in the presence of distinct flanking sequences. Thus, Egr-1 may induce additional markers of the mature MSN.
Egr-1 is a ubiquitously expressed immediate early gene, usually associated with neuronal plasticity after stimuli that alter intracellular calcium stores (Knapska and Kaczmarek, 2004; Thiel et al., 2010). Despite its widespread expression, many examples exist of regulation of cell-specific markers by Egr-1, both within and outside the nervous system (Boyle et al., 2009). Tissue-specific expression of proteins may be mediated by cell-specific TFs but also by unique combinations of ubiquitously expressed TFs (Hobert et al., 2010). For example, Egr-1 acts synergistically with NFATc to regulate expression of the IL-2 gene in T lymphocytes by binding independently to adjacent sites within the IL-2 gene promoter (Decker et al., 1998). Thus, it is unlikely that Egr-1 alone can induce the DARPP-32 phenotype in precursors that have not yet committed to the MSN lineage but may be sufficient once the neurons are committed. In both cases, therefore, Egr-1 almost certainly acts in concert with other TFs, because induction of Egr-1 in most cell subtypes does not induce DARPP-32. For example, Egr-1 is required in part for BDNF induction of the tyrosine hydroxylase promoter (Fukuchi et al., 2010), but DARPP-32 is not expressed in aminergic neurons.
Conversely, the adult Egr-1-null mouse has a normal level of striatal DARPP-32 (Valjent et al., 2006), indicating that Egr-1 is ultimately not necessary for expression of DARPP-32 in vivo. Importantly, development of the striatum in the absence of Egr-1 has not been examined, and we know that, in the BDNF-null mouse, DARPP-32 expression is reduced early but normalizes during the second postnatal week (Ivkovic et al., 1997). Also, because we show that Egr-2 binds to the same H10 binding site as does Egr-1, Egr-2 may substitute for Egr-1. An Egr-2 regulated transcription network has been suggested to exist in the striatum, downstream of the adenosine A2A receptor. However, the involvement of Egr-2 in regulating DARPP-32 transcription in vivo remains to be determined.
Our data are highly relevant to the pathophysiology of diseases in which the striatum plays a role and their potential therapies. Notably, Egr-1 levels are specifically decreased in the dorsal subregion of the striatum in R6/2 mouse model of HD (Spektor et al., 2002) and are elevated in the striatum after exposure to drugs of abuse (Valjent et al., 2000, 2006). More important perhaps is the potential contribution to cell replacement therapy and to the use of induced pluripotent stem (iPS) cells for the study of disease mechanisms. At this juncture, these approaches are hampered by the inability to induce large numbers of striatal subtype-specific neurons that are physiologically active and able to properly integrate into the CNS. Evidence is accumulating that the forced expression of neuronal subtype-specific TFs is able to re-program cells, e.g., fibroblasts, and to convert embryonic stem cells and iPS to a specific phenotype (Caiazzo et al., 2011; Son et al., 2011; Sánchez-Danés et al., 2012). Perhaps the most successful MSN differentiation program to date is that of Aubry et al. (2008) in which neural precursors are treated with BDNF and then with BDNF and valproic acid for terminal differentiation into MSN-like neurons. Multiple markers were assayed, although not their colocalization, and only extrinsic signals were used to induce differentiation. In addition to our findings, Egr-1 was recently identified in a novel SOX2-positive neuronal progenitor cell population in developing NeuN-positive striatal neurons, indicating a role for Egr-1 in both intermediate and terminal differentiation of MSNs and perhaps in maintenance of the differentiated phenotype (Wells et al., 2011).
In summary, our study is the first to identify a TF, Egr-1, that directly interacts with and regulates Ppp1r1b gene expression in MSNs. In addition, we demonstrate that it acts downstream of BDNF, a major regulator of DARPP-32 and other markers of the differentiated MSN. As we continue to identify specific TFs that can directly induce DARPP-32, it will be critical to test them for their utility in driving an MSN phenotype, the identification of which should include multiple, colocalized markers and physiological properties.
Footnotes
This work was supported by the National Institutes of Health Grants NS-059936 and NS-045942 (M.E.E.), P30 NS061777 and S10 RR022415 (R.W.), and HL069908, HL103575, and EY014856 (G.N.R.).
References
- Alder J, Thakker-Varia S, Bangasser DA, Kuroiwa M, Plummer MR, Shors TJ, Black IB. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J Neurosci. 2003;23:10800–10808. doi: 10.1523/JNEUROSCI.23-34-10800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Molyneaux BJ, Jabaudon D, Yoshida Y, Macklis JD. Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. J Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry L, Bugi A, Lefort N, Rousseau F, Peschanski M, Perrier AL. Striatal progenitors derived from human ES cells mature into DARPP-32 neurons in vitro and in quinolinic acid-lesioned rats. Proc Natl Acad Sci U S A. 2008;105:16707–16712. doi: 10.1073/pnas.0808488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck H, Semisch M, Culmsee C, Plesnila N, Hatzopoulos AK. Egr-1 regulates expression of the glial scar component phosphacan in astrocytes after experimental stroke. Am J Pathol. 2008;173:77–92. doi: 10.2353/ajpath.2008.070648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckwith JR, Signer ER, Epstein W. Transposition of the Lac region of E. coli. Cold Spring Harb Symp Quant Biol. 1966;31:393–401. doi: 10.1101/sqb.1966.031.01.051. [DOI] [PubMed] [Google Scholar]
- Blau S, Daly L, Fienberg A, Teitelman G, Ehrlich ME. DARPP-32 promoter directs transgene expression to renal thick ascending limb of loop of Henle. Am J Physiol. 1995;269:F564–F570. doi: 10.1152/ajprenal.1995.269.4.F564. [DOI] [PubMed] [Google Scholar]
- Bogush AI, McCarthy LE, Tian C, Olm V, Gieringer T, Ivkovic S, Ehrlich ME. DARPP-32 genomic fragments drive Cre expression in postnatal striatum. Genesis. 2005;42:37–46. doi: 10.1002/gene.20118. [DOI] [PubMed] [Google Scholar]
- Bogush A, Pedrini S, Pelta-Heller J, Chan T, Yang Q, Mao Z, Sluzas E, Gieringer T, Ehrlich ME. AKT and CDK5/p35 mediate brain-derived neurotrophic factor induction of DARPP-32 in medium size spiny neurons in vitro. J Biol Chem. 2007;282:7352–7359. doi: 10.1074/jbc.M606508200. [DOI] [PubMed] [Google Scholar]
- Boyle KB, Hadaschik D, Virtue S, Cawthorn WP, Ridley SH, O'Rahilly S, Siddle K. The transcription factors Egr1 and Egr2 have opposing influences on adipocyte differentiation. Cell Death Differ. 2009;16:782–789. doi: 10.1038/cdd.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TB, Bogush AI, Ehrlich ME. Neocortical expression of mutant huntingtin is not required for alterations in striatal gene expression or motor dysfunction in a transgenic mouse. Hum Mol Genet. 2008;17:3095–3104. doi: 10.1093/hmg/ddn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiazzo M, Dell'Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Calella AM, Nerlov C, Lopez RG, Sciarretta C, von Bohlen und Halbach O, Bereshchenko O, Minichiello L. Neurotrophin/Trk receptor signaling mediates C/EBPalpha, -beta and NeuroD recruitment to immediate-early gene promoters in neuronal cells and requires C/EBPs to induce immediate-early gene transcription. Neural Dev. 2007;2:4. doi: 10.1186/1749-8104-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CW, Tsai CW, Wang HF, Tsai HC, Chen HY, Tsai TF, Takahashi H, Li HY, Fann MJ, Yang CW, Hayashizaki Y, Saito T, Liu FC. Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc Natl Acad Sci U S A. 2004;101:2613–2618. doi: 10.1073/pnas.0308645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W, Jr, Chen ZJ, Ile KE, Tew KD. Reciprocal regulation of expression of the human adenosine 5′-triphosphate binding cassette, sub-family A, transporter 2 (ABCA2) promoter by the early growth response-1 (EGR-1) and Sp-family transcription factors. Nucleic Acids Res. 2003;31:1097–1107. doi: 10.1093/nar/gkg192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker EL, Skerka C, Zipfel PF. The early growth response protein (EGR-1) regulates interleukin-2 transcription by synergistic interaction with the nuclear factor of activated T cells. J Biol Chem. 1998;273:26923–26930. doi: 10.1074/jbc.273.41.26923. [DOI] [PubMed] [Google Scholar]
- Desplats PA, Kass KE, Gilmartin T, Stanwood GD, Woodward EL, Head SR, Sutcliffe JG, Thomas EA. Selective deficits in the expression of striatal-enriched mRNAs in Huntington's disease. J Neurochem. 2006;96:743–757. doi: 10.1111/j.1471-4159.2005.03588.x. [DOI] [PubMed] [Google Scholar]
- Desplats PA, Lambert JR, Thomas EA. Functional roles for the striatal-enriched transcription factor, Bcl11b, in the control of striatal gene expression and transcriptional dysregulation in Huntington's disease. Neurobiol Dis. 2008;31:298–308. doi: 10.1016/j.nbd.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich ME, Rosen NL, Kurihara T, Shalaby IA, Greengard P. DARPP-32 development in the caudate nucleus is independent of afferent input from the substantia nigra. Brain Res Dev Brain Res. 1990;54:257–263. doi: 10.1016/0165-3806(90)90148-r. [DOI] [PubMed] [Google Scholar]
- Flames N, Hobert O. Transcriptional control of the terminal fate of monoaminergic neurons. Annu Rev Neurosci. 2011;34:153–184. doi: 10.1146/annurev-neuro-061010-113824. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi M, Fujii H, Takachi H, Ichinose H, Kuwana Y, Tabuchi A, Tsuda M. Activation of tyrosine hydroxylase (TH) gene transcription induced by brain-derived neurotrophic factor (BDNF) and its selective inhibition through Ca2+ signals evoked via the N-methyl-d-aspartate (NMDA) receptor. Brain Res. 2010;1366:18–26. doi: 10.1016/j.brainres.2010.10.034. [DOI] [PubMed] [Google Scholar]
- Gadgil H, Jurado LA, Jarrett HW. DNA affinity chromatography of transcription factors. Anal Biochem. 2001;290:147–178. doi: 10.1006/abio.2000.4912. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Annu Rev Neurosci. 1992;15:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- Gerfen CR, Young WS., 3rd Distribution of striatonigral and striatopallidal peptidergic neurons in both patch and matrix compartments: an in situ hybridization histochemistry and fluorescent retrograde tracing study. Brain Res. 1988;460:161–167. doi: 10.1016/0006-8993(88)91217-6. [DOI] [PubMed] [Google Scholar]
- Gokce O, Runne H, Kuhn A, Luthi-Carter R. Short-term striatal gene expression responses to brain-derived neurotrophic factor are dependent on MEK and ERK activation. PLoS One. 2009;4:e5292. doi: 10.1371/journal.pone.0005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM. The basal ganglia: learning new tricks and loving it. Curr Opin Neurobiol. 2005;15:638–644. doi: 10.1016/j.conb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Grebb JA, Girault JA, Ehrlich M, Greengard P. Chronic treatment of rats with SCH-23390 or raclopride does not affect the concentrations of DARPP-32 or its mRNA in dopamine-innervated brain regions. J Neurochem. 1990;55:204–207. doi: 10.1111/j.1471-4159.1990.tb08839.x. [DOI] [PubMed] [Google Scholar]
- Greengard P, Allen PB, Nairn AC. Beyond the dopamine receptor: the DARPP-32/protein phosphatase-1 cascade. Neuron. 1999;23:435–447. doi: 10.1016/s0896-6273(00)80798-9. [DOI] [PubMed] [Google Scholar]
- Gustafson EL, Ehrlich ME, Trivedi P, Greengard P. In situ hybridization analysis of the development of DARPP-32 and ARPP-21 in rat striatum. Neuroscience. 1992;51:65–75. doi: 10.1016/0306-4522(92)90471-d. [DOI] [PubMed] [Google Scholar]
- Handen JS, Rosenberg HF. An improved method for Southwestern blotting. Front Biosci. 1997;2:c9–c11. doi: 10.2741/A166. [DOI] [PubMed] [Google Scholar]
- Hobert O, Carrera I, Stefanakis N. The molecular and gene regulatory signature of a neuron. Trends Neurosci. 2010;33:435–445. doi: 10.1016/j.tins.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A, Strand AD, Aragaki AK, Kuhn A, Sengstag T, Hughes G, Elliston LA, Hartog C, Goldstein DR, Thu D, Hollingsworth ZR, Collin F, Synek B, Holmans PA, Young AB, Wexler NS, Delorenzi M, Kooperberg C, Augood SJ, Faull RL, Olson JM, Jones L, Luthi-Carter R. Regional and cellular gene expression changes in human Huntington's disease brain. Hum Mol Genet. 2006;15:965–977. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- Huang RP, Fan Y, Ni Z, Mercola D, Adamson ED. Reciprocal modulation between Sp1 and Egr-1. J Cell Biochem. 1997;66:489–499. [PubMed] [Google Scholar]
- Ivkovic S, Ehrlich ME. Expression of the striatal DARPP-32/ARPP-21 phenotype in GABAergic neurons requires neurotrophins in vivo and in vitro. J Neurosci. 1999;19:5409–5419. doi: 10.1523/JNEUROSCI.19-13-05409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivkovic S, Polonskaia O, Fariñas I, Ehrlich ME. Brain-derived neurotrophic factor regulates maturation of the DARPP-32 phenotype in striatal medium spiny neurons: studies in vivo and in vitro. Neuroscience. 1997;79:509–516. doi: 10.1016/s0306-4522(96)00684-7. [DOI] [PubMed] [Google Scholar]
- Jain M, Armstrong RJ, Barker RA, Rosser AE. Cellular and molecular aspects of striatal development. Brain Res Bull. 2001;55:533–540. doi: 10.1016/s0361-9230(01)00555-x. [DOI] [PubMed] [Google Scholar]
- Jiang D, Jia Y, Zhou Y, Jarrett HW. Two-dimensional southwestern blotting and characterization of transcription factors on-blot. J Proteome Res. 2009;8:3693–3701. doi: 10.1021/pr900214p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser UB, Halvorson LM, Chen MT. Sp1, steroidogenic factor 1 (SF-1), and early growth response protein 1 (egr-1) binding sites form a tripartite gonadotropin-releasing hormone response element in the rat luteinizing hormone-beta gene promoter: an integral role for SF-1. Mol Endocrinol. 2000;14:1235–1245. doi: 10.1210/mend.14.8.0507. [DOI] [PubMed] [Google Scholar]
- Keilani S, Sugaya K. Reelin induces a radial glial phenotype in human neural progenitor cells by activation of Notch-1. BMC Dev Biol. 2008;8:69. doi: 10.1186/1471-213X-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapska E, Kaczmarek L. gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Kundumani-Sridharan V, Niu J, Wang D, Van Quyen D, Zhang Q, Singh NK, Subramani J, Karri S, Rao GN. 15(S)-hydroxyeicosatetraenoic acid-induced angiogenesis requires Src-mediated Egr-1-dependent rapid induction of FGF-2 expression. Blood. 2010;115:2105–2116. doi: 10.1182/blood-2009-09-241802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JX, Leonard WJ. The immediate-early gene product Egr-1 regulates the human interleukin-2 receptor beta-chain promoter through noncanonical Egr and Sp1 binding sites. Mol Cell Biol. 1997;17:3714–3722. doi: 10.1128/mcb.17.7.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci. 2006;9:443–452. doi: 10.1038/nn1654. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Strand A, Peters NL, Solano SM, Hollingsworth ZR, Menon AS, Frey AS, Spektor BS, Penney EB, Schilling G, Ross CA, Borchelt DR, Tapscott SJ, Young AB, Cha JH, Olson JM. Decreased expression of striatal signaling genes in a mouse model of Huntington's disease. Hum Mol Genet. 2000;9:1259–1271. doi: 10.1093/hmg/9.9.1259. [DOI] [PubMed] [Google Scholar]
- Luthi-Carter R, Hanson SA, Strand AD, Bergstrom DA, Chun W, Peters NL, Woods AM, Chan EY, Kooperberg C, Krainc D, Young AB, Tapscott SJ, Olson JM. Dysregulation of gene expression in the R6/2 model of polyglutamine disease: parallel changes in muscle and brain. Hum Mol Genet. 2002;11:1911–1926. doi: 10.1093/hmg/11.17.1911. [DOI] [PubMed] [Google Scholar]
- Minichiello L, Calella AM, Medina DL, Bonhoeffer T, Klein R, Korte M. Mechanism of TrkB mediated hippocampal long-term potentiation. Neuron. 2002;36:121–137. doi: 10.1016/s0896-6273(02)00942-x. [DOI] [PubMed] [Google Scholar]
- Minig V, Kattan Z, van Beeumen J, Brunner E, Becuwe P. Identification of DDB2 protein as a transcriptional regulator of constitutive SOD2 gene expression in human breast cancer cells. J Biol Chem. 2009;284:14165–14176. doi: 10.1074/jbc.M808208200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet CC, Langley-Gullion KC, Greengard P. Quantitative immunocytochemistry of DARPP-32-expressing neurons in the rat caudatoputamen. Brain Res. 1998;808:8–12. doi: 10.1016/s0006-8993(98)00724-0. [DOI] [PubMed] [Google Scholar]
- Papanikolaou NA, Sabban EL. Ability of Egr1 to activate tyrosine hydroxylase transcription in PC12 cells. Cross-talk with AP-1 factors. J Biol Chem. 2000;275:26683–26689. doi: 10.1074/jbc.M000049200. [DOI] [PubMed] [Google Scholar]
- Parent A, Hazrati LN. Functional anatomy of the basal ganglia. I. The cortico-basal ganglia-thalamo-cortical loop. Brain Res Brain Res Rev. 1995;20:91–127. doi: 10.1016/0165-0173(94)00007-c. [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Sánchez-Danés A, Consiglio A, Richaud Y, Rodríguez-Pizà I, Dehay B, Edel M, Bové J, Memo M, Vila M, Raya A, Izpisua Belmonte JC. Efficient generation of A9 midbrain dopaminergic neurons by lentiviral delivery of LMX1A in human embryonic stem cells and iPS cell. Hum Gene Ther. 2012;23:56–69. doi: 10.1089/hum.2011.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini-extracts,” prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomics studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- Snyder-Keller A, Chandra R, Lin Y, Mitchell ES. Basal EGR-1 (zif268, NGFI-A, Krox-24) expression in developing striatal patches: role of dopamine and glutamate. Brain Res. 2002;958:297–304. doi: 10.1016/s0006-8993(02)03602-8. [DOI] [PubMed] [Google Scholar]
- Son EY, Ichida JK, Wainger BJ, Toma JS, Rafuse VF, Woolf CJ, Eggan K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell. 2011;9:205–218. doi: 10.1016/j.stem.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spektor BS, Miller DW, Hollingsworth ZR, Kaneko YA, Solano SM, Johnson JM, Penney JB, Jr, Young AB, Luthi-Carter R. Differential D1 and D2 receptor-mediated effects on immediate early gene induction in a transgenic mouse model of Huntington's disease. Brain Res Mol Brain Res. 2002;102:118–128. doi: 10.1016/s0169-328x(02)00216-4. [DOI] [PubMed] [Google Scholar]
- Stroppolo A, Guinea B, Tian C, Sommer J, Ehrlich ME. Role of phosphatidylinositide 3-kinase in brain-derived neurotrophic factor-induced DARPP-32 expression in medium size spiny neurons in vitro. J Neurochem. 2001;79:1027–1032. doi: 10.1046/j.1471-4159.2001.00651.x. [DOI] [PubMed] [Google Scholar]
- Svenningsson P, Nishi A, Fisone G, Girault JA, Nairn AC, Greengard P. DARPP-32: an integrator of neurotransmission. Annu Rev Pharmacol Toxicol. 2004;44:269–296. doi: 10.1146/annurev.pharmtox.44.101802.121415. [DOI] [PubMed] [Google Scholar]
- Tai TC, Wong DL. Protein kinase A and protein kinase C signaling pathway interaction in phenylethanolamine N-methyltransferase gene regulation. J Neurochem. 2003;85:816–829. doi: 10.1046/j.1471-4159.2003.01728.x. [DOI] [PubMed] [Google Scholar]
- Tan L, Peng H, Osaki M, Choy BK, Auron PE, Sandell LJ, Goldring MB. Egr-1 mediates transcriptional repression of COL2A1 promoter activity by interleukin-1beta. J Biol Chem. 2003;278:17688–17700. doi: 10.1074/jbc.M301676200. [DOI] [PubMed] [Google Scholar]
- Thiel G, Mayer SI, Müller I, Stefano L, Rössler OG. Egr-1-A Ca2+-regulated transcription factor. Cell Calcium. 2010;47:397–403. doi: 10.1016/j.ceca.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Thomas EA, Coppola G, Desplats PA, Tang B, Soragni E, Burnett R, Gao F, Fitzgerald KM, Borok JF, Herman D, Geschwind DH, Gottesfeld JM. The HDAC inhibitor 4b ameliorates the disease phenotype and transcriptional abnormalities in Huntington's disease transgenic mice. Proc Natl Acad Sci U S A. 2008;105:15564–15569. doi: 10.1073/pnas.0804249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EA, Coppola G, Tang B, Kuhn A, Kim S, Geschwind DH, Brown TB, Luthi-Carter R, Ehrlich ME. In vivo cell-autonomous transcriptional abnormalities revealed in mice expressing mutant huntingtin in striatal but not cortical neurons. Hum Mol Genet. 2011;20:1049–1060. doi: 10.1093/hmg/ddq548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Corvol JC, Pages C, Besson MJ, Maldonado R, Caboche J. Involvement of the extracellular signal-regulated kinase cascade for cocaine-rewarding properties. J Neurosci. 2000;20:8701–8709. doi: 10.1523/JNEUROSCI.20-23-08701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valjent E, Aubier B, Corbillé AG, Brami-Cherrier K, Caboche J, Topilko P, Girault JA, Hervé D. Plasticity-associated gene Krox24/Zif268 is required for long-lasting behavioral effects of cocaine. J Neurosci. 2006;26:4956–4960. doi: 10.1523/JNEUROSCI.4601-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooy D, Fishell G. Neuronal birthdate underlies the development of striatal compartments. Brain Res. 1987;401:155–161. doi: 10.1016/0006-8993(87)91176-0. [DOI] [PubMed] [Google Scholar]
- Wells T, Rough K, Carter DA. Transcription mapping of embryonic rat brain reveals EGR-1 induction in SOX2 neural progenitor cells. Front Mol Neurosci. 2011;4:6. doi: 10.3389/fnmol.2011.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker B, Luthi-Carter R, Kama JA, Dunah AW, Stern EA, Fox JH, Standaert DG, Young AB, Augood SJ. Transcriptional dysregulation in striatal projection- and interneurons in a mouse model of Huntington's disease: neuronal selectivity and potential neuroprotective role of HAP1. Hum Mol Genet. 2005;14:179–189. doi: 10.1093/hmg/ddi014. [DOI] [PubMed] [Google Scholar]