Abstract
Midbrain dopaminergic (mDA) neurons control movement and emotion, and their degeneration leads to motor and cognitive defects in Parkinson's disease (PD). miR-133b is a conserved microRNA that is thought to regulate mDA neuron differentiation by targeting Pitx3, a transcription factor required for appropriate development of mDA substantia nigra neurons. Moreover, miR-133b has been found to be depleted in the midbrain of PD patients. However, the function of miR-133b in the intact midbrain has not been determined. Here we show that miR-133b null mice have normal numbers of mDA neurons during development and aging. Dopamine levels are unchanged in the striatum, while expression of dopaminergic genes, including Pitx3, is also unaffected. Finally, motor coordination and both spontaneous and psychostimulant-induced locomotion are unaltered in miR-133b null mice, suggesting that miR-133b does not play a significant role in mDA neuron development and maintenance in vivo.
Introduction
Midbrain dopaminergic (mDA) neurons of the substantia nigra pars compacta (SNpc) and ventral tegmental area (VTA) are critical for the regulation of motor function and emotion-related behaviors. mDA neurons are affected in several debilitating neurological disorders including Parkinson's disease (PD), which results from selective loss of SNpc DA neurons (Moore et al., 2005). While much progress has been made (Smidt and Burbach, 2007; Andressoo and Saarma, 2008; Cookson and Bandmann, 2010), the molecular mechanisms underlying the development and maintenance of mDA neurons and their loss in PD have not been fully elucidated.
Recently, microRNAs (miRNAs) have been implicated in mDA neuron differentiation and PD (Harraz et al., 2011). miRNAs are small, noncoding RNAs with diverse roles in nervous system development, function, and disease (Cao et al., 2006; Im and Kenny, 2012). miRNAs regulate gene expression by binding to the 3′-UTR of mRNA targets, decreasing mRNA stability and protein translation (Bushati and Cohen, 2007). Deletion of the miRNA processing enzyme Dicer in postmitotic mDA neurons in mice causes extensive cell death, suggesting that miRNAs are critical for maintaining the mDA neuron population (Kim et al., 2007). One miRNA, miR-133b, was found to be enriched in the midbrain and specifically depleted in PD patients (Kim et al., 2007). In a mouse embryonic stem (ES) cell culture system, miR-133b negatively regulated ES cell differentiation into dopamine (DA) neurons. miR-133b has been proposed to mediate these effects by targeting Pitx3, a transcription factor required for differentiation of SNpc DA neurons; Pitx3 mutant mice lack SNpc DA neurons and display severe motor defects (van den Munckhof et al., 2003). miR-133b is a promising candidate in regulating mDA neuron differentiation and degeneration, but its role in vivo has not been determined.
Here we show that the mDA system develops normally in miR-133b null mice. mDA neuron morphology, number, and gene expression are unaltered early in development and in adult and aged miR-133b mutant mice. Whole-tissue levels of DA, norepinephrine (NE), and 5-hydroxytryptamine (5-HT) monoamine neurotransmitters in knock-out (KO) striatum are unchanged, while baseline and amphetamine-induced extracellular DA release are also unaffected. Furthermore, motor and anxiety-like behaviors in miR-133b null mice are equivalent to those of wild-type controls. From these data we conclude that miR-133b is not critical for mDA neuron development or function in vivo. In situ hybridization for mature miR-133b using locked nucleic acid (LNA) probes revealed modest expression in the SNpc as well as other brain regions, suggesting that miR-133b may have as yet undetermined functions in the mammalian nervous system.
Materials and Methods
miR-133b knock-out mouse generation
miR-133b conditional KO mice were generated by homologous recombination of a targeting construct containing the floxed miR-133b stem loop in R1 129/SvJ mouse ES cells using standard procedures (Nagy et al., 1993; Heyer et al., 2011). A targeting vector containing a loxP-flanked miR-133b stem loop and a FRT-flanked pGK-Neo cassette was used to replace the endogenous miR-133b stem loop in ES cells. The floxed stem loop sequence was 5′·loxP·AGGCTTGGACAAGTGGTGCTCAAACTCCAAGGACTTCACAAGCTTCAGGCTGCAGTCACCTCCAAAGGGAGTGGCCCCCTGCTCTGGCTGGTCAAACGGAACCAAGTCCGTCTTCCTGAGAGGTTTGGTCCCCTTCAACCAGCTACAGCAGGGCTGGCAAAGCTCAATATTTGGAGAAAGAGAAGAGAAGAGAAAATAGCTGCTACAGCCTTGCTTATGAAAAGTAATGCTGGTTTTGCCATCATAGCCTT·XhoI·FRT·loxP·3′ (mature miR-133b in bold). Correctly targeted ES cells were identified by PCR screening and confirmed by Southern blot of genomic DNA. A correctly targeted clone was injected into C57BL/6J blastocysts, and chimeras mated to C57BL/6J mice. Germline transmission was confirmed by PCR genotyping of tail DNA. miR-133bNeo-fl offspring were mated to FLPe deleter mice (The Jackson Laboratory) (Farley et al., 2000) to remove the neomycin resistance cassette, and miR-133bfl/+ offspring were confirmed by PCR genotyping. miR-133bfl/+ mice were crossed with a Cre deleter strain (The Jackson Laboratory) (Tallquist and Soriano, 2000) to create a miR-133-null allele, confirmed by PCR genotyping. miR-133b heterozygous mice were backcrossed to C57BL/6 for at least 8 generations, and all data were from wild-type (WT) and KO mice bred from miR-133b heterozygotes. Genotyping primers for detecting the floxed, WT, and null alleles were as follows: 133b-F, CAAGCTCTGTGAGAGGTTAGTCAGG; and 133b-R, CTCGACTGCATTTCCATTGTACTG (where F is forward and R is reverse). WT and KO bands were sequenced using the 133b-F and 133b-R primers to confirm deletion and correct orientation following loxP-mediated excision. Mice were housed 2–5 per cage in an Association for Assessment and Accreditation of Laboratory Animal Care-approved vivarium on a 12 h light–dark cycle. All experiments were carried out according to approved protocols from the Duke University and The Scripps Research Institute Florida Institutional Animal Care and Use Committees.
Antibodies
Mouse monoclonal anti-tyrosine hydroxylase (TH) antibody (1:10,000, Immunostar) and rabbit anti-Pitx3 antibody (1:400, a gift from Dr. Marten Smidt, University Medical Center Utrecht, Utrecht, The Netherlands) were used for immunostaining and Western blot. Cy3-conjugated anti-rabbit antibody (1:400, Jackson ImmunoResearch) and Alexa Fluor 488-conjugated anti-mouse antibody (1:400, Invitrogen) were used for immunostaining. Mouse anti-GAD1 (1:1000, Millipore), goat anti-ChAT (1:1000, Millipore), anti-β-tubulin (1:5000, Sigma), anti-β-actin (1:5000, Sigma), anti-GAPDH (1:2000,Santa Cruz Biotechnology), and HRP-conjugated goat anti-mouse and anti-rabbit (1:5000, Jackson ImmunoResearch) antibodies were used for Western blot. Western blots of midbrain protein samples were performed using infra-red dye-conjugated secondary antibodies (1:10000, Li-Cor) and a Li-Cor Odyssey infrared imager.
Tyrosine hydroxylase immunohistochemistry
Two-month-old male WT and miR-133b KO mice were deeply anesthetized with isoflurane (Butler Animal Health Supply) and transcardially perfused with physiological Ringer's solution followed by fixative (4% paraformaldehyde in PBS). Whole brain was fixed overnight, equilibrated in 30% sucrose, and embedded in O.C.T. (Sakura). Thirty micrometer frozen coronal serial sections were collected using a cryostat (Leica CM1850). Heat-mediated antigen retrieval was performed by autoclaving in 1.8 mm citric acid and 8.2 mm sodium citrate. TH immunohistochemistry was performed as previously described (Johnson et al., 2010). Montage images were acquired using an AxioImager microscope fitted with a motorized stage controlled by the MosaiX functionality within AxioVision 5.1 software (Carl Zeiss MicroImaging).
Unbiased stereology and cell counting
Unbiased stereological estimation of the total number of TH immunoreactive (TH-IR) cell bodies in the SNpc and VTA was performed using the optical fractionator method, as described previously (Johnson et al., 2010). Every second section was collected through the midbrain and every fourth section was analyzed for cell counting, totaling 10 sections per animal. Cells at least 10 μm in diameter stained for TH were classified as TH-IR. Embryonic day (E)18.5 embryos were fixed in 4% PFA overnight at 4°C, equilibrated in 30% sucrose, and embedded in O.C.T. Frozen 20 μm coronal serial sections were collected throughout the midbrain region. After heat-mediated antigen retrieval, sections were incubated with anti-TH and anti-Pitx3 primary antibodies in 5% normal goat serum, 2% BSA, and 0.1% Triton X-100 in PBS overnight at 4°C. Sections were incubated with Cy3 anti-rabbit and Alexa Fluor 488 anti-mouse antibodies and mounted with Fluoro-Gel (Electron Microscopy Sciences). Montage images were acquired of every second section throughout the rostral region of the SNpc, totaling five images per animal. All SNpc TH-IR, and Pitx3-IR cells were counted in each image using the counter tool in Photoshop (Adobe). All analyses were performed blind to genotype.
Western blotting
Three-month-old male and postnatal day (P)0 WT and KO mice were killed and whole brain or ventral midbrain dissected and flash frozen in liquid nitrogen. Tissue was homogenized in RIPA buffer (Sigma) supplemented with protease inhibitors (Complete, Roche). Twenty micrograms of lysate were run on 10–12% SDS-polyacrylamide gels, and Western blotting was carried out according to standard procedures. Bands of whole brain Western blots were visualized using ECL Plus (GE Healthcare Life Sciences) and a LAS-3000 Intelligent Dark Box (Fujifilm). Bands of midbrain Western blots were visualized using a Li-Cor Odyssey infrared imager. Bands were quantified by densitometry analysis using ImageJ software and normalized to β-actin (for TH and GAD1), β-tubulin (for Pitx3) or GAPDH (midbrain samples).
RNA extraction and real-time PCR
Three-month-old male and P0 WT and KO mice were sacrificed and whole brain was flash frozen in liquid nitrogen. Total RNA was extracted using TRIzol reagent (Invitrogen) and treated with Turbo DNase (Ambion). One microgram of RNA was used for reverse transcription with oligo-dT and random primers and AffinityScript enzyme (Stratagene). cDNA was used for quantitative PCR (Power SYBR Green, ABI 7300 real-time PCR machine, Applied Biosystems). The following intron-spanning primers were used: Gapdh-F, CATGGCCTTCCGTGTTCCT; Gapdh-R, TGATGTCATCATACTTGGCAGGTT; TH-F, CTCTCCACGGTGTACTGGTTCAC; TH-R, GGCTGGTAGGTTTGATCTTGGTAG; DAT-F, GTATGTGGTCGTGGTCAGCATT; DAT-R, CTTTCTCAGGTGTGATGGCATAG; GAD1-F, CAACCAGATGTGTGCAGGCTAC; GAD1-R, GCGTAGAGGTAATCAGCCAGCTC; ChAT-F, CTTTTGTGCAAGCCATGACTGAC; ChAT-R, CATCCATGAACATCTCAGGTGG; Pitx3-F, CAGAGGAATCGCTACCCTGACA; Pitx3-R, AAGCCACCTTTGCACAGCTC. Primers amplifying the miR-133a1 stem loop were as follows: miR-133a1-F, GGTTGACAGTTGCTAGGTATTTGC; and miR-133a1-R, GCTTTGCTAAAGCTGGTAAAATGG. Whole brain cDNA was also used for standard PCR amplification and quantitative PCR to confirm loss of the miR-133b stem loop in KO samples using the following primers: miR-133b-F, AGGCTTGGACAAGTGGTGCTCAA; and miR-133b-R, AAGGCTATGATGGCAAAACCAGC. Each sample was measured in triplicate and normalized to expression of Gapdh.
Neurochemical analysis of whole tissue
Two-month-old male WT and miR-133b KO mice were deeply anesthetized with isoflurane and sacrificed by decapitation. Frontal cortex, nucleus accumbens, dorsal striatum, and thalamus were rapidly dissected on a cold plate, flash-frozen in liquid nitrogen, and stored at −80°C for high performance liquid chromatography with electrochemical detection (HPLC-ECD) analysis. A modified chromatographic procedure was employed, as described previously (Pani, 2009). All procedures were performed blind to genotype.
In vivo microdialysis
Ten-week-old male WT and miR-133b KO mice were deeply anesthetized with tribromoethanol and a stereotaxic apparatus (David Kopf Instruments) used to intracranially implant microdialysis guide cannulae (CMA7, CMA Microdialysis) in dorsal striatum (coordinates in mm: 0.5 anteroposterior, 2.0 mediolateral, −1.5 dorsoventral from bregma). Following recovery for 3–5 days, a microdialysis probe (CMA7, 2 mm membrane length, CMA Microdialysis) was inserted into the guide cannula and perfused overnight with artificial CSF (aCSF perfusion fluid: 145 mm NaCl, 3.5 mm KCl, 2 mm Na2HPO4, 1 mm CaCl2, 1.2 mm MgCl2, pH 7.4, CMA Microdialysis) at a flow rate of 0.45 μl/min (CMA Microdialysis 400 syringe pump). The following day, the flow rate was increased to 1 μl/min for 1 hour before sample collection. Dialysate samples were collected at 20 min intervals into vials containing 10% 0.1 m perchloric acid to prevent oxidation of monoamines. Three baseline samples were collected, 100 μm amphetamine in aCSF was delivered to the probe by reverse microdialysis, and four more samples were collected to assess amphetamine-induced neurotransmitter release. Animals were kept on a reverse light–dark cycle, and all microdialysis samples were collected during the dark–active phase from freely moving animals. Mice were deeply anesthetized with isoflurane and transcardially perfused, brains were removed and embedded in O.C.T., and frozen coronal sections were collected from striatum to confirm probe placement.
Liquid chromatography and electrochemical detection
Dopamine contents from dialysates were quantified by HPLC-ECD. Ten microliters of each sample was automatically injected using an Agilent 1100 liquid chromatography apparatus and run on an MD-150 HPLC column (3 × 150 mm, 3 μm particle size). MD-TM mobile phase (ESA) was delivered at a flow rate of 0.6 ml/min. Following column separation, monoamines were detected using an ESA 5021A guard cell, an ESA 5011A analytical cell, and an ESA Coulochem III detector (EGC = +350 mV, E1 = −150 mV, E2 = +220 mV; filter time constant, 5 s). Characteristic retention times of dihydroxyphenylacetic acid, dopamine, and homovanillic acid were 3.5, 4.3, and 7.2 min, respectively. Analyte peak areas were integrated and interpolated from known standards using Agilent ChemStation software and GraphPad software. All procedures were carried out blind to genotype.
Behavioral analyses
Open field.
The open field test was performed as described previously (Peça et al., 2011). Briefly, 3- to 4-month-old WT and KO mice were placed in automated 16 × 16 inch chambers (AccuScan Instruments) for 60 min. Total distance traveled (centimeters) and time spent in the center were detected in 5 min bins. Two weeks later, the same mice were placed in 8 × 8 inch chambers, and distance traveled was similarly detected for 60 min. Three milligrams per kilogram of amphetamine was immediately administered by i.p. injection, the mice were returned to their chambers, and locomotor activity was recorded for a further 120 min.
Zero maze.
Five-month-old WT and KO mice were placed in the closed arm of an elevated zero maze indirectly illuminated at 100 lux, and their behavior was videotaped over 5 min (Peça et al., 2011). The time spent in the closed and open arms was scored blind to genotype using The Observer software (Noldus).
Dark–light emergence.
The dark–light emergence task was carried out as described previously (Peça et al., 2011). Three- to four-month-old WT and KO mice were tested in an automated dual chamber apparatus (Med Associates) containing a gate between a “light” (1000 lux) and “dark” chamber. The mice were placed in the dark chamber and the gate lifted, allowing the mice to freely move between both chambers. The duration in the light and dark chambers was recorded over 5 min.
Marble burying.
Marble burying behavior of 5-month-old male WT and KO mice was assessed (Deacon, 2006). Twenty-four 2 cm diameter marbles were evenly spaced on the surface in an 8.5 inch × 16 inch chamber filled 5 inches deep with aspen shavings. A mouse was placed in one corner of the chamber and allowed to move freely. After 30 min, the mouse was removed and an image taken with a digital camera (Canon). The number of visible marbles was counted, and the remainder recorded as the number of marbles buried.
Rotarod.
Five-month-old male WT and KO mice were tested on the rotarod task (Jones and Roberts, 1968). Mice were trained for four trials per day over 3 days on a rotarod device (Med Associates) at a steady speed of 24 rpm. The latency to fall was recorded as the time at which the mouse fell completely off the rod or failed to stay atop the rod. The intertrial interval was 5 min. On the fourth day the mice underwent an accelerating rotarod task, where rotation velocity increased from 4 to 40 rpm over 5 min. Performance on the task was assessed by measuring the average latency to fall off the rod over three trials.
miRNA fluorescence in situ hybridization
Fluorescence in situ hybridization for miR-133b was performed on 16 μm sagittal sections of 3-month-old WT and KO mouse brains according to a previously described method using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide fixative to prevent loss of miRNAs (Pena et al., 2009). LNA oligonucleotides (Exiqon) were labeled with digoxigenin using a 3′-end labeling kit (Roche). In situ hybridization was carried out with a miR-133b antisense probe, a miR-124 antisense probe as a positive control, and a scrambled miR-133b probe as a negative control. Images were acquired with an Olympus BX51 microscope and the Virtual Tissue module of Stereo Investigator software.
Statistical analyses
All data were analyzed using Prism software (GraphPad Software). Stereological cell counts, mRNA and protein expression data, zero maze data, dark–light emergence test data, and rotarod data were analyzed by an unpaired two-tailed t test. Tissue monoamine levels were analyzed by one-way ANOVA, and microdialysis and open field data were analyzed by repeated-measures ANOVA followed by Bonferroni post hoc test. Data are presented as mean plus SEM.
Results
Generation of miR-133b mutant mice
miR-133b is embedded in an intergenic noncoding gene, 7H4, located on chromosome 1 (Velleca et al., 1994). loxP sites were placed around the miR-133b stem loop precursor region, and a FRT-flanked pGK-Neo cassette was inserted 3′ to the floxed stem loop (Fig. 1A,B). The targeting vector was introduced into ES cells, and homologous recombination at the miR-133b locus was detected by Southern blot (Fig. 1C). Correctly targeted ES cells were used to generate chimeric mice, and germline transmission of the targeted allele was confirmed by PCR screening. The Neo cassette was excised by mating miR-133bflox-Neo/+ mice with a FLPe deleter strain, and subsequent miR-133bflox/+ mice were mated with a Cre deleter strain to remove the miR-133b stem loop. Floxed, WT, heterozygous, and KO mouse genotypes were detected by PCR of tail DNA (Fig. 1D), and WT and KO PCR products were sequenced to detect deletion of miR-133b and presence of a single loxP site in the KO (Fig. 1F,G). Loss of miR-133b was confirmed by RT-PCR and quantitative RT-PCR for the miR-133b stem loop region (Fig. 1E,H). Heterozygous mice were backcrossed at least eight generations to C57BL/6 mice and maintained on a C57BL/6 background. miR-133b KO mice were viable, fertile, and born at a Mendelian ratio. The KO mice appeared grossly normal, had similar weights and lifespan as their WT littermates, and did not display any overt behavioral defects.
Figure 1.
Generation of a miR-133b knock-out mouse. A, Schematic of miR-133b stem-loop targeting strategy. The box containing miR-133b represents the miR-133b stem-loop region. FLP, Mouse expressing germline FLP; Cre, mouse expressing germline Cre. Arrows show locations of PCR primers for genotyping. B, Sequence of the miR-133b precursor region flanked by loxP and Neo cassette. Mature miR-133b sequence is in large letters. C, Southern blot analysis following EcoRI digestion of WT and correctly targeted ES cell genomic DNA. The 5′ probe binding site is indicated in A. D, PCR genotyping of miR-133b floxed, wild-type, heterozygous, and knock-out mice. E, RT-PCR demonstrates loss of miR-133b stem-loop expression in brain and muscle of miR-133b KO mice. F, Sequencing chromatogram of WT allele genotyping PCR product. miR-133b is underlined in black. G, Sequencing chromatogram of KO allele genotyping PCR product. The miR-133b precursor has been deleted following Cre/loxP-mediated recombination. H, Quantitative pre-miR-133b RNA expression in whole brain and tibialis muscle of 3-month-old WT and KO mice. Expression was normalized to Gapdh and results were scaled to an average WT value of 1 (muscle); n = 3 WT and 3 KO mice. Average CT values were 28.80 (WT brain), 35.27 (KO brain), 23.46 (WT muscle), and 34.70 (KO muscle). Error bars indicate SEM.
Detection of miR-133b in brain by fluorescence in situ hybridization
miR-133b has previously been found to be enriched in midbrain; however, its precise expression pattern has not been determined. We performed fluorescence in situ hybridization (FISH) for mature miR-133b in adult brain using LNA modified, digoxigenin (DIG)-labeled antisense and scrambled probes (Fig. 2). miR-133b signal could be observed in many different brain regions, particularly cortex, hippocampus, and cerebellum (Fig. 2A,B), as well as regions neighboring the midbrain such as hypothalamic and pontine nuclei (Fig. 2A,D). Lack of signal from a miR-133b scrambled probe suggested that these regions were not stained due to nonspecific binding (Fig. 2C). Consistent with previous reports of biochemical enrichment in midbrain, we did detect moderate miR-133b expression in the SNpc region of ventral midbrain (Fig. 2E,F). However, some signal from the miR-133b antisense probe could still be detected in miR-133b KO brain, including the SNpc (Fig. 2G,H). The orthologs of miR-133b, miR-133a1, and miR-133a2 only differ by one base from miR-133b and, although not found to be enriched, have also been biochemically detected in brain and midbrain. Thus, signal from the miR-133b antisense probe in various regions of miR-133b KO brain is likely due to binding to the remaining miR-133 family members.
Figure 2.

Detection of miR-133 expression in brain. A–H, Fluorescence in situ hybridization using antisense or scrambled LNA-DIG probes for mature miR-133b. Blue, DAPI staining of cell nuclei; Red, DIG detected by Cy3 signal following tyramide amplification. A, miR-133b antisense probe signal in sagittal section of 3-month-old WT brain. B, D, Enlarged regions in A showing miR-133b signal in cortex and hippocampus (B) and ventral midbrain (arrow) and neighboring regions (hypothalamus, pons) (D). C, Signal from scrambled miR-133b LNA-DIG probe. E, F, Coronal section through midbrain region showing miR-133 signal in substantia nigra pars compacta region, indicated by arrow in inset (F). G, H, Coronal section through 3-month-old miR-133b KO midbrain region with detectable miR-133 signal in SNpc, indicated by arrow in inset (H), likely due to miR-133a1 and miR-133a2 expression. Scale bars, 1 mm.
Detection of miR-133b expression in SNpc by FISH, together with previous findings that miR-133b is enriched in midbrain and expressed in ES cell-derived DA neurons in vitro, warranted further investigation into its function in the midbrain dopaminergic system in vivo.
The midbrain is morphologically unchanged in miR-133b mutant mice
miR-133b has been shown to negatively regulate dopaminergic neuron differentiation in cultured mouse ES cells (Kim et al., 2007); therefore, miR-133b KO mice might be expected to have altered numbers of mDA neurons. The general morphology of the midbrain and striatum of 2-month-old male KO mice was indistinguishable from those of WT mice (Fig. 3A,B), while morphology and unbiased stereological counts of TH-IR cells in VTA and SNpc were also unchanged (Fig. 3A,D). miR-133b is selectively depleted in the midbrain of PD patients; if loss of miR-133b contributes to PD pathogenesis, deletion of miR-133b could lead to decreased numbers of DA neurons in aged mice. We found no difference in midbrain TH-IR cell number in 12-month-old WT and KO animals (Fig. 3C,F), suggesting that loss of miR-133b does not cause degeneration of mDA neurons. To determine whether KO mice have abnormal DA neuron development that is corrected postnatally, TH-IR cells were counted in E18.5 WT and KO SNpc. No difference was observed in number or morphology of TH-IR cells (Fig. 3C,F), consistent with normal development of DA neurons. A small increase in the number of Pitx3-IR cells was observed in the KO (Fig. 3F), suggesting that miR-133b may target Pitx3 in vivo. However, the number of cells immunoreactive for both TH and Pitx3 was equivalent in WT and KO (Fig. 3F), suggesting that the increased number of Pitx3-expressing cells in KOs does not have a major influence on mDA cell fate.
Figure 3.

Normal midbrain DA neuron morphology and number in miR-133b KO mice. A, B, TH immunohistochemistry and Nissl staining of midbrain (A) and dorsal striatum (B) of 2-month-old WT and KO littermates. C, TH and Pitx3 immunofluorescence in E18.5 SNpc of WT and KO mice. D, Unbiased stereological counts of TH-IR cells in SNpc and VTA of 2-month-old male mice; n = 4 WT and 5 KO mice. E, Unbiased stereological counts of TH-IR cells in SNpc of 12-month-old mice; n = 7 WT and 5 KO mice. F, Cell count of TH-IR, Pitx3-IR, and TH + Pitx3 coexpressing cells in SNpc of E18.5 WT and KO mice. *p < 0.05, Student's t test; n = 3 WT and 3 KO embryos. Scale bars: 0.5 mm in A, 1 mm in B, and 100 μm in C. Error bars indicate SEM.
Midbrain dopaminergic neuron gene expression is unaffected in miR-133b mutant mice
While miR-133b does not appear to regulate the differentiation of DA neurons in vivo, it could act by regulating expression of genes that affect neuron function but not cell number. Therefore, we quantified the expression levels of several genes important for mDA neuron function. TH, DAT, and Pitx3 transcripts were at WT levels in neonatal and adult KO brain, and levels of GAD1 and ChAT, expressed in GABAergic and cholinergic midbrain neurons, respectively, were also unchanged (Fig. 4A,B). Furthermore, expression of the related miRNA, miR-133a1, was unaltered in KO brain. Since many miRNAs act post-transcriptionally, miR-133b deletion could affect protein expression without altering transcript levels. However, no difference in neonatal or adult brain TH, GAD1, or Pitx3 protein levels was observed between WT and KO animals (Fig. 4C–E). To more specifically determine whether dopaminergic, GABAergic or cholinergic neurons had altered protein expression in midbrain, TH, GAD1, ChAT, and Pitx3 levels were quantified in WT and KO ventral midbrain, and no difference was observed (Fig. 4F,G), suggesting that global expression patterns in brain and midbrain are not affected by loss of miR-133b.
Figure 4.
Unaltered dopaminergic gene expression in miR-133b KO mice. A, B, TH, DAT, GAD1, ChAT, Pitx3, and miR-133a1 mRNA expression in whole brain of 3-month-old male (A) and P0 (B) WT and miR-133b KO mice. Gene expression is normalized to Gapdh and results are scaled to the average value of the WT samples (WT average, 1; n = 4 WT and 4 KO mice). C, TH, GAD1, and Pitx3 protein expression in whole brain of 3-month-old male (quantified in D) and P0 (quantified in E) WT and miR-133b KO mice. Western blot bands were quantified by densitometry and normalized to β-actin (for TH and GAD1) or β-tubulin (for Pitx3). Results were scaled to the average value of the WT samples (WT average, 1; n = 4 WT and 4 KO mice). F, TH, ChAT, GAD1, and Pitx3 protein expression in ventral midbrain of WT and miR-133b KO mice. G, Western blot bands in F were quantified in by densitometry and normalized to GAPDH. Results were scaled to the average value of the WT samples (WT average, 1; n = 5 WT and 6 KO mice). Error bars indicate SEM.
Monoamine neurotransmitters are unaltered in miR-133b mutant mice
To evaluate whether the activity of mDA neurons was altered in miR-133b KO mice, whole tissue monoamine neurotransmitter levels were quantified by HPLC-ECD. Levels of DA, NE, 5-HT, and their metabolites were unchanged in dorsal striatum (Fig. 5A), nucleus accumbens (Fig. 5B), frontal cortex (Fig. 5C), thalamus (Fig. 5D), and hypothalamus (Fig. 5E) of KO mice. To determine whether extracellular DA release was altered following miR-133b deletion, baseline and amphetamine-stimulated dorsal striatal microdialysis perfusates were collected from 3-month-old WT and KO male mice, and DA levels were quantified by HPLC-ECD. Baseline and amphetamine-induced DA release was equivalent between WT and KO samples, with both exhibiting a similar increase in extracellular DA upon administration of amphetamine (∼20-fold above baseline), followed by a gradual decline (Fig. 5F). These data suggest that miR-133b does not regulate total DA neurotransmitter production in mDA neurons or DA release in dorsal striatum.
Figure 5.
Unchanged monoamine neurotransmitter levels in miR-133b KO mice. A–E, Neurochemical analysis of whole-tissue monoamine contents in dorsal striatum (A), nucleus accumbens (B), frontal cortex (C), thalamus (D), and hypothalamus (E) from 2-month-old male WT and miR-133b KO mice; n = 8 WT and 7 KO mice. F, Neurochemical analysis of baseline and amphetamine-stimulated extracellular DA levels in dorsal striatum of 3-month-old male WT and KO mice. Amphetamine at 100 μm was continually infused via reverse microdialysis after 40 min (arrow); n = 9 WT and 8 KO mice. Error bars indicate SEM.
Locomotor and anxiety-like behaviors in miR-133b mutants are similar to those of wild-type mice
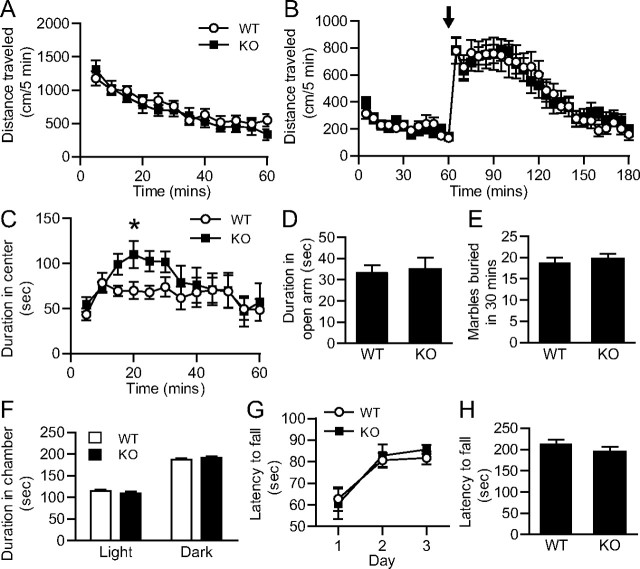
Movement and motivation-related behaviors are directly linked to mDA neuron function. To determine whether miR-133b KO mice have an underlying defect in the midbrain dopaminergic system, mice underwent a battery of behavioral tests. KO mice exhibited spontaneous locomotor behavior in the open field test that was indistinguishable from that of WT mice (Fig. 6A). The response of KO mice after injection of the psychostimulant amphetamine, which elevates extracellular DA levels, was also similar to that of WT mice (Fig. 6B). KO mice spent more time in the center of the open field during the first 30 min of the test as compared to WT mice (Fig. 6C), suggesting a decrease in anxiety-like behavior. mDA neuron dysfunction in mice has been found to cause generalized anxiety (Zweifel et al., 2011); to test whether KO mice were less anxious, behavior was observed in the elevated zero maze test (Fig. 6D). KO mice exhibited anxiety-like behavior equivalent to that of WT, preferring to spend less time in the open arms of the maze. The behavior of KO mice did not differ from that of WT in the marble burying test (Fig. 6E) or dark-light emergence test (Fig. 6F), which are both tests of anxiety-like behavior. Finally, motor coordination and learning were evaluated in the rotarod test. Mice were trained over three days to stay on top of a rod rotating at a fixed velocity, and KO mice learned the task as easily as their WT counterparts (Fig. 6G). Motor coordination was tested further on the fourth day as the rod accelerated to its maximum velocity. The performance of KO mice was equivalent to that of WT mice, as assessed by the duration of their attempt to stay on the rod (Fig. 6H). Therefore, miR-133b KO mice appear to have normal motor and anxiety-like behaviors, consistent with appropriate midbrain dopaminergic neuron function.
Figure 6.
Motor and anxiety-like behaviors are similar in miR-133b WT and KO mice. A, Spontaneous locomotor activity of 3- to 4-month-old WT and KO mice in the open field test; n = 12 WT and 12 KO mice. B, Amphetamine-induced locomotor activity of 3- to 4-month-old WT and KO mice in the open field test. Three milligrams per kilogram of amphetamine was administered by i.p. injection at 60 min (arrow); n = 9 WT and 11 KO mice. C, miR-133b KO mice spend more time in the center of the open field chamber during the first 30 min of the open field test; *p < 0.05, Student's t test. n = 12 WT and 12 KO mice. D, Time spent by 3- to 4-month-old WT and KO mice in the open arm of the elevated plus zero maze over 5 min; n = 18 WT and 13 KO mice. E, Number of marbles buried beneath bedding by 5-month-old male WT and KO mice after 30 min; n = 13 WT and 9 KO mice. F, Time spent in the light versus dark chamber by 3- to 4-month-old WT and KO mice in the dark–light emergence test over 5 min; n = 15 WT and 10 KO mice. G, Average latency to fall of 5-month-old male WT and KO trained on a rotarod at a fixed speed of 24 rpm for 4 trials per day over 3 days. H, Latency to fall from an accelerating rotarod (4–40 rpm) on the fourth day, averaged over 3 trials; n = 13 WT and 9 KO mice. Error bars indicate SEM.
Discussion
Here we show that mice with a deletion of miR-133b, a miRNA previously proposed to regulate differentiation and survival of mDA neurons (Kim et al., 2007), display no differences in development or maintenance of mDA neurons. We could detect no significant abnormalities in mDA neuron morphology, cell number, gene expression, monoamine levels, or motor and anxiety-like behaviors in these mice. These data suggest that the function of miR-133b in an in vitro model of DA neuron differentiation does not translate to a similar role in the intact animal. There are several explanations for this discrepancy. First, miR-133b expression in the midbrain may not contribute significantly to mDA neuron function. Indeed, miR-133b is part of the miR-133 family of miRNAs that is highly expressed in muscle and has been more extensively characterized in skeletal muscle development (Townley-Tilson et al., 2010). Overexpression of muscle miRNAs in heterologous cells globally inhibits brain-specific gene expression (Lim et al., 2005); introduction of miR-133b in a neuron culture system could be expected to inhibit neuronal differentiation in a broad manner without having a specific role in DA neurons.
Second, while not found to be enriched, expression of the miR-133b paralogs miR-133a1 and miR-133a2 has also been detected in the midbrain (Kim et al., 2007) and could compensate for the loss of miR-133b. miR-133a1 expression was not altered in miR-133b KO brain, while miR-133a2 expression was below our detection limits. Therefore, the lack of miR-133b does not appear to feed back to increase expression of its paralogs, although we cannot rule out the possibility of compensation by existing levels of miR-133a in midbrain. miR-133a1 and miR-133a2 double KO mice have recently been reported (Liu et al., 2008). It will now be possible to generate mice with a triple deletion of miR-133a1, miR-133a2, and miR-133b to determine whether the miR-133 family of miRNAs is important for brain development and function.
We have found that miR-133b is expressed in the SNpc region of midbrain, in addition to many other brain regions, including cortex, hippocampus, and cerebellum. Recently, cell type-specific miRNA biochemical purification combined with deep sequencing has shown that miR-133b is expressed in GABAergic neurons of cortex and cerebellum (He et al., 2012). This method will be critical for teasing apart the specific expression patterns of miR-133b, miR-133a1, and miR-133a2 in various neuronal subtypes and brain regions. The specific functions of miR-133b in these cell types may subsequently be explored using the constitutive or conditional miR-133b knock-out mouse lines described here.
There is growing evidence that miRNAs are implicated in neurodegenerative disorders (Hébert and De Strooper, 2009), presenting new possibilities for therapeutic intervention (Seto, 2010). miR-133b depletion in the midbrain of PD patients led to the hypothesis that loss or dysfunction of miR-133b could promote PD progression (Kim et al., 2007). However, we did not detect mDA neuron degeneration or impaired motor function in aged miR-133b KO animals. Moreover, no association has been found between miR-133b genetic variants and PD (de Mena et al., 2010). These results suggest that miR-133b depletion may not be causal in PD neurodegeneration; rather, loss of mDA neurons during disease progression would necessarily lead to a reduction in levels of miR-133b, along with other genes expressed in mDA neurons. However, we cannot exclude the possibility that a decrease in miR-133b may enhance the susceptibility of mDA neurons to neurodegeneration resulting from PD or toxic insults. This could be tested in the future by comparing PD-like disease progression in WT and miR-133b KO mice following MPTP or 6-OHDA treatment. Although miR-133b does not appear to play a causal role in midbrain neurodegeneration, loss of all miRNAs in mDA neurons in vivo does cause profound neuronal cell death (Kim et al., 2007). Therefore, other miRNAs expressed in mDA neurons may act alone or in concert to promote neuronal development and survival. In vivo analyses of these miRNAs may pave the way for novel therapeutic strategies in PD and other disorders of the mDA system.
Footnotes
This research was supported by a Ruth K. Broad Foundation Fellowship and a NINDS NRSA to M.P.H. G.F. was supported by NIH/NINDS and by The Poitras Center for Affective Disorders Research. We are grateful to Professor Marten Smidt of University Medical Center Utrecht for donation of the Pitx3 antibody. We thank Dr. Jacob Jacobsen of Duke University and Dr. Michael Cameron of The Scripps Research Institute Florida for advice related to in vivo microdialysis and HPLC-ECD.
The authors declare no competing financial interests.
References
- Andressoo JO, Saarma M. Signalling mechanisms underlying development and maintenance of dopamine neurons. Curr Opin Neurobiol. 2008;18:297–306. doi: 10.1016/j.conb.2008.07.005. [DOI] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cookson MR, Bandmann O. Parkinson's disease: insights from pathways. Hum Mol Genet. 2010;19:R21–R27. doi: 10.1093/hmg/ddq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon RM. Digging and marble burying in mice: simple methods for in vivo identification of biological impacts. Nat Protoc. 2006;1:122–124. doi: 10.1038/nprot.2006.20. [DOI] [PubMed] [Google Scholar]
- de Mena L, Coto E, Cardo LF, Díaz M, Blázquez M, Ribacoba R, Salvador C, Pastor P, Samaranch L, Moris G, Menéndez M, Corao AI, Alvarez V. Analysis of the Micro-RNA-133 and PITX3 genes in Parkinson's disease. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1234–1239. doi: 10.1002/ajmg.b.31086. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson's disease. J Chem Neuroanat. 2011;42:127–130. doi: 10.1016/j.jchemneu.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Liu Y, Wang X, Zhang MQ, Hannon GJ, Huang ZJ. Cell-type-based analysis of microRNA profiles in the mouse brain. Neuron. 2012;73:35–48. doi: 10.1016/j.neuron.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert SS, De Strooper B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009;32:199–206. doi: 10.1016/j.tins.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Heyer MP, Feliciano C, Peça J, Feng G. Elucidating gene function through use of genetically engineered mice. In: Starkey MP, Elaswarapu R, editors. Genomics: essential methods. Oxford: Wiley-Blackwell; 2011. pp. 211–248. [Google Scholar]
- Im HI, Kenny PJ. MicroRNAs in neuronal function and dysfunction. Trends Neurosci. 2012;35:325–334. doi: 10.1016/j.tins.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ML, Ho CC, Day AE, Walker QD, Francis R, Kuhn CM. Oestrogen receptors enhance dopamine neurone survival in rat midbrain. J Neuroendocrinol. 2010;22:226–237. doi: 10.1111/j.1365-2826.2010.01964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BJ, Roberts DJ. The quantiative measurement of motor in co-ordination in naive mice using an acelerating rotarod. J Pharm Pharmacol. 1968;20:302–304. doi: 10.1111/j.2042-7158.1968.tb09743.x. [DOI] [PubMed] [Google Scholar]
- Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes Dev. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci U S A. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani AK. Influence of intrastriatal infusion of dynorphin fragments on overflow of acetylcholine and dopamine in the rat brain. Int J Neurosci. 2009;119:1362–1383. doi: 10.1080/00207450902955770. [DOI] [PubMed] [Google Scholar]
- Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena JT, Sohn-Lee C, Rouhanifard SH, Ludwig J, Hafner M, Mihailovic A, Lim C, Holoch D, Berninger P, Zavolan M, Tuschl T. miRNA in situ hybridization in formaldehyde and EDC-fixed tissues. Nat Methods. 2009;6:139–141. doi: 10.1038/nmeth.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG. The road toward microRNA therapeutics. Int J Biochem Cell Biol. 2010;42:1298–1305. doi: 10.1016/j.biocel.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Smidt MP, Burbach JP. How to make a mesodiencephalic dopaminergic neuron. Nat Rev Neurosci. 2007;8:21–32. doi: 10.1038/nrn2039. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Townley-Tilson WH, Callis TE, Wang D. MicroRNAs 1, 133, and 206: critical factors of skeletal and cardiac muscle development, function, and disease. Int J Biochem Cell Biol. 2010;42:1252–1255. doi: 10.1016/j.biocel.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Munckhof P, Luk KC, Ste-Marie L, Montgomery J, Blanchet PJ, Sadikot AF, Drouin J. Pitx3 is required for motor activity and for survival of a subset of midbrain dopaminergic neurons. Development. 2003;130:2535–2542. doi: 10.1242/dev.00464. [DOI] [PubMed] [Google Scholar]
- Velleca MA, Wallace MC, Merlie JP. A novel synapse-associated noncoding RNA. Mol Cell Biol. 1994;14:7095–7104. doi: 10.1128/mcb.14.11.7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifel LS, Fadok JP, Argilli E, Garelick MG, Jones GL, Dickerson TM, Allen JM, Mizumori SJ, Bonci A, Palmiter RD. Activation of dopamine neurons is critical for aversive conditioning and prevention of generalized anxiety. Nat Neurosci. 2011;14:620–626. doi: 10.1038/nn.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]