Abstract
Elevated levels of reactive oxygen species (ROS) play a critical role in cardiac myocyte signaling in both healthy and diseased cells. Mitochondria represent the predominant cellular source of ROS, specifically the activity of complexes I and III. The model presented here explores the modulation of electron transport chain ROS production for state 3 and state 4 respiration and the role of substrates and respiratory inhibitors. Model simulations show that ROS production from complex III increases exponentially with membrane potential (ΔΨm) when in state 4. Complex I ROS release in the model can occur in the presence of NADH and succinate (reverse electron flow), leading to a highly reduced ubiquinone pool, displaying the highest ROS production flux in state 4. In the presence of ample ROS scavenging, total ROS production is moderate in state 3 and increases substantially under state 4 conditions. The ROS production model was extended by combining it with a minimal model of ROS scavenging. When the mitochondrial redox status was oxidized by increasing the proton permeability of the inner mitochondrial membrane, simulations with the combined model show that ROS levels initially decline as production drops off with decreasing ΔΨm and then increase as scavenging capacity is exhausted. Hence, this mechanistic model of ROS production demonstrates how ROS levels are controlled by mitochondrial redox balance.
Introduction
Experimental evidence suggests that low levels of reactive oxygen species (ROS) play a critical role in the cellular signaling pathways of healthy cardiac myocytes (1), including the protective effects of ischemic preconditioning (2). However, ROS contribute to pathophysiology at higher concentrations. Oxidative stress can sensitize the mitochondria to permeability transition pore opening (3) and can also modulate the activity of many key ion channels and excitation-contraction coupling proteins in the cardiac myocyte. High levels of ROS contribute to contractile dysfunction and fatal arrhythmias in diseased hearts (4) and have been implicated in the progression of heart failure (5–7), particularly ROS derived from the mitochondrial electron transport chain (ETC) (8).
Cellular ROS levels depend not only on the rate of ROS production, but on the rate of ROS scavenging. The redox-optimized ROS balance (R-ORB) hypothesis (9) posits that ROS levels are minimized in an intermediate cellular redox environment, neither too reduced nor too oxidized, when energy output is maximal, but increase when the system deviates toward a more oxidized or more reduced state. This property arises from the dual control of ROS production and scavenging, respectively, by NADH redox potential, which determines the electron transport rate, and the glutathione (GSH) and nicotinamide adenine dinucleotide phosphate (NADPH) redox potentials, which drive antioxidant enzyme reactions. As the redox environment becomes more reduced than that corresponding to minimal ROS levels, ROS scavenging rates are maintained, but higher mitochondrial NADH concentrations lead to a high level of reduction of the ETC complexes, which increases ROS production above the rate of scavenging. On the other hand, as the redox environment becomes more oxidized than that corresponding to minimal ROS levels, mitochondrial ROS production is low, but decreased availability of reducing equivalents for ROS scavenging can also result in accumulation of cellular ROS. ROS balance is maintained in an intermediate redox environment that is reduced enough to support high scavenging rates but oxidized enough to prevent high ROS production.
In ventricular myocytes isolated from failing hearts, oxidation of NAD(P)H occurs during increased workload (10), leading to increased cellular ROS accumulation (11), likely stemming from decreased scavenging capacity in an increasingly oxidized redox environment. In contrast, hypoxia, which reduces the cellular redox environment, can also lead to increased ROS and activation of hypoxia-inducible factor-dependent signaling (12). Furthermore, experiments in isolated mitochondria have shown that ROS production varies with NADH redox potential (13–15), the concentration (16) and type of respiratory substrate used (9,15), mitochondrial membrane potential (ΔΨm) (17,18), and direction of electron transport through complex I (9,15). Understanding how mitochondrial ROS production varies under different metabolic conditions will help elucidate how redox homeostasis is maintained in healthy cells and how metabolic changes in diseases like heart failure can lead to pathological levels of ROS production.
NADH-ubiquinone oxidoreductase (complex I of the ETC), is one proposed source of mitochondrial ROS production (13–16,19–22). Its role in respiration is to transfer electrons from NADH, a product of the tricarboxylic acid (TCA) cycle or β-oxidation, to ubiquinone (Q). Complex I is a large protein, consisting of up to 45 subunits (23) depending on species. Its structure was not fully resolved until recently (24), and its mechanism of proton pumping is still a source of controversy (25,26). A variety of conditions have been shown to elicit ROS production from complex I, including rotenone inhibition in the presence of NADH (13–15,19,20) and reverse electron transport (RET) in the presence of NADH, succinate, and high ΔΨm (15,16,19,21,22). However, the source of the electrons forming superoxide is still under debate (27). It has been identified by some (14,15) as the flavin mononucleotide (FMN) cluster in the hydrophilic arm of the protein and by others (22,28) as the Q-site at the interface of the protein’s matrix and membrane domains.
Mitochondrial ubiquinol-cytochrome-c oxidoreductase (complex III) transfers electrons from ubiquinol to cytochrome c by means of the Q cycle. By virtue of the fact that the high- and low-potential b-hemes (bL and bH) of complex III are situated in the membrane, electron transfers to and from those centers are dependent on the electric field imposed by ΔΨm. As ΔΨm increases, the rates of electron transfer through the b-hemes decrease and the concentration of the upstream ubisemiquinone on the cytoplasm-facing side increases. One proposed mechanism for ΔΨm-dependent ROS production from complex III attributes superoxide formation to the escape of an electron from the unstable ubisemiquinone to reduce oxygen. Experimental data have confirmed the existence of a semiquinone on the cytoplasm-facing side (29,30), suggesting a mechanism for ROS production by complex III (20,31,32). Additional experiments have further elaborated on this mechanism by proposing complex III ROS release to both sides of the IMM (20,31). In contrast to complex I, high ROS production from complex III occurs during forward electron transport (FET).
In this work, we formulate a biophysically derived computational model of the ETC to investigate how changes in the mitochondrial environment control the rate of ROS production. This model is constructed with a minimal number of states to facilitate its future incorporation into multiscale models of mitochondria and cardiac myocytes. We extend this model to include a previously published minimal model of mitochondrial ROS scavenging (33) to show how, as the redox environment varies from highly reduced to highly oxidized, opposite but complementary changes in the rate of ROS production and the ROS scavenging capacity produce a minimal level of mitochondrial ROS at intermediate mitochondrial redox potentials, in agreement with the R-ORB hypothesis (9).
Methods
Model description
Fig. 1 A schematically depicts protein structures and redox centers of the respiratory complexes represented in this electron transport chain model. Briefly, forward and reverse rate constants for electron-transfer models of complexes II–IV were modified from Demin et al. (34,35) (see Fig. 1, C and D) to better match data on the redox state of the ETC from Kim et al. (36). A thermodynamic model of complex I was constructed (see Fig. 1 B) based on the whole ETC model of Magnus and Keizer (37), and its rate constants were parameterized to match respiration and ROS production data from Aon et al. (9). Enzyme concentrations and thermodynamic quantities remained unchanged with the exception of complex I protein concentration. Overall, 23 parameters were obtained from experimental results or previous models and 39 parameters were adjusted based on experimental data. More complete methods, equations, and parameters can be found in the Supporting Material.
Figure 1.
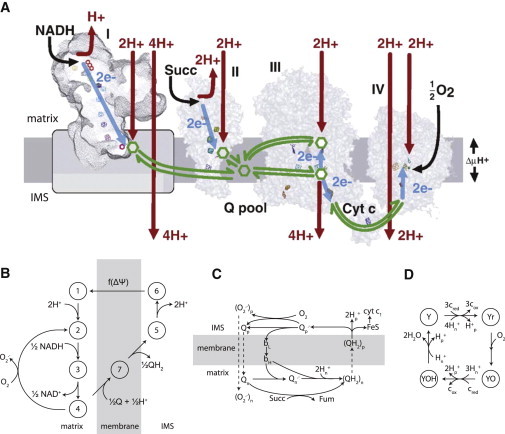
Overview of the mitochondrial ETC and model representation. (A) Mitochondrial respiratory complexes I–IV from Moser et al. ((81), reproduced with permission from Elsevier). (B) State diagram for the complex I model, modified from Magnus and Keizer (37). (C) Reaction scheme for complexes II and III, from Demin et al. (35). (D) State diagram describing the reactions of complex IV, from Demin et al. (34). Matrix and intermembrane space (IMS) compartments are indicated for topologically explicit diagrams.
Results
In the following sections, we demonstrate the ability of the model to simulate data from which model parameters were estimated, as well as to predict experimental data not included in the optimization process.
Model reconstructions
Redox state of the ETC
ROS can only be produced from reduced redox centers, since those centers act as electron donors to reduce oxygen to superoxide. As such, the oxidation state of the redox components of the ETC can offer important information about likely sites of ROS production and how those sites are modulated by changes in the mitochondrial environment. The ETC model presented here includes variables describing the concentrations of ubiquinone, ubiquinol, and ubisemiquinone, along with the oxidation states of cytochrome c and the redox centers in complex III, i.e., the high- and low-potential b-hemes (bH and bL) and cytochrome c1. Modeling results were obtained for a range of ΔΨm, from the high ΔΨm associated with state 4 respiration, through the state 3 ΔΨm range to lower values of ΔΨm found after the addition of protonophoric uncouplers. These computational results are compared with experimental data from the literature in Fig. 2, A–C. Fig. 2 A shows the reduction in the ubiquinone redox potential that occurs at higher ΔΨm as the concentration of ubiquinone (Q) falls and ubiquinol (QH2) increases. It is important to note that different tissue types show different mitochondrial NAD+/NADH redox potentials at rest. Estimations using ratios of other metabolites indicate that rat heart maintains a lower NAD+/NADH ratio from 1 to 16 (38,39), liver attains ratios from 30 to 100 (38), and alveolar macrophages have a ratio near 30 (40). The agreement of the redox potential of these upstream electron carriers in Fig. 2 A helps to validate the oxidation-state results of the downstream electron carriers. Fig. 2 B shows the typical ΔΨm-dependent reduction of the b-hemes. This is expected, because these redox centers are embedded in the membrane, and electron transfers from the cytosolic to the matrix face of the membrane are subject to the opposing force of the membrane potential. The increased reduction of bL plays a critical role in the mechanism of complex III ROS production in the model presented here, in which the buildup of electrons on the cytosolic side of the Q cycle causes an increase in the ubisemiquinone concentration and subsequent release of its unpaired electron to form superoxide. In the model, superoxide resulting from this process is released to either the matrix or the intermembrane space with equal probability, according to the mechanisms proposed by Muller et al. (31). Fig. 2 C validates the ΔΨm dependence of the redox states of cytochromes c1 and c, which play a role in the transfer of electrons between complex III and complex IV. The agreement of model results and experimental data from Brown and Brand (41) in isolated mitochondria is reasonably good over the range of physiological ΔΨm (120–180 mV). In contrast, data from Kim et al. (36) show that in whole macrophage cells, oxidation of cytochrome c1 and c increases with ΔΨ over the range 150–180 mV. This may be related to the high [ATP]/[ADP]-ratio-induced inhibition of complex IV (42), which is common in cells but not in isolated mitochondria (43). Given that the model presented here represents isolated mitochondria, the disparity between the model results and the experimental data from cells may be due to a control mechanism that is not represented in the model. It is also important to note that studies using metabolic control analysis, such as those by Rossignol et al. (44), have revealed that metabolic control can vary widely between tissue types, which could potentially explain the difference between the Brown and Brand (41) liver data and the Kim et al. (36) macrophage data.
Figure 2.
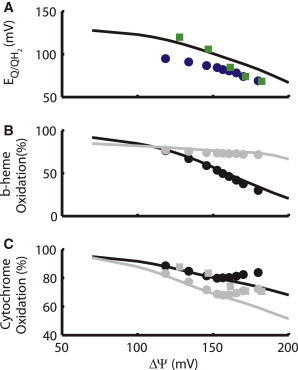
Electron carrier redox state versus mitochondrial membrane potential (ΔΨm). (A) Model results for redox potential of Q/QH2 (black line) follow a decrease in redox potential with increasing ΔΨm similar to the experimental data of Kim et al. (36) (circles) and Brown and Brand (41) (squares). (B) Model results (lines) are in good agreement with the increasing reduction of the bL (black) and bH (gray) redox centers shown by experimental measurements from Kim et al. (36) (circles). (C) Model results (lines) show increasing reduction of cytochrome c1 (black) and c (gray), similar to the trend in cytochrome c redox state shown by Brown and Brand (41) (squares). Experiments of Kim et al. (36) (circles) show a slight reoxidation at potentials exceeding 160 mV.
Control of respiration
See the Supporting Material for results validating the dependence of respiratory flux on ΔΨ and NADH.
Control of ROS production
When complex I rates are fitted to guinea pig cardiac mitochondrial data from Aon et al. (9), the model reproduces the experimental patterns of ROS production in the presence of glutamate/malate or succinate in states 3 and 4 (Fig. 3). ROS production in complex III exhibits higher rates for state 4 than for state 3. The conditions for the highest levels of total ROS production correspond to state 4 respiration in the presence of complex II substrates. For the RET conditions in state 4 in the presence of succinate, 72% of ROS production is derived from complex I, though the contribution from the ubisemiquinone site of complex III is significant at 28%. After the addition of ADP, ROS production by both complexes, I and III, decreases substantially. For complex I substrates, complex I makes a minimal contribution to ROS production in state 3. Given that mitochondrial succinate levels have been measured at 0.3 mM (45), much lower than the 4–10 mM used to induce RET (9,15,22) in isolated mitochondria, and that complex I substrates are always present in cells, it is unlikely that physiological conditions favor RET in vivo under nonischemic conditions (46). This suggests that FET ROS production would predominate physiologically. In the absence of pathological conditions leading to complex I inhibition, model simulations therefore support complex III as the primary contributor to superoxide production in normal cells.
Figure 3.
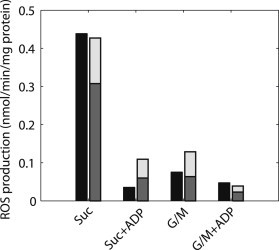
ROS production as a function of different substrates and mitochondrial respiratory states. Simulated state 4 ROS production in succinate (Suc; dark gray/light gray bar), predominated by RET-induced complex I ROS, quantitatively matches the experimental data (black). The bottom segment of each bar (dark gray) for the simulations depicts the ROS contribution from complex I, whereas the upper segment (light gray) depicts the contribution from complex III ROS production. During maximal rates of oxidative phosphorylation, stimulated by ADP (state 3), ROS production with succinate is minimized (Suc+ADP). ROS production with the complex I substrates (G/M) is split between complex I and complex III and is diminished when oxidative phosphorylation flux is maximal (G/M+ADP).
Model predictions
After the model parameters were adjusted to reproduce the experimental data featured previously in the Model reconstructions section, the final parameter set was used to simulate additional experimental protocols. The model results using these protocols are able to reproduce data that were not included in the model parameterization process.
Control of ROS production
For mitochondria respiring on succinate in state 4, ROS production has been shown to be highly dependent on ΔΨm. Fig. 4 compares the model prediction of the normalized rate of ROS production from complex III as a function of ΔΨm to experimental data from rat heart mitochondria (18) and isolated complex III (17). In the model, this property arises from the reduction in the rate of Q-cycle reactions involving the b-hemes as membrane potential increases. As the b-hemes become more reduced, the ability to accept electrons from ubisemiquinone on the side of the membrane facing the cytoplasm decreases, leading to a buildup of that species. Electrons from the ubisemiquinone are transferred to oxygen to produce superoxide. It is important to note that the relationship between respiratory rate and ROS production shown in Fig. 4 is derived by modulating ΔΨm through increased membrane proton leak. Under these conditions, the increased permeability of the membrane to ions leads to higher rates of electron flux, and thus respiration (VO2), as membrane potential decreases. However, if membrane potential is modulated by inhibition of the ETC either by using malonate (18,47) or by modulating the concentration of cytochrome c as an electron acceptor (48), then VO2 will decrease with membrane potential, thus paralleling the trend in ROS production (not shown). Decreasing ΔΨm using an uncoupler would also be expected to oxidize the NAD+/NADH couple, which is evident in experiments (9,49) but is not represented in the model due to the exclusion of the TCA cycle. Given that uncoupling oxidizes the cellular redox environment, the decrease in ROS shown in Fig. 4 is consistent with the R-ORB hypothesis over the range of intermediate to highly reduced redox environments.
Figure 4.
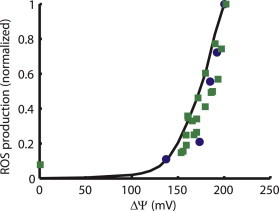
ΔΨm dependence of complex-III-derived ROS. Model ROS production from complex III (black) is minimal for ΔΨm < 150 mV, then increases exponentially as ΔΨm increases to >150 mV. Experimental data from Korshusnov et al. (18) (circles) and from Rottenberg et al. (17) (squares) follow a similar trend. The normalized ΔΨm reported by Korshunov et al. (18) were scaled to a maximum of 200 mV for comparison.
Model simulations in which matrix pH is increased (Fig. S2 in the Supporting Material) show that ROS production from complexes I and III during FET increases with matrix alkalinization, in agreement with the results of Selivanov et al. (50). From a control matrix pH of 7.3, the model results show a 3.7-fold increase in net ROS production with 0.6 pH units of alkalinization and a 30% decrease with 0.2 units of acidification. As Fig. 5 shows, RET-derived ROS production from complex I also exhibits matrix pH dependence. Under state 4 conditions with high succinate, matrix alkalinization causes an increase in complex I ROS production, qualitatively similar to data shown in Lambert and Brand (22) (reproduced in Fig. 5 A), though the experimental ROS production rates are around sevenfold greater. The results of Figs. 5 B and S2 show that complex I ROS production under glutamate/malate (G/M) substrates is consistently low, even for high matrix pH. However, a notable prediction of the model is that under state 3 respiration, in the presence of succinate, an increase in matrix pH will elevate complex-I-derived ROS from the minimal levels observed at control pH to the high levels observed in state 4 RET (Fig. 5 B).
Figure 5.
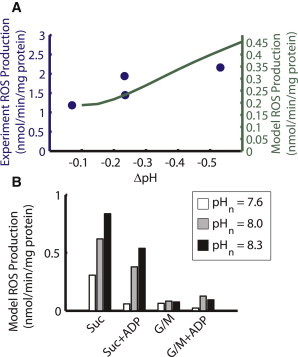
Effect of matrix alkalinization on complex I ROS production. (A) Comparison of experimental complex I H2O2 production data (circles) from Lambert and Brand (22) and model ROS production for the same ΔpH (line). Model cytosolic pH was held constant at 7.0 and model ΔΨm was clamped to 200 mV. Experimental data were given with the effect due to ΔΨm subtracted (22). (B) Model complex I ROS production for an increase in matrix pH from 7.6 (white) to 8.0 (gray) to 8.3 (black). Not only does matrix alkalinization increase complex I ROS for succinate respiration without ADP, it increases ROS for succinate respiration with ADP fivefold at pH 8 and nearly sevenfold at pH 8.3.
The highest levels of ROS production are typically measured in the presence of various ETC inhibitors that promote high levels of reduction at sites favoring direct electron transfer to molecular oxygen. The simulations shown in Fig. 6 predict the effects of these inhibitors on ROS production under different respiratory conditions. Rat heart mitochondria data by St. Pierre et al. (20) show that complex I ROS production is limited to the matrix side, in agreement with the ROS production mechanism chosen for the model (Fig. 6 A). Complex-III-derived ROS production rates in the experiments and the model are low for succinate respiration, except in the presence of antimycin A, which blocks the Qi site, preventing the b-hemes from being oxidized by matrix-facing Q (Fig. 6 B). This results in an accumulation of cytosol-facing ubisemiquinone and subsequent electron transfer to O2 and superoxide formation. In a similar way, block of bL heme reduction by ubisemiquinone at the Qo site also prevents electrons from crossing the membrane via the b-hemes (not shown). The result is a buildup of cytosol-facing ubisemiquinone and elevation of ROS production. Additional simulations show that inhibition of oxygen binding to complex IV by cyanide decreases ROS production from complex III (not shown) by causing nearly complete reduction of cytochrome c and the FeS cluster, preventing the oxidation of QH2 and formation of semiquinone. These results are in agreement with experimental data demonstrating a cyanide-dependent decrease in ROS production in heart mitochondria (19,51). Each inhibition simulation is the result of drastically reducing or eliminating the reaction in the model analogous to that inhibited by the drug intervention.
Figure 6.
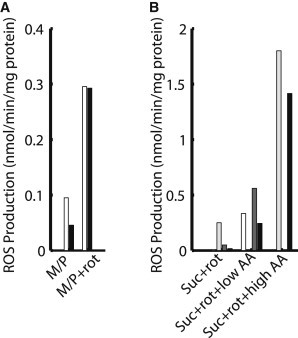
Control of ROS production by respiratory blockers. (A) ROS release in the presence of complex I substrates. For mitochondria respiring on malate plus pyruvate (M/P), experimental H2O2 recordings (white) from St. Pierre et al. (20) were measured in the absence and presence of rotenone (rot) and compared to model ROS production under similar conditions (black). (B) ROS release from complex III. Experimental results from St. Pierre et al. (20) (white), Turrens et al. (48) (light gray), and Starkov and Fiskum (82) (dark gray) show ROS release for rat heart mitochondria respiring on succinate (Suc). St. Pierre et al., Turrens et al. and simulations results included rotenone to eliminate RET-induced complex I ROS release. Antimycin A (AA) was added to observe ROS production resulting from the disruption of the complex III Q cycle. Similar results are obtained from the model (black). St. Pierre et al. used 0.625 nmol/mg protein AA, Turrens et al. used 2 nmol/mg protein AA, and Starkov and Fiskum used 2 μM AA. Specific model simulation conditions are given in the Supporting Material.
Fig. S1 shows the NADH dependence of complex I ROS production in the absence of Q and QH2, as in Pryde and Hirst (15). This was predicted by fitting the rate constants of complex I to the data from Aon et al. (9), as shown in Fig. 3. The midpoint potential of this curve matches the two-electron reduction potential of complex I’s FMN, given by Sled et al. (53) as −0.38 V at pH 7.5. This prediction supports the validity of the proposed mechanism of ROS production from the FMN (14,15).
Control of ROS levels
Many experiments infer matrix ROS levels from data obtained using indicators of hydrogen peroxide (H2O2) emission into the extramitochondrial space (e.g., the Amplex Red method). This technique thus reports only the excess H2O2 diffusing across the inner mitochondrial membrane (IMM) at steady state, which reflects the net balance of superoxide production and its removal via superoxide dismutase and glutathione/thioredoxin-dependent H2O2 scavenging systems. Thus, the ROS production rates depicted in Fig. 3 provide only one-half of the picture. To more accurately represent mitochondrial ROS dynamics, both ROS production and scavenging must be taken into account. Hence, we combined the ETC ROS production model with a recent minimal model of ROS scavenging (33). Model NADH levels were derived from an earlier mitochondrial model (54) subjected to the same uncoupling protocol. NADPH levels were set to follow the same decreasing function as NADH, but with a minimum of 3 μM and a maximum of 8 μM, spanning the critical ROS overflow range given in Aon et al. (33). The mitochondrial redox environment was calculated as the sum of redox potentials for NADH, NADPH, and GSH weighted by their respective concentrations, as described in Schafer and Buettner (55). In the simulation, mitochondrial respiratory uncoupling from state 4 is represented by increasing membrane proton leak conductance, leading to oxidation of the mitochondrial redox environment (Fig. 7). The model’s rate of H2O2 emission across the IMM decreases from initial levels near 10 nmol/min/mg protein to a minimum of ∼7 nmol/min/mg protein as the redox environment is oxidized from −1300 to −1150 mV-mM. As uncoupling progresses, the scavenging systems are compromised by further oxidation of the mitochondrial redox environment, and H2O2 emission begins to increase with further uncoupling. At −400 mV-mM redox environment, NADH is 2% reduced, ΔΨm has decreased to 135 mM and the rate of H2O2 emission reaches nearly 40 nmol/min/mg protein. This fourfold increase in H2O2 emission at −400 mV-mM redox environment compared to the initial H2O2 emission rate is in good agreement with the experimental data given in Aon et al. (9).
Figure 7.
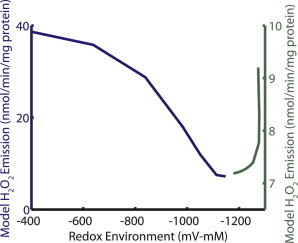
Interplay of ROS production and scavenging systems. Simulations were performed with progressive respiratory uncoupling simulated by increasing proton leak conductance in the model. Steady-state H2O2 emission rates are plotted against the model’s redox environment, incorporating NADH, NADPH, and GSH redox couples as described by Schafer and Buettner (55). The U-shaped dependence of H2O2 emission on ΔΨm arises from the combined effects of decreased ROS production and decreased ROS scavenging capacity on net ROS levels. Model NADH levels were derived from a previously published mitochondria model (54) subjected to the same uncoupling protocol. NADPH levels were set to follow the same decreasing function as NADH, but with a minimum of 3 μM and a maximum of 8 μM, spanning the critical ROS overflow range given in Aon et al. (33). See the Supporting Material for scavenging equations and parameters.
The model mechanism that produces the U-shaped dependence of ROS levels on redox environment follows closely that proposed in the R-ORB hypothesis. In the model simulations, state 4 conditions are modulated with increasing respiratory uncoupling. Increasing leak causes a decrease in ΔΨm, which drives a subsequent decrease in ROS production, similar to the behavior depicted in Fig. 4. As membrane potential declines from the initial state 4 value, the NAD+/NADH redox couple becomes more oxidized as more NADH is consumed to pump protons in a futile attempt to counteract the increased membrane leak. ROS scavenging systems remain largely intact for small oxidative perturbations in the redox environment (not shown). Thus, for the initial increase in uncoupling, comprising the right side of the U-shape in Fig. 7, model ROS production decreases 60%, whereas GSH and the associated scavenging capacity are almost unchanged, leading to the ROS minimum at the so-called redox-optimum. Model ΔΨm at this redox optimum is 152 mV, similar to the nearly minimal ROS production at 153 mV observed by Korshunov et al. (18) (Fig. 4, green squares) Further uncoupling continues to decrease ΔΨm, but ROS production only decreases by another 13% from control levels. The NAD+/NADH redox couple continues to oxidize, leading to lower NADPH levels and a decreased ability to recharge the oxidized glutathione and thioredoxin scavenger pools. As a result, GSH levels drop by 72%, compromising the H2O2 scavenging capacity of the mitochondria. The decrease in GSH levels exceeds the reduction in ROS production, causing an increase in H2O2 emission as the redox environment is further oxidized from the optimum.
Discussion
In this work, we have used a mechanistic formulation to develop what is to our knowledge a novel model of the mitochondrial ETC, which we then used to study control of ROS production in cardiac mitochondria. This model is shown to reproduce 1), experimental data measuring the ΔΨ dependence of the redox potential of the Q pool and the oxidation states of the b-hemes and cytochromes c1 and c; 2), experimental results describing the dependence of the respiratory flux from complex I or complex II substrates on ΔΨm; 3), measurements of ROS production as a function of substrate concentration and ΔΨm. The model predicts the dependence of ROS production on ΔΨ, matrix pH, NAD+/NADH redox potential, and inhibition of complexes I and III. With the addition of a minimal ROS scavenging model, the computational work presented here also permits analysis of the relative roles of ROS production and scavenging on mitochondrial H2O2 emission and ROS balance, as well as the impact of alterations in the redox environment on oxidative stress.
The detailed description of redox-dependent electron transport processes in the model is the basis for its predictive power. Because the ΔΨ dependence of the electron transfer between hemes bL and bH is included explicitly, suppression of this flux (e.g., by antimycin A) leads to a cytosolic-side accumulation of ubisemiquinone, producing the ΔΨ-dependent ROS production behavior shown in Fig. 4. The complex III inhibitor effects shown in Fig. 6 also arise from the explicit representation of electron transfers of the Q cycle. The antimycin A block increases ROS production by a mechanism similar to the increase produced due to high ΔΨ. Although the model of complex I does not include explicit electron transfer reactions like the complex III model, each state transition does have a mechanistic basis (Fig. 1 B). As a result, the complex I model is able to predict the influence of rotenone (Fig. 6 A) and NAD+/NADH redox potential (Fig. S1) on ROS production.
The explicit inclusion of reactions requiring protons leads to some interesting predictions regarding the pH dependence of ROS production. As shown in Fig. S2, ROS production by complex I and complex III increases under conditions of matrix alkalinization, in agreement with data from Selivanov et al. (50). In the model, the reduction of semiquinone and the binding of two protons are lumped into a single reaction. As matrix-side protons become more scarce, matrix-side ubisemiquinone is converted to ubiquinol more slowly because of the scarcity of protons to form QH2. An increasing proportion of the Q pool stagnates on the matrix side without enough protons to become the charge-neutral QH2 form that is able to diffuse across the membrane, reducing the amount of ubiquinone able to accept electrons from heme bH. Through these mechanisms, both hemes become more reduced, preventing heme bL from accepting electrons from the cytosol-side semiquinone. The buildup of this ubisemiquinone species is directly proportional to ROS production from complex III. The model shows that complex I ROS production is also increased under more alkaline matrix conditions. The pH dependence of complex I ROS production is compared to RET data from Lambert and Brand (22) in Fig. 5 A. The model is able to qualitatively reproduce the experimental data. However, a quantitative comparison of model results and experimental data for the pH dependence of complex I shows that the rate of ROS production in the experimental data is around sevenfold higher. ROS production rates vary widely depending on the species used, the age of the animal, and the preparation of the sample. The complex I model was constrained using the data from Aon et al. (9), which show a sixfold-lower rate of ROS production compared with the Lambert and Brand data (22) when mitochondria are respiring on succinate in the absence of ADP. A sixfold increase in the rate of ROS production from complex I (represented by the rate constant between complex I reaction states 4 and 2) can match the magnitude of the Lambert and Brand (22) data without changing the model response to the VO2 versus ΔΨ protocol from Fig. S1 A (not shown), but at the expense of the quantitative agreement shown in Fig. 3.The effects of matrix alkalinization on complex I ROS production were also tested for complex I and complex II substrates in states 3 and 4 (Fig. 5 B). An increase in matrix pH leaves G/M ROS production largely unchanged in state 3 and state 4, but large increases are predicted for succinate respiration. The high levels of complex I ROS production occurring during RET conditions at control pH increase by 87% as pH increases from 7.6 to 8.3. An even larger increase is shown for state 3 succinate respiration, with complex I ROS production increasing almost sevenfold. To our knowledge, this property of state 3 succinate respiration has not been tested experimentally, but this prediction illustrates the utility of the cycle of experiment-based modeling and model-derived predictions as the basis of further experiments.
The most complete description of mitochondrial ROS levels includes not only ROS production but the native mitochondrial ROS scavenging capabilities. A previously published, minimal model of ROS scavenging (33) was combined with the ROS production model developed here to examine the overall control of mitochondrial ROS levels by both production and scavenging systems. To simulate the oxidation of the mitochondrial environment, respiration was uncoupled from ΔΨ with increasing membrane proton leak conductance. In vitro, this has the effect of decreasing NADH levels and subsequently NADPH levels, as the two are coupled via the nicotinamide nucleotide transhydrogenase. Model simulations show that whereas ROS production drops off dramatically as ΔΨ decreases (Fig. 4), the depletion of scavenging systems with increased uncoupling results in an increase in H2O2 levels as the GSSG/GSH redox couple is oxidized. This is in agreement with the hypothesis put forth by Aon et al. (9) that the interplay between ROS production and ROS scavenging leads to a minimum of mitochondrial ROS levels at intermediate redox potentials. The simulations presented using this model make it possible to explore a range of redox environments both more reduced and more oxidized than that of the ROS minimum.
Validation of the redox-optimized ROS balance hypothesis
By combining the model of ROS production by the ETC, presented here, and the previously published minimal scavenging model (33), we are able to computationally validate the R-ORB hypothesis. In simulations, the mitochondrial redox environment was modulated by increasing respiratory uncoupling, starting from state 4 conditions. The presence of a minimum in H2O2 emission at an intermediate redox environment and the 1:4 ratio of H2O2 emission for the maximally reduced environment in this protocol compared to the most oxidized environment is in good agreement with the experimental data of Aon et al. (9) H2O2 emission reaches a minimum at levels of uncoupling corresponding to a ΔΨm between 150 and 155 mV, similar to state 3 ΔΨm values for isolated mitochondria (9) and to the ΔΨm value shown by Korshunov et al. (18) to strongly suppress H2O2 emission. However, this ΔΨm range is higher than the typical mitochondrial operating conditions in vivo, which are closer to 100–130 mV (43). Therefore, the expectation is that intact cells operate at the optimal conditions for minimizing ROS levels during high rates of oxidative phosphorylation. It is important to note that the model simulates the behavior of isolated mitochondria, which can be pushed to redox extremes and utilize substrate concentrations not normally encountered in cells. The difference in operating conditions between isolated myocytes and isolated mitochondria was a key point addressed by the R-ORB hypothesis (9). Moreover, a whole-cell ROS model would also need to take into account other nonmitochondrial ROS sources (e.g., NADPH and xanthine oxidases) and cytoplasmic ROS scavenging pathways. Although the model ΔΨm value at the redox optimum is reasonable, the state of the model redox environment at that optimum is far more reduced than that of the experimentally-observed optimum (9). This discrepancy may be due to the approximation of NADH and NADPH levels used in the model. Specifically, the simplifying assumption that NADPH decreases in parallel with NADH may not be an accurate approximation. However, given that the work presented here represents ROS production and scavenging networks in the absence of key NADH and NADPH production mechanisms such as the TCA cycle or the nicotinamide nucleotide transhydrogenase, the qualitative agreement between the model responses to redox environment changes and the experimental data are still highly informative with regard to how ROS production is modulated and how ROS production contributes to matrix ROS levels. The ROS production results shown in this protocol incorporate changes in both ΔΨ and NADH redox state that occur with uncoupling. To continue the cycle of experiments and modeling, the ROS production results shown here could be supported by experiments using antioxidant-depleted mitochondria and valinomycin or nigericin to investigate changes in ROS production with ΔΨ and ΔpH and to validate the role of ΔΨ versus NADH redox state in controlling ROS production, as suggested by the model. Once the model has been validated by additional experiments, it can be used to investigate the impact of ΔΨ and NADH redox state on scavenging and ROS production, respectively.
This model, by extension of the R-ORB hypothesis, shows the importance of the interplay between ROS production and scavenging. The uncoupling protocols reproduced with the model (Fig. 7) show the effects of oxidizing the mitochondrial redox environment. ROS production decreases as NADH is oxidized and ΔΨ decreases, but net ROS levels begin to increase as GSH becomes oxidized. An interesting extension of this hypothesis would be to investigate changes in ROS production and scavenging that result from a more reduced redox environment, as might occur during inhibition with cyanide. Although the model presented here does not include a dynamically varying redox environment, preliminary studies show that, with ΔΨ and NADH redox potential clamped, application of submaximal cyanide block increases ROS production (not shown), in agreement with data from the literature, which also show reduced complex IV function and incomplete block of respiration (48,56,57). In contrast to the oxidation associated with uncoupling, the inhibition of respiration by cyanide will serve to reduce the electron carriers of the ETC. In the uncoupling protocol, oxidation of the redox environment depletes the mitochondrial scavenging capacity and leads to ROS overflow. Anoxia protocols would explore reduction of the redox environment, in which case the R-ORB hypothesis suggests that ROS production will play a larger role than scavenging in ROS overflow.
Mechanism of ROS production from complex I
Complex I ROS production was modeled as originating from a redox center in the hydrophilic arm of the protein. Without the explicit inclusion of complex I redox centers, this is a generalized implementation of the hypothesis, proposed by Turrens and Boveris (19) and further investigated in recent years by the Hirst group (14,15), that complex I ROS is produced from the FMN site. Under this assumption of complex I ROS production, the model can reproduce experimental data describing ROS production using complex I and complex II substrates under state 3 and state 4. In the absence of ubiquinone or ubiquinol, the model predicts a dependence of ROS production on NADH redox potential, closely matching experimental data from Pryde and Hirst (15) (see Fig. S1). In the work by those authors, it is noted that the midpoint potential of this NADH dependence closely matches the two-electron reduction potential of the complex I FMN. This serves as validation of the mechanism chosen in the model to represent complex I ROS production. Other groups (22,28,58) have proposed complex I’s quinone-binding site to be the primary source of ROS production in the absence of rotenone. The model described here was modified to test this Q-linked ROS production hypothesis by removing the superoxide-generating reaction connecting states 4 and 2 of the complex I model (see Fig. 1 B) and replacing it with a reaction connecting state 7 (reduced enzyme bound to Q) to state 2, such that the ROS production cycle of the revised model connects states 2, 3, 4, 7, and 2 again (see the Supporting Material for details). When the modified model was refitted to the ROS production data from Aon et al. (9), as for the original model, the ability of the Q-linked ROS production model to reproduce the data were very similar (not shown). None of the parameters for the Q-site ROS production model varied by more than 10 times the original model parameters, with the largest differences occurring in the rates for NADH and NAD+ binding. Given that the modeling results for the two different hypotheses were identical, this modeling experiment was unable to distinguish between the two mechanisms.
Mechanism of ROS production from complex III
The model presented here assumes that the mechanism of ROS production from complex III involves the reduction of oxygen by the cytosol-side semiquinone radical and is associated with a highly reduced ubiquinone pool and high proton motive force. Since the Demin et al. models (34,35) were developed, providing the basis for the model presented here, more recent experimental data have led to alternative proposals for complex-III-derived ROS production. Dröse and Brandt (59) found that in submitochondrial particles, ROS production is maximized when the Q pool is 25% oxidized, in contrast to the mechanism in the model presented here, which requires a highly reduced Q pool. These data led Dröse and Brandt to propose a mechanism of ROS production derived from RET from the reduced heme bL to oxidized ubiquinol to form a transient semiquinone, which reduces oxygen to form superoxide. Both this mechanism and the one used in the model depend on highly reduced heme bL derived from high ΔΨ. The difference arises from whether the electron that forms superoxide is released from the forward reaction from the QH2 bifurcation with FeS or in the reverse reaction from heme bL. Another recently proposed mechanism comes from Borek et al. (60), who used mutant, bacterial complex III proteins to study the impact of the position of the FeS head domain on ROS production. Their results show that ROS production occurs when the FeS head is in the distal position, when the diffusion of oxygen to a position near enough for electron transfer from the Qo site is not sterically hindered. Those authors also suggest that the highest ROS production rates may occur when a semiquinone is formed at the Qo site via RET from heme bL. Both of these mechanisms, in contrast to the mechanism used in this model, emphasize ROS production without a significant accumulation of semiquinone at the Qo site. All of the mechanisms described rely on reduced heme bL to produce ROS. However, the differences in the distribution of ubiquinone forms and origin of the electron used to generate the semiquinone at Qi may lead to different ΔΨ dependence of the electron carrier redox states. The ROS production mechanism of the model is subject to further refinements as consensus is achieved in the bioenergetics community with regard to the mechanism of ROS production in complex III.
Critique of the model
The model presented here was constructed with the goal of incorporating a high level of mechanistic detail while limiting the number of equations necessary. This will be an asset in future work, when the model of respiration is to be incorporated into a full description of mitochondrial dynamics and eventually further nested into a cardiac myocyte model. Such a multiscale model will require a conservative number of states and a limited range of timescales for computational efficiency.
The model presented here only accounts for superoxide release from the Qo site. Given that the difference in redox potential between the ubiquinone/semiquinone and semiquinone/ubiquinol couples is significantly smaller at the Qi site than at the Qo site (61), the semiquinone at Qi is considered thermodynamically stable (30,62). As such, we have not accounted for superoxide release from that radical. However, it is worth noting that the concentration of semiquinone at Qi does increase with membrane potential as well as matrix alkalinization (not shown), following a trend similar to that observed for the superoxide-producing semiquinone at Qo.
The complex IV model included here is a relatively simplistic description of an enzyme with a wide variety of properties, from allosteric ATP inhibition to variable Km for O2 to intrinsic uncoupling at high ΔΨm. A recent model by Krab et al. (63) incorporates more mechanistic detail to model the changing Km. This could be a potentially valuable addition to the model described here, facilitating investigation of ROS production in conditions of varying oxygen content, such as in a spatial model or under an ischemia-reperfusion protocol.
As described above, mitochondrial and cellular ROS levels are determined by both ROS production and ROS scavenging. Only the simulations of Fig. 7 include the contribution of mitochondrial ROS scavenging. As such, our model underreports the amount of ROS produced in most protocols. The most comprehensive estimates would require a full mitochondrial model that includes NADH and NADPH production, which is outside the scope of this work.
The mitochondrial ROS production and scavenging pathways represent a highly coupled and nonlinear system. The goal of this modeling effort was to reproduce key experimental protocols with a minimal model of mitochondrial regulation and control. Although the model’s ability to reproduce some of the experimental data falls short, as shown by the disparity in cytochrome c oxidation in Fig. 2 C, these disparities should not be seen as failures, but instead as clues to critical control mechanisms that are not represented in this version of the model.
Comparison to existing models
Several models reported in the literature describe the ETC. Some are detailed representations of electron transfer reactions (64–67), with the advantage of mechanistic accuracy at the cost of computational intensity. Others use a simpler thermodynamic approach (37,68) that requires fewer equations and state variables, but may capture less detail in the model results. For a full comparison of the work presented here with previously published modeling efforts, please refer to the Supporting Material.
Conclusion
A mechanistic approach to modeling the mitochondrial electron transport chain led to the construction of a model that displays a strong qualitative agreement with experimental data describing the control of respiration of ROS production and good quantitative agreement with many of those protocols. An additional asset of the model afforded by its mechanistic nature is the ability to reproduce experimental data describing the influence of inhibitors on ROS production. Furthermore, the model offers predictions regarding complex I ROS generation mechanisms and the interplay between ROS production and scavenging that together set mitochondrial and cellular ROS levels. This model has not only strong explanatory value but the ability to predict properties of the ETC in the protocols simulated in this study and potentially in further studies.
Acknowledgments
This work was supported by National Institutes of Health grants R21HL106054 (SC), R01HL052160 (RLW), R33HL87345 (RLW), and R37HL54598 (BO’R). LDG was supported by NDSEG and NSF graduate fellowships.
Supporting Material
References
- 1.Dröge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 2.Baines C.P., Goto M., Downey J.M. Oxygen radicals released during ischemic preconditioning contribute to cardioprotection in the rabbit myocardium. J. Mol. Cell. Cardiol. 1997;29:207–216. doi: 10.1006/jmcc.1996.0265. [DOI] [PubMed] [Google Scholar]
- 3.Petronilli V., Costantini P., Bernardi P. The voltage sensor of the mitochondrial permeability transition pore is tuned by the oxidation-reduction state of vicinal thiols. Increase of the gating potential by oxidants and its reversal by reducing agents. J. Biol. Chem. 1994;269:16638–16642. [PubMed] [Google Scholar]
- 4.Xie L.-H., Chen F., Weiss J.N. Oxidative-stress-induced afterdepolarizations and calmodulin kinase II signaling. Circ. Res. 2009;104:79–86. doi: 10.1161/CIRCRESAHA.108.183475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordano F.J. Oxygen, oxidative stress, hypoxia, and heart failure. J. Clin. Invest. 2005;115:500–508. doi: 10.1172/JCI200524408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura R., Egashira K., Takeshita A. Probucol attenuates left ventricular dysfunction and remodeling in tachycardia-induced heart failure: roles of oxidative stress and inflammation. Circulation. 2002;106:362–367. doi: 10.1161/01.cir.0000021430.04195.51. [DOI] [PubMed] [Google Scholar]
- 7.Murdoch C.E., Zhang M., Shah A.M. NADPH oxidase-dependent redox signalling in cardiac hypertrophy, remodelling and failure. Cardiovasc. Res. 2006;71:208–215. doi: 10.1016/j.cardiores.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Ide T., Tsutsui H., Takeshita A. Mitochondrial electron transport complex I is a potential source of oxygen free radicals in the failing myocardium. Circ. Res. 1999;85:357–363. doi: 10.1161/01.res.85.4.357. [DOI] [PubMed] [Google Scholar]
- 9.Aon M.A., Cortassa S., O'Rourke B. Redox-optimized ROS balance: a unifying hypothesis. Biochim. Biophys. Acta. 2010;1797:865–877. doi: 10.1016/j.bbabio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T., O’Rourke B. Enhancing mitochondrial Ca2+ uptake in myocytes from failing hearts restores energy supply and demand matching. Circ. Res. 2008;103:279–288. doi: 10.1161/CIRCRESAHA.108.175919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohlhaas M., Liu T., Maack C. Elevated cytosolic Na+ increases mitochondrial formation of reactive oxygen species in failing cardiac myocytes. Circulation. 2010;121:1606–1613. doi: 10.1161/CIRCULATIONAHA.109.914911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guzy R.D., Hoyos B., Schumacker P.T. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Kushnareva Y., Murphy A.N., Andreyev A. Complex I-mediated reactive oxygen species generation: modulation by cytochrome c and NAD(P)+ oxidation-reduction state. Biochem. J. 2002;368:545–553. doi: 10.1042/BJ20021121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kussmaul L., Hirst J. The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proc. Natl. Acad. Sci. USA. 2006;103:7607–7612. doi: 10.1073/pnas.0510977103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pryde K.R., Hirst J. Superoxide is produced by the reduced flavin in mitochondrial complex I: a single, unified mechanism that applies during both forward and reverse electron transfer. J. Biol. Chem. 2011;286:18056–18065. doi: 10.1074/jbc.M110.186841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grivennikova V.G., Vinogradov A.D. Generation of superoxide by the mitochondrial Complex I. Biochim. Biophys. Acta. 2006;1757:553–561. doi: 10.1016/j.bbabio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Rottenberg H., Covian R., Trumpower B.L. Membrane potential greatly enhances superoxide generation by the cytochrome bc1 complex reconstituted into phospholipid vesicles. J. Biol. Chem. 2009;284:19203–19210. doi: 10.1074/jbc.M109.017376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korshunov S.S., Skulachev V.P., Starkov A.A. High protonic potential actuates a mechanism of production of reactive oxygen species in mitochondria. FEBS Lett. 1997;416:15–18. doi: 10.1016/s0014-5793(97)01159-9. [DOI] [PubMed] [Google Scholar]
- 19.Turrens J.F., Boveris A. Generation of superoxide anion by the NADH dehydrogenase of bovine heart mitochondria. Biochem. J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.St-Pierre J., Buckingham J.A., Brand M.D. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- 21.Zoccarato F., Cavallini L., Alexandre A. Succinate is the controller of O2−/H2O2 release at mitochondrial complex I : negative modulation by malate, positive by cyanide. J. Bioenerg. Biomembr. 2009;41:387–393. doi: 10.1007/s10863-009-9238-2. [DOI] [PubMed] [Google Scholar]
- 22.Lambert A.J., Brand M.D. Superoxide production by NADH:ubiquinone oxidoreductase (complex I) depends on the pH gradient across the mitochondrial inner membrane. Biochem. J. 2004;382:511–517. doi: 10.1042/BJ20040485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carroll J., Fearnley I.M., Walker J.E. Bovine complex I is a complex of 45 different subunits. J. Biol. Chem. 2006;281:32724–32727. doi: 10.1074/jbc.M607135200. [DOI] [PubMed] [Google Scholar]
- 24.Efremov R.G., Sazanov L.A. Structure of the membrane domain of respiratory complex I. Nature. 2011;476:414–420. doi: 10.1038/nature10330. [DOI] [PubMed] [Google Scholar]
- 25.Mourier A., Larsson N.-G. Tracing the trail of protons through complex I of the mitochondrial respiratory chain. PLoS Biol. 2011;9:e1001129. doi: 10.1371/journal.pbio.1001129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandt U. A two-state stabilization-change mechanism for proton-pumping complex I. Biochim. Biophys. Acta. 2011;1807:1364–1369. doi: 10.1016/j.bbabio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Dröse S., Brandt U. Molecular mechanisms of superoxide production by the mitochondrial respiratory chain. In: Kadenbach B., editor. Mitochondrial Oxidative Phosphorylation. Springer; New York: 2012. pp. 145–169. [DOI] [PubMed] [Google Scholar]
- 28.Lambert A.J., Brand M.D. Inhibitors of the quinone-binding site allow rapid superoxide production from mitochondrial NADH:ubiquinone oxidoreductase (complex I) J. Biol. Chem. 2004;279:39414–39420. doi: 10.1074/jbc.M406576200. [DOI] [PubMed] [Google Scholar]
- 29.Cape J.L., Bowman M.K., Kramer D.M. A semiquinone intermediate generated at the Qo site of the cytochrome bc1 complex: importance for the Q-cycle and superoxide production. Proc. Natl. Acad. Sci. USA. 2007;104:7887–7892. doi: 10.1073/pnas.0702621104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H., Osyczka A., Moser C.C. Exposing the complex III Qo semiquinone radical. Biochim. Biophys. Acta. 2007;1767:883–887. doi: 10.1016/j.bbabio.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller F.L., Liu Y., Van Remmen H. Complex III releases superoxide to both sides of the inner mitochondrial membrane. J. Biol. Chem. 2004;279:49064–49073. doi: 10.1074/jbc.M407715200. [DOI] [PubMed] [Google Scholar]
- 32.Han D., Antunes F., Cadenas E. Voltage-dependent anion channels control the release of the superoxide anion from mitochondria to cytosol. J. Biol. Chem. 2003;278:5557–5563. doi: 10.1074/jbc.M210269200. [DOI] [PubMed] [Google Scholar]
- 33.Aon M.A., Stanley B.A., Cortassa S. Glutathione/thioredoxin systems modulate mitochondrial H2O2 emission: an experimental-computational study. J. Gen. Physiol. 2012;139:479–491. doi: 10.1085/jgp.201210772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demin O.V., Goryanin I.I., Westerhoff H.V. Kinetic modeling of energy metabolism and superoxide generation in hepatocyte mitochondria. Mol. Biol. (Mosk.) 2001;35:940–949. [PubMed] [Google Scholar]
- 35.Demin O.V., Kholodenko B.N., Skulachev V.P. A model of O2.− generation in the complex III of the electron transport chain. Mol. Cell. Biochem. 1998;184:21–33. [PubMed] [Google Scholar]
- 36.Kim N., Ripple M.O., Springett R. Measurement of the mitochondrial membrane potential and pH gradient from the redox poise of the hemes of the bc1 complex. Biophys. J. 2012;102:1194–1203. doi: 10.1016/j.bpj.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magnus G., Keizer J. Minimal model of β-cell mitochondrial Ca2+ handling. Am. J. Physiol. 1997;273:C717–C733. doi: 10.1152/ajpcell.1997.273.2.C717. [DOI] [PubMed] [Google Scholar]
- 38.Erecińska M., Wilson D.F. Regulation of cellular energy metabolism. J. Membr. Biol. 1982;70:1–14. doi: 10.1007/BF01871584. [DOI] [PubMed] [Google Scholar]
- 39.Sato K., Kashiwaya Y., Veech R.L. Insulin, ketone bodies, and mitochondrial energy transduction. FASEB J. 1995;9:651–658. doi: 10.1096/fasebj.9.8.7768357. [DOI] [PubMed] [Google Scholar]
- 40.Mintz S., Robin E.D. Redox state of free nicotinamide-adenine nucleotides in the cytoplasm and mitochondria of alveolar macrophages. J. Clin. Invest. 1971;50:1181–1186. doi: 10.1172/JCI106595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown G.C., Brand M.D. Thermodynamic control of electron flux through mitochondrial cytochrome bc1 complex. Biochem. J. 1985;225:399–405. doi: 10.1042/bj2250399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kadenbach B. Intrinsic and extrinsic uncoupling of oxidative phosphorylation. Biochim. Biophys. Acta. 2003;1604:77–94. doi: 10.1016/s0005-2728(03)00027-6. [DOI] [PubMed] [Google Scholar]
- 43.Kadenbach B., Ramzan R., Vogt S. The role of mitochondrial membrane potential in ischemic heart failure. Mitochondrion. 2011;11:700–706. doi: 10.1016/j.mito.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Cramer W.A., Knaff D. Springer-Verlag; New York: 1991. Energy Transduction in Biological Membranes. A Textbook of Bioenergetics. [Google Scholar]
- 45.Randle P., Tubbs P. Carbohydrate and fatty acid metabolism. In: Berne R.M., Sperelakis N., Geiger R., editors. Handbook of Physiology. American Physiological Society; Bethesda, MD: 1979. pp. 805–844. [Google Scholar]
- 46.Adam-Vizi V., Chinopoulos C. Bioenergetics and the formation of mitochondrial reactive oxygen species. Trends Pharmacol. Sci. 2006;27:639–645. doi: 10.1016/j.tips.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 47.Liu S.S. Cooperation of a “reactive oxygen cycle” with the Q cycle and the proton cycle in the respiratory chain—superoxide generating and cycling mechanisms in mitochondria. J. Bioenerg. Biomembr. 1999;31:367–376. doi: 10.1023/a:1018650103259. [DOI] [PubMed] [Google Scholar]
- 48.Turrens J.F., Alexandre A., Lehninger A.L. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 49.Wei A.-C., Aon M.A., Cortassa S. Mitochondrial energetics, pH regulation, and ion dynamics: a computational-experimental approach. Biophys. J. 2011;100:2894–2903. doi: 10.1016/j.bpj.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selivanov V.A., Zeak J.A., Votyakova T.V. The role of external and matrix pH in mitochondrial reactive oxygen species generation. J. Biol. Chem. 2008;283:29292–29300. doi: 10.1074/jbc.M801019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cadenas E., Boveris A. Enhancement of hydrogen peroxide formation by protophores and ionophores in antimycin-supplemented mitochondria. Biochem. J. 1980;188:31–37. doi: 10.1042/bj1880031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reference deleted in proof.
- 53.Sled V.D., Rudnitzky N.I., Ohnishi T. Thermodynamic analysis of flavin in mitochondrial NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 1994;33:10069–10075. doi: 10.1021/bi00199a034. [DOI] [PubMed] [Google Scholar]
- 54.Cortassa S., Aon M.A., Winslow R.L. A computational model integrating electrophysiology, contraction, and mitochondrial bioenergetics in the ventricular myocyte. Biophys. J. 2006;91:1564–1589. doi: 10.1529/biophysj.105.076174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schafer F.Q., Buettner G.R. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic. Biol. Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 56.Dawson T.L., Gores G.J., Lemasters J.J. Mitochondria as a source of reactive oxygen species during reductive stress in rat hepatocytes. Am. J. Physiol. 1993;264:C961–C967. doi: 10.1152/ajpcell.1993.264.4.C961. [DOI] [PubMed] [Google Scholar]
- 57.Zuckerbraun B.S., Chin B.Y., Otterbein L.E. Carbon monoxide signals via inhibition of cytochrome c oxidase and generation of mitochondrial reactive oxygen species. FASEB J. 2007;21:1099–1106. doi: 10.1096/fj.06-6644com. [DOI] [PubMed] [Google Scholar]
- 58.Turrens J.F. Mitochondrial formation of reactive oxygen species. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dröse S., Brandt U. The mechanism of mitochondrial superoxide production by the cytochrome bc1 complex. J. Biol. Chem. 2008;283:21649–21654. doi: 10.1074/jbc.M803236200. [DOI] [PubMed] [Google Scholar]
- 60.Borek A., Sarewicz M., Osyczka A. Movement of the iron-sulfur head domain of cytochrome bc1 transiently opens the catalytic Qo site for reaction with oxygen. Biochemistry. 2008;47:12365–12370. doi: 10.1021/bi801207f. [DOI] [PubMed] [Google Scholar]
- 61.Zhang H., Chobot S.E., Moser C.C. Quinone and non-quinone redox couples in Complex III. J. Bioenerg. Biomembr. 2008;40:493–499. doi: 10.1007/s10863-008-9174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meinhardt S.W., Ohnishi T. Determination of the position of the Qi.- quinone binding site from the protein surface of the cytochrome bc1 complex in Rhodobacter capsulates chromatophores. Biochim. Biophys. Acta. 1992;1100:67–74. doi: 10.1016/0005-2728(92)90127-n. [DOI] [PubMed] [Google Scholar]
- 63.Krab K., Kempe H., Wikström M. Explaining the enigmatic KM for oxygen in cytochrome c oxidase: a kinetic model. Biochim. Biophys. Acta. 2011;1807:348–358. doi: 10.1016/j.bbabio.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 64.Orii Y., Miki T. Oxidation process of bovine heart ubiquinol-cytochrome c reductase as studied by stopped-flow rapid-scan spectrophotometry and simulations based on the mechanistic Q cycle model. J. Biol. Chem. 1997;272:17594–17604. doi: 10.1074/jbc.272.28.17594. [DOI] [PubMed] [Google Scholar]
- 65.Selivanov V.A., Cascante M., Votyakova T.V. Multistationary and oscillatory modes of free radicals generation by the mitochondrial respiratory chain revealed by a bifurcation analysis. PLOS Comput. Biol. 2012;8:e1002700. doi: 10.1371/journal.pcbi.1002700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Selivanov V.A., Votyakova T.V., Cascante M. Reactive oxygen species production by forward and reverse electron fluxes in the mitochondrial respiratory chain. PLOS Comput. Biol. 2011;7:e1001115. doi: 10.1371/journal.pcbi.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Selivanov V.A., Votyakova T.V., Cascante M. Bistability of mitochondrial respiration underlies paradoxical reactive oxygen species generation induced by anoxia. PLOS Comput. Biol. 2009;5:e1000619. doi: 10.1371/journal.pcbi.1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin Q., Bethke C.M. Kinetics of electron transfer through the respiratory chain. Biophys. J. 2002;83:1797–1808. doi: 10.1016/S0006-3495(02)73945-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cortassa S., Aon M.A., O’Rourke B. A mitochondrial oscillator dependent on reactive oxygen species. Biophys. J. 2004;87:2060–2073. doi: 10.1529/biophysj.104.041749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cortassa S., Aon M.A., O’Rourke B. An integrated model of cardiac mitochondrial energy metabolism and calcium dynamics. Biophys. J. 2003;84:2734–2755. doi: 10.1016/S0006-3495(03)75079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lenaz G., Genova M.L. Kinetics of integrated electron transfer in the mitochondrial respiratory chain: random collisions vs. solid state electron channeling. American Journal of Physiology - Cell Physiology. 2007;292:C1221–C1239. doi: 10.1152/ajpcell.00263.2006. [DOI] [PubMed] [Google Scholar]
- 72.Zhou L., Cortassa S., O’Rourke B. Modeling cardiac action potential shortening driven by oxidative stress-induced mitochondrial oscillations in guinea pig cardiomyocytes. Biophys. J. 2009;97:1843–1852. doi: 10.1016/j.bpj.2009.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X., Qi F., Beard D.A. Kinetics and regulation of mammalian NADH-ubiquinone oxidoreductase (Complex I) Biophys. J. 2010;99:1426–1436. doi: 10.1016/j.bpj.2010.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Genova M.L., Ventura B., Lenaz G. The site of production of superoxide radical in mitochondrial Complex I is not a bound ubisemiquinone but presumably iron-sulfur cluster N2. FEBS Lett. 2001;505:364–368. doi: 10.1016/s0014-5793(01)02850-2. [DOI] [PubMed] [Google Scholar]
- 75.Qi F., Dash R.K., Beard D.A. Generating rate equations for complex enzyme systems by a computer-assisted systematic method. BMC Bioinformatics. 2009;10:238. doi: 10.1186/1471-2105-10-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Muraoka S., Slater E.C. The redox states of respiratory-chain components in rat-liver mitochondria II. The “crossover” on the transition from State 3 to State 4. Biochim. Biophys. Acta. 1969;180:227–236. doi: 10.1016/0005-2728(69)90109-1. [DOI] [PubMed] [Google Scholar]
- 77.Brzezinski P., Ädelroth P. Pathways of proton transfer in cytochrome c oxidase. J. Bioenerg. Biomembr. 1998;30:99–107. doi: 10.1023/a:1020567729941. [DOI] [PubMed] [Google Scholar]
- 78.Hafner R.P., Brown G.C., Brand M.D. Analysis of the control of respiration rate, phosphorylation rate, proton leak rate and protonmotive force in isolated mitochondria using the “top-down” approach of metabolic control theory. Eur. J. Biochem. 1990;188:313–319. doi: 10.1111/j.1432-1033.1990.tb15405.x. [DOI] [PubMed] [Google Scholar]
- 79.Lionetti L., Iossa S., Liverini G. Relationship between membrane potential and respiration rate in isolated liver mitochondria from rats fed an energy dense diet. Mol. Cell. Biochem. 1996;158:133–138. doi: 10.1007/BF00225839. [DOI] [PubMed] [Google Scholar]
- 80.Marcinkeviciute A., Mildaziene V., Kholodenko B. Kinetics and control of oxidative phosphorylation in rat liver mitochondria after chronic ethanol feeding. Biochem. J. 2000;349:519–526. doi: 10.1042/0264-6021:3490519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moser C.C., Farid T.A., Dutton P.L. Electron tunneling chains of mitochondria. Biochim. Biophys. Acta. 2006;1757:1096–1109. doi: 10.1016/j.bbabio.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 82.Starkov A.A., Fiskum G. Myxothiazol induces H(2)O(2) production from mitochondrial respiratory chain. Biochem. Biophys. Res. Commun. 2001;281:645–650. doi: 10.1006/bbrc.2001.4409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


