Abstract
Pertussis toxin is transported across the outer membrane of Bordetella pertussis by the type IV secretion system known as the Ptl transporter, which is composed of nine different proteins. In order to determine the relative levels of production of pertussis toxin subunits and Ptl proteins in B. pertussis, we constructed translational fusions of the gene for alkaline phosphatase, phoA, with various ptx and ptl genes. Comparison of the alkaline phosphatase activity of strains containing ptx′- or ptl′-phoA fusions indicated that pertussis toxin subunits are produced at higher levels than Ptl proteins, which are encoded by genes located toward the 3′ end of the ptx-ptl operon. We also engineered strains of B. pertussis by introducing multiple copies of the ptl genes or subsets of these genes and then examined the ability of each of these strains to secrete pertussis toxin. From these studies, we determined that certain Ptl proteins appear to be limiting in the secretion of pertussis toxin from the bacteria. These results represent an important first step in assessing the stoichiometric relationship of pertussis toxin and its transporter within the bacterial cell.
Pertussis toxin (PT) is an important virulence factor produced by Bordetella pertussis, the causative agent of the disease pertussis (whooping cough). The toxin is composed of five different subunits, S1, S2, S3, S4, and S5, that are found in a 1:1:1:2:1 ratio (32). The S1 subunit is enzymatically active and sits atop a ring formed by the remaining subunits, which are collectively known as the B oligomer (29).
PT exits the bacterial cell by a complex pathway which is not completely understood. Each of the toxin subunits is believed to be individually secreted across the inner membrane by a Sec-like system. After the subunits have traversed the inner membrane, they assemble to form the holotoxin, which is then transported across the outer membrane by an apparatus known as the Ptl transporter, comprising the nine proteins PtlA to PtlI (5, 8, 33). The ptl genes encoding these proteins are located on the bacterial chromosome directly downstream from the ptx genes that encode the toxin structural subunits (33). The ptx and ptl genes are cotranscribed (1, 17).
The Ptl apparatus is a member of the type IV family of transporters. Other members of this family include the VirB systems of Agrobacterium tumefaciens, Brucella spp., and Bartonella henselae, which are involved in the transport of pathogenic factors across the outer membranes of these gram-negative bacteria (4, 22, 23, 27). Studies from a number of laboratories indicate that the VirB proteins of A. tumefaciens form a structure that spans a region from the inner surface of the inner membrane through the periplasmic space to the outer membrane, and finally extending outward from the surface of the bacterial cell (6, 11, 14, 18, 25, 26). The striking homologies between the VirB proteins of A. tumefaciens and the Ptl proteins suggest that the Ptl apparatus likely has a similar architecture.
The mechanistic details of PT secretion remain elusive, including the specific architecture of the Ptl transport apparatus and the mode of interaction of the toxin with the Ptl proteins. Also unknown are the relative amounts of PT subunits and Ptl proteins produced within the cell. Knowledge of the ratio of PT to the Ptl proteins produced within the cell would contribute to our general understanding of the mechanism of secretion of the toxin from the cell.
Since the ptx and ptl genes are located within the same operon and transcription is controlled by a single promoter (1, 17), the possibility exists that PT subunits and Ptl proteins might be produced in comparable amounts. However, differential transcription and/or translation of regions of the ptx-ptl operon or differential stability of the various Ptx and Ptl proteins could result in markedly different levels of PT and its transporter.
In order to begin to understand the stoichiometry of PT and its transporter, we have constructed strains containing translational fusions consisting of phoA, devoid of its initiation codon, fused to the initiation codons of various ptx and ptl genes. By comparing the alkaline phosphatase activities of these strains, we estimated the relative levels of production of the PT subunits and Ptl proteins encoded by these ptx and ptl genes. In addition, we examined the effects of overexpression of the ptl genes, as well as subsets of these genes, on the secretion of the toxin from the bacterial cell. We found that genes located at the 5′ end of the operon are transcribed and/or translated at levels higher than those of genes located at the 3′ end of the operon. Moreover, we found that the Ptl proteins are produced in amounts that are limiting for the secretion of the toxin from the bacterium.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The strains of B. pertussis and Escherichia coli and the plasmids used in this study are listed in Table 1. B. pertussis strains were grown at 37°C on Bordet-Gengou (BG) agar or in Stainer-Scholte liquid medium. For liquid cultures, cells that had been grown on BG agar plates were suspended in medium to yield an A550 of 0.2 to 0.5. The strains were then grown in shaking culture for 48 h to an A550 of 1.2 to 1.5.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | F− ϕ80d lacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44λ−thi-1 gyrA96 relA1 | GIBCO BRL |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpirR6K | 28 |
| B. pertussis | ||
| BP536 | Wild-type, nalidixic acid-resistant, streptomycin-resistant derivative of Tohama I | 31 |
| BP536Δptxptl936-8003 | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 936 to 8003 | This study |
| BP536Δptxptl936-3941 | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 936 to 3941 | This study |
| BP536Δptxptl3822-8003 | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 3822 to 8003 | This study |
| BP536::pAMC48 | BP536 containing a ptxS1′-phoA fusion | This study |
| BP536::pAMC17 | BP536 containing a ptxS2′-phoA fusion | This study |
| BP536::pAMC67 | BP536 containing a ptlA′-phoA fusion | This study |
| BP536::pAMC107 | BP536 containing a ptlB′-phoA fusion | This study |
| BP536::pAMC121 | BP536 containing a ptlD′-phoA fusion | This study |
| BP536::pAMC28 | BP536 containing a ptlF′-phoA fusion | This study |
| BP536Δptxptl936-8003::pAMC28 | BP536Δptxptl936-8003::pAMC28 containing a ptlF′-phoA fusion | This study |
| BP536Δptxptl936-3941::pAMC28 | BP536Δptxptl936-3941::pAMC28 containing a ptlF′-phoA fusion | This study |
| BP536Δptxptl3821-8003::pAMC28 | BP536Δptxptl3821-8003::pAMC28 containing a ptlF′-phoA fusion | This study |
| Plasmids | ||
| pUFR047 | Broad-host-range (IncW) vector; Mob+lacZα+; gentamicin-resistant | 7 |
| pGEM11Zf(+) | Ampicillin-resistant (Ampr) cloning vector | Promega |
| pALTER-1 | Tetr cloning vector | Promega |
| pUC19 | Ampr cloning vector | GIBCO BRL |
| pZErO-1 | Zeocin-resistant cloning vector | Invitrogen |
| pSZH4 | pUC19 containing the entire ptx-ptl region | 12 |
| pSS1129 | Genr Ampr Sms allelic exhange vector | 30 |
| pAMC48 | pSS1129 containing nucleotides 1 to 509 of the ptx-ptl region followed by phoA devoid of its initiation codon | This study |
| pAMC17 | pSS1129 containing nucleotides 1 to 1358 of the ptx-ptl region followed by phoA devoid of its initiation codon | This study |
| pAMC67 | pSS1129 containing nucleotides 1312 to 3686 of the ptx-ptl region followed by phoA devoid of its initiation codon | This study |
| pAMC107 | pSS1129 containing nucleotides 2831 to 4013 of the ptx-ptl region followed by phoA devoid of its initiation codon | This study |
| pAMC121 | pSS1129 containing nucleotides 5738 to 6806 of the ptx-ptl region followed by phoA devoid of its initiation codon | This study |
| pAMC28 | pSS1129 containing nucleotides 5007 to 9056 of the ptx-ptl region followed by phoA devoid of its initiation codon | This study |
| pAMC111 | pUFR047 containing the nine ptl genes under the control of the ptx-ptl promoter | This study |
| pAMC147 | pUFR047 containing ptlA, ptlB, and ptlC under the control of the ptx-ptl promoter | This study |
| pTC11 | pUFR047 containing ptlD, ptlI, ptlE, and ptlF under the control of the lac promoter | This study |
| pAMC151 | pUFR047 containing ptlI-ptlH under the control of the lac promoter | This study |
| pAMC171 | pUFR047 containing ptlA under the control of the ptx-ptl promoter | This study |
| pAMC176 | pUFR047 containing ptlB and ptlC under the control of the ptx-ptl promoter | This study |
Construction of in-frame deletions. (i) In-frame deletion from ptxS1 to ptlD.
A 7.1-kb segment of the ptx-ptl operon (diagramed in Fig. 1A), extending from nucleotide 936 through nucleotide 8003 (according to the numbering system described in references 21 and 33), was deleted from the B. pertussis chromosome as follows. Plasmid pTH14 [pGEM11Zf(+) containing nucleotides 1 to 935 of the ptx-ptl region followed by nucleotides 3515 to 4574 of the same region], which has been described previously (9), was digested with EcoRI and SalI to yield a 0.9-kb fragment. Using PCR essentially as previously described (13), we amplified nucleotides 8004 to 8965 of the ptl region by using primers 1a and 1b. (All primers used in PCR are listed in Table 2). The 0.9-kb fragment from pTH14 and the amplified DNA fragment were inserted into pSS1129. The resulting plasmid was transformed into E. coli SM10 and transferred into B. pertussis BP536 by conjugation as described previously (8). The BP536 cells in which pAMC159 had integrated into the chromosome by homologous recombination were selected on BG agar containing gentamicin (10 μg/ml) and nalidixic acid (50 μg/ml). A second homologous recombination was achieved by selection on BG agar containing streptomycin (100 μg/ml), resulting in loss of the plasmid as previously described (8) to yield BP536Δptxptl936-8003. The 7.1-kb deletion in the ptx-ptl region was verified by PCR.
FIG. 1.
In-frame deletions constructed in the ptx-ptl region. (A) The entire ptx-ptl region is depicted along with a diagram of the ptx-ptl region containing an in-frame deletion from nucleotide 936 to nucleotide 8003. The nucleotide numbering system of the ptx-ptl region has been described previously (21, 33). (B) In-frame deletions of the ptx-ptl region in which the phoA gene devoid of its initiation codon was fused to the ATG initiation codon of ptlF.
TABLE 2.
Primers used in this study
| Primer | Primer sequencea | Relevant characteristics |
|---|---|---|
| 1a | 5′-CTCCAGGTCGACGCCGGTACGTACGCAGCCTCGGC | SalI site followed by nucleotides 8004-8026 |
| 1b | 5′-CTCCAGAAGCTTGCTGATCGTAGGCGAATGCCACGG | HindIII site followed by nucleotides 8965-8942 |
| 2a | 5′-CTCCAGGTCGACGGACTGCTGATCGGCGCATCGGCC | SalI site followed by nucleotides 3842-3965 |
| 2b | 5′-CTCCAGAAGCTTGGTGAGCAGGAATTCGAGCAGGGTC | HindIII site followed by nucleotides 4945-4921 |
| 3a | 5′-CTCCAGGAATTCGCCAGGCATCGTCATCCCGCCGAAGG | EcoRI site followed by nucleotides 3034-3059 |
| 3b | 5′-CTCCAGGTCGACGCTCGCCATGAAGTGGTTGACGCG | SalI site followed by nucleotides 3821-3798 |
| 4a | 5′-CTCCAGGTCGACGCCGGTACGTACGCAGCCTCGGC | SalI site followed by nucleotides 8004-8026 |
| 4b | 5′-CTCCAGAAGCTTGCTGATCGTAGGCGAATGCCACGG | HindIII site followed by nucleotides 8965-8942 |
| 5a | 5′-CTCCAGTCTAGAGAAGACGGGATGAAACAAAGCACTATTGCACTGG | XbaI site followed by nucleotides 498-509 and first 22 nucleotides of phoA |
| 5b | 5′-CTCCAGAAGCTTTTATTTCAGCCCCAGAGCGGCTTT | HindIII followed by the last 24 nucleotides of phoA |
| 6a | 5′-CTCCAGTCTAGACCTGGCCCAGCCCCGCCCAACTCCGGTAATTGAACAG CATGAAACAAAGCACTATTGCACTGG | Nucleotides 1312-1358 followed by the first 22 nucleotides of phoA |
| 7a | 5′-CTCCAGTCTAGAGCTGAGCCGCCGGCTCGGATCTGTTCGCCTGTCCATGT TTTTCCTTGACGGATACCGCGAATGAAACAAAGCACTATTGCACTGG | XbaI site followed by nucleotides 3624-3686 and the first 22 nucleotides of phoA |
| 7b | 5′-CTCCAGAAGCTTGGATCCTTATTTCAGCCCCAGAGCGGCTTTCATG | HindIII site, a BamHI site, the last 24 nucleotides of phoA |
| 8a | 5′-CAGTCTAGACGGCCGAAATCGCTCGTTATCTGCTGACCTGAATCCTGGACG TATCGAACATGAAACAAAGCACTATTGCACTGG | XbaI site followed by nucleotides 3961-4013 and the first 22 nucleotides of phoA |
| 9a | 5′-CTCCAGCCTCAGGTGAAACCAGCATGAAACAAAGCACTATTGCACTGG | Nucleotides 6788-6806 followed by the first 22 nucleotides of phoA |
| 10a | 5′-CTCCAGGTCGACGCGGAGGCTGGACAGCCATGAAACAAAGCACTATTGCACTGG | Nucleotides 9031-9056 followed by the first 22 nucleotides of phoA |
| 11a | 5′-CGCGAATTCTAATTAATTAAAGGAAACAGCGATGGCCGGCCTGTCACGAATCCTGCTG | EcoRI site, stop codons in three reading frames, ribosomal binding site, and nucleotides 6804-6830 |
| 11b | 5′-CGCTCTAGATCATGGCAGGTTGGGTTTGCGGGGAAGAACCCG | XbaI site followed by nucleotides 8195-8163 |
| 12a | 5′-CTGGCGGCCGCCGCCGCGGGACCGCACCAGGCCGGTACG | Nucleotides 7977-8012 |
| 12b | 5′-CGCAAGCTTCTAGAGGGCCGGTTCAGCATGGCATTC | HindIII followed by nucleotides 9875-9849 |
| 13a | 5′-CGCTCTAGATAATTAATTAAAGGAAACAGCGATGGCGGCCGCCGCCGCGGGACCGCAC | XbaI site, stop codons in three reading frames, ribosomal binding site, and nucleotides 7977-8000 |
| 13b | 5′-CAGAAGCTTGTGACGACCGAGGCCGCGCCGCTGCTCTTTTCG | HindIII site followed by nucleotides 8606-8574 |
| 14a | 5′-CTCCAGGTCGACGTACGCGTCCACGTCAGCAAGGAAG | SalI site followed by nucleotides 3515-3539 |
| 14b | 5′-CTCCAGAAGCTTTCAGGTCAGCAGATAACGAGCGATTTC | HindIII site followed by nucleotides 3992-3966 |
| 15a | 5′-CTCCAGGTCGACGGACTGCTGATCGGCGCATCGGCCGAAATC | SalI site followed by nucleotides 3842-3971 |
| 15b | 5′-CGAGATGGCGCGCCACAGGCCGTTGAGCTG | Nucleotides 4549-4520 containing an AscI site |
Underlined sequences are restriction enzyme sites described under “Relevant characteristics.”
(ii) In-frame deletion from ptxS1 to ptlA.
A 3.0-kb segment of the ptx-ptl region, extending from nucleotide 936 through nucleotide 3941, was deleted from the B. pertussis chromosome as follows. Plasmid pTH14 was digested with EcoRI and SalI to yield a 0.9-kb fragment. Using PCR, we amplified nucleotides 3942 to 4945 of the ptx-ptl region with primers 2a and 2b. The 0.9-kb fragment from pTH14 and the amplified DNA fragment containing nucleotides 3942 to 4945 were inserted into pSS1129. The resulting plasmid was transferred into B. pertussis BP536 and used to create the in-frame deletion as described above.
(iii) In-frame deletion from ptlA to ptlD.
A 4.2-kb segment of the ptx-ptl region, extending from nucleotide 3822 through nucleotide 8003, was deleted from the B. pertussis chromosome as follows. First, we PCR amplified a DNA fragment containing nucleotides 3034 to 3821 of the ptl region by using primers 3a and 3b. We then amplified a second DNA fragment containing nucleotides 8004 to 8965 of the ptl region by using primers 4a and 4b. Both segments were inserted into pSS1129. The resulting plasmid was transferred into B. pertussis BP536 and used to create the in-frame deletion as described above.
Construction of phoA fusions. (i) ptxS1′-phoA fusion.
A DNA fragment which included nucleotides 498 to 509 of the ptx region followed by phoA devoid of its initiation codon was generated by using primers 5a and 5b. Chromosomal DNA derived from E. coli HB101 was used as the template for generation of phoA. To minimize the possibility of PCR-generated errors in this sequence (and all other PCR-generated phoA sequences described in this study), a 1.2-kb BsgI-SphI fragment of the amplified DNA was replaced with the corresponding BsgI-SphI fragment from plasmid p2959, which contains phoA (kindly provided by Scott Stibitz, Food and Drug Administration). The phoA gene contained in p2959 has silent point mutations in the two EcoRI sites of phoA engineered into it, resulting in a change of codon 266 from AAT to AAC and of codon 376 from GAA to GAG, but retains the ability to express active alkaline phosphatase. The 5′ and 3′ ends of the DNA fragment that were not derived from p2929 were sequenced.
pSZH4, a plasmid containing the entire ptx-ptl region from B. pertussis (12), was digested with EcoRI and XbaI, and the 1.3-kb fragment that was generated was inserted into pUC19. The resulting plasmid was digested with BbsI and HindIII, and the PCR-derived fragment containing ptxS1′-phoA, generated as described above, was inserted. The resulting plasmid was digested with EcoRI and HindIII, and the 1.8-kb fragment was inserted into pSS1129 to generate pAMC48, which was then transformed into E. coli SM10 and transferred into B. pertussis by conjugation as described previously (30). Exconjugants, in which pAMC48 had integrated into the chromosome by homologous recombination, were selected on BG agar containing gentamicin (10 μg/ml) and nalidixic acid (50 μg/ml).
(ii) ptxS2′-phoA fusion.
Primers 6a and 5b were used to generate a DNA fragment by PCR. pSZH4 was then digested with XbaI and HindIII to yield a 4.0-kb fragment containing the vector and nucleotides 1 to 1317 of the ptx-ptl region. The PCR fragment containing the ptxS2′-phoA fusion was ligated with the digested plasmid. The EcoRI-HindIII fragment from the resultant plasmid was then inserted into pSS1129, generating pAMC17, which was then integrated into the chromosome of B. pertussis BP536 as described above.
(iii) ptlA′-phoA fusion.
A DNA fragment was generated by using primers 7a and 7b. Next, pSZH4 was digested with XbaI and BamHI, and the resulting 3.4-kb fragment was inserted into the pGEM7Zf(+) vector. The resulting plasmid, pAMC35, was then ligated to the PCR fragment containing the ptlA′-phoA fusion which had been digested with BlpI and BamHI. This plasmid was digested with XbaI and BamHI, and the resulting 3.8-kb fragment was inserted into pUC19. This plasmid was then digested with BamHI and HindIII, and the 3.8-kb fragment was inserted into pSS1129, generating pAMC67, which was then integrated into the chromosome of B. pertussis BP536.
(iv) ptlB′-phoA fusion.
An in-frame gene fusion between ptlB and phoA was constructed as follows. A DNA fragment was generated by using primers 8a and 7b. pAMC35, described above, was digested with BglII and BamHI, and the resulting 1.7-kb fragment was inserted into the BamHI site of the pGEM3Zf(+) vector. This plasmid was then digested with EagI and BamHI and ligated to the PCR fragment containing ptlB′-phoA. The resultant plasmid was digested with EcoRI and HindIII, and the 2.6-kb fragment was inserted into pSS1129, generating pAMC107, which was integrated into the chromosome of B. pertussis BP536 as described above.
(v) ptlD′-phoA fusion.
A DNA fragment was generated by PCR using primers 9a and 5b. Next, pSZH4 was digested with ClaI, and the 5.4-kb fragment was inserted into the ClaI site of the pGEM7Zf(+) vector, generating pAMC5. The PCR-generated DNA fragment containing ptlD′-phoA was then inserted into the Bsu36I-HindIII site of this plasmid. The resultant plasmid was digested with PstI and HindIII, and the 2.4-kb fragment was inserted into the pGEM3Zf(+) vector. Next, this plasmid was digested with EcoRI and HindIII, and the 2.4-kb fragment was inserted into pSS1129, generating pAMC121, which was then integrated into the chromosome of B. pertussis BP536 as described above.
(vi) ptlF′-phoA fusion.
A DNA fragment was generated by PCR using primers 10a and 5b. The DNA fragment was then inserted into the SalI-HindIII site of pAMC5 (described above). Digestion of the resultant plasmid with NotI and HindIII yielded a 2.5-kb fragment which was inserted into the NotI-HindIII site of pSZH4. This plasmid was digested with BamHI and HindIII, and the 5.5-kb fragment was inserted into pSS1129, generating pAMC28, which was then integrated into the chromosome of either B. pertussis BP536, BP536Δptxptl936-8003, BP536Δptxptl936-3941, or BP536Δptlptl3821-8003 as described above.
Plasmid construction. (i) Construction of pAMC111.
Plasmid pAMC111, which carries all nine ptl genes under the control of the ptx-ptl promoter, was constructed as follows. Plasmid pTH14 (described above) was digested with SacI and HindIII. The 2.0-kb fragment was inserted into pZErO-1. The resultant plasmid was digested with SapI and HindIII, and the 3.7-kb fragment containing the vector backbone was isolated. Plasmid pSZH4 was digested with SapI and HindIII, and the 9.5-kb fragment was inserted into the 3.7-kb fragment described above. The resulting plasmid, pAMC110, was digested with SacI and HindIII, and the 10.4-kb fragment was inserted into pUFR047, a broad-host-range vector capable of replicating in B. pertussis, which had been digested with the same enzymes, to yield pAMC111.
After the introduction of pAMC111 into B. pertussis as described below, we were not able to detect extrachromosomal copies of the plasmid. However, we were able to determine that the plasmid had integrated into the chromosome by isolating genomic DNA from B. pertussis BP536::pAMC111 and the wild-type strain B. pertussis BP536 and examining the DNA for copies of the ptx-ptl region by Southern blot analysis. By using MfeI, which cut the integrated plasmid at a single site, and by probing with a labeled oligonucleotide corresponding to nucleotides 3515 to 4574 from the ptx-ptl operon, we observed a single band in B. pertussis BP536, as expected, and two bands in B. pertussis BP536::pAMC111, one of which corresponded to the band seen in the wild-type strain (data not shown). Thus, the plasmid had integrated at only a single site in B. pertussis BP536::pAMC111.
(ii) Construction of pAMC147.
Plasmid pAMC147, which carries ptlA, ptlB, and ptlC under the control of the ptx-ptl promoter, was constructed as follows. Plasmid pAMC110 (described above) was digested with SacI and SmaI, and the 4.3-kb fragment containing nucleotides 1 to 935 and nucleotides 3515 to 6895 of the ptx-ptl region was isolated and inserted into pUFR047 to yield pAMC147.
(iii) Construction of pTC11.
Plasmid pTC11, which carries ptlD, ptlI, ptlE, and ptlF under the control of the lac promoter, was constructed as follows. A DNA fragment consisting of the coding sequence for ptlD, preceded by the sequence for a ribosomal binding site, was generated by PCR using primers 11a and 11b and was inserted into the EcoRI-XbaI site of pALTER-1, yielding pTC9. Next, a DNA fragment extending from the NotI site in ptlD through the end of ptlF was generated by PCR using primers 12a and 12b and was inserted into the NotI-HindIII fragment of pTC9, generating pTC11A. The EcoRI-HindIII fragment from pTC11A was then inserted into pUFR047, generating pTC11.
(iv) Construction of pAMC151.
Plasmid pAMC151 which carries ptlI through ptlH under the control of the lac promoter, was constructed as follows. A DNA fragment was generated by using primers 13a and 13b. The amplified DNA fragment was digested with XbaI and HindIII and was inserted into pUC19, which had been digested with the same enzymes, to yield pAMC177. Plasmid pSZH4 was digested with NotI and HindIII to yield a 5.0-kb fragment containing the 3′ end of ptlD through ptlH. This fragment was inserted into pAMC177, which had been digested with NotI and HindIII. The resultant plasmid was digested with SacI and HindIII, and the 5.0-kb fragment containing the 3′ end of ptlD through ptlH was isolated and inserted into pUFR047, which had been digested with the same enzymes, to yield pAMC151.
(v) Construction of pAMC171.
Plasmid pAMC171, which carries ptlA behind the ptx-ptl promoter in pUFR047, was constructed as follows. Plasmid pTH14 (described above) was digested with EcoRI and SalI to yield a 0.9-kb fragment. A DNA fragment was generated by PCR using primers 14a and 14b. The 0.9-kb fragment and the amplified DNA were inserted into pUFR047 to yield pAMC171.
(vi) Construction of pAMC176.
Plasmid pAMC176, which carries ptlB and ptlC behind the ptx-ptl promoter in pUFR047, was constructed as follows. Plasmid pTH14 (described above) was digested with SacI and SalI to yield a 0.9-kb fragment. A DNA fragment was generated by PCR using primers 15a and 15b. The 0.9-kb fragment and the purified PCR fragment were inserted into the 11.0-kb fragment of pAMC147 (described above) that was isolated after digestion of pAMC147 with SacI and AscI to yield pAMC176.
Introduction of plasmids into B. pertussis.
The introduction of plasmids with a pUFR047 backbone into strains of B. pertussis by conjugation has been described previously (9).
Immunoblot analysis of cell extracts and culture supernatants.
B. pertussis cells from plate cultures were collected by suspension in 5 ml of phosphate-buffered saline (pH 7.4) to an A550 of 2.0. B. pertussis cells and culture supernatant fractions from liquid cultures were collected by centrifugation of 5 ml of the culture at 17,000 × g for 10 min. Cells from liquid cultures were suspended in 5 ml of phosphate-buffered saline (pH 7.4). Samples of cells from plate cultures (50 μl), cells from liquid cultures (100 μl or as otherwise specified), and culture supernatants (100 μl or as otherwise specified) were precipitated with an equal volume of 20% trichloroacetic acid or with 2.3 times the volume of 100% ethanol by incubation in a dry-ice-ethanol bath for 30 min. After centrifugation, the precipitates were suspended in 15 μl of sodium dodecyl sulfate (SDS) sample buffer.
Samples were subjected to SDS-polyacrylamide gel electrophoresis, performed essentially as described by Laemmli (19), using 4 to 20% gradient polyacrylamide gels obtained from Bio-Rad Laboratories (Hercules, Calif.). Immunoblot analysis was performed essentially as described previously (3); monoclonal antibody 3CX4 (15, 16) was used to visualize the S1 subunit, and monoclonal antibody P11B10 (10) was used to visualize the S2 subunit. Polyclonal antibodies raised in mice that have been described previously (13) were used to visualize PtlF. Where indicated, blots were scanned with a Bio-Rad GS-800 densitometer and analyzed by using Quality One software. The densities of the bands representing the portions of the S1 subunit that had been secreted and that remained cell associated were determined. The percentage of the S1 subunit that was secreted was calculated as the amount of S1 subunit found in the supernatant divided by the total amount of S1 subunit (supernatant plus cell associated), multiplied by 100.
Alkaline phosphatase assay.
B. pertussis strains were grown at 37°C in Stainer-Scholte liquid medium for 48 h to an optical density of 1.2 to 1.5 at 550 nm. B. pertussis cells from liquid cultures were collected by centrifugation of 5 ml of the culture at 17,000 × g for 10 min and were suspended in 5 ml of 1.0 M Tris-HCl (pH 8.0) buffer. Alternatively, cells were grown on BG agar and suspended in 1.0 M Tris-HCl (pH 8.0) to give an A550 of 2. A serial dilution from 1:5 to 1:320 of the cell suspensions was made by using the same buffer. A 1-ml volume of the diluted cell suspensions was permeabilized by the addition of 30 μl of chloroform and 30 μl of 0.1% SDS, followed by vortexing. The assay for alkaline phosphatase was performed essentially as described by Brickman and Beckwith (2). After the addition of 0.1 ml of 0.4% para-nitrophenylphosphate, samples were incubated at room temperature for 15 min, at which time 0.1 ml of 1 M K2HPO4 was added to stop the reactions. The optical density of each sample was read at 420 and 550 nm. Readings in the linear portion of the output curve were used to calculate alkaline phosphatase activity.
Southern blot analysis.
Chromosomal DNA was isolated from BP536 and BP536::pAMC111 by using the MasterPure DNA Purification kit (Epicentre). DNA was digested with MfeI and was transferred to Hybond-N+ membranes (Amersham Pharmacia Biotech). DNA was probed by using a gel-purified 1.0-kb SalI-BamHI restriction fragment (nucleotides 3515 to 4574) from the ptx-ptl operon that was labeled with alkaline phosphatase.
Statistical analysis.
Data are presented as means and standard deviations from triplicate experiments. Statistical significance was determined by using Student's t test. Differences were considered significant when the P value was less than 0.05.
RESULTS
Since the ptx and ptl genes are cotranscribed, PT and its transporter might be produced at comparable levels. Alternatively, the PT subunits and Ptl proteins could be produced in different amounts for a number of possible reasons. For example, regulatory signals, such as attenuators, might exist at points along the operon that could alter the transcriptional efficiency of specific regions of the operon. Or the levels of translation of specific regions of the mRNA might differ such that the quantity of the PT subunits produced would differ from that of the Ptl proteins.
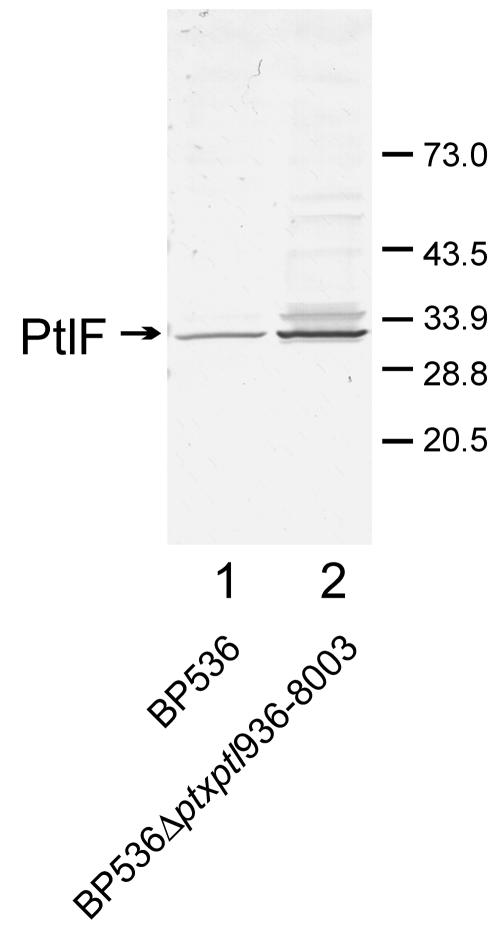
If the ptx and ptl genes are transcribed and translated at comparable levels, then introduction of a large in-frame deletion of the DNA within the region should not affect the production of Ptl proteins encoded by genes located downstream of the deletion. If, on the other hand, the ptx and ptl genes are transcribed or translated at different levels because, for example, either specific regulatory signals exist along the DNA and/or mRNA, different portions of the mRNA exhibit different stabilities, or ribosomes are released from the mRNA after translation of a specific gene and reinitiation of translation at downstream genes fails to occur, then introduction of a large in-frame deletion within the operon should affect the production of Ptl proteins encoded by downstream genes. In order to test whether a large in-frame deletion in this region of the chromosome results in altered production of Ptl proteins from downstream genes, we generated a 7.1-kb deletion, from nucleotide 936 to nucleotide 8003 of the ptx-ptl region, that retained the reading frames of the two partial genes spanning the deletion (Fig. 1A). With this deletion, the 5′ end of ptxS1 was fused to the 3′ end of ptlD, resulting in a gene that encoded a fusion protein consisting of the N-terminal 50% of the S1 subunit of PT and the C-terminal 15% of PtlD. The seven genes between ptxS1 and ptlD (ptxS2, ptxS4, ptxS5, ptxS3, ptlA, ptlB, and ptlC) were deleted in their entirety. Such a deletion would move ptlF approximately 7.1 kb closer to the promoter region of the operon (Fig. 1A). We then compared the amount of PtlF produced by this strain (BP536Δptxptl936-8003) with the amount produced by the wild-type strain (BP536). As seen in Fig. 2, significantly more PtlF was produced by the strain with the 7.1-kb deletion than by the wild-type strain.
FIG. 2.
Immunoblot analysis of PtlF in a wild-type strain and a deletion strain. Cell extracts from BP536 (lane 1) or BP536Δptxptl936-8003 (lane 2) were prepared as described in Materials and Methods. Samples (50 μl) were subjected to SDS-polyacrylamide gel electrophoresis and immunoblot analysis with a polyclonal antibody to visualize PtlF. Arrow indicates PtlF. The minor band migrating slightly more slowly than PtlF has been identified as the PtlF-PtlI complex (8). Positions of molecular size markers are shown on the right. The results observed for the bands seen in this immunoblot are typical of those seen in replicate experiments. Densitometry of the PtlF band in triplicate experiments yielded values of 0.19 ± 0.021 for BP536 and of 0.44 ± 0.006 for BP536Δptxptl936-8003 (P < 0.05). Densitometry values are expressed in arbitrary units.
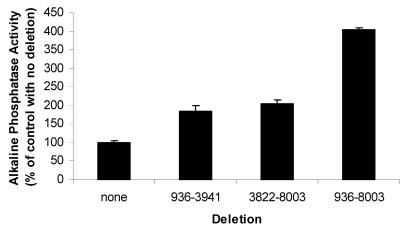
In order to determine whether a single regulatory signal along the operon or message might account for the increase in the production of PtlF observed in the deletion strain described above, we constructed additional deletion strains. In each of these deletion strains, we also replaced the ptlF gene with the phoA gene so that protein production could easily be quantified simply by measuring alkaline phosphatase activity (Fig. 1B). In all of these strains phoA was fused to ptlF such that the fusion gene contained the initiation codon of ptlF followed by phoA devoid of its start codon. We compared the alkaline phosphatase activities of strains with no deletion or with a deletion from nucleotide 936 to 8003, from nucleotide 936 to 3941, or from nucleotide 3822 to 8003. As shown in Fig. 3, the level of alkaline phosphatase activity increased in proportion to the size of the deletion. Moreover, the two strains containing deletions from nucleotide 936 to 3941 or from nucleotide 3822 to 8003 exhibited similar alkaline phosphatase activities, neither of which reached the level of alkaline phosphatase activity produced by the strain with the largest deletion, spanning nucleotides 936 to 8003 (P < 0.05). These data are not compatible with the idea that only a single regulatory signal exists along the operon and/or message, deletion of which results in an increase in the production of proteins encoded by downstream genes.
FIG. 3.
Alkaline phosphatase activities of strains containing in-frame deletions and ptlF′-phoA fusions. The strains containing the indicated deletions in the ptx-ptl region were analyzed for alkaline phosphatase activity. Values are averages from three independent assays. Error bars, standard deviations.
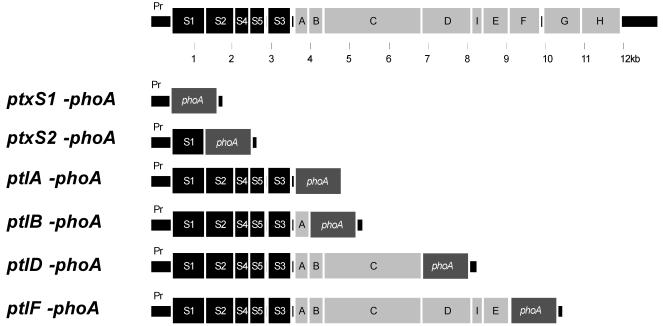
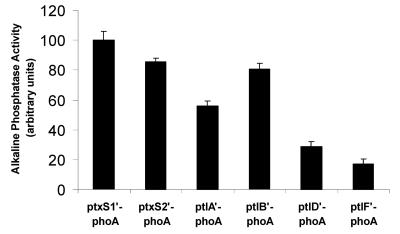
In order to further examine the relative levels of production of proteins encoded by genes in the ptx-ptl operon, we constructed strains in which the phoA gene (devoid of its initiation codon) was fused to the initiation codon of either ptxS1, ptxS2, ptlA, ptlB, ptlD, or ptlF (Fig. 4), and then we compared the alkaline phosphatase activities of these strains. As shown in Fig. 5, we found that alkaline phosphatase activity decreased in a relatively linear fashion as the phoA gene was moved from the 5′ to the 3′ end of the operon. The amounts of alkaline phosphatase activity produced by the strains containing the ptlD′-phoA fusion and the ptlF′-phoA fusion were significantly smaller than that produced by the strain containing the ptxS1′-phoA fusion (P < 0.05). These data suggest that production of the Ptl proteins located at the 3′ end of the operon is lower than that of the PT subunits. In fact, PT subunits may be produced in quantities as much as five times greater than those of Ptl proteins encoded by genes located at the 3′ end of the operon (Fig. 5).
FIG. 4.
phoA gene fusions. phoA devoid of its initiation codon was fused to the ATG initiation codon of the indicated ptx and ptl genes as described in Materials and Methods. The ptx-ptl regions containing these fusions are depicted.
FIG. 5.
Alkaline phosphatase activities of strains containing ptx′- or ptl′-phoA gene fusions. Alkaline phosphatase activities of strains containing the indicated phoA fusions and grown in liquid culture were measured as described in Materials and Methods. Values are averages from three independent assays. Error bars, standard deviations.
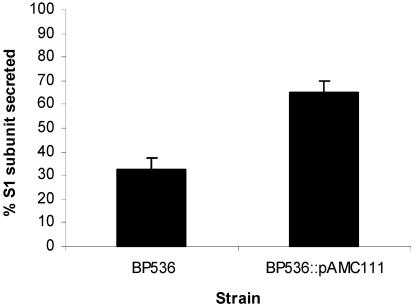
Since our data indicate that certain Ptl proteins might be produced in quantities smaller than those of the PT subunits, we wanted to determine whether the Ptl proteins might be limiting in the secretion of the toxin from the bacteria. We first attempted to introduce a broad-host-range vector, pAMC111, containing the ptx-ptl promoter followed by the ptl region, into a wild-type strain. However, we were unable to isolate a strain in which the plasmid containing the ptl region replicated extrachromosomally. Such a finding could be explained if overproduction of Ptl proteins were toxic to the cell. However, we were able to isolate a strain in which the plasmid had integrated into the chromosome (see Materials and Methods for details) such that the strain contained a single extra copy of the ptl region. We examined the production of PtlF in this strain and found that more PtlF was produced in this strain than in the wild-type strain (data not shown), as would be expected if both copies of the ptlF gene within the chromosome of this strain were expressed. We then compared the secretion of PT in this strain containing 2 copies of the ptl region to that in the wild-type strain. As shown in Fig. 6, we observed that secretion of PT from the wild-type strain (BP536) was inefficient, as reported previously (9). When we examined the secretion of PT from the strain that contains 2 copies of the ptl region (BP536::pAMC111), we found that significantly more of the S1 subunit was observed in the supernatant of BP536::pAMC111 than in that of BP536. BP536::pAMC111 and BP536 produced approximately the same amount of PT, as determined by densitometry of the amount of PT found in the culture supernatant and the amount that remained associated with the cell (data not shown). These results suggest that the Ptl proteins are limiting in the secretion of the toxin.
FIG. 6.
Secretion of PT from BP536 and BP536::pAMC111. Samples of culture supernatants (100 μl) and cell extracts (100 μl) of the indicated strains were prepared as described in Materials and Methods. Samples were subjected to SDS-polyacrylamide gel electrophoresis and immunoblot analysis with monoclonal antibody 3CX4 to visualize the S1 subunit of PT. Immunoblots were then subjected to densitometry to quantify the amount of S1 found in the supernatant and cell fractions as described in Materials and Methods. The percentage of the S1 subunit secreted by each strain was calculated as the amount of the S1 subunit found in the supernatant fraction divided by the total amount of the S1 subunit (supernatant plus cell associated), multiplied by 100. Values shown are means ± standard deviations of values determined from three independent experiments. The percentage of the S1 subunit secreted by BP536::pAMC111 was determined to be significantly different from that secreted by BP536 (P < 0.05).
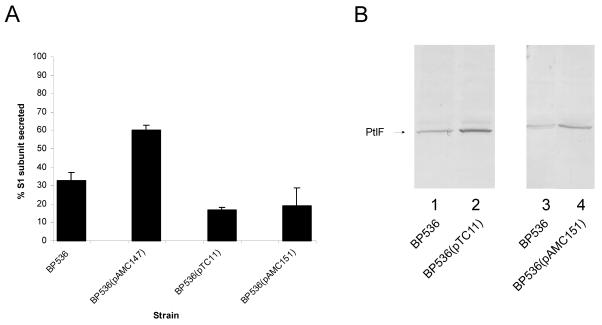
As a first step in determining which Ptl proteins might be limiting in the secretion of the toxin, we introduced plasmids containing either ptlA-ptlC controlled by the ptx-ptl promoter, ptlD-ptlF controlled by the lac promoter, or ptlI-ptlH controlled by the lac promoter into BP536 to yield strains BP536(pAMC147), BP536(pTC11), and BP536(pAMC151), respectively. As shown in Fig. 7A, an increase in the gene dosage of ptlA-ptlC resulted in a significant increase in the proportion of S1 secreted over that for the wild-type strain. The total amount of PT produced by BP536(pAMC147) was similar to that produced by BP536, as determined by densitometry of the amount of PT found in the supernatant and that associated with the cell (data not shown). Overexpression of either ptlD-ptlF or ptlI-ptlH did not increase secretion. We examined the amounts of PtlF produced by BP536(pTC11) and BP536(pAMC151) and found that they were greater than that produced by the wild-type strain, thus ensuring that the ptl genes on the plasmids of both strains were being expressed (Fig. 7B).
FIG. 7.
Secretion of PT from strains overexpressing subsets of the ptl genes. (A) Samples of culture supernatants (100 μl) and cell extracts (100 μl) of the indicated strains were prepared as described in Materials and Methods. Samples were subjected to SDS-polyacrylamide gel electrophoresis and immunoblot analysis using monoclonal antibody 3CX4 to visualize the S1 subunit of PT. Immunoblots were then subjected to densitometry to quantify the amounts of S1 found in the supernatant and cell fractions as described in Materials and Methods. The percentage of the S1 subunit secreted by each strain was calculated as the amount of the S1 subunit found in the supernatant fraction divided by the total amount of the S1 subunit (supernatant plus cell associated), multiplied by 100. Values shown are means ± standard deviations of values determined from three independent experiments. The percentage of the S1 subunit secreted by BP536(pAMC147), the strain which carries extra copies of ptlA-ptlC, was determined to be significantly different from that secreted by BP536, the wild-type strain (P < 0.05). (B) Samples of cells from plate cultures of the indicated strains were prepared as described in Materials and Methods. Samples were subjected to immunoblot analysis using polyclonal antibodies to visualize PtlF. Results are typical of those seen in replicate experiments. Densitometry of the PtlF band in triplicate experiments yielded values of 0.21 ± 0.04 for BP536, 0.52 ± 0.03 for BP536(pTC11), and 0.37 ± 0.05 for BP536(pAMC151). Densitometry values are expressed in arbitrary units.
Since an increase in the gene dosage of ptlA-ptlC increased the secretion of PT, we next wanted to determine which of these genes might be responsible for the increased secretion. Therefore, we introduced plasmids containing either ptlA controlled by the ptx-ptl promoter or ptlB-ptlC controlled by the ptx-ptl promoter into BP536 to yield strains BP536(pAMC171) and BP536(pAMC176), respectively. We found that neither of these strains exhibited increased secretion of PT relative to that of the wild-type strain, indicating that a combination of ptlA, ptlB, and/or ptlC is needed for increased secretion (data not shown).
DISCUSSION
In this study, we generated translational fusions between phoA and various ptx and ptl genes in order to examine the relative levels of production of PT subunits and Ptl proteins. Because we used translational fusions to examine gene expression, our results for the expression of these genes reflect a combination of both transcription and translation of the genes that were examined. We found that genes located at the 5′ end of the ptx-ptl operon are transcribed and/or translated more efficiently than those at the 3′ end; in fact, proteins encoded by genes located at the 5′ end of the operon may be produced at levels more than five times higher than those of proteins encoded by genes located at the 3′ end of the operon.
Previously, Nicosia et al. (21) noted the existence of a potential stem-loop structure located in the intergenic region between the ptx genes and the ptl genes and suggested that this region resembled a rho-independent termination signal of transcription. Ricci et al. (24) investigated the role that this region might play in regulation of ptx/ptl expression. They found that deletion of this region resulted in at least a 30% higher accumulation of ptl transcripts. They concluded that this intergenic region contains the information for a weak transcriptional termination that occurs after the transcription of the ptx region and suggested that such a signal might be responsible for the correct equilibrium between the available amount of PT and the amounts of the Ptl proteins that are needed for the transport of PT.
Our results suggest that a single region that regulates the level of transcription of the ptx versus the ptl region is not solely responsible for the differences in the levels of production of PT subunits and Ptl proteins. Additional factors appear to come into play in determining the relative levels of PT and Ptl proteins that are produced within the bacterium, since we found that transcription and/or translation of genes generally decreases across the entire length of the ptx-ptl operon. One possible explanation for differential production of PT subunits versus Ptl proteins might be variation in the stability of portions of the transcribed message. For example, our results can be explained if degradation of the message is initiated more often at the 3′ end of the message than at the 5′ end. Another likely explanation for the observation that proteins encoded by genes located at the 5′ end of the operon are produced at higher levels than those encoded by genes located at the 3′ end of the operon is that translation may not be reinitiated after the release of ribosomes from upstream genes. Such a phenomenon has been reported to be at least partially responsible for the differential translation of the three genes found within the E. coli lac operon: lacZ, lacY, and lacA (20). A single ribosomal binding site exists at the 5′ end of the lac message, and all ribosomes attach to the mRNA at this site. At each stop codon, some of the ribosomes detach. Therefore, the number of ribosomes translating a gene decreases from the 5′ to the 3′ end of the message. The ribosomal binding sites of the ptx-ptl operon are not well defined. A sequence resembling the consensus Shine-Dalgarno sequence was identified 9 bp before the ATG initiation codon of the ptxS1 gene (21). Other such sites have not as yet been identified within the ptx-ptl operon. A number of the ptx-ptl genes may not have an associated Shine-Dalgarno sequence; rather, translational coupling may occur between genes, since the stop codon of the upstream gene overlaps with the coding region of the downstream gene. Such is the case for ptxS4, ptxS5, ptlC, ptlI, ptlE, ptlF, and ptlH. Thus, a limited number of ribosomal binding sites may exist within the operon. Release of ribosomes at the stop codon of an upstream gene and lack of reinitiation of translation of the message at the start codon of the downstream gene would then lead to a decrease in the production of downstream genes.
Our finding that genes located at the 3′ end of the operon are transcribed and/or translated at levels lower than those of genes located at the 5′ end of the operon suggested the possibility that certain of the Ptl proteins might be produced in quantities that are limiting for secretion of the toxin. Indeed, we did find that expression of a second copy of the ptl region resulted in more-efficient secretion. In order to determine which Ptl proteins are limiting, we expressed subsets of the ptl genes. We found that introduction of a plasmid containing ptlA, ptlB, and ptlC under the control of the ptx-ptl promoter resulted in increased levels of secretion of the toxin. In contrast, increases in the gene dosage of ptlD, ptlI, ptlE, ptlF, ptlG, and ptlH had little effect on secretion of the toxin. These results suggest that PtlA, PtlB, and/or PtlC may be limiting in the transport process. A combination of these proteins may be limiting, since increases in the gene dosage of either ptlA or a combination of ptlB and ptlC were not sufficient to increase the secretion of the toxin. The finding that PtlA, PtlB, and/or PtlC appear to be limiting in the secretion of PT was somewhat surprising, since it is the Ptl proteins encoded by genes located at the extreme 3′ end of the operon that appear to be produced at the lowest levels. Several possible explanations exist for these results. First, PtlA, PtlB, and/or PtlC may be the most labile of the Ptl proteins, and they may be rapidly degraded after synthesis. Second, the possibility exists that multiple copies of one or more of these proteins may be required for the intact transporter apparatus.
In summary, the results presented here suggest that the Ptl proteins, especially those encoded by genes located at the 3′ end of the operon, are produced at levels lower than those of the PT subunits. In our studies, certain of the Ptl proteins appear to be limiting in the secretion process, since introduction of multiple copies of the genes expressing these proteins results in more-efficient secretion of the toxin. This information represents an important step toward obtaining a solid understanding of the stoichiometry of the PT subunits and the Ptl proteins within the bacteria and the dynamics that exist between PT and its transporter.
Acknowledgments
We are grateful to Scott Stibitz, who kindly provided strains and plasmids for this study, and to Thomas Cafarella for technical assistance.
Editor: J. T. Barbieri
REFERENCES
- 1.Baker, S. M., A. Masi, D.-F. Liu, B. K. Novitsky, and R. A. Deich. 1995. Pertussis toxin export genes are regulated by the ptx promoter and may be required for efficient translation of ptx mRNA in Bordetella pertussis. Infect. Immun. 63:3920-3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brickman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J. Mol. Biol. 96:307-316. [DOI] [PubMed] [Google Scholar]
- 3.Burnette, W. N. 1981. “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal. Biochem. 112:195-203. [DOI] [PubMed] [Google Scholar]
- 4.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional tranporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Covacci, A., and R. Rappuoli. 1993. Pertussis toxin export requires accessory genes located downstream from the pertussis toxin operon. Mol. Microbiol. 8:429-434. [DOI] [PubMed] [Google Scholar]
- 6.Das, A., and Y.-H. Xie. 2000. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J. Bacteriol. 182:758-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeFeyter, R., Y. Yang, and D. W. Gabriel. 1993. Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant-Microbe Interact. 6:225-237. [DOI] [PubMed] [Google Scholar]
- 8.Farizo, K. M., T. G. Cafarella, and D. L. Burns. 1996. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J. Biol. Chem. 271:31643-31649. [DOI] [PubMed] [Google Scholar]
- 9.Farizo, K. M., T. Huang, and D. L. Burns. 2000. Importance of holotoxin assembly in Ptl-mediated secretion of pertussis toxin from Bordetella pertussis. Infect. Immun. 68:4049-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, D. W., and C. D. Parker. 1984. Interaction of monoclonal antibodies with pertussis toxin and its subunits. Infect. Immun. 46:195-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hapfelmeier, S., N. Domke, P. C. Zambryski, and C. Baron. 2000. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J. Bacteriol. 182:4505-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hausman, S. Z., J. D. Cherry, U. Heininger, C. H. Wirsing von Konig, and D. L. Burns. 1996. Analysis of proteins encoded by the ptx and ptl genes of Bordetella bronchiseptica and Bordetella parapertussis. Infect. Immun. 64:4020-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, F. D., and D. L. Burns. 1994. Detection and subcellular localization of three Ptl proteins involved in the secretion of pertussis toxin from Bordetella pertussis. J. Bacteriol. 176:5350-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones, A. L., E.-M. Lai, K. Shirasu, and C. I. Kado. 1996. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 178:5706-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenimer, J. G., K. J. Kim, P. G. Probst, C. R. Manclark, D. G. Burstyn, and J. L. Cowell. 1989. Monoclonal antibodies to pertussis toxin: utilization as probes of toxin function. Hybridoma 8:37-51. [DOI] [PubMed] [Google Scholar]
- 16.Kim, K. J., W. N. Burnette, R. D. Sublett, C. R. Manclark, and J. G. Kenimer. 1989. Epitopes on the S1 subunit of pertussis toxin recognized by monoclonal antibodies. Infect. Immun. 57:944-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kotob, S. I., S. Z. Hausman, and D. L. Burns. 1995. Localization of the promoter for the ptl genes of Bordetella pertussis, which encode proteins essential for secretion of pertussis toxin. Infect. Immun. 63:3227-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Maloy, S. R., J. E. Cronan, Jr., and D. Freifelder. 1994. Microbial genetics. Jones and Bartlett Publishers, Boston, Mass.
- 21.Nicosia, A., M. Perugini, C. Franzini, M. C. Casagli, M. G. Borri, G. Antoni, M. Almoni, P. Neri, G. Ratti, and R. Rappuoli. 1986. Cloning and sequencing of the pertussis toxin genes: operon structure and gene duplication. Proc. Natl. Acad. Sci. USA 83:4631-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Callaghan, D., C. Cazevieille, A. Allardet-Servent, M. L. Boschiroli, G. Bourg, V. Foulongne, P. Frutos, Y. Kulakov, and M. Ramuz. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210-1220. [DOI] [PubMed] [Google Scholar]
- 23.Padmalayam, I., K. Karem, B. Baumstark, and R. Massung. 2000. The gene encoding the 17-kDa antigen of Bartonella henselae is located within a cluster of genes homologous to the virB virulence operon. DNA Cell Biol. 19:377-382. [DOI] [PubMed] [Google Scholar]
- 24.Ricci, S., R. Rappouli, and V. Scarlato. 1996. The pertussis toxin liberation genes of Bordetella pertussis are transcriptionally linked to the pertussis toxin operon. Infect. Immun. 64:1458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagulenko, V., E. Sagulenko, S. Jakubowski, E. Spudich, and P. J. Christie. 2001. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J. Bacteriol. 183:3642-3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt-Eisenlohr, H., N. Domke, C. Angerer, G. Wanner, P. C. Zambryski, and C. Baron. 1999. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J. Bacteriol. 181:7485-7492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmiederer, M., and B. Anderson. 2000. Cloning, sequencing and expression of three Bartonella henselae genes homologous to the Agrobacterium tumefaciens virB region. DNA Cell Biol. 19:141-147. [DOI] [PubMed] [Google Scholar]
- 28.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 29.Stein, P. E., A. Boodhoo, G. D. Armstrong, S. A. Cockle, M. H. Klein, and R. J. Read. 1994. The crystal structure of pertussis toxin. Structure 2:45-57. [DOI] [PubMed] [Google Scholar]
- 30.Stibitz, S. 1994. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 235:458-465. [DOI] [PubMed] [Google Scholar]
- 31.Stibitz, S., and M.-S. Yang. 1991. Subcellular localization and immunochemical detection of proteins encoded by the vir locus of Bordetella pertussis. J. Bacteriol. 173:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamura, M., K. Nogimori, S. Murai, M. Yajima, K. Ito, T. Katada, M. Ui, and S. Ishii. 1982. Subunit structure of islet-activating protein, pertussis toxin, in conformity with the A-B model. Biochemistry 21:5516-5522. [DOI] [PubMed] [Google Scholar]
- 33.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. USA 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]