Abstract
Unisexual sperm-dependent vertebrates are of hybrid origins, rare, and predicted to be short-lived as a result of several challenges arising from their mode of reproduction. In particular, because of a lack of recombination, clonal species are predicted to have a low potential to respond to natural selection. However, many unisexual sperm-dependent species persist, and assessing the genetic diversity present in these species is fundamental to understanding how they avoid extinction. We used population genomic methods to assess genotypic variation within the unisexual fish Poecilia formosa. Measures of admixture and population differentiation, as well as clustering analyses, indicate that the genomes of individuals of P. formosa are admixed and intermediate between Poecilia latipinna and Poecilia mexicana, consistent with the hypothesis of their hybrid origins. Bayesian genomic cline analyses indicate that about 12% of sampled loci exhibit patterns consistent with inheritance from only one parent. The estimation of observed heterozygosity clearly suggests that P. formosa is not comprised of direct descendants of a single nonrecombining asexual F1 hybrid individual. Additionally, the estimation of observed heterozygosity provides support for the hypothesis that the history of this unisexual species has included backcrossing with the parent species before the onset of gynogenesis. We also document high levels of variation among asexual individuals, which is attributable to recombination (historical or ongoing) and the accumulation of mutations. The high genetic variation suggests that this unisexual vertebrate has more potential to respond to natural selection than if they were frozen F1 hybrids.
The maintenance of sex presents a conundrum for evolutionary biology because the costs of sexual reproduction (cost of producing males, energy expenditure to find a mate, exposure to diseases, and segregation of alleles) appear to be immediate and substantial, whereas its benefits (facilitation of adaptations, elimination of deleterious mutations) are postponed (reviewed in ref. 1). The long-term maintenance of unisexual organisms is of interest to evolutionary biologists as well because the advantages of asexual reproduction are all immediate (no cost of producing males and, therefore, exponential population growth), but the long-term costs are substantial (accumulation of deleterious mutations and lack of genetic recombination to respond to environmental changes). Asexual vertebrate species are, therefore, predicted to be short-lived compared with sexually reproducing species (2–4). However, recent work focused on nonvertebrate species has challenged the view that recombination is absent in asexual lineages and that, therefore, those species are doomed to extinction. Asexual aphids, fungi, and microcrustaceans have all been shown to be genetically variable [aphids (5), fungi (6), Daphnia (7)] and, in some cases, mitotic recombination facilitates the spread of beneficial mutations (8). Therefore, understanding how much genetic variation is present in asexual lineages, and whether the presence of this variation and the mechanisms that facilitate it are shared among taxa, is an essential step toward understanding the evolution of sexual and asexual reproduction and, perhaps, challenging existing paradigms.
Asexuality is common in many phyla (reviewed in ref. 7), but it is relatively rare in vertebrates (1). All known unisexual (all-female) vertebrates are products of hybridization events between sexually reproducing species (ref. 9 and references therein), constitute only 0.1% of extant vertebrate species (1, 9). One type of asexual reproduction found in unisexual vertebrates is gynogenesis, where females must mate with males of a closely related species (but refer to ref. 10 for exceptions), but the nonrecombinant embryos do not inherit any genetic information from the sperm donor (9). Because gynogens require sperm to initiate development of offspring, but no paternal genes are expressed, they are considered “sexual parasites” (11).
The maintenance of a gynogenetic species is paradoxical because gynogens face the costs of both sexual and asexual reproduction: the cost of finding a mate, exposure to diseases, accumulation of deleterious mutations, and lack of genetic recombination to facilitate adaptation. In addition, because male sperm donors do not gain a fitness advantage from mating with gynogens, selection should favor males that avoid mating with them.
Given the extensive and diverse list of challenges faced by gynogenetic species, they are predicted to be short-lived with a limited potential to respond to natural selection. Nevertheless, gynogenetic species persist, and some have origins in the distant past (12, 13). This suggests that gynogenetic species might be able to avoid or ameliorate some of the costs associated with their reproductive mode. One question that arises is how much genetic variation persists in gynogenetic species?
The Amazon molly (Poecilia formosa) is an excellent system to explore this question. P. formosa is the first vertebrate recognized as asexual (11) and is a gynogenetic species that uses Poecilia mexicana (Atlantic molly), Poecilia latipinna (sailfin molly), and Poecilia latipunctata (Tamesi molly) as sexual hosts (14). Like every other known unisexual vertebrate, P. formosa is thought to be a hybrid lineage (9, 13, 15–17). P. mexicana is recognized to be the maternal species of P. formosa (13, 15–17), whereas P. latipinna (or an extinct ancestor of P. latipinna) is the putative paternal species (17). P. formosa lives in sympatry with at least one of the two parent species throughout its range from the Tampico region in Mexico to the southeastern United States (Fig. 1A). Although recent studies suggest that P. formosa is a species that consists of “frozen” F1 hybrid clones (i.e., individuals with ancestry from P. mexicana and P. latipinna at all loci) (15–19), this result is based on limited genetic data. Additionally, it is still not clear whether P. formosa is the product of a single or multiple hybridization events, although a recent investigation supports the hypothesis of a single event possibly giving rise to several clonal lineages (19).
Fig. 1.
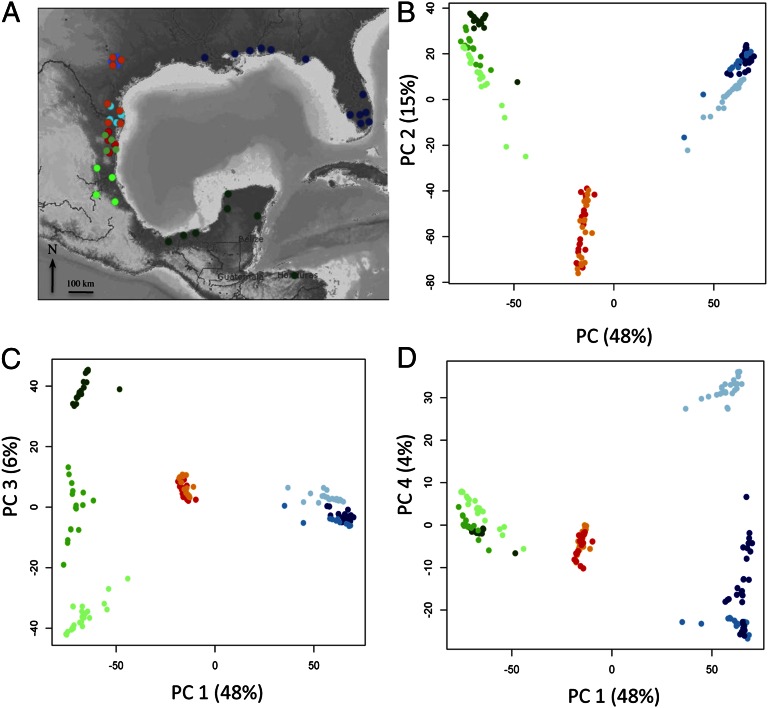
Sampled populations (A) and PCA plots for the 192 individuals based on the genotype probabilities at each locus (B–D). Dark blue, north P. latipinna; blue, central P. latipinna; light blue, south P. latipinna; orange, north P. formosa sympatric with P. latipinna; red, south P. formosa sympatric with P. mexicana; light green, north P. mexicana; green, central P. mexicana; dark green, south P. mexicana.
The overall objective of this research was to examine genotypic variation in P. formosa at a genomic scale. We generated thousands of DNA sequence markers to address three main questions. (i) Is P. formosa the product of hybridization between P. latipinna and P. mexicana? This is the conclusion of several previous studies (11, 13, 15–19), and, here, we attempt to confirm this result with a genome-wide survey of P. formosa and its putative parent species. (ii) Is P. formosa composed of frozen F1 hybrid clones, or is there evidence of a more complicated history of nonclonal reproduction or recombination? Previous genetic analyses (13, 15–19), and the fact that no laboratory has been able to synthetize P. formosa from artificial hybridization experiments (15, 19–21), suggest that the genome of this species is more complicated than that of a simple F1 hybrid. If P. formosa is not composed of clones of F1 hybrid lineages, then, what is the genetic contribution of each parent species? (iii) How much genotypic diversity exists within P. formosa and is this genotypic diversity consistent with P. formosa having evolved from a single or multiple independently formed hybrid individuals? Quantifying genetic variation will provide some estimate of this species’ potential to respond to selection.
Results and Discussion
We used a next-generation sequencing population genetics approach to collect information on variation from across the genomes of P. formosa, P. latipinna, and P. mexicana and to understand the genomic composition of the gynogenetic species. The methodology used herein allowed us to obtain genotype information from thousands of variable sites dispersed across the entire genomes of these fishes, with which we achieved a higher level of resolution of patterns of genetic variation than previous studies. We generated DNA-sequence data using the Illumina GAII platform for 192 fish: 41 P. formosa (5 localities where P. formosa is sympatric with P. mexicana and 6 localities where it is sympatric with P. latipinna), 82 P. latipinna (from 22 localities across Louisiana, Texas, and Mexico), and 69 P. mexicana (from 13 localities across Mexico and Honduras; Fig. 1A). We identified 26,313 single-nucleotide polymorphism (SNP) markers that were used for the analyses.
Our first goal was to confirm the hybrid origin of P. formosa and determine the genomic contribution from its parent species. Our results are in agreement with the findings of several previous studies: P. formosa is a hybrid between P. mexicana and P. latipinna (11, 14–19). A principal components analysis (PCA) of the genotype estimates of each individual at each locus (Fig. 1 B–D), an admixture analysis performed in STRUCTURE with K = 2 (forcing individuals to be assigned to one of only two clusters) (Fig. 2A), calculation of genome-average pairwise GST (Fig. S1), and the hybrid index for each P. formosa (Fig. 3) all suggest that gynogenetic individuals have a genome that is intermediate between P. latipinna and P. mexicana. Principal component axes 1 (PC1) and 2 (PC2) collectively explained 63% of the genetic variation (Fig. 1B). PC1 appears to divide the three species into three distinct clusters, with P. formosa in an intermediate position between the parent species, consistent with the hypothesis of a hybrid origin for P. formosa (Fig. 1B). PC2 separates the asexual species from the sexual species and illustrates the variation in genotypes among individuals (Fig. 1B). Interestingly, the genotypic variation within P. formosa was high: individuals of P. formosa did not all cluster together but were differentiated along the second principal component just as much as the sexual individuals. PC3 explained 5.7% of the variation and separated the populations of P. mexicana into three groups, which corresponded to three geographic regions (north, central, and south) (Fig. 1C). PC4 explained 4.0% of the variation and divided P. latipinna into two geographic regions: north plus central (the central group is comprised of populations in central Texas that were introduced from the north group, including population in Florida and the Gulf Coast of the United States) and south (Fig. 1D).
Fig. 2.
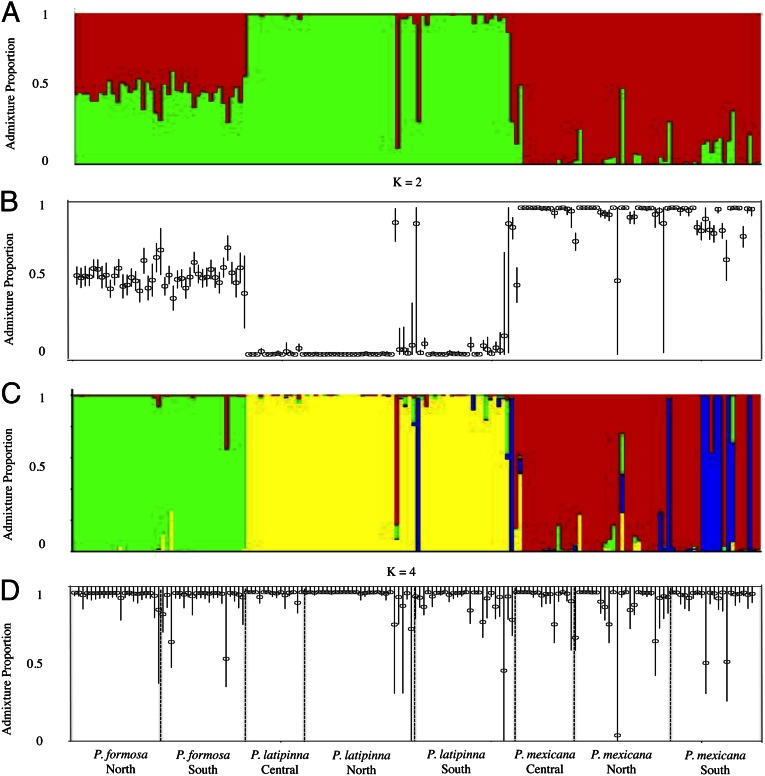
Results from STRUCTURE clustering analyses. Admixture proportion for K = 2 (A) and mean assignment probabilities to cluster 1 ±95% credible intervals for K = 2 (B). Admixture proportions for K = 4 (C) and mean assignment probability ±95% credible intervals for K = 4 for cluster individuals were assigned to D. K = 2 and K = 4 were chosen as the appropriate number of groups after examination of the log of the marginal likelihood and the ad hoc delta(K) statistic (41) for each K (Fig. S6).
Fig. 3.
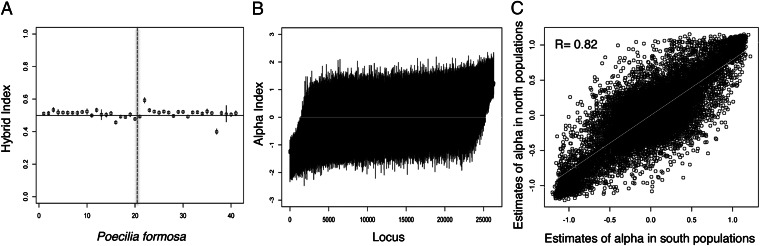
Posterior probability of estimates of hybrid indices and cline parameter α for P. formosa. The assigned putative parent populations were P. mexicana found in the northern part of its range and P. latipinna found in the southern part of its range. A depicts the posterior probability distribution of the hybrid index estimates (±95% credible intervals) for the 41 P. formosa used in this study. The vertical dotted line in A divides P. formosa sympatric with P. latipinna (Left) from P. formosa sympatric with P. mexicana (Right). Each black line in B represents the 95% credible interval for the estimate of the cline parameter α for each of the 26,313 SNPs, for all 41 P. formosa. C shows the correlation between the alpha estimates at each locus in the populations of P. formosa in the south vs. populations in the north.
Estimates of pairwise GST also confirmed the genomic intermediacy of P. formosa (Fig. S1 and Table 1). Differentiation between P. latipinna and P. mexicana was approximately twice that observed between P. formosa and either parent species (Table 1). Within-species (among-population) differentiation was an order of magnitude lower, and lowest in P. formosa, although distinctly nonzero. Estimation of hybrid indices for the asexual individuals was also consistent with the hypothesis of hybrid origin: for the 41 P. formosa examined, hybrid index ranged from 0.37 to 0.56 (mean, 0.49; Fig. 3A). These estimates are in agreement with the estimates of admixture proportion calculated using STRUCTURE (Fig. 2A).
Table 1.
Summary of genome-wide GST calculation
| Pairwise comparisons | n | Mean GST | Maximum GST | Minimum GST |
| Intraspecific comparisons | ||||
| P. latipinna vs. P. latipinna | 3 | 0.06 ± 0.02 | 0.043 | 0.081 |
| P. mexicana vs. P. mexicana | 3 | 0.096 ± 0.02 | 0.077 | 0.125 |
| P. formosa vs. P. formosa | 1 | 0.028 | Upper bound: 0.0278 | Lower bound: 0.0288 |
| Interspecific comparisons | ||||
| P. latipinna vs. P. mexicana | 9 | 0.361 ± 0.01 | 0.341 | 0.380 |
| P. mexicana vs. P. formosa | 6 | 0.155 ± 0.02 | 0.132 | 0.180 |
| P. formosa vs. P. latipinna | 6 | 0.163 ± 0.01 | 0.144 | 0.170 |
Mean and range of GST for comparisons between populations of P. latipinna and P. mexicana and for comparison within species among populations of P. latipinna (North, Central, and South; Fig. 1) and P. mexicana (North, Central, and South; Fig. 1). Mean and upper and lower bounds provided for the comparison between the two regions (North and South) of P. formosa.
Given the patterns and diversity captured by both the estimates of hybrid index and the PCA, we calculated the probability of ancestry for each locus in P. formosa given the two parent populations and the individual’s hybrid index. This estimate is calculated using the Bayesian genomic cline model (22, 23) and is summarized in the α parameter. The α parameter is a population-level parameter that specified whether, on average, individuals are less or more likely to have either parent species’ ancestry given their hybrid index (which is a genome-wide measure of ancestry). The parameter α provides information about ancestry for a locus across individuals and not allelic state; thus, we used α to examine the organization of the genome of P. formosa with respect to ancestry. For frozen F1 hybrids obtained from parent species that are differentiated at all loci, we expect hybrid indexes of 0.5 (consistent with our mean estimates for P. formosa) and values of the cline parameter α to be zero across all loci, indicating no excess ancestry. However, our results suggest that P. formosa is either not a frozen F1 hybrid derived from a single F1 individual or substantial evolution has occurred in P. latipinna and P. mexicana since they gave rise to P. formosa. As shown in Fig. 3B, most loci in P. formosa have parameter α estimates not different from zero, indicating no excess ancestry for either parent species at those loci. However, about 12% of the loci have excess ancestry from one or the other parent species, consistent with the conclusion that genetic recombination might have occurred in P. formosa. An alternative explanation for these results could be that the allele frequencies of the parent species at the time of hybridization were much more similar than they are now. Interestingly 15% of the SNPs analyzed in this study are polymorphic in all three species, whereas 12.5% are shared only by P. formosa and P. latipinna and 21.6% are shared by P. formosa and P. mexicana. These results suggest that possibly P. latipinna and P. mexicana were more similar at the time of hybridization but have since diverged because of drift, mutations, and selection. In fact, only 1.8% of SNPs are now shared exclusively between the two sexual species, indicating that they are now clearly differentiated (as also suggested from the estimates of GST).
The possibility that our results are attributable only to the fact that the parent populations were more similar at the time of hybridization and differentiated since is not a trivial one. To estimate both the hybrid index and α parameter we must assign putative parent species to P. formosa. We chose populations of P. mexicana found in the northern part of the species’ range and populations of P. latipinna found in the southern part of its range because previous work has suggested that the region of Tampico, Mexico, is where P. formosa has originated (13, 17). However, this ad hoc decision might have influenced the estimates of α, given that the allele frequencies of the sexual individuals found in those populations now might be significantly different from the allele frequencies during the time of hybridization. To address this possibility, we also estimated α for all of the other parent population combinations and found that the locus-specific α estimates did not correlate between different parent population combinations (Table S1 and Fig. S2). The patterns found during the estimation of α across the genome can be explained by (i) recombination in the gynogenetic species coupled with differentiation between the parent populations or (ii) high differentiation in the parent species (and populations within each parent species) after the hybridization event. The estimation of linkage and Hardy–Weinberg (HW) disequilibria (Fig. S3) provide evidence to support the hypothesis that some recombination has occurred in P. formosa. In fact, calculations of a Burrow’s composite measure of linkage disequilibrium demonstrate that P. formosa exhibits less disequilibrium than expected for an asexual lineage of hybrid origin descending from a single F1 individual, which suggests that recombination has reduced the linkage disequilibria created by admixture (SI Results and Discussion and Fig. S3).
To rule out the hypothesis that the excess ancestry recorded was attributable to recent gene flow with either of the parent species, we compared the estimations of the α parameter for the northern and southern populations of P. formosa, where P. formosa is sympatric with P. latipinna and P. mexicana, respectively. This analysis revealed no difference in the pattern of parent contribution (Fig. 3C). Thus, the excess ancestry for ∼12% of the loci surveyed does not vary with geography across P. formosa and does not appear to be a function of recent gene flow from the parent species. Although previous studies have found that microchromosomes might be inherited by P. formosa (24), we found no evidence to support their results. However, given that we only used SNPs that had a minimum coverage of five reads per population, and microchromosome inheritance has been recorded only in a handful of populations not sampled extensively in this study, it is possible that microchromosomes were not included in our analysis at all because they are not present in every population.
Together with investigating the ancestry of P. formosa, we tried to shed light on the allelic state of the loci used for the analyses. The estimation of observed heterozygosity across loci revealed patterns that are inconsistent with the hypothesis that P. formosa is a frozen F1 hybrid descended from a single F1 individual (Fig. S4). Specifically, loci with observed heterozygosities different from 0 (for loci at which the parents are not differentiated) or 1 (for loci at which the parents are differentiated) are not expected in F1 hybrids descended from a single F1 individual (which would be consistent with the hypothesis of a single hybridization forming the mother of all P. formosa). The variation in heterozygosity across loci (Fig. S4) is, instead, consistent with a short history of backcrossing, perhaps before the onset of gynogenesis or with the hypothesis that P. formosa is comprised of descendants of multiple, independently formed F1 gynogenetic individuals. However, the data gathered for the present study and work performed in other laboratories suggest that a short history of recombination is more likely than the hypothesis of independent origins of clonally reproducing F1 hybrids. The ancestral P. formosa might have been a sexually reproducing hybrid for some time before becoming gynogenetic, as hypothesized by Turner et al. (16), and later by Stöck et al. (19). Unisexual sperm-dependent organisms might be rare because certain genetic compositions (and possibly specific epistatic interactions) are necessary for the evolution of gynogenesis. Stöck et al. (19) referred to this idea as the “rare formation hypothesis.” Perhaps the specific combination of alleles required for gynogenesis only occurred after some cycles of recombination and independent assortment of alleles. This possibility could explain why no one has been able to reproduce P. formosa in the laboratory, even after extensive attempts to do so (13–16, 19–21).
A recent study also found some loci in P. formosa that were homozygous for one of the parent species, and the authors suggested that mitotic gene conversion (or mitotic recombination) during gamete formation might explain the pattern (18, 19). Our results are consistent with the hypothesis of mitotic gene conversion. When this particular type of recombination occurs, some loci become homozygous for one of the alleles (25), causing a loss of heterozygosity and an increase in linkage disequilibrium. The probability of the occurrence and success of gene conversion varies across the genome (26). This mechanism causes genomes to vary among individuals and causes a decay of admixture linkage disequilibrium because recombination within admixed individuals and between chromosomes of different ancestry occurs. Thus, mitotic gene conversion could potentially explain the observed variation in ancestry across loci in P. formosa and the variation among individuals of P. formosa.
The last goal of our study was to determine the amount of genotypic variation present within P. formosa. The GST calculations and the PCA suggest relatively high genotypic diversity in P. formosa. This observation is in agreement with previously published results (17, 27, 28), which all found that P. formosa was genotypically variable. Stöck et al. (19) suggest that the high genetic diversity in P. formosa is attributable to high mutation rates because the phylogenetic analyses of mtDNA variation suggested a monophyletic origin of P. formosa (19). Turner et al. (27) also suggested that high mutation rates are more probable than multiple hybrid origins based on allozyme data. However, some of the P. formosa studied by Turner et al. (27) were collected in the population where triploid individuals are present, and, therefore, the high clonal diversity found in this population in the Rio Purification might have been caused by the presence of triploids. We did not include triploids in this study. To address the possibility suggested in previous studies, that high mutation rates within P. formosa contribute to high genetic variation, we calculated the percentage of variable SNPs private to P. formosa and found that 3% of the SNPs used in this study are only found in the gynogenetic species. Thus, mutation accumulation within P. formosa has been moderate and does not fully explain the genotypic diversity in this species.
Our analyses using more than 25,000 SNPs from across the genome of P. formosa document considerable genotypic variation within this gynogenetic species. Given the complexity of the genomic patterns across loci and among individuals, it is not currently possible to make definitive inferences about the number of clonal lineages or the number of hybridization events involved in the origin of P. formosa. We can conclude, however, that our results are consistent with the hypothesis that the formation of P. formosa as a unisexual species is the result of hybridization and possible subsequent recombination. Recombination might have occurred following hybridization and the resulting genotypic variation has obscured our ability to discern distinct lineages. Results from calculation of genetic distances between individuals across loci illustrates that individual P. formosa are not identical to one another (Fig. S5). However, differentiation between P. formosa from different geographic regions appears to be less than among populations of either parent species (Fig. S5). Recombination following hybridization (i.e., recombination via sexual reproduction) could have occurred before the onset of the gynogenetic reproductive mode, resulting in numerous genotypes (as suggested from the calculation of observed heterozygosity). Alternatively, some form of asexual recombination, most likely mitotic gene conversion or automixis (29, 30, 31), might have occurred (or might still be occurring). Similar mechanisms for the production of genetic diversity have been proposed for the unisexual lizards in the genus Darevskia [formerly Lacerta, Lacertidae (32, 33)] and have been shown to possibly play a significant role in the maintenance of asexual species (8).
Regardless of the origins of genetic variation in P. formosa, it is clear that this variation could contribute to the persistence of this species. Coexistence between a unisexual sperm-dependent species, and its host can be achieved and maintained if genetic variation is present in a population because natural selection can select against the clones that overlap extensively in resource use with their host species (frozen niche variation) (34, 35). Intriguingly, some form of asexual recombination, such as mitotic gene conversion or automixis, might also facilitate a reduction in the rate of accumulation of deleterious mutations (2) and increase the longevity of P. formosa beyond what is predicted by theoretical models (36).
Materials and Methods
In-depth descriptions of the protocols, models, and calculations used are in SI Materials and Methods. This work was performed under Institutional Animal Care and Use Committee no. 0818_0325_18.
Next-generation DNA sequence data from 192 fish were generated with the Illumina GAII platform following recently developed methods (for more details see, refs. 37 and 38 and SI Materials and Methods). A total of 32,492 variable sites were identified using custom Perl scripts together with samtools and bcftools (39). Because of the low numbers of individuals sampled from each locality, we pooled individuals across localities into eight geographical regions to obtain adequate sample sizes to perform all of our analyses (Fig. 1A).
We trimmed data to only those SNPs with a minimum of five reads per marker per region (population grouping), which produced 26,313 SNPs, and then used Bayesian hierarchical models to estimate allele frequencies for each locus based on the observed data by using the allele frequency Bayesian model presented in Gompert and Buerkle (37). We summarized population genetic structure at the individual level using both a PCA and STRUCTURE 2.2 (40, 41). We also summarized population genetic structure at the population level by calculating pairwise GST statistics (42) for all combinations of regional groupings (Fig. 3).
To investigate the genomic composition of P. formosa, we used a Bayesian approach to estimate hybrid index for all P. formosa individuals and to asses ancestry (relative to the putative parental species) at all SNP loci for all individuals. To obtain a clearer picture about the robustness of the pattern obtained using the putative parent populations (P. latipinna and P. mexicana in northern Mexico), we repeated the estimation of both the hybrid index (Fig. S2) and α for all combinations of possible parent populations and calculated the correlation coefficient between these new estimates vs. the estimates obtained with the putative parent populations (Table S1).
Results from the genomic clines analysis (Results and Discussion) provided possible evidence of a history of recombination in P. formosa. This was unexpected given the hybrid origin of P. formosa and the presumed lack of recombination in this asexual species. Consequently, we predicted substantially higher linkage disequilibrium in this species compared with the parental species. We, therefore, calculated Burrow’s composite measure of linkage disequilibrium (Δ) between all pairs of variable loci (43, 44) and performed a set of simulations to untangle the effects of linkage disequilibrium and HW disequilibrium on Δ.
To further investigate the results of the linkage disequilibrium estimation and to better understand the allelic state of the loci analyzed, we calculated the observed heterozygosity for each locus in each population (Fig. S4).
To address the question of whether mutation accumulation only can explain the variation present in P. formosa (as suggested by previous studies), we calculated the proportion of variable SNPs private to P. formosa, P. mexicana, and P. latipinna, as well as the proportion of SNPs shared by all species and by only two species.
As an alternative means of illustrating the genotypic variation observed within P. formosa (Fig. 1B), we calculated the “genotypic distance” between each pair of individuals at each locus as a measure of genotypic dissimilarity among individuals (Fig. S5).
Supplementary Material
Acknowledgments
We thank Michael Sandel, Ryan Earley, Margaret Ptacek, José Jaime Zúñiga-Vega, Kristen Epp, and Celeste Espinedo for collecting fish; Alex Buerkle for computational help and suggestions; Tom Parchman for help with scripts; Kate Bell, Lauren Lucas, Darrin Hulsey, and Mariana Mateos for suggestions; the Mexican government for collection permit DGOPA.07011.210612.1750; the Texas State University computer cluster; and the University of Wyoming Seismic computer cluster. This work was funded by the National Science Foundation (Integrative Organismal Systems Grant IOS-1021873 to C.R.G., A.S.A., and C.C.N.; and Division of Environmental Biology Grant DEB-1050355 to C.C.N.). Additional funding was provided by the Raney Fund Award (American Society for Ichthyologists and Herpetologists), the Theodore Roosevelt Memorial Grant (American Museum of Natural History), the Howard McCarley Student Research Award (Southwestern Association of Naturalists), the Vern Parish Grant (American Livebearer Association), and the Texas Academy of Science Student Research Grant (Texas Academy of Science) (to L.A.d.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Dryad Digital Repository, www.datadryad.org (Dryad data package DOI: 10.5061/dryad.h6p35).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303730110/-/DCSupplemental.
References
- 1.Avise JC. Clonality: The Genetics, Ecology and Evolution of Sexual Abstinence in Vertebrate Animals. New York: Oxford Univ Press; 2008. [Google Scholar]
- 2.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106(1):2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 3.Maynard Smith J. Evolution in sexual and asexual populations. Am Nat. 1968;102(197):469–473. [Google Scholar]
- 4.Beukeboom LW, Vrijenhoek RC. Evolutionary genetics and ecology of sperm-dependent parthenogenesis. J Evol Biol. 1998;11(6):755–782. [Google Scholar]
- 5.Normark BB. Evolution in a putatively ancient asexual aphid lineage: Recombination and rapid karyotype change. Evolution. 1999;53(5):1458–1469. doi: 10.1111/j.1558-5646.1999.tb05410.x. [DOI] [PubMed] [Google Scholar]
- 6.Souza-Júnior SA, Becker TCA, Castro-Prado MAA. Asexual recombination in a uvsH mutant of Aspergillus nidulans. Biol Res. 2007;40(1):65–71. doi: 10.4067/s0716-97602007000100007. [DOI] [PubMed] [Google Scholar]
- 7.Omilian AR, Cristescu MEA, Dudycha JL, Lynch M. Ameiotic recombination in asexual lineages of Daphnia. Proc Natl Acad Sci USA. 2006;103(49):18638–18643. doi: 10.1073/pnas.0606435103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mandegar MA, Otto SP. Mitotic recombination counteracts the benefits of genetic segregation. Proc Biol Sci. 2007;274(1615):1301–1307. doi: 10.1098/rspb.2007.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawley RM, Bogart JP. In: Evolution and Ecology of Unisexual Vertebrates. Dawley RM, Bogart JP, editors. New York: New York State Museum; 1989. [Google Scholar]
- 10.Choleva L, Apostolou A, Rab P, Janko K. Making it on their own: Sperm-dependent hybrid fishes (Cobitis) switch the sexual hosts and expand beyond the ranges of their original sperm donors. Philos Trans R Soc Lond B Biol Sci. 2008;363(1505):2911–2919. doi: 10.1098/rstb.2008.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hubbs CL, Hubbs LC (1932) Apparent parthenogenesis in nature in a form of fish of hybrid origin. Science 76(1983):628–630. [DOI] [PubMed]
- 12.Bi K, Bogart JP. Time and time again: Unisexual salamanders (genus Ambystoma) are the oldest unisexual vertebrates. BMC Evol Biol. 2010;10:238. doi: 10.1186/1471-2148-10-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avise JC, Trexler JC, Travis J, Nelson W. Poecilia mexicana is the recent female parent of the unisexual fish P. formosa. Evolution. 1991;45(6):1530–1533. doi: 10.1111/j.1558-5646.1991.tb02657.x. [DOI] [PubMed] [Google Scholar]
- 14.Niemeitz A, Kreutzfeldt R, Schartl M, Parzefall J, Schlupp I. Male mating behavior of a molly, Poecilia latipunctata: A third host for the sperm-dependent Amazon molly, Poecilia formosa. Acta Ethol. 2002;5(1):45–49. [Google Scholar]
- 15.Abramoff P, Darnell RM, Balsano JS. Electrophoretic demonstration of the hybrid origin of the gynogenetic Teleost Poecilia formosa. Am Nat. 1968;102(928):555–558. [Google Scholar]
- 16.Turner BJ, Brett BH, Miller RR. Interspecific hybridization and the evolutionary origin of a gynogenetic fish, Poecilia formosa. Evolution. 1980;34(5):917–922. doi: 10.1111/j.1558-5646.1980.tb04029.x. [DOI] [PubMed] [Google Scholar]
- 17.Schartl M, Wilde B, Schlupp I, Parzefall J. Evolutionary origin of a parthenoform, the Amazon molly, Poecilia formosa, on the basis of a molecular genealogy. Evolution. 1995;49(5):827–835. doi: 10.1111/j.1558-5646.1995.tb02319.x. [DOI] [PubMed] [Google Scholar]
- 18.Tiedemann R, Moll K, Paulus KB, Schlupp I. New microsatellite loci confirm hybrid origin, parthenogenetic inheritance, and mitotic gene conversion in the gynogentic Amazon molly (Poecilia formosa) Mol Ecol Notes. 2005;5(3):586–589. [Google Scholar]
- 19.Stöck M, Lampert KP, Möller D, Schlupp I, Schartl M. Monophyletic origin of multiple clonal lineages in an asexual fish (Poecilia formosa) Mol Ecol. 2010;19(23):5204–5215. doi: 10.1111/j.1365-294X.2010.04869.x. [DOI] [PubMed] [Google Scholar]
- 20.Dries LA. Peering through the looking glass at a sexual parasite: Are Amazon mollies red queens? Evolution. 2003;57(6):1387–1396. doi: 10.1111/j.0014-3820.2003.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 21.Ptacek MB. Patterns of inheritance of mating signals in interspecific hybrids between sailfin and shortfin mollies (Poeciliidae: Poecilia: Mollienesia) Genetica. 2002;116(2-3):329–342. [PubMed] [Google Scholar]
- 22.Gompert Z, Buerkle CA. Bayesian estimation of genomic clines. Mol Ecol. 2011;20(10):2111–2127. doi: 10.1111/j.1365-294X.2011.05074.x. [DOI] [PubMed] [Google Scholar]
- 23.Gompert Z, et al. Genomic regions with a history of divergent selection affect fitness of hybrids between two butterfly species. Evolution. 2012;66(7):2167–2181. doi: 10.1111/j.1558-5646.2012.01587.x. [DOI] [PubMed] [Google Scholar]
- 24.Nanda I, et al. Stable inheritance of host species-derived microchromosomes in the gynogenetic fish Poecilia formosa. Genetics. 2007;177(2):917–926. doi: 10.1534/genetics.107.076893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen JM, Cooper DN, Chuzhanova N, Férec C, Patrinos GP. Gene conversion: Mechanisms, evolution and human disease. Nat Rev Genet. 2007;8(10):762–775. doi: 10.1038/nrg2193. [DOI] [PubMed] [Google Scholar]
- 26.Jeffreys AJ, May CA. Intense and highly localized gene conversion activity in human meiotic crossover hot spots. Nat Genet. 2004;36(2):151–156. doi: 10.1038/ng1287. [DOI] [PubMed] [Google Scholar]
- 27.Turner BJ, Elder JF, Jr, Laughlin TF, Davis WP. Genetic variation in clonal vertebrates detected by simple-sequence DNA fingerprinting. Proc Natl Acad Sci USA. 1990;87(15):5653–5657. doi: 10.1073/pnas.87.15.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaschl H, Tobler M, Plath M, Penn DJ, Schlupp I. Polymorphic MHC loci in an asexual fish, the amazon molly (Poecilia formosa; Poeciliidae) Mol Ecol. 2008;17(24):5220–5230. doi: 10.1111/j.1365-294X.2008.03997.x. [DOI] [PubMed] [Google Scholar]
- 29.Lutes AA, Neaves WB, Baumann DP, Wiegraebe W, Baumann P. Sister chromosome pairing maintains heterozygosity in parthenogenetic lizards. Nature. 2010;464(7286):283–286. doi: 10.1038/nature08818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasch EM, Monaco PJ, Balsano JS. Cytophotometric and autoradiographic evidence for functional apomixis in a gynogenetic fish, Poecilia formosa and its related, triploid unisexuals. Histochemistry. 1982;73(4):515–533. doi: 10.1007/BF00493366. [DOI] [PubMed] [Google Scholar]
- 31.Lampert KP, et al. Automictic reproduction in interspecific hybrids of poeciliid fish. Curr Biol. 2007;17(22):1948–1953. doi: 10.1016/j.cub.2007.09.064. [DOI] [PubMed] [Google Scholar]
- 32.Murphy RW, et al. Old age, multiple formations or genetic plasticity? Clonal diversity in the uniparental Caucasian rock lizard, Lacerta dahli. Genetica. 1997;101(2):125–130. doi: 10.1023/A:1018392603062. [DOI] [PubMed] [Google Scholar]
- 33.Kupriyanova L. Cytogenetic and genetic trends in the evolution of unisexual lizards. Cytogenet Genome Res. 2009;127(2-4):273–279. doi: 10.1159/000303325. [DOI] [PubMed] [Google Scholar]
- 34.Vrijenhoek RC, Angus RA, Schultz RJ. Variation and clonal structure in unisexual fish. Am Nat. 1978;112(983):41–55. [Google Scholar]
- 35.Vrijenhoek RC, Parker ED. Lost Sex. The Evolutionary Biology of Parthenogenesis. New York: Springer; 2009. pp. 99–131. [Google Scholar]
- 36.Loewe L, Lamatsch DK. Quantifying the threat of extinction from Muller’s ratchet in the diploid Amazon molly (Poecilia formosa) BMC Evol Biol. 2008;8:88–108. doi: 10.1186/1471-2148-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gompert Z, Buerkle CA. A hierarchical Bayesian model for next-generation population genomics. Genetics. 2011;187(3):903–917. doi: 10.1534/genetics.110.124693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parchman TL, et al. Genome-wide association genetics of an adaptive trait in lodgepole pine. Mol Ecol. 2012;21(12):2991–3005. doi: 10.1111/j.1365-294X.2012.05513.x. [DOI] [PubMed] [Google Scholar]
- 39.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 42.Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA. 1973;70(12):3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weir BS. Inferences about linkage disequilibrium. Biometrics. 1979;35(1):235–254. [PubMed] [Google Scholar]
- 44.Zaykin DV. Bounds and normalization of the composite linkage disequilibrium coefficient. Genet Epidemiol. 2004;27(3):252–257. doi: 10.1002/gepi.20015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.