Significance
We describe conditional by inversion (COIN), a new design for conditional alleles that uses an optimized conditional gene trap module (COIN module) inserted into the target gene in an orientation opposite to the gene’s direction of transcription. Activation by Cre recombinase inverts the COIN module, resulting in expression of a reporter and termination of transcription, thereby inactivating the target gene while marking the cells where the conditional event has occurred. Creation of COIN alleles for more than 20 genes showed that it is a robust and universal method—applicable to any gene regardless of exon–intron structure—that overcomes the limitations of previous conditional approaches.
Keywords: genome engineering, conditional-null
Abstract
Conditional mutagenesis is becoming a method of choice for studying gene function, but constructing conditional alleles is often laborious, limited by target gene structure, and at times, prone to incomplete conditional ablation. To address these issues, we developed a technology termed conditionals by inversion (COIN). Before activation, COINs contain an inverted module (COIN module) that lies inertly within the antisense strand of a resident gene. When inverted into the sense strand by a site-specific recombinase, the COIN module causes termination of the target gene’s transcription and simultaneously provides a reporter for tracking this event. COIN modules can be inserted into natural introns (intronic COINs) or directly into coding exons as part of an artificial intron (exonic COINs), greatly simplifying allele design and increasing flexibility over previous conditional KO approaches. Detailed analysis of over 20 COIN alleles establishes the reliability of the method and its broad applicability to any gene, regardless of exon–intron structure. Our extensive testing provides rules that help ensure success of this approach and also explains why other currently available conditional approaches often fail to function optimally. Finally, the ability to split exons using the COIN’s artificial intron opens up engineering modalities for the generation of multifunctional alleles.
Conditional alleles are rapidly becoming a method of choice for the analysis of gene function, particularly for mutations that result in embryonic lethality or when it is desirable to study the role of a gene in a specific tissue or developmental stage. The most commonly used conditional alleles are those alleles where part of the gene of interest is flanked by site-specific recombinase recognition sites (SRSs) in a manner such that deletion or inversion of the SRS-flanked sequence—induced by the action of a cognate recombinase—will result in the generation of a null allele. Combined with the ability to spatiotemporally control the expression and/or the activity of the recombinase, this methodology enables induction of the null state in a cell type-selective as well as temporally regulated manner. As a result, recombinase-regulated conditional-null alleles have provided a powerful tool to query the function of a target gene in specific cell types or tissues and/or at different time points during the life of a model organism (1).
Conditional alleles can, however, be difficult to design and engineer, and they may require laborious vector construction as well as multiple manipulations in ES cells (example in ref. 2). Approaches that involve flanking the entire exon–intron region of the target gene with SSR sites while attempting to avoid disruption of associated transcriptional control elements are practical only for genes with compact exon–intron structure and well-mapped regulatory regions. Therefore, alternative approaches have been developed in which either only exons that are deemed critical for a gene’s function are flanked by loxP sites (3) or deletion is avoided altogether by inserting invertible elements into introns and antisense to the target gene, with the intent that these elements remain inert in their initial orientation but disable the gene on conditional inversion to the sense strand (4) (SI Appendix, Fig. S17). Both of these approaches suffer from the limitation that they are not applicable to single exon genes. Furthermore, although the latter method is, in theory, more amenable to broader and less customized use, in practice, it is limited by the availability and location of targetable introns, and it may yield hypomorphs before inversion and incomplete nulls after inversion (4, 5). Finally, the majority of currently available conditional approaches do not incorporate a reporter that can mark individual cells where the conditional allele has been activated, making it difficult to assess the efficiency and cell-type specificity of the target gene’s inactivation in complex tissues.
In an attempt to rectify the deficiencies inherent in the currently available conditional-null allele methods, we sought to develop a unique allele design that combines four properties:
i) Simple, modular, and standardized design that can be engineered in a single bacterial homologous recombination (BHR) step and a single targeting event in ES cells.
ii) General applicability to the vast majority of genes, regardless of a gene’s exon–intron structure, to minimize the need to define transcriptional control elements or functional domains in the target gene most importantly and uniquely by allowing for insertion into an exon by the creation of an artificial intron.
iii) Optimal properties of conditional-null alleles (silent before induction and null after induction by the recombinase).
iv) Ability to mark cells in which the targeted gene has been inactivated by turning on expression of a reporter simultaneously with induction of the conditional allele.
We present here conditional by inversion (COIN), a method that incorporates these properties. By applying rigorous tests of conditionality to a set of well-studied genes, we establish the COIN conditional-null reporter allele as a reliable and versatile gene modification method that opens up unique design modalities.
Results
Structure of a COIN Conditional Allele.
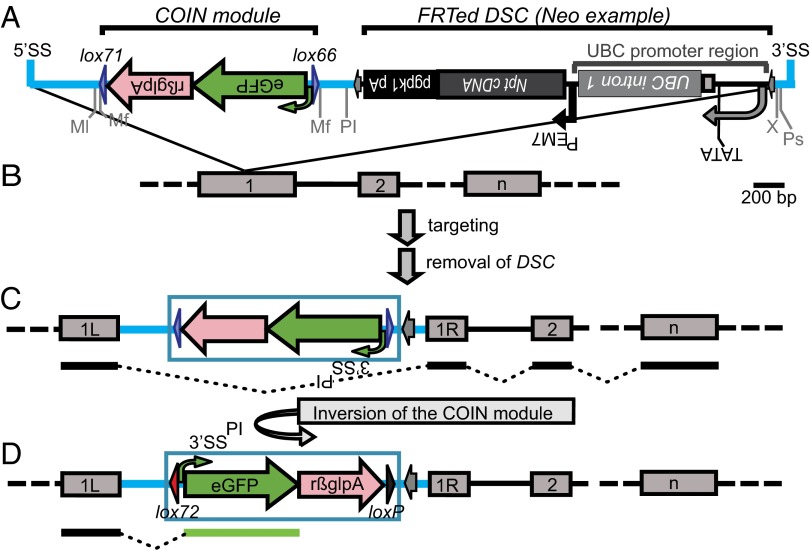
Like all conditional alleles, COIN alleles are designed to modify a target gene in such a way that the gene’s expression and function remain normal until the allele is inactivated by a site-specific recombinase, which induces a physical rearrangement that renders the gene inactive or alters the function of the gene product. The COIN design (Fig. 1A) consists of two linked elements. The first element is a conventional FRT-flanked drug selection cassette (DSC), which is removed by Flp recombinase before use of the COIN. The second element is the COIN module, the functional part of the allele, which consists of initially inverted sequences that comprise a terminal exon encoding a reporter protein, such as enhanced GFP (eGFP), preceded by a 3′ splice site (3′SSPI) and followed by a polyadenylation region (pA). Extensive testing of alternative sequences for the 3′SSPI and pA and their positioning was performed to optimize the COIN module (SI Appendix, Fig. S1). To enable its inversion by Cre recombinase, the COIN module is flanked by lox66 and lox71 sites in a head-to-head orientation (6).
Fig. 1.
Design of COIN alleles. (A) Detailed schematic of the COIN intron (version eGFP, phase 0; neo). The COIN module was inserted into the MfeI site (Mf) of HBB2 intron 2, and it is comprised of a 3′SSPI-eGFP-rßglpA flanked by lox71 and lox66 in a mirror image configuration to enable inversion of 3′SSPI-eGFP-rßglpA; 3′SSPI is the 3′ splice region of HBB2 intron 2 (SI Appendix, Fig. S14). Postinversion (PI) indicates that this 3′SS participates in trapping the message of the COIN allele after inversion of the COIN module. rßglpA is mainly the 3′UTR region of HBB2 (SI Appendix, Fig. S15). FRTed DSC is an FRT-flanked drug selection minigene [e.g., PUBC-driven neomycin phosphotransferase (Npt)] (SI Appendix, Fig. S16); FTR sites are indicated as gray arrows. The 5′ splice site (5′SS) and 3′ splice site (3′SS) of rßgl i2 mark the boundaries of the COIN intron (blue line). Restriction sites are shown below the sequence line. Ml, MluI; Ps, PspOMI; X, XmaI. All elements are drawn to scale. (Scale bar: 200 bp.) (B) A hypothetical gene comprised of n number of exons is rendered conditional-null by insertion of the COIN intron (elements not drawn to scale) into exon 1, (C) splitting exon 1 into a left exon (1L) and a right exon (1R). After BHR, targeting, and removal of the DSC, the resulting exCOIN allele encodes a WT transcript, because the COIN module is in the antisense strand and therefore, stealth to transcription. (D) Inversion of the COIN module results in a null allele encoding for a transcript that incorporates exon 1L and eGFP. In addition, Cre-mediated inversion of the COIN module through lox71 and lox66 gives rise to a double mutant lox site, lox72 (red arrowhead), and loxP (black arrowhead). The COIN module is framed by a blue box in C and D to facilitate visualization of the region being inverted.
Although COINs can be engineered into native introns (SI Appendix, Fig. S6), it is their ability to be introduced into exons that renders COINs a general strategy applicable to the majority of genes. Insertion into an exon is enabled by embedding the COIN module (and DSC) into an exogenous artificial intron (Fig. 1A) (hereafter referred to as the COIN intron). This strategy requires that (i) the COIN intron is efficiently spliced so that the COIN allele remains silent in its original orientation and (ii) the COIN module operates as a terminal exon when inverted into the sense orientation. We based the COIN intron on rabbit β-hemoglobin (HBB2) intron 2, because it has been shown to function in the context of several genes (7). To maximize the chances of efficient splicing when the COIN module is operating in different genomic contexts, we chose an efficient (8, 9) (SI Appendix, Fig. S1) and coevolved 3′SSPI–pA pair from HBB2 (10). A protocol detailing where to place the COIN intron can be found in Materials and Methods.
COIN allele targeting vectors are engineered by BHR to insert the linked DSC and COIN module elements into a defined site within the target gene (Fig. 1B), such that the COIN module’s protein coding, splicing, and pA sequences are antisense to the direction of transcription of the target gene, thereby ensuring that the COIN module is not incorporated into the target gene’s transcript (Fig. 1C). After targeting in ES cells, we use FLPe recombinase (11) to excise the DSC, resulting in a silent COIN allele ready for induction (Fig. 1C). Our experience is consistent with observations reported by others (12, 13) (i.e., that removal of the DSC is essential to avoid the generation of partially defective hypomorphic alleles).
Upon Cre-mediated inversion of the COIN module, its reporter-coding exon is brought in position to trap the target gene’s transcript by splicing to the 3′SS of the module and terminating transcription at the pA of the module, simultaneously abrogating transcription of the target gene’s downstream exons (Fig. 1D). Moreover, the inverted COIN is effectively fixed into the sense orientation, because Cre-mediated inversion converts the lox66-lox71 pair into an lox72-loxP pair, which does not support reinversion (14); in fact, we never observed a reinversion event in the course of these studies.
To validate the COIN method, we engineered COIN alleles for 26 protein-coding genes whose inactivation would result in known or predicted phenotypes and scored them (Table 1) using the following criteria: (i) normal Mendelian inheritance and wild type (WT) phenotype in mice either homozygous for the noninverted and thus, silent COIN allele (COIN/COIN) or compound heterozygous with the corresponding null allele (COIN/null); (ii) phenotypes and inheritance patterns identical to those of KO mice (null/null) in mice either homozygous for the inverted COIN allele (COIN-INV/COIN-INV) or compound heterozygous with the corresponding null allele (COIN-INV/null); (iii) normal target gene expression for the COIN allele compared with the unmodified gene; and (iv) COIN module reporter expression on inversion of the COIN. Where noted, phenotypes of some COIN alleles were studied in ES cells. Based on these criteria, nearly all of the COIN alleles were found to be completely WT before inversion and completely null postinversion (Table 1); our extensive testing revealed COIN design rules to help prevent rare failures (i.e., to prevent perturbation of WT functionality before inversion and ensure complete nullness as opposed to hypomorphism after inversion). The following sections describe COIN allele examples that show the efficacy of the design and illustrate features and properties that can affect the performance of the alleles.
Table 1.
List of COIN alleles and their pre- and postinversion phenotypes
| Gene | VG no. | Placement of COIN module | Initial exon or intron size | Exon L or intron L size | Exon R or intron R size | Preinversion phenotype | Postinversion phenotype |
| Acvr1b | 1601 | Exon 2 | 240 | 125 | 115 | WT | NULL |
| Chrd | n.a. | Intron 2 | 372 | 206 | 105 | WT | HYPO |
| Ctgf | 1511 | Exon 2 | 223 | 120 | 103 | WT | NULL |
| Dicer1 | 1399 | Intron 6 | 4,162 | 3,770 | 446 | WT | NULL |
| Dkk4 | 4003 | Intron 1 | 953 | 730 | 223 | WT | n.d. |
| Dll4 | 1407, 1513 | Intron 3 | 808 | 631 | 144 | WT | NULL |
| Exosc3 | 1788 | Intron 3 | 649 | 235 | 327 | WT | NULL |
| Exosc10 | 1790 | Intron 2 | 1,328 | 412 | 1,018 | WT | NULL |
| Gdf11 | 1422 | Exon 1 | 472 | 297 | 175 | WT | NULL |
| Gpr124 | 1402 | Exon 1 | 371 | 133 | 238 | WT | NULL |
| Gt(ROSA)26Sor | 2154, 2234 | Intron 1 | 5,632 | 1,036 | 4,595 | WT (Tg) | NULL (Tg) |
| Hprt1 | 1272 | Exon 3 | 184 | 85 | 99 | WT | NULL |
| Il2rg | 1253 | Exon 1 | 201 | 89 | 112 | WT | NULL |
| Il2rg | 1452 | Exon 1 | 201 | 89 | 112 | NULL* | n.d. |
| Mstn | 1427 | Exon 1 | 478 | 185 | 293 | NULL† | n.d. |
| Plxnd1 | 1618 | Exon 2 | 177 | 97 | 80 | WT | NULL |
| Ret | 1541 | Exon 2 | 264 | 121 | 143 | WT | NULL |
| Drosha | 1390 | Intron 3 | 6,536 | 5,548 | 925 | WT | HYPO |
| Drosha | 1483 | Exon 4 | 834 | 625 | 209 | WT | NULL |
| Scn9a | 1589 | Exon 2 | 308 | 105 | 203 | WT | n.d. |
| Sirt1 | n.a. | Exon 3 | 242 | 141 | 101 | WT | n.d. |
| Sost | 1445 | Intron 2 | 2,492 | 938 | 1,504 | WT | NULL |
| Sox2 | 1413 | Exon 1 | 2,457 | 441 | 2,016 | WT | NULL |
| Sox10 | n.a. | Intron 3 | 2,493 | 94 | 2,358 | WT | DN |
| Tek | 1379 | Exon 1 | 393 | 354 | 40 | WT | NULL |
| Tgfbr1 | 1602 | Exon 2 | 246 | 90 | 156 | WT | NULL |
| Tie1 | 1260 | Exon 1 | 424 | 380 | 34 | WT | NULL |
| VegfR1 | 1308 | Exon 1 | 347 | 291 | 56 | WT | n.d. |
| VegfR2 | 1345 | Exon 1 | 327 | 272 | 55 | WT | NULL |
Gene names are those names used in the public genome servers. VG no. is the VelociGene number of each COIN allele after removal of the drug selection cassette. Placement of COIN module denotes the number of the exon or intron where the COIN module was introduced, whereas initial exon or intron size lists the size of the respective exon or intron. In the case of genes that are alternatively spiced, the numbering of the exon or intron corresponds to the longest form. Exon L or intron L size denotes the size of the 5′ part of the split exon or intron, whereas exon R or intron R size denotes the size of the 3′ part of the split exon or intron. For intronic COINs, the left and right parts of the modified intron are defined by the boundaries with the COIN module, thereby defining the position of the COIN module within the intron. Preinversion phenotype indicates the phenotype observed before inversion of the COIN allele in either homozygosis (COIN/COIN) or hemizygosis with a null allele (COIN/null), whereas for X-linked genes, such as Hprt1 and Il2rg, it is the phenotype associated with the COIN/Y genotype. A WT designation indicates that, before inversion of the COIN module, the phenotype observed in COIN/COIN, COIN/null, or COIN/Y cells or mice matches the WT cells or mice. Postinversion phenotype indicates the phenotype observed after inversion of the COIN allele in either homozygosis (COIN-INV/COIN-INV) or hemizygosis with a null allele (COIN-INV/null), whereas for X-linked genes, it is the phenotype associated with the COIN-INV/Y genotype. A NULL designation indicates that the phenotype observed in COIN-INV/COIN-INV, COIN-INV/null, or COIN-INV/Y cells or mice phenocopies COIN-INV/COIN-INV, COIN-INV/null, or COIN-INV/Y cells or mice. The Tg designation appended to the two Gt(ROSA)26Sor alleles (VG2154 and VG2234) indicates that the WT and NULL designations are based on the analysis of artificial COIN transgenes built into the Gt(ROSA)26Sor locus rather than the biological function of the locus itself. The DN designation for SRY-box containing gene 10 (Sox10) indicates that the corresponding inverted COIN allele is dominant negative as intended by design. For the i2COIN allele of chordin (Chrd) and the i3COIN allele of Drosha (VG1390), the HYPO designation indicates that the allele resulting from inversion of the COIN module is hypomorphic and does not phenocopy the null allele (in homozygosis). For Dll4, two different COIN alleles where generated; VG1513 replaced VG1407 because of improved reporter function. The Ctgf COIN allele has been presented elsewhere (27). Data on Acvr1b, Chrd, Dkk4, Exosc3, Exosc10, Ret, Scn9a, Sirt1, SRY-box containing gene 2 (Sox2) (28), Sox10, endothelial-specific receptor tyrosine kinase (Tek), Tgfbr1, vascular endothelial growth receptor 1 (Vegfr1), and vascular endothelial growth receptor 2 (Vegfr2) COINs will be presented elsewhere. n.a., not applicable; n.d., not determined; WT, wild type; Tg, transgene; HYPO, hypomorphic allele.
For Il2rg, line VG1452, the NULL designation reflects the fact that VG1452 is a reverse COIN, where the COIN module is placed in the sense orientation with respect to Il2rg, thereby deliberately generating a null allele before inversion of the COIN module.
For the COIN allele of myostatin (Mstn), the listed NULL designation indicates that Mstnex1COIN/LacZ mice phenocopy MstnLacZ/LacZ mice.
Hprt1ex3COIN: A Stringent Test for a Cellular Null Phenotype.
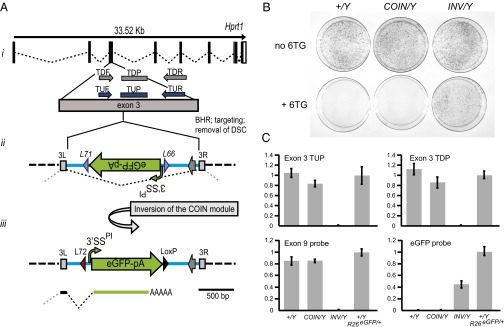
The COIN method was initially tested using the X-linked hypoxanthine guanine phosphoribosyl transferase 1 (Hprt1) gene, because it provides a rapid and stringent functional test in targeted XY ES cells in culture: the toxicity of 6-thioguanine (6-TG) is dependent on expression of Hprt1 (15), with as little as 5% of WT Hprt1 protein levels being adequate for 6-TG–mediated cell death. The Hprt1ex3COIN allele was engineered by inserting the COIN intron into exon 3 of Hprt1, thereby dividing this 184-nt exon into two new exons, 3L and 3R, of 85 and 99 nt. Exon 3 was chosen over exons 1 and 2, because both of them are too short to accommodate insertion of the COIN intron. As would be expected if the COIN allele was functionally equivalent to WT before inversion and completely null postinversion, the preinversion Hprt1ex3COIN/Y ES cells died on treatment with 6-TG just as the unmodified parental ES cells did, whereas the postinversion Hprt1ex3COIN-INV/Y cells survived (Fig. 2B).
Fig. 2.
Hprt1ex3COIN validates COIN approach in ES cells. (A) Engineering of Hprt1ex3COIN. (i) Schematic of Hprt1ex3COIN allele. The exon–intron region of Hprt1 adapted from Ensembl.org. Exon 3 (ENSMUSE00000491684) is highlighted together with the loss-of-allele (LOA) probes (TUP, TDP) and primers (TUF, TUR; TDF, TDR). The same probes and primers are used for RT-PCR to quantitate Hprt1 mRNA levels. (ii) The COIN intron was placed after the 85th nucleotide of exon 3 (SI Appendix, Fig. S2), splitting exon 3 into exons 3L and 3R. (iii) Before inversion, the Hprt1ex3COIN allele generates a normal message as the COIN intron is spliced out. (iv) After inversion, the COIN module becomes the terminal exon of the modified gene (Hprt1ex3COIN-INV), abrogating transcription of the downstream exons and resulting in a functional null allele. Exons 3L and 3R of Hprt1ex3COIN allele are shown as light gray boxes. Blue line denotes the COIN intron sequence. L66, lox66; L71, lox71; L72, lox72. lox and FRT sites are not drawn to scale. (Scale bar: ii–iv, 500 bp.) (B) Hprt1ex3COIN-INV/Y cells are Hprt1-null. ES cells were cultured in either the absence (Upper) or presence (Lower) of 10 µM 6-TG for 10 d, and then, they were fixed and stained with Giemsa. Hprt1+/Y and Hprt1ex3COIN/Y cells die on treatment with 6-TG, whereas Hprt1ex3COIN-INV/Y cells survive. (C) Insertion of the COIN element does not alter the expression level of Hprt1 before inversion but ablates it after inversion. Quantitative RT-PCR analysis of Hprt1 and eGFP-encoding mRNA from Hprt1+/Y (+/Y), Hprt1ex3COIN/Y (COIN/Y), Hprt1ex3COIN-INV/Y (INV/Y), and Hprt1+/Y; Gt(ROSA)26SoreGFP/+ (+/Y; R26eGFP/+) ES cells. (Upper Left) Level of Hprt1 mRNA relative to Hprt1+/Y; Gt(ROSA)26SoreGFP/+ detected using probe TUP. (Upper Right) Level of Hprt1 mRNA relative to Hprt1+/Y; Gt(ROSA)26SoreGFP/+ detected using probe TDP. (Lower Left) Level of Hprt1 mRNA relative to Hprt1+/Y; Gt(ROSA)26SoreGFP/+ detected using a probe that detects exon 9 of Hprt1. (Lower Right) Level of eGFP-encoding mRNAs relative to Hprt1+/Y; Gt(ROSA)26SoreGFP/+ detected using a probe for eGFP.
Quantitative RT-PCR analysis showed that, preinversion, the artificial intron is correctly spliced at the exon 3L/3R junction and insertion of the COIN module did not alter Hprt1 mRNA levels in the Hprt1ex3COIN/Y ES cells (Fig. 2C and SI Appendix, Fig. S2). In contrast, after inversion of the COIN module, the Hprt1 mRNA is entirely ablated and replaced with an eGFP-encoding mRNA; the latter is absent in Hprt1+/Y and Hprt1ex3COIN/Y ES cells. Therefore, by two different criteria—functionality (resistance to 6-TG) and Hprt1 mRNA expression—Hprt1ex3COIN/Y ES cells are identical to WT, whereas their postinversion counterparts, Hprt1ex3COIN-INV/Y ES cells, exhibit a phenotype identical to Hprt1-null ES cells.
Il2rgex1COIN/Y: A Stringent Test for a Complete Null Phenotype in Mice.
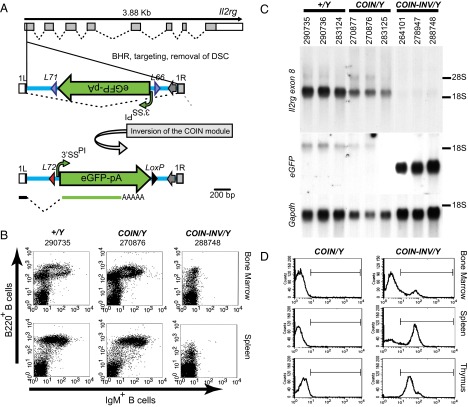
To extend the in vitro observations made with the Hprt1 COIN allele to the organismal level, we created a COIN allele for the gene encoding the common receptor γ-chain used by IL-2 and related cytokines [interleukin 2 receptor, gamma chain (Il2rg)] because of the well-established immunological phenotypes associated with loss of this X-linked gene (16–19). The Il2rgex1COIN allele was engineered by inserting the COIN intron into exon 1 immediately after the start codon (initiating ATG; Fig. 3A and SI Appendix, Fig. S3). Il2rgex1COIN/Y mice were identical to WT mice for every immune parameter examined; in contrast, the immune properties of postinversion Il2rgex1COIN-INV/Y mice matched the published phenotypes of Il2rg-null lines (SI Appendix, Table S1). For example, the B220+, IgM+ compartments of bone marrow and splenic lymphocytes in preinversion Il2rgex1COIN/Y mice were indistinguishable from the corresponding compartments in WT Il2rg+/Y mice, but this lymphocyte class was nearly absent in postinversion Il2rgex1COIN-INV/Y mice (Fig. 3B and SI Appendix, Fig. S4). As expected from the phenotypes, the expression of Il2rg mRNA in the spleens of Il2rgex1COIN/Y mice was largely unperturbed (Fig. 3C), whereas in Il2rgex1COIN-INV/Y mice, the Ilr2g mRNA was replaced by an mRNA-expressing eGFP, which was evidenced by the presence of green fluorescent cells in lymphocyte populations isolated from Il2rgex1COIN-INV/Y mice (Fig. 3D). Therefore, by phenotype, mRNA size and quantity, and expression of the reporter after inversion of the COIN module, the Il2rg COIN allele functioned as intended. Because even a few WT cells can reconstitute the immune system, the severe immune-deficient phenotype of the Il2rgex1COIN-INV/Y mice stringently confirms that the inverted COIN allele confers a completely null phenotype in all cells.
Fig. 3.
Il2rgex1COIN-INV provides in vivo validation of COIN and shows the functionality of the reporter. (A) Schematic of Il2rgex1COIN allele. The exon–intron region of Il2rg (isoform Il2rg-001, CCDS30312) is shown as adapted from Ensembl.org. The COIN intron was placed after the 90th nucleotide of exon 1 (SI Appendix, Fig. S3), splitting exon 1 into exons 1L and 1R. Before inversion, the Il2rgex1COIN allele generates a normal message as the COIN intron is spliced out. After inversion, the COIN module becomes the terminal exon of the modified gene, abrogating expression of the downstream exons and resulting in a functional null allele incorporating eGFP. Naming conventions, abbreviations, and markings are as noted in Fig. 2. (Scale bar: 200 bp.) (B) IgM+, B220+ B-cell population is largely absent in bone marrow and spleen cells of Il2rgex1COIN-INV/Y (288748) but unaffected in Il2rgex1COIN/Y (270876) compared with Il2rg+/Y (290735) mice. Numbers denote mouse identity. (C) Insertion of the COIN element does not alter the expression level of Il2rg before inversion but ablates it after inversion. Northern analysis of RNA isolated from spleens of Il2rg+/Y (290736, 290735, and 283124), Il2rgex1COIN/Y (270877, 270876, and 283125), and Il2rgex1COIN-INV/Y (278947, 288748, and 283126) mice. Probes are (Top) Il2rg, (Middle) eGFP, and (Bottom) Gapdh. The positions of 18S and 28S rRNAs are marked. Numbers denote the identification of each mouse belonging to each genotypic class; the same mice were analyzed phenotypically (B) (SI Appendix, Fig. S4). (D) Bone Marrow, thymic, and splenic lymphocyte populations from Il2rgex1COIN-INV/Y mice express eGFP. eGFP expression is absent from Il2rgex1COIN/Y lymphocyte populations.
Dll4i3COIN: A Sensitive Test for Conditionality.
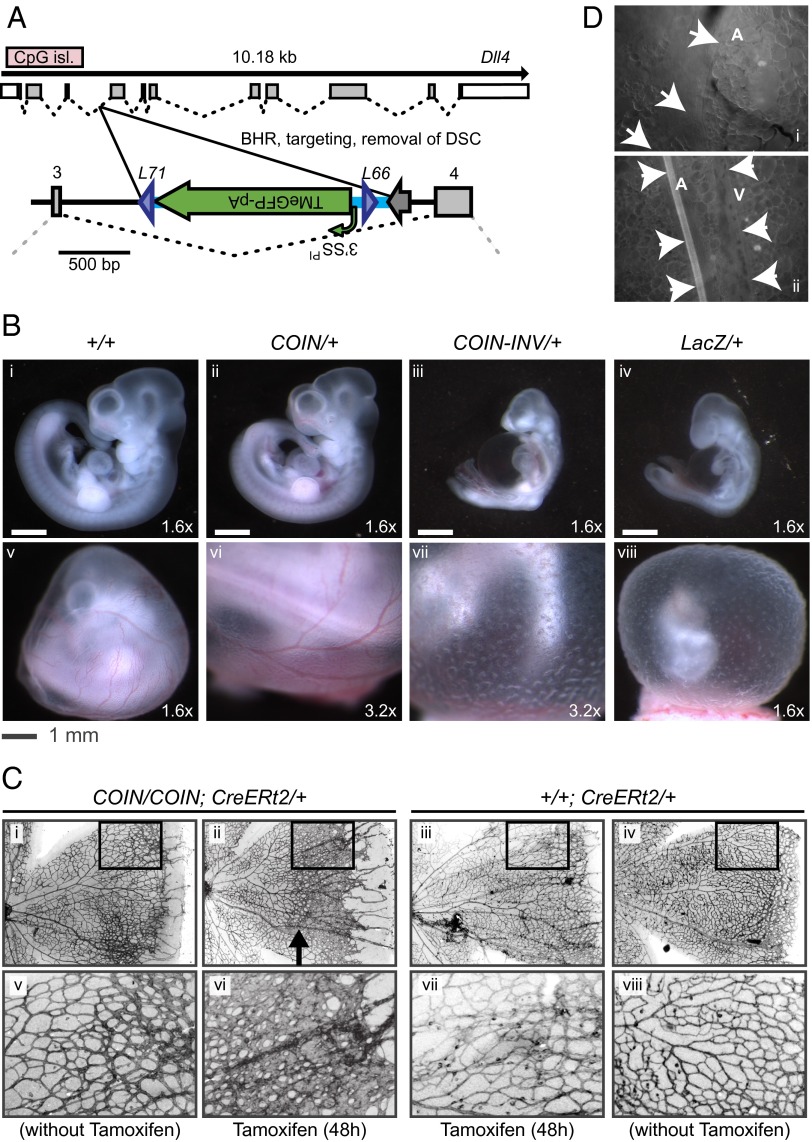
To further establish that preinversion COIN alleles are innocuous, we created a COIN for delta-like 4 (Dll4). Embryos heterozygous-null for this autosomal gene display a severe angiogenesis phenotype resulting in death during gestation (20–22). Therefore, if the preinversion Dll4i3COIN allele were to cause even a small reduction in Dll4 function, then it might be expected to produce an observable angiogenesis phenotype in embryos. To avoid disruption of a conserved CpG island, we opted to place the COIN module within intron 3 (Fig. 4A). As would be expected if the preinversion COIN allele of Dll4 functioned at WT levels, embryonic day 10.5 (E10.5) Dll4i3COIN/+ embryos appeared normal (Fig. 4B). Furthermore, E10.5 Dll4i3COIN-INV/+ embryos (i.e., embryos heterozygous for the postinversion allele generated by breeding to an Nanog-Cre transgenic line) displayed a severe angiogenesis phenotype identical to the phenotype observed in Dll4LacZ/+ embryos heterozygous for a conventional null allele (Fig. 4B, iii and iv and SI Appendix, Table S2). This COIN allele also functioned postnatally when conditionally inverted by tamoxifen/CreERt2-mediated recombination, because such inversion reproduced the angiogenic abnormalities seen with pharmacological Dll4 blockade in the postnatal developing eye (23) (Fig. 4C). Lastly, eGFP fluorescence was detected in the arterial vasculature of the skin, which is analogous to the pattern previously seen with conventional reporter alleles (21) (Fig. 4D), indicating correct expression of the COIN module’s reporter postinversion.
Fig. 4.
Dll4i3COIN provides a stringent test for engineering COIN alleles that are WT before inversion. (A) Schematic of Dll4i3COIN allele. The exon–intron region of Dll4 was adapted from Ensembl.org. The COIN module was inserted into intron 3. Before inversion, the Dll4i3COIN allele generates a normal message as the COIN intron is spliced out. For brevity, the Dll4i3COIN-INV allele is not depicted schematically; however, as depicted in Fig. 1, after inversion, the COIN module becomes the terminal exon of the modified gene, abrogating expression of the downstream exons and resulting in a functional null allele incorporating TMeGFP. The Dll4i3T2ACOIN allele is identical to the Dll4i3COIN allele, except for the incorporation of a T2A peptide leading into the marker (TMT2AeGFP). Naming conventions, abbreviations, and markings for different elements are as noted in Fig. 2. (Scale bar: 500 bp.) (B) The Dll4i3COIN-INV allele corresponds to a null allele. Embryos from a Dll4i3COIN × Nanog-Cre cross were collected at E10.5, visualized by light microscopy, genotyped, and compared with Dll4LacZ/+ E10.5 embryos. Embryos and yolk sacs with genotypes (i and v) Dll4+/+; LocXNanog-Cre/+ (+/+); (ii and vi) Dll4i3COIN/+ (COIN/+); (iii and vii) Dll4i3COIN-INV/+LocXNanog-Cre/+ (COIN-INV/+); and (iv and vii) Dll4LacZ/+ (LacZ/+) are shown. Dll4i3COIN-INV/+ embryos phenocopy Dll4LacZ/+ embryos. (C) Inversion of the COIN module in Dll4i3COIN mice at postnatal day 5 (P5) results in abrogated maturation of the retinal vasculature. P5 mice were treated with tamoxifen to activate CreERt2 or vehicle; 48 h later, retinas were collected, and their vasculatures were visualized. Neither (i and v) the COIN allele in homozygosis [Dll4i3COIN/i3COIN; Gt(ROSA)26SOR+/+, denoted as COIN/COIN; CreERt2/+] nor (iii and vii) the activation of CreERt2 by tamoxifen (+/+; CreERt2/+) has any impact on retinal angiogenesis and (iv and viii) phenocopy of DII4+/+; Gt(ROSA)26SORCreERt2/+. (ii and vi) Activation of CreERt2 in Dll4i3COIN/i3COIN; Gt(ROSA)26SORCreERt2/+ results in abrogated maturation of the retinal vasculature. Treatment with tamoxifen is indicated. (D) Inclusion of T2A in Dll4i3T2ACOIN restores function of the reporter (TMT2AeGFP). Fluorescence photomicrographs of skin from Dll4i3T2ACOIN/+; Gt(ROSA)26SORCreERt2/+ mice (i) before and (ii) after treatment with tamoxifen. Note the expression of eGFP in the arteries (A) but not veins (V) only in ii.
Failure of an Intronic COIN Allele to Function as a KO Postinversion Is Corrected by Reengineering the Allele as an Exonic COIN.
The vast majority of conditional-null alleles engineered as COINs and analyzed to date have functioned as intended: they are WT preinversion and null postinversion (Table 1). Exceptions to this experience are two intronic COINs, Chrdi2COIN and Droshai3COIN, that both function as incomplete nulls postinversion. For example, mice carrying the postinversion Droshai3COIN allele in conjunction with a conventional null allele (Droshai3COIN-INV/LacZ) are born and survive (although runted), whereas this same null allele, DroshaLacZ, results in embryonic lethality when homozygous (DroshaLacZ/LacZ) (SI Appendix, Table S3). The reason the postinversion Droshai3COIN-INV allele is not a complete null is that it still produces low levels of WT mRNA, apparently because of alternative splicing that removes the inverted COIN module (SI Appendix, Fig. S19); similar observations have been made with conventional gene traps, accounting for the hypomorphic character observed with many of them (24, 25) and highlighting one of the major weaknesses of gene trapping.
We hypothesized that the hypomorphic phenotype of an intronic COIN postinversion could be corrected by placing the COIN module contained within a discrete artificial intron (intronic COIN) into a nearby exon. This hypothesis was tested by engineering a COIN allele in exon 4 of Drosha (Droshaex4COIN) (Fig. 5A and SI Appendix, Fig. S5); postinversion, this allele resulted in early embryonic lethality when paired with a conventional null allele (Droshaex4COIN-INV/LacZ), indicating that the exonic placement of the COIN module corrects the problems observed with intronic placement (SI Appendix, Table S4). Unlike the intronic COIN allele of Drosha, the postinversion exonic COIN completely lacks WT Drosha mRNA (Fig. 5B) and abrogates the function of Drosha (26) in pri-miR processing (Fig. 5 C and D). These data indicate that exonic placement of the COIN module more reliably generates completely null alleles postinversion compared with placement of the COIN module into native introns.
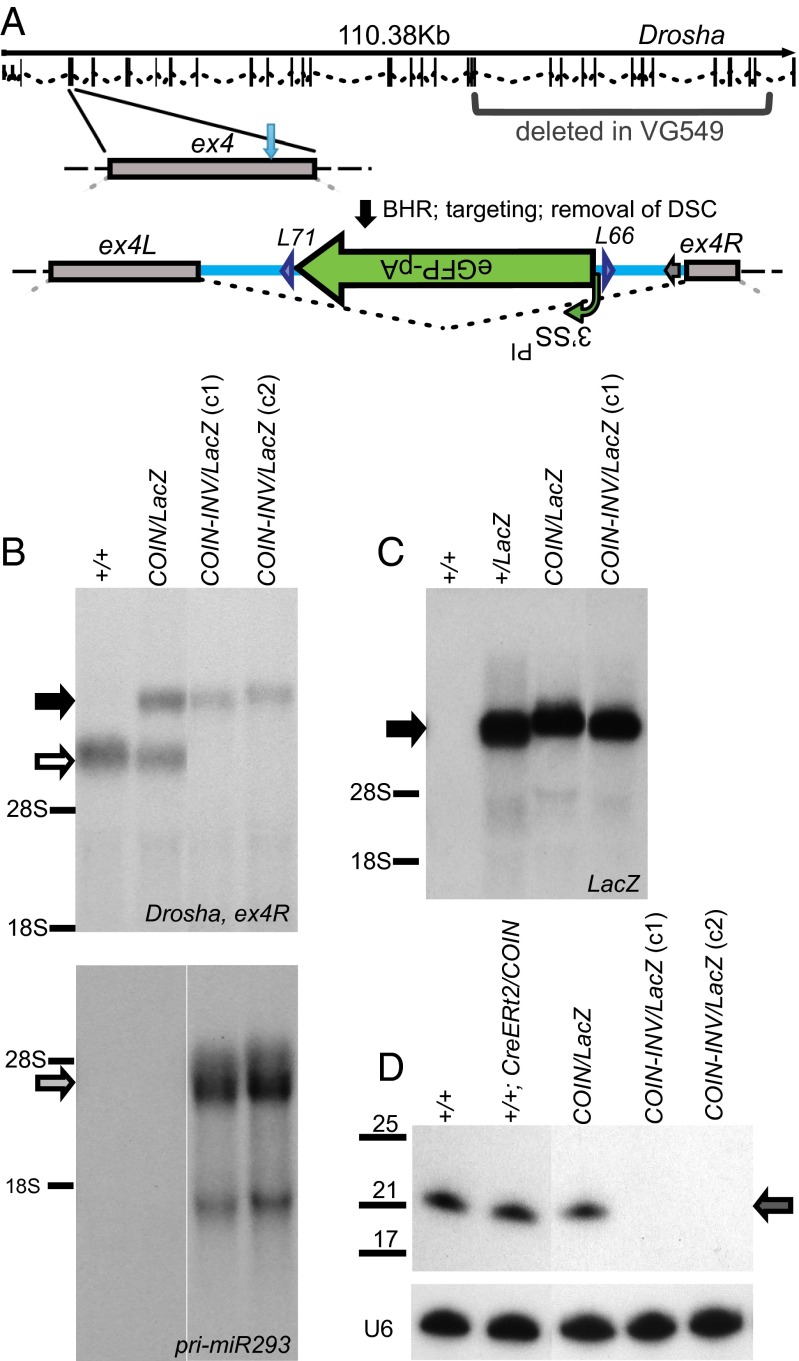
Fig. 5.
The exonic COIN of Drosha is a postinversion functional null. (A) Schematic of Droshaex4COIN allele. The exon–intron region of Drosha (splice variant 001, transcript ID ENSMUST00000090292) was adapted from Ensembl.org. Light blue vertical arrow indicates the point of insertion of the COIN intron within exon 4 (SI Appendix, Fig. S5). Before inversion, Droshaex4COIN generates a normal message as the COIN intron is spliced out. For brevity, Droshaex4COIN-INV is not depicted schematically; however, as shown in Fig. 1, after inversion, the COIN module becomes the terminal exon of the modified gene, abrogating expression of the downstream exons and resulting in a functional null allele incorporating eGFP. The region replaced in the DroshaLacZ allele VG549 is marked by brackets. Naming conventions, abbreviations, and markings for different elements are as noted in Fig. 2. (Scale bar: 100 bp.) (B–D) Inversion of the COIN module results in abrogation of expression of Drosha and microRNA maturation and concomitant accumulation of pri-miRs. Northern analysis of Drosha using exon 4 (ENSMUSE00000563117) as a probe reveals lack of Drosha mRNA (B, Upper, white arrow) in Droshaex4COIN-INV/LacZ ES cells and the presence of a hybrid/fusion message encoding exons 1–21 of Drosha plus LacZ (B, Upper, black arrow), which is also detected with a LacZ probe (C, black arrow). Loss of Drosha expression results in accumulation of pri-miR293 (B, Lower, gray arrow) and loss of the mature miR-293 (D, Upper, gray arrow) in Droshaex4COIN-INV/LacZ ES cells. Maturation of the miR is not affected by induction of Cre activity or expression of eGFP (D, lane 2). The positions of 18S and 28S rRNAs (B and C) as well as the positions of U6, miR-293, and a small RNA ladder (D) are marked. Genotype key: +/+, Drosha+/+; +/LacZ, Drosha+/LacZ; CreERt2/COIN-INV, Drosha+/+; Gt(ROSA)26SorCreERt2/COIN-INV; COIN-INV/LacZ, Droshaex4COIN-INV/LacZ; Gt(ROSA)26SorCreERt2/+, +/+; COIN/LacZ, Droshaex4COIN/LacZ. c1 and c2 denote clones 1 and 2 of Droshaex4COIN-INV/LacZ; Gt(ROSA)26SorCreERt2/+ derived from treatment with tamoxifen.
COIN Method Is Applicable to a Wide Variety of Genes.
We further validated the COIN method by analyzing COIN alleles for an additional 23 genes (Table 1). Representative data from a subset of these genes are provided in SI Appendix or as indicated: Ctgf (27), Dicer1 (SI Appendix, Fig. S7 and Table S5), Gdf11 (SI Appendix, Fig. S8 and Table S6), Gpr124 (SI Appendix, Fig. S9 and Table S7), Gt(ROSA)26Sor (SI Appendix, Fig. S10), Plxnd1 (SI Appendix, Fig. S11 and Table S8), Sost (SI Appendix, Fig. S12 and Table S9), Sox2 (28), and Tie1 (SI Appendix, Fig. S13 and Table S10). With the exception of Mstnex1COIN, which was hypomorphic before inversion for reasons that are not fully understood, all of the COIN alleles were shown to function as intended (Table 1)—WT before inversion and null after inversion.
Discussion
COINs Provide a State-of-the-Art Method for Engineering Conditional-Null Alleles.
As a flexible and widely applicable method for generating conditional alleles, the COIN allele addresses the limitations inherent in other conditional methods while opening up unique design modalities. COINs are subject to few design constraints; they use an optimized gene trap-like cassette—the COIN module—that is modular and adaptable to high-throughput schemes while overcoming the limitations and deficiencies of existing gene trap and conditional-null allele approaches. The COIN method is applicable to the majority of genes as it is independent of the target gene's intron–exon structure, eliminates the need to flank entire genes with recombinase recognition elements or define critical regions, and also, reduces concerns about disrupting or deleting essential regulatory elements. Although most of genes where COIN has been tested are protein-coding genes, the method is applicable to noncoding genes, as shown by the COIN allele of Gt(ROSA26)Sor.
In addition to its broad applicability, the COIN allele design includes a reporter (e.g., eGFP) that enables visualization of the cells in which the conditional induction of the allele has occurred. The COIN reporter gene can be easily replaced by any transgene or engineered exon of choice. This latter capability is being exploited to generate conditional mutant alleles to model human genetic disorders (29). However, the most unique technological advance introduced by COINs is the engineering modality of splitting exons and exploiting artificial introns. Exon splitting enables the application of COIN technology even to single exon genes, which comprise ∼15% of the protein-coding genes in the mouse genome (30). This property is being used by EUCOMMTOOLS to generate conditional-null alleles for single exon genes (www.knockoutmouse.org/about/eucommtools/vectors).
Previously, conditional allele design methods, such as FlEx (5) and KO-first (3), have attempted to address some of the problems associated with traditional conditional-null methodology (SI Appendix, Fig. S17). Of these two methods, FlEx is most similar to COIN in that it relies on an invertible conditional gene trap cassette that incorporates a reporter, but it retains the limitation of gene traps in that it must be inserted into introns; with one exception (5), it has been used solely in gene trapping (4). Although the value of FlEx has yet to be realized, KO-first is one of the methods adopted by large-scale mouse KO projects, such as EUCOMM and KOMP (31). KO-first alleles are targeted gene traps that can be converted, posttargeting, into either null or conditional-null alleles (SI Appendix, Fig. S17). KO-first alleles are, however, limited to genes for which a critical exon (i.e., an exon with deletion that places the coding sequence of downstream exons out of frame; thus, they are predicted to result in a null through the process of nonsense-mediated decay) (32) can be identified, and they lack a reporter that can mark the cells where Cre-mediated inactivation of the conditional allele has taken place. In overcoming the limitations of both of these methods, COINs provide a reliable alternative conditional allele design strategy.
Exonic COINs Provide Unique and Reliable Engineering Modalities for Engineering Conditional Alleles.
The majority of both exonic and intronic COIN alleles was shown to be WT before inversion of the COIN module and null after inversion (Table 1). One instructive exception was the postinversion hypomorphism encountered with two intronic COINs, Chrdi2COIN and Droshai3COIN. For the latter, this problem was corrected by engineering an exonic version (Droshaex4COIN) in the immediate downstream exon, indicating that exonic COINs reduce the possibility of regenerating WT mRNA by alternative splicing postinversion, thus more reliably generating completely null alleles compared with intronic COINs. Although this property of exonic COINs is not fully understood, the 3′SSPI of the COIN module and the 3′SSPI of the COIN intron are identical and matched to the 5′SS and pA site in a tried-and-true arrangement in which the COIN module 3′SSPI and pA site dominate, resulting in full use of the COIN module’s exon as a terminal exon and concomitant termination of transcription after inversion. Regardless of mechanism, our discovery that even an optimized gene trap cassette, such as the COIN module (SI Appendix, Fig. S1), can be spliced out when placed into some dominant native introns provides a cautionary note for the use of gene traps as null alleles (24, 25) and indicates that exonic COINs present a more reliable method for generating conditional-null alleles.
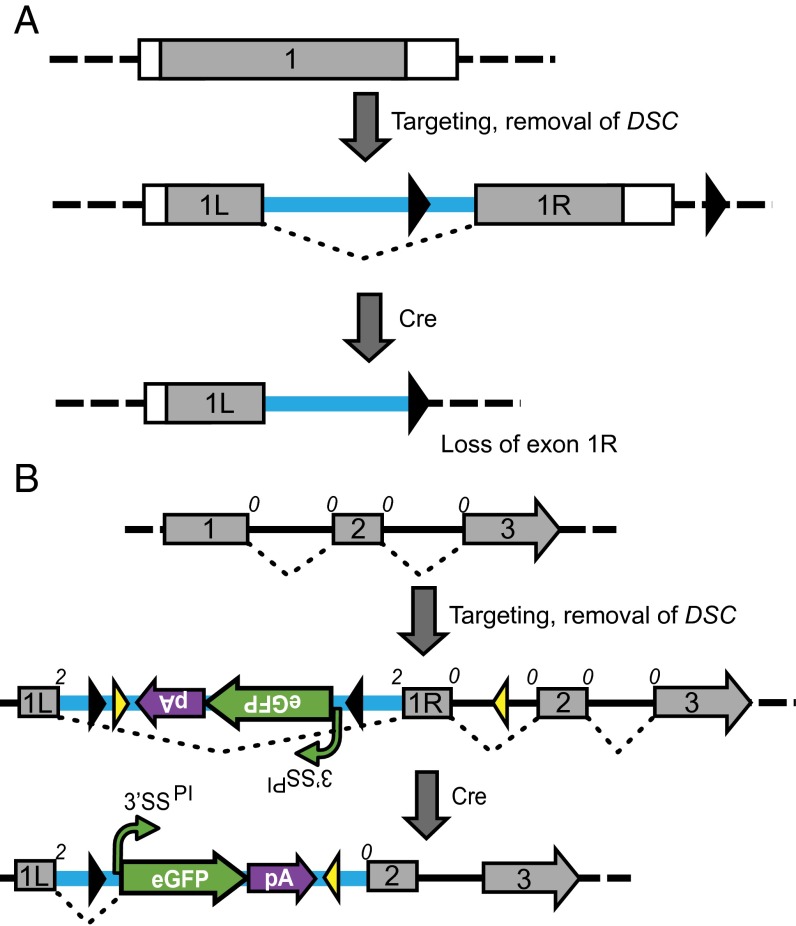
Without exception, the alleles described here have been designed as conditional-nulls. It should be apparent, however, that the COIN method expands the repertoire of possible allele designs. For example, the COIN intron can be used for simple splitting of an exon (Fig. 6A) in cases where deletion of part of the exon is a desirable outcome and particularly, enabling the design of simple, deletion-based floxed alleles for single exon genes. More complex designs, such as hybrid COIN–FlEx alleles (Fig. 6B and SI Appendix, Fig. S19), are equally achievable and useful where both the COIN module functionality and the ability to delete part of the floxed sequence is desired. Other possibilities include rescue COIN alleles, where the starting allele is mutant and the WT state is induced after inversion (29) (for example, VG1452, the reverse COIN allele of Il2rg) (Table 1). Given that the reporter present in the COIN module can be replaced with a variety of functional sequences, the use of COINs to generate conditional alleles of point mutations, exon swaps, domain swaps, and fusion proteins and even coexpress these engineered proteins or reporters with small noncoding RNAs are variations on the COIN theme currently being used.
Fig. 6.
COIN methodology enables unique engineering modalities. (A) Exon splitting enables floxing of single exon genes as well as genes lacking critical exons. A hypothetical protein-coding gene comprised of a single exon (1) is depicted. This exon is split into exons 1L and 1R using an intron that contains an loxP site. Another loxP site is placed downstream of exon 1R in a parallel configuration, rendering exon 1R amenable to deletion by Cre. Black triangles denote loxP sites, protein-coding sequence is denoted by gray color, and splicing is denoted by dotted black lines. All other elements are as described in Fig. 1. (B) Converting a noncritical exon to critical using a FlEx–COIN hybrid design. A hypothetical protein-coding gene comprised of three exons is depicted, where the exons are in the same phase (phase 0). Exons are shown as gray boxes, splicing is denoted by dotted black lines, the starting and end phases of each exon are denoted by numbers above the corresponding spot on each exon, and black and yellow triangles denote loxP and lox2372 sites, respectively. Exon 1 is rendered critical (3) by splitting it using the COIN intron in a manner such that exon 1L ends in phase 2. Recombination by Cre results in inversion of the COIN module and simultaneous deletion of exon 1R. (A version where exon 1R is preserved is shown in SI Appendix, Fig. S18.) Consequently, exons 1L and 2 are out of phase; thus, even if the COIN module is spliced out, the resulting mRNA will encode for a truncated, nonfunctional protein.
Designing reliable conditional alleles remains a challenging art. Any approach, including the currently described COIN approach, will have some failure rate. We report probably the most extensive testing and characterization of a conditional allele approach and find perhaps the lowest failure rate ever documented. Thus, the data that we provide suggest the COIN approach to be exceptionally reliable, generally applicable, and flexible enough to allow broad adoption and exploitation for designing alleles beyond conditional nulls.
Materials and Methods
Engineering COIN Modification Cassettes.
The COIN module is comprised of a 3′SSPI (denoted as such to indicate that it is intended to function as a 3′SS after inversion of the COIN module to the sense strand of the modified gene) with sequence 5′-CGG GCC CCT CTG CTA ACC ATG TTC ATG CCT TCT TCT TTT TCC TAC AG-3′ derived from the second intron of rabbit HBB2 (GeneID 100009084) (SI Appendix, Fig. S14) (9) followed by the ORF of eGFP (33) and the pA region of HBB2 (rßglpA; coordinates 32041:32560 in gb|M18818.1|RABBGLOB) (SI Appendix, Fig. S15). The 3′SSPI-eGFP-rßglpA module is flanked by left element/right element (LE/RE) mutant lox sites, lox66 and lox71 (6), to generate the complete COIN module, [lox66]-3′SSPI-eGFP-rßglpA-lox71, where lox66 and lox71 are in a head-to-head configuration with respect to each other and this relationship is denoted by showing lox66 within brackets ([…]) (Fig. 1).
The COIN module was inserted into the antisense strand of intron 2 of HBB2 at the unique MfeI site. Placement of the COIN module in the antisense strand allows introduction of the COIN intron into exons, thereby rendering possible the engineering of COIN alleles even for genes comprised of only a single exon. In addition, to accommodate in-frame insertions into any particular location within a gene of interest, COIN modules were generated in all three different reading frames by altering the phase of the eGFP-coding sequence. Furthermore, in cases where the gene product of the gene of interest is a protein that bears a secretion signal, a transmembrane version of eGFP (TMeGFP) was used as the reporter, and later, it also incorporated the T2A peptide (34) between the transmembrane domain of TMeGFP and eGFP (TM-T2A-eGFP) to enable expression of a minimally modified eGFP.
To facilitate BHR in Escherichia coli and targeting in ES cells, an antibiotic/DSC minigene was placed within the COIN intron in its antisense strand and between the unique PleI and PspOMI sites. The DSCs used were either a neomycin phosphotransferase artificial minigene (neo) or hygromycin B phosphotransferase artificial minigene (hyg) flanked by flippase recognition target (FRT) sites to allow removal of neo or hyg by Flp recombinase or its derivatives (35). Other than the ORF encoding neomycin phosphotransferase (npt) or hygromycin B phosphotransferase (hph), the elements of both minigenes are identical. Expression of npt or hph in mammalian cells was driven by the promoter region of the human ubiquitin C (UBC) gene (coordinates 125398319:125399530 in the antisense strand of human chromosome 12; Human CCDS set: CCDS9260) (36), whereas expression in E. coli was driven by the EM7 promoter (Invitrogen). To ensure polydenylation of the neo or hyg messages, the 3′ region of the mouse Pgk1 gene containing a pA and associated sequence (37) (coordinates 103398979–103399440 of chromosome X) was cloned past the stop codon of the npt or hph ORFs. Details on the engineering of an optimal COIN module are provided in SI Appendix.
Genetic Engineering of Alleles, ES cell lines, and Mouse Lines.
Velocigene technology was used to generate the genetically modified ES cells and mice used in this study (38). Briefly, targeting vectors were generated by modifying BACs using BHR (39). Targeting was performed into F1H4 ES cells, a 129S6/ScEvTac-C57BL/6NTac hybrid line, or ES lines derived from F1H4 (through gene targeting). Gene names and ch coordinates and chromosome numbers, where noted, correspond to nucleotide coordinates in mouse genome Ensembl release 58—May 2010; we have provided these coordinates to allow precise definition of the alleles being engineered and facilitate future annotation of these alleles into the genome. Gene names are also as they appeared in Ensembl release 58, May 2010. Details of the sets of primers and probes used for genotyping as well as the targeting frequencies obtained for each allele are listed in Dataset S1. Unless otherwise noted, all genotyping was performed using loss of allele (LOA) assay as described (38). Mouse lines were generated either by microinjection or using the VelociMouse method (40) as noted. The design of each allele used in this study along with detailed protocols for ES cell targeting and genotyping of COIN alleles are described in SI Appendix.
Genetic Engineering of COIN Alleles: Guidelines and Practices.
A simple set of rules guides the design of COINs:
i) Splitting of exons shorter than 100 bp should be generally avoided, because exons shorter than 50 bp are rare in the mouse genome (41).
ii) After splitting of the original exon, each of the resulting new exons [left (L) and right (R)] should preferably be no less than 50 bp. (However, for the four COIN alleles generated with exon 1L that are shorter than 50 bp—Tekex1COIN, Tie1ex1COIN, Vegfr1ex1COIN, and Vegfr2ex1COIN (Table 1)—there were no apparent negative consequences, and all of these COIN alleles functioned as intended.)
iii) Splitting of the exon by the COIN intron is preferably rendered after an MAG trinucleotide motif in the exon, hence preserving the MAG/gtragt 5′SS consensus (where M = A or C, r = a or g, and “/” denotes the cleavage site; nucleotides in uppercase are exonic, and nucleotides in lowercase are intronic) (42). If an MAG is not available in the target exon, one may be engineered by introducing appropriate silent mutations; the Gpr124ex1COIN allele provides such an example (SI Appendix, Fig. S9).
iv) Insertions within the first exon or intron are preferred to render as much as possible of the gene’s sequence inaccessible to transcription.
v) Disruption of CpG islands and conserved regulatory regions is avoided whenever possible. (However, we found that, in two alleles where the CpG island was disrupted by insertion of the COIN intron—Gdf11ex1COIN and Sox2exCOIN—there were minimal and no apparent consequences, respectively.)
vi) Lastly, a cautionary note regarding use is necessary. Before use of COIN alleles, the DSC must be removed to avoid aberrations in the splicing of the modified gene as well as its level of expression (13) or avoid even effects on neighboring genes (12, 43, 44).
For all COIN alleles presented in this paper, the DSC was placed in the antisense strand with respect to the direction of the COIN module, and for exonic COINs, it was contained within the COIN intron (Fig. 1). Additionally, all COIN alleles were studied only after excision of the DSC with FLPe (11). The types of COIN alleles—exonic (exCOIN) or intronic (iCOIN)—generated for each gene are listed in Table 1. The choice of COIN cassette was determined by placement requirements: in the case of exCOINs, the modification cassette was the full COIN intron plus DSC, whereas in the case of iCOINs, the modification cassette is the COIN module plus DSC without COIN intron sequence 5′ to the COIN intron’s MluI and 3′ to its XmaI site (Fig. 1 and SI Appendix, Fig. S6). In addition, the phase of the reporter (eGFP, TMeGFP, or TM-T2A-eGFP) was chosen so that after inversion of the COIN module into the strand encoding the target gene, the reporter will be in frame with the target’s gene-coding sequence upstream of the COIN module insertion point. The particular version of COIN modification cassette used for each allele is indicated as “reporter; phase number; DSC”. Details regarding the placement of the COIN modification cassette for each allele are provided in Dataset S1.
Supplementary Material
Acknowledgments
We would like to acknowledge Drs. Klaus Rajewsky, Francis Stewart, Konstantinos Anastasiadis, Barry Rosen, Daniel Graf, George Kollias, Douglas P. Mortclock, and Triantafyllos Chavakis for their encouragement and helpful discussions during the course of this work; and Drs. Michelle Southard-Smith, Stavroula Kousteni, and Anne Calof for sharing their data prepublication.
Footnotes
Conflict of interest statement: All authors of this manuscript are employed by Regeneron Pharmaceuticals, Inc., and some of the authors own a substantial amount of Regeneron stock.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217812110/-/DCSupplemental.
References
- 1.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6(1):7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 2.Voehringer D, Wu D, Liang HE, Locksley RM. Efficient generation of long-distance conditional alleles using recombineering and a dual selection strategy in replicate plates. BMC Biotechnol. 2009;9:69. doi: 10.1186/1472-6750-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Testa G, et al. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38(3):151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 4.Schnütgen F, et al. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci USA. 2005;102(20):7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schnütgen F, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21(5):562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- 6.Albert H, Dale EC, Lee E, Ow DW. Site-specific integration of DNA into wild-type and mutant lox sites placed in the plant genome. Plant J. 1995;7(4):649–659. doi: 10.1046/j.1365-313x.1995.7040649.x. [DOI] [PubMed] [Google Scholar]
- 7.Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8(10):4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil A, Proudfoot NJ. A sequence downstream of AAUAAA is required for rabbit beta-globin mRNA 3′-end formation. Nature. 1984;312(5993):473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- 9.Wieringa B, Hofer E, Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- 10.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108(4):501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 11.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16(7):657–662. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 12.Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci USA. 1996;93(23):13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meyers EN, Lewandoski M, Martin GR. An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet. 1998;18(2):136–141. doi: 10.1038/ng0298-136. [DOI] [PubMed] [Google Scholar]
- 14.Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K. Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res. 2003;31(22):e140. doi: 10.1093/nar/gng140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doetschman T, et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330(6148):576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- 16.Cao X, et al. Defective lymphoid development in mice lacking expression of the common cytokine receptor gamma chain. Immunity. 1995;2(3):223–238. doi: 10.1016/1074-7613(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 17.DiSanto JP, Müller W, Guy-Grand D, Fischer A, Rajewsky K. Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor gamma chain. Proc Natl Acad Sci USA. 1995;92(2):377–381. doi: 10.1073/pnas.92.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohbo K, et al. Modulation of hematopoiesis in mice with a truncated mutant of the interleukin-2 receptor gamma chain. Blood. 1996;87(3):956–967. [PubMed] [Google Scholar]
- 19.Ikebe M, et al. Lymphohaematopoietic abnormalities and systemic lymphoproliferative disorder in interleukin-2 receptor gamma chain-deficient mice. Int J Exp Pathol. 1997;78(3):133–148. doi: 10.1046/j.1365-2613.1997.230356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duarte A, et al. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18(20):2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gale NW, et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci USA. 2004;101(45):15949–15954. doi: 10.1073/pnas.0407290101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krebs LT, et al. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18(20):2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lobov IB, et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci USA. 2007;104(9):3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abuin A, Hansen GM, Zambrowicz B. Gene trap mutagenesis. Handb Exp Pharmacol. 2007;178(2007):129–147. doi: 10.1007/978-3-540-35109-2_6. [DOI] [PubMed] [Google Scholar]
- 25.De-Zolt S, et al. High-throughput trapping of secretory pathway genes in mouse embryonic stem cells. Nucleic Acids Res. 2006;34(3):e25. doi: 10.1093/nar/gnj026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43(6):892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canalis E, Zanotti S, Beamer WG, Economides AN, Smerdel-Ramoya A. Connective tissue growth factor is required for skeletal development and postnatal skeletal homeostasis in male mice. Endocrinology. 2010;151(8):3490–3501. doi: 10.1210/en.2010-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mandalos N, et al. Application of a novel strategy of engineering conditional alleles to a single exon gene, Sox2. PLoS ONE. 2012;7(9):e45768. doi: 10.1371/journal.pone.0045768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lutz CM, et al. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J Clin Invest. 2011;121(8):3029–3041. doi: 10.1172/JCI57291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakharkar KR, Sakharkar MK, Culiat CT, Chow VT, Pervaiz S. Functional and evolutionary analyses on expressed intronless genes in the mouse genome. FEBS Lett. 2006;580(5):1472–1478. doi: 10.1016/j.febslet.2006.01.070. [DOI] [PubMed] [Google Scholar]
- 31.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474(7351):337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 33.Zhang G, Gurtu V, Kain SR. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem Biophys Res Commun. 1996;227(3):707–711. doi: 10.1006/bbrc.1996.1573. [DOI] [PubMed] [Google Scholar]
- 34.Donnelly ML, et al. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol. 2001;82(Pt 5):1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- 35.Andrews BJ, Proteau GA, Beatty LG, Sadowski PD. The FLP recombinase of the 2 micron circle DNA of yeast: Interaction with its target sequences. Cell. 1985;40(4):795–803. doi: 10.1016/0092-8674(85)90339-3. [DOI] [PubMed] [Google Scholar]
- 36.Schorpp M, et al. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res. 1996;24(9):1787–1788. doi: 10.1093/nar/24.9.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adra CN, Boer PH, McBurney MW. Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene. 1987;60(1):65–74. doi: 10.1016/0378-1119(87)90214-9. [DOI] [PubMed] [Google Scholar]
- 38.Valenzuela DM, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21(6):652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20(2):123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 40.Poueymirou WT, et al. F0 generation mice fully derived from gene-targeted embryonic stem cells allowing immediate phenotypic analyses. Nat Biotechnol. 2007;25(1):91–99. doi: 10.1038/nbt1263. [DOI] [PubMed] [Google Scholar]
- 41.Sakharkar MK, Perumal BS, Sakharkar KR, Kangueane P. An analysis on gene architecture in human and mouse genomes. In Silico Biol. 2005;5(4):347–365. [PubMed] [Google Scholar]
- 42.Horowitz DS, Krainer AR. Mechanisms for selecting 5′ splice sites in mammalian pre-mRNA splicing. Trends Genet. 1994;10(3):100–106. doi: 10.1016/0168-9525(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 43.Olson EN, Arnold HH, Rigby PW, Wold BJ. Know your neighbors: Three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85(1):1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 44.Meier ID, et al. Short DNA sequences inserted for gene targeting can accidentally interfere with off-target gene expression. FASEB J. 2010;24(6):1714–1724. doi: 10.1096/fj.09-140749. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.