Abstract
The melanocortin 1 receptor (MC1R) mediates the tanning response through induction of cAMP and downstream pigmentary enzymes. Diminished function alleles of MC1R are associated with decreased tanning and increased melanoma risk, which has been attributed to increased rates of mutation. We have found that MC1R or cAMP signaling also directly decreases proliferation in melanoma cell lines. MC1R overexpression, treatment with the MC1R ligand, or treatment with small-molecule activators of cAMP signaling causes delayed progression from G2 into mitosis. This delay is caused by phosphorylation and inhibition of cdc25B, a cyclin dependent kinase 1-activating phosphatase, and is rescued by expression of a cdc25B mutant that cannot be phosphorylated at the serine 323 residue. These results show that MC1R and cAMP signaling can directly inhibit melanoma growth through regulation of the G2/M checkpoint.
Keywords: skin cancer, genetic risk factors, g-protein coupled receptors
Melanoma is an extremely aggressive skin tumor of the pigment-producing melanocyte lineage. As with other skin tumors, there is a close link between melanoma risk, sun exposure, skin tone and the skin’s ability to respond to sun exposure by inducing the pigmentation response (1, 2). This response is mediated by melanocytes following α-melanocyte–stimulating hormone (MSH) binding and activation of the melanocortin 1 receptor (MC1R) and the cAMP cascade (3, 4). MC1R and cAMP activation leads to induction of pigment producing enzymes such as tyrosinase, and a switch from the production of the red pigment phaeomelanin to the black pigment eumelanin.
MC1R is a highly polymorphic gene with over 50 variants found in humans (5). A subset of these variants, termed red hair color (RHC) alleles, are closely associated with a phenotype that includes red hair, freckling, and decreased tanning response (6). These RHC alleles have diminished function with respect to cAMP signaling, although it is not clear whether this loss of function is complete or partial (7–9). The red hair phenotype is a strong risk factor for skin cancers of both the keratinocyte and melanocyte lineages (10). Because the red hair phenotype is marked by decreased tanning and associated UV protection, it has been hypothesized that the risk associated with the RHC phenotype is linked to increased UV damage and mutation. In mice, rescuing defective MC1R activity by small molecule induction of cAMP signaling restores tanning and confers protection against UV-induced epithelial tumors (11). However, in melanoma, it is less clear what role the loss of pigment and UV-protection play in the increased risk associated with RHC alleles of MC1R. RHC variants of MC1R are associated with melanoma risk (12, 13); however, individuals who possess RHC alleles, but do not show an RHC phenotype, have equal or increased susceptibility to melanoma compared with individuals with RHC alleles and RHC phenotype (14, 15). These findings suggest that MC1R’s protective effect against melanoma extends beyond the induction of tanning and the prevention of UV-induced photo damage.
Adding to this complexity, a recent report has shown that phaeomelanin, which is up-regulated in MC1R RHC individuals, can directly drive oxidative damage and can contribute to melanoma development independent from UV damage (16). Although there is strong evidence that active MC1R and cAMP signaling can have a protective effect against mutation through pigmentary and nonpigmentary mechanisms (17), the role of cAMP signaling as a direct modulator of cancer-related phenotypes in relation to MC1R and melanoma is still not clear. Depending on cell type, cAMP can act as an inducer or an inhibitor of proliferation (18). Of particular interest in relation to melanoma, which has a high frequency of up-regulated MAPK signaling through mutation of the upstream kinase BRaf (60–70%) and small GTPase NRas (15–20%), cAMP blocks MAPK signaling through Raf- and Ras-dependent mechanisms (19).
Alongside its role in inhibiting proliferation at the level of S-phase entry, cAMP signaling may influence other stages of the cell cycle. In frogs and mice, protein kinase A (PKA), which is a key transducer of cAMP signals, inhibits meiotic resumption at the G2/M checkpoint. In mitosis and meiosis, the G2/M checkpoint is regulated by cyclin B/cyclin dependent kinase 1 (CDK1) complexes. Cyclin B accumulates during late S phase and G2 and forms complexes with CDK1 which are held inactive by phosphorylation at residues Thr14 and Tyr15 on CDK1 (20). CDK1 dephosphorylation allows cyclin B-associated CDK1 to become active and phosphorylate various targets, leading to mitotic entry. These phosphates are removed by the dual-specificity phosphatases cdc25B and C (21). Cdc25C, which is thought to dephosphorylate the majority of CDK1, is phosphorylated and inhibited by PKA during Xenopus meiosis (22). Cdc25B, which is thought to act as a trigger for CDK1 activation has also been reported to be phosphorylated and inhibited by PKA during mouse meiosis (23, 24).
Because of the genetic evidence linking decreased MC1R function and cAMP signaling to melanoma risk independently from pigmentary effects, and the well documented roles for cAMP signaling in inhibiting tumor growth, we sought to investigate the role of MC1R and cAMP signaling in the proliferation of melanoma cell lines. We show that expression of active MC1R or induction of cAMP slows the growth of melanoma cell lines in culture. cAMP signaling can inhibit MAPK signaling in NRas, but not BRaf mutant melanoma lines; however, even when the MAPK pathway is inhibited, there is no effect on S-phase entry. In all melanoma lines tested, we identified a delay in G2/M progression following activation of cAMP signaling. This G2/M delay is caused by increased inhibitory phosphorylation of cdc25B and can be rescued by expression of a PKA insensitive mutant of cdc25B. These findings describe a method of cell cycle regulation by cAMP that is distinct from previously described mechanisms of G1/S phase regulation. Additionally, they raise the possibility that, alongside the pathway’s roles in modulating mutation rates, MC1R and cAMP signaling may directly regulate proliferation in developing melanomas.
Results
MC1R and cAMP Signaling Inhibit Melanoma Cell Cycle Progression.
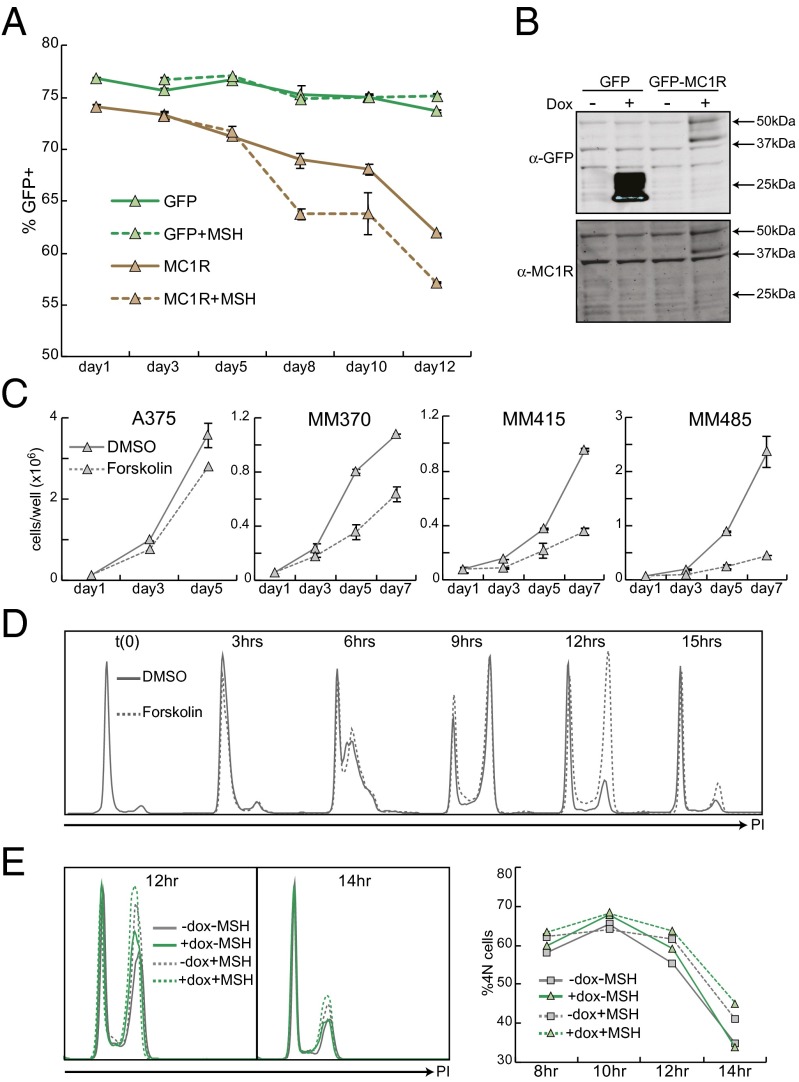
To test whether MC1R and cAMP signaling could influence melanoma susceptibility independently from their effects on pigmentation and mutation, we assessed the roles of MC1R and the adenylyl cyclase agonist forskolin on the growth of melanoma cells in culture. We first tested the effects of restoration of wild-type MC1R in an MC1R RHC cell line. MM485 cells stably expressing inducible GFP or GFP-tagged MC1R were diluted with the parental non-GFP–expressing cell line and treated with doxycycline or doxycycline plus the MC1R ligand MSH. Fig. 1A shows that, during 12 d of growth in the presence of doxycycline, the percentage of GFP-MC1R–expressing cells decreased with each time point and this effect was enhanced by treatment with MSH (−12.1% and −16.9% respectively), suggesting that MC1R activity inhibited the proliferation of these cells relative to the nontransduced cells. The percentage of cells expressing GFP alone decreased only slightly, and showed no MSH dependence. For the first four time points, cells were counted before flow cytometry and the total numbers of GFP+ and GFP− cells were calculated. Growth curves showed no significant changes in proliferation in uninfected and GFP+ cells following MSH treatment. However, GFP-MC1R–expressing cells had decreased cell number relative to GFP cells, and this effect was significantly enhanced by MSH treatment (Fig. S1). Expression of the GFP-tagged receptor is shown in Fig. 1B and the activity of the GFP-tagged receptor was confirmed by glosensor cAMP assay (Fig. S2).
Fig. 1.
MC1R and cAMP signaling decrease proliferation and delay cell cycle progression. (A) MM485 cells were lentivirally transduced and selected for expression of pLVX tet on and pLVX tight puro GFP or GFP-MC1R. GFP-expressing cells were diluted with the parental (pLVX tet on) cells, plated at 50,000 cells per well and treated daily with doxycycline (1 μg/mL) with or without MSH (100 nM). Every other day, duplicate wells were trypsinized and the percentage of GFP-positive cells was measured by flow cytometry. (B) MM485 tet-on GFP or GFP-MC1R cells were grown for 48 h with or without doxycycline and lysed. Protein levels were assessed by Western blot. (C) A panel of melanoma cell lines was plated at 50,000 cells per well in six-well plates. Starting 24 h after plating, cells were treated daily with DMSO or forskolin (50 μM). Wells were counted in duplicate every two days. (D) MM370 cells were synchronized by thymidine double block and released in the presence of DMSO or forskolin (50 μM). Cells were harvested and fixed every 3 h. Cells were stained with propidium iodide (PI), and cell cycle was assessed by DNA content analysis. All PI experiments were replicated multiple times. Shown is one representative experiment. (E) MM485 pLVX tet on GFP-MC1R cells were synchronized by thymidine double block and released in the presence or absence of doxycycline. MSH was added 6 h postrelease where indicated. Cells were collected and fixed every 2 h beginning 6 h postrelease. Cell cycle was analyzed by PI staining. The percentage of 4N cells was determined by gating and is quantified in Right.
To determine whether this effect could be recapitulated by cAMP signaling alone, the nontransduced parental MM485 cells were plated at low density and treated daily with DMSO or forskolin. Fig. 1C shows that forskolin dramatically reduced cell number. This experiment was repeated in several different melanoma lines including A375 (RHC), MM370 (non-RHC), and MM415 (non-RHC). In all of the lines tested, we found decreased cell number relative to unstimulated controls following forskolin treatment, although the magnitude of this effect varied from cell line to cell line. The cell lines tested had a range of MC1R, NRAS, and BRAF genotypes, but there was no clear correlation between genotype and forskolin response. Instead, baseline proliferative rate was the greatest predictor of forskolin response, with faster growing cell lines showing diminished sensitivity to growth inhibition.
To inhibit the growth kinetics of the melanoma cultures, forskolin had to be replaced in the media on a daily basis. Additionally, there was no visual evidence of apoptosis or cell death in the treated wells so we investigated the possibility that the forskolin effect on cell number resulted from changes in proliferation and the cell cycle. MM370 cells were synchronized in G1 by thymidine double block, released in the presence of DMSO or forskolin, and progression through the cell cycle was determined by DNA content analysis. There was no difference between DMSO- and forskolin-treated cells with respect to S-phase entry and the cells accumulated equally with 4N DNA content. At 12 h postrelease, the majority of the DMSO-treated cells had completed the cell cycle and returned to a 2N DNA content; however, the forskolin-treated cells maintained a 4N DNA content and did not complete the cell cycle until 15 h postrelease (Fig. 1D). A similar, although attenuated effect was seen following activation of MC1R. MM485 GFP-MC1R–inducible cells (Fig. 1A) were synchronized in G1 by thymidine double block and released in the presence or absence of doxycycline and MSH, and progression through the cell cycle was determined by DNA content analysis. GFP-MC1R–expressing cells accumulated with a 4N DNA content similar to uninduced cells, but showed a 5% increase in 4N cells at 12 h, suggesting that they were delayed in progression through mitosis (Fig. 1E). MSH treatment also resulted in delayed progression through the cell cycle in both uninduced and GFP-MC1R–expressing cells, although to a greater extent in the MC1R-overexpressing cells.
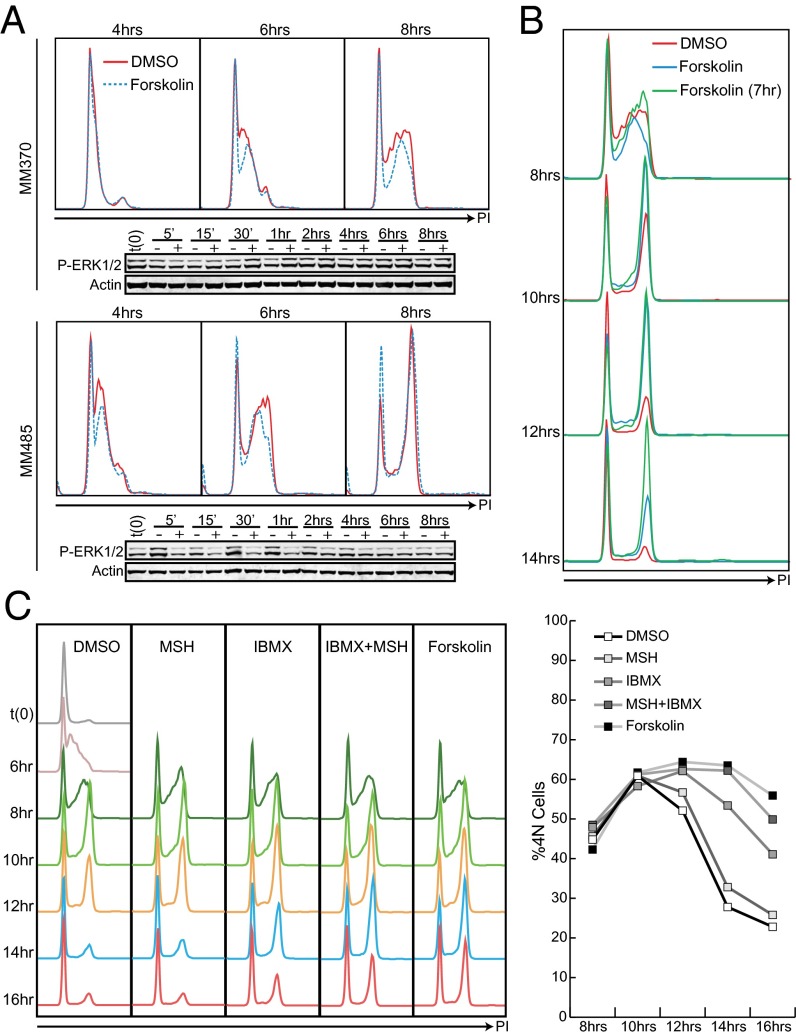
In other systems, cAMP signaling can delay entry into S phase, in part through inhibition of the MAPK cascade (18, 25, 26). In all of the melanoma lines tested, delayed progression through the cell cycle was not associated with delayed rates of S-phase entry (Fig. 2A). Forskolin treatment decreased MAPK signaling in NRas mutant but not BRaf mutant cell lines, however, this change in MAPK pathway activity was not correlated with delayed entry into S phase. To confirm that the cAMP-induced cell cycle delay was not caused by inhibition of S-phase entry, synchronized cells were treated with forskolin 7 h postrelease, when the cells were reaching the end of S phase. A similar but more pronounced delay in cell cycle progression was seen following delayed stimulation, and the cell cycle delay extended to 4 h or longer (Fig. 2B). These experiments were repeated with several different inducers of cAMP signaling, including physiological activators of MC1R and cAMP signaling (MSH) and a broad-spectrum inhibitor of phosphodiesterases, the enzymes that catalyze the breakdown of cAMP (IBMX). All of these cAMP-enhancing agents caused a delay in cell cycle progression that did not require interference with S phase. Of note, and confirming previous reports that cAMP levels can be uncoupled from receptor signaling by the activity of phosphodiesterases in melanoma cell lines (25, 27), activation of endogenous signaling through MSH treatment had only a modest effect on cell cycle delay. However, this delay was comparable to that seen with forskolin treatment when MSH was combined with the phosphodiesterase inhibitor IBMX (Fig. 2C).
Fig. 2.
cAMP-induced delay in cell cycle progression is not S-phase dependent. (A) MM370 (BRaf V600E) and MM485 (NRas Q61R) were synchronized by thymidine double block and released in the presence of DMSO or forskolin. Cells were fixed and lysed at the indicated times. Cell cycle progression was assessed by DNA content analysis and protein levels were assessed by Western blot with indicated antibodies. (B) MM370 cells were synchronized as described and released in the presence of DMSO, forskolin, or no treatment. Untreated cells were treated with forskolin 7 h postrelease toward the end of S phase. Cells were harvested and fixed at indicated time points and stained with PI for DNA content analysis. (C) MM370 cells were synchronized by thymidine double block and released. Seven hours postrelease, cells were treated with indicated MC1R and cAMP agonists. Cells were fixed at the indicated time points, and the cell cycle was assessed by PI staining. 4N fraction was quantified in the graph to the right.
cAMP Signaling Causes a Defect in Mitotic Entry.
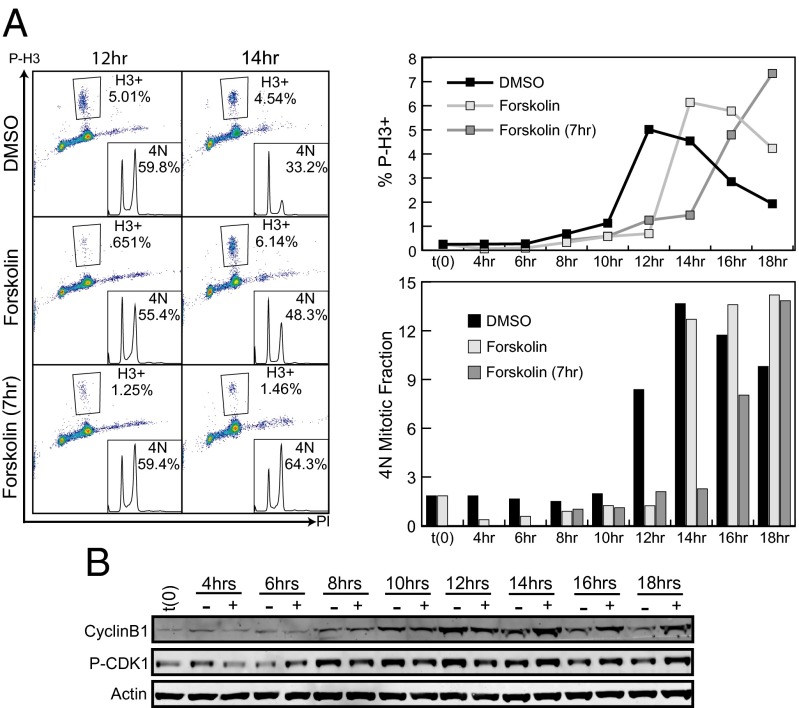
Because the delay in cell cycle progression manifested itself as an accumulation of cells with 4N DNA content and was independent of the presence of cAMP agonists during G0, G1, or S phases, we reasoned that the delay was the result of a defect in mitotic entry, mitosis or cytokinesis. To determine whether the accumulation of cells with 4N DNA was the result of a defect in mitotic entry, cells were synchronized in G1 and released in the presence of DMSO or forskolin, or treated with forskolin 7 h postrelease (forskolin 7hrs). Fixed cells were stained for phospho-histone H3 S10 (P-H3: a marker of condensed chromatin and mitosis) and DNA content. Twelve hours postrelease, DMSO-, forskolin-, and forskolin 7hrs-treated cells had similar percentages of 4N cells; however, DMSO-treated cells had a 4- to 7.6-fold higher percentage of P-H3–positive cells (Fig. 3A). At 14 h, nearly half of the DMSO-treated cells had completed the cell cycle and a high percentage of the remaining 4N cells were mitotic (4.5% P-H3/33.2% 4N). The cells that were released in the presence of forskolin also showed a high number of mitotic cells and had begun to complete the cell cycle; however, the cells that were treated with forskolin at the completion of S phase still had a low percentage of mitotic cells (1.5% H3/64.3% 4N), and none of the cells had completed the cell cycle. The 4N mitotic fraction was determined by dividing the number of mitotic cells by the total number of cells with 4N DNA content. Relative to DMSO-treated wells, the 4N mitotic fraction increased and peaked with a 2-h delay in cells released in the presence of forskolin and a 4-h delay in cells treated with forskolin at the G2/M transition. This delay in phospho-H3 staining strongly suggests that the delay in cell cycle progression seen upon forskolin treatment was caused by delayed entry into mitosis.
Fig. 3.
Forskolin delays mitotic entry. (A) MM485 cells were synchronized, released and treated with DMSO or forskolin at the time of release or with forskolin 7 h postrelease. Cells were fixed at indicated time points and stained for P-H3 and DNA content. The P-H3–positive population was defined by the indicated gate. P-H3–positive fraction and the 4N mitotic fraction (P-H3/4N DNA) are quantified in the graphs at Right. (B) Duplicate wells from A were lysed, separated by SDS/PAGE, and proteins were visualized by immunoblot using indicated antibodies.
Entry into mitosis is controlled primarily by the G2/M checkpoint proteins cyclin B and CDK1. Like other checkpoint kinases, CDK1 activity requires binding to its cyclin partner; however, CDK1 must also be dephosphorylated at two key inhibitory residues (T14 and Y15) to become fully active. To test whether this checkpoint complex was inhibited by forskolin treatment, lysates from synchronized cells were probed for levels of phospho-CDK1 Y15 and cyclin B (Fig. 3B). Forskolin treatment delayed dephosphorylation of CDK1 relative to DMSO-treated cells by more than 4 h. Cyclin B expression was stronger and sustained in forskolin-treated relative to DMSO-treated cells. Cyclin B is degraded after the metaphase to anaphase transition, and these increased levels of cyclin B serve as another marker of delayed mitosis.
Cdc25B Is Phosphorylated and Mislocalized by Forskolin Treatment and PKA.
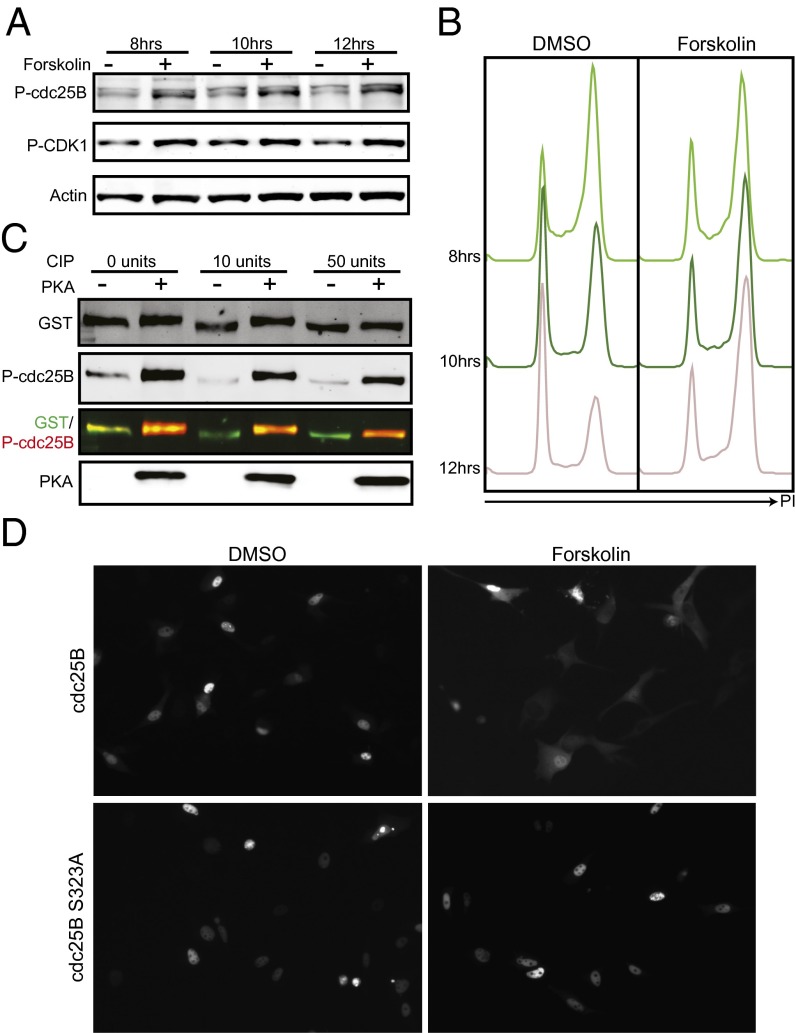
Cdc25B and C are responsible for dephosphorylating and activating CDK1. Cdc25C is thought to dephosphorylate and activate the majority of CDK1, however, it requires phosphorylation by CDK1 to become active itself. Cdc25B dephosphorylates and activates this initial pool of CDK1 and serves as a trigger for mitotic entry. Both cdc25s have an inhibitory phosphorylation site (S323 and S216, respectively) that causes binding to 14-3-3 proteins, is a target of CHK1 and has been demonstrated to be phosphorylated and inhibited by PKA during meiosis in mice and frogs respectively (23, 24). We first tested whether cdc25C could be the target of cAMP signaling causing delayed mitotic entry following forskolin treatment (Fig. S3). We found that forskolin treatment delayed the activation of cdc25C (Fig. S3B) and caused decreased protein and mRNA expression of cdc25C (Fig. S3 C and D). In contrast to what has been reported in Xenopus meiosis, we did not detect increased inhibitory phosphorylation (Fig. S3 C, F, and G). Replacement of cdc25C did not rescue the forskolin effect, indicating that cdc25C inhibition was not mediating the cell cycle delay (Fig. S3E). On the other hand, we found that forskolin treatment of synchronized cells led to increased cdc25B S323 inhibitory phosphorylation. This increase was correlated with increased P-CDK1 (Y15) levels and delayed entry into mitosis (Fig. 4 A and B).
Fig. 4.
cdc25B is phosphorylated and inhibited in response to forskolin treatment. (A) MM485 cells were synchronized and released. DMSO or forskolin was added to media 7 h postrelease. Cells were lysed at indicated time points and phospho-protein levels were assessed by Western blot with indicated antibodies. (B) Duplicate wells from A were fixed and stained with PI for DNA content analysis. (C) GST-tagged cdc25B was purified from transiently transfected 293FT cells by Glutathione pulldown. Purified protein was treated with varying amounts of calf intestinal phosphatase and then with purified recombinant PKA. Treated protein was visualized by Western blot for total cdc25B (GST), P-cdc25B (S323), and PKA. (D) MM370 tet on cells were transfected with pLVX GFP-cdc25B or GFP-cdc25B S323A, synchronized and released in the presence of doxycycline and DMSO or forskolin. Protein localization was assessed by live cell fluorescence microscopy. Images were taken 10 h postrelease.
To determine whether cdc25B is a direct target of PKA, an in vitro kinase assay was performed on GST-tagged cdc25B purified from 293FT cells. Basal cdc25B phosphorylation was removed by calf intestinal phosphatase treatment, and purified protein was incubated with recombinant PKA for 1 h. Western blots with phospho-specific antibodies showed a massive increase in cdc25B S323 phosphorylation following PKA treatment, indicating that PKA can directly phosphorylate cdc25B on this inhibitory residue (Fig. 4C). Furthermore, we tested the ability of the PKA inhibitor H-89 to block the phosphorylation of cdc25B in the presence of forskolin and found that PKA inhibition does reduce the amount of phosphorylated cdc25B in cell synchronization experiments (Fig. S4).
Phosphorylation at S323 leads to 14-3-3 binding and cytoplasmic sequestration (28). To test the effects of cAMP on subcellular localization, cells were transfected with doxycycline-inducible GFP-tagged cdc25B or GFP-tagged cdc25B S323A. Cells were synchronized and released with doxycycline and DMSO or forskolin, and subcellular localization was determined by live-cell fluorescence imaging (Fig. 4D). By 10 h postrelease, the DMSO-treated wild-type cdc25B showed predominantly nuclear fluorescence; however, the forskolin-treated cells showed primarily cytoplasmic cdc25B. Like wild type, a S323A mutant of cdc25B showed primarily nuclear fluorescence following DMSO treatment; however, forskolin treatment had no effect on localization of the mutant. Together, these data show that cdc25B becomes phosphorylated at S323 in the presence of cAMP signaling, that this phosphorylation can be carried out directly by PKA, and that this results in localization of cdc25B to the cytoplasm during the G2/M transition.
Restoring cdc25B Activity Rescues Forskolin-Induced Delay in Mitotic Entry.
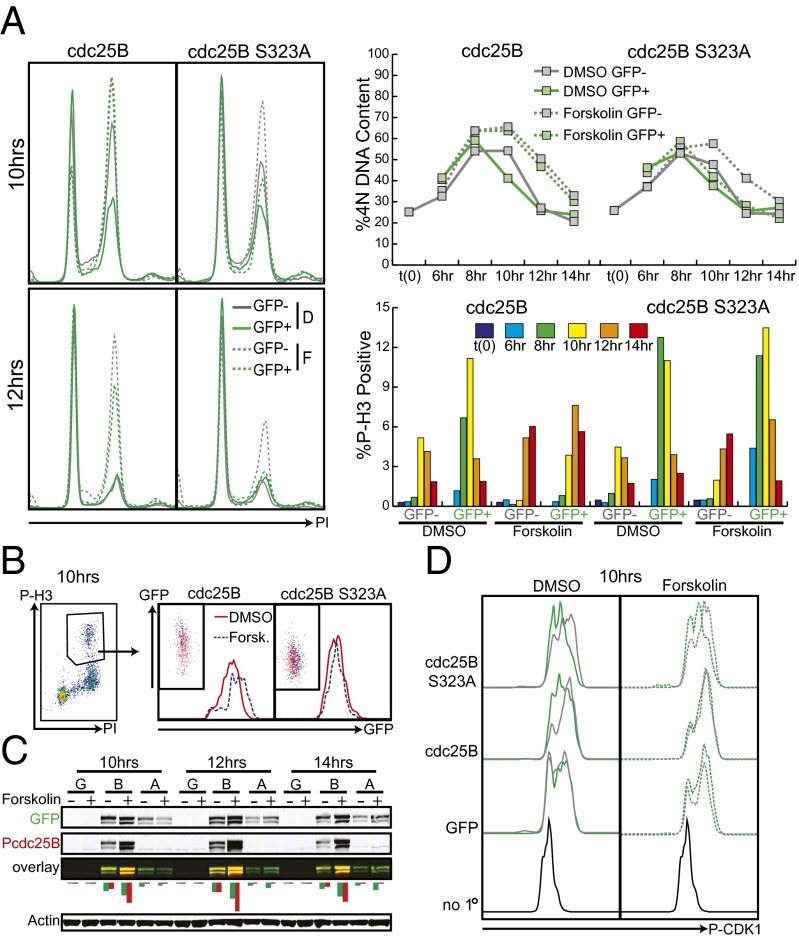
To determine whether forskolin’s effect on mitotic entry was mediated by inhibition of cdc25B activity, we overexpressed GFP-tagged cdc25B and cdc25B S323A and assessed mitotic entry following forskolin stimulation. Prolonged overexpression of cdc25B or its mutant caused mitotic defects, so a doxycycline-inducible system was used to control the levels and timing of cdc25B expression. Mixed populations of MM370 tet-on and MM370 tet-on pLVX GFP-cdc25B or -cdc25B S323A were synchronized by thymidine double block, released in the presence of doxycycline and treated at 5 h postrelease with DMSO or forskolin to a final concentration of 50 μM. Because these cells were a mixed population we were able to compare the cell cycles of cdc25B overexpressing (GFP+) and normal (GFP−) cells growing in the same well. Cdc25B wt and S323A caused rapid progression through the cell cycle in the presence of DMSO, with fewer 4N cells at 10 h postrelease than uninfected counterparts (Fig. 5A). However, cdc25B wild-type was unable to rescue the forskolin-induced cell cycle delay, and following forskolin treatment, cdc25B-expressing cells showed a similar percentage of 4N cells to the nontransduced cells at 10, 12 and 14 h postrelease. Cdc25B S323A, on the other hand, was able to rescue the forskolin effect, and cdc25B S323A-expressing cells did not show delayed cell cycle progression following forskolin treatment. The effects of cdc25B wild-type and S323A on 4N DNA content were mirrored by their effects on mitotic entry. Following DMSO treatment, cdc25B wild-type or mutant expression caused P-H3 levels to begin to peak 2 h earlier than in uninfected cells. Following forskolin treatment, wild-type cdc25B-expressing cells lost this premature mitotic entry; however, they did show enhanced mitosis relative to the uninfected, forskolin-treated cells (Fig. 5A). We analyzed the levels of GFP in this population of P-H3–positive cells and found that in the cdc25B wild-type, but not S323A mutant, there was a twofold increase in mean fluorescence intensity of the forskolin treated cells relative to the DMSO treated cells (Fig. 5B and Fig. S5). This finding suggests cdc25B’s ability to drive mitosis is subject to regulation by cAMP and PKA but, at very high levels of cdc25B expression, PKA is not able to inhibit all of the cdc25B and this remaining pool is able to drive mitotic entry. Cdc25B S323A was able to induce premature mitotic entry equally well in the presence or absence of forskolin treatment as seen in the P-H3 graph in Fig. 5A.
Fig. 5.
Forskolin-induced cell cycle delay is rescued by expression of active cdc25B. (A) MM370 pLVX tet on/pLVX tight puro GFP-cdc25B or GFP-cdc25B 323A cells were synchronized, released into doxycycline and treated with DMSO or forskolin 5 h postrelease. Cells were fixed at indicated time points and stained for DNA content and P-H3. The percentages of 4N cells and P-H3+ cells are quantified in Right. (B) GFP levels were assessed in the P-H3–positive population of cells from A. In the cdc25B wild type, but not S323A mutant, there was a nearly twofold increase in median fluorescence intensity in the forskolin treated wells compared with the DMSO-treated wells. There was no change in median fluorescence intensity in GFP-positive cells that were ungated for P-H3 (Fig. S1). (C) Duplicate wells from the above experiment were lysed and protein was visualized by immunoblot. Fluorescence intensities of the green (GFP/total cdc25B) and red (P-cdc25B S323) bands were quantified and graphed below the overlay blot. Transfection conditions are identified as: G = GFP, B = cdc25B, A = cdc25B S323A. (D) Duplicate wells from the above experiment were fixed in formaldehyde, methanol-permeabilized, stained for P-CDK1 (Y15), and analyzed by flow cytometry.
Western blots on lysates from these experiments showed enhanced S323 phosphorylation of the GFP-tagged wild-type cdc25B in the presence of forskolin relative to DMSO and no phosphorylation of the S323A mutant regardless of the stimulation (Fig. 5C). We used flow cytometry to compare the levels of CDK1 inhibitory phosphorylation in normal cells and cells overexpressing the wild-type or mutant forms of cdc25B. Cdc25B-expressing cells showed a shift toward lower CDK1 phosphorylation in DMSO-treated, but not forskolin-treated conditions (Fig. 5D). In cdc25B S323A-expressing cells, there was an even more pronounced effect on CDK1 dephosphorylation, and this was not altered by treatment with forskolin. We attempted to determine whether cdc25B activity could reverse the forskolin-induced decrease in long-term proliferation of melanoma cell cultures; however, prolonged and deregulated cdc25B activity caused cell death, and these experiments could not adequately address whether the inhibition of cdc25B by forskolin is responsible for the decreased proliferation of melanoma cell lines (Fig. S6). Together, these experiments have shown a role for cAMP signaling in delaying mitotic entry and cell cycle progression. This effect results from phosphorylation and inhibition of cdc25B, and a mutant form of cdc25B that cannot be phosphorylated allows cells to progress through the cell cycle normally in the presence of cAMP signaling.
Discussion
We have investigated the role of MC1R and cAMP signaling in the proliferation and cell cycle of melanoma. We found that sustained activation of MC1R or up-regulation of cAMP levels by forskolin slows the proliferation of melanoma cell lines. Experiments testing the effects of sustained up-regulation of MC1R activity showed that cells ectopically expressing wild type MC1R were at a selective disadvantage relative to cells expressing only endogenous RHC alleles of MC1R, and that this effect was enhanced by stimulation of the receptor by MSH. This finding provides evidence that wild-type MC1R can inhibit melanoma cell growth directly. Loss of function MC1R RHC alleles confer an increased risk to melanoma that has been associated with increased UV-associated mutagenesis or increased oxidative DNA damage; however, these results suggest that the activity of functional MC1R could directly inhibit the proliferation and growth of melanomas.
Besides suggesting a potential role and mechanism for MC1R signaling in directly inhibiting melanoma proliferation, this work also elucidates a role for cAMP signaling in the regulation of the mitotic G2/M checkpoint. cAMP signaling has been linked to regulation of the G1/S checkpoint through inhibition of the Ras/MAPK pathway and has been implicated in regulation of meiotic resumption through similar mechanisms to those presented in this paper. Here we show that this mechanism is also operative in the somatic cell cycle. The question remains whether this mechanism is generalizable to primary melanocytes and other cell types, or if the cAMP-dependent inhibition of the cell cycle is specific to cancer. We attempted to address this question in primary melanocytes using the approaches described above. However, because cultured melanocytes are not primed for proliferation, the synchronization experiments that reveal the cell cycle effects of MC1R and cAMP signaling were not feasible. An additional confounding factor that has been previously published by other groups and observed in our own laboratory, is the fact that MC1R and cAMP signaling induce transient MAPK activation and are proproliferative in cultured primary melanocytes (29, 30). It is possible that the cdc25B response is present in these cells, but that the mitogenic MAPK signal has a greater impact on the overall proliferation of these slow-growing cultures. Alternatively, oncogene activation may cause global or specific signaling changes that make cells susceptible to cAMP-induced cdc25B inhibition and mitotic entry delay. High MAPK activity can decrease the levels of cdc25B (31); and in developing melanomas, MC1R and cAMP signaling could cooperate with BRaf or NRas mutation and MAPK activation to inhibit the activity of cdc25B, causing decreased proliferation. Going forward, it will be important to determine whether cAMP’s inhibition of cdc25B activity is specific to cells experiencing oncogenic stress, or if this represents a mechanism of cell cycle regulation in response to physiological stress signals in other cell and tissue types.
In conclusion, we have found evidence that MC1R signaling can have a direct inhibitory role on melanoma cell line proliferation, and that MC1R and cAMP signaling can regulate progression from G2 into mitosis through a cdc25B-dependent mechanism. This work helps further our understanding of the role of MC1R and cAMP signaling in proliferation and cell cycle arrest, and its potential as a target of therapeutic intervention.
Materials and Methods
Cell Culture, Synchronization, and Drug Treatment.
MM200, MM370, MM485, MM415, and A375 cell lines were grown in RPMI +10% (vol/vol) FBS. Cells were synchronized by thymidine double block. Cells were grown in media containing 2 mM thymidine for 14 h, washed with PBS and grown in normal media for 10 h, returned to 2 mM thymidine for 14 h and then washed with PBS and released into thymidine-free media with various drug treatments. Dox induction was carried out in 1 μg/mL doxycycline. Forskolin was used in concentrations ranging from 20 to 50 μM. IBMX was used at 100 μM. MSH was used at a concentration of 100 nM. DMSO was added to wells where necessary to bring the concentration of DMSO up to that of the treated wells.
Flow Cytometry.
Cells were fixed by one of three different protocols depending on the cells and application (for details, see SI Materials and Methods). For DNA content analysis, cells were stained with 10 μg/mL propidium iodide with RNaseA. P-H3 staining was carried out with anti P-H3 S10-Alexa 647 (molecular probes) at 1:100 dilution for 1 h on ice. All cell cycle experiments were replicated a minimum of three times. Because of variability in synchronization, we have shown one representative experiment in each case. P-CDK1 staining was performed with anti-CDK1 Y15 (Cell Signaling) and an anti-rabbit–Alexa647 secondary from molecular probes. Cells were collected on the Beckman Dickson LSRII flow cytometer and analyzed in FlowJo.
Pull Downs and Western Blot.
Cells were lysed in TNM lysis buffer (TNM+protease and phosphatase inhibitors, DTT, and 1% triton). For GST pulldown, lysates were added to Glutathione-conjugated Sepharose beads and agitated for 2 h at 4 °C. Following incubation, beads were washed three times with ice cold TNM + 0.3% triton, drained and resuspended in sample buffer. Blots were visualized and quantified using the licor fluorescent detection system and Alexa 680-conjugated anti-rabbit and Alexa 800-conjugated anti-mouse secondary antibodies.
Supplementary Material
Acknowledgments
We thank past and present members of F.M.’s laboratory for invaluable technical help and discussion, and, in particular, Sang Hyun-Lee for sharing his cell cycle expertise. Additionally, we thank Kevin Haigis and Doug Lauffenburger for providing space and feedback for the completion of this manuscript, and Jeane Ann Grisso, Mark Lyons, and Sherry Stolberg for critical reading and editing of the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201917110/-/DCSupplemental.
References
- 1.Gandini S, et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. Eur J Cancer. 2005;41(14):2040–2059. doi: 10.1016/j.ejca.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 2.Elwood JM, Jopson J. Melanoma and sun exposure: An overview of published studies. Int J Cancer. 1997;73(2):198–203. doi: 10.1002/(sici)1097-0215(19971009)73:2<198::aid-ijc6>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 3.Abdel-Malek Z, et al. The melanocortin-1 receptor is a key regulator of human cutaneous pigmentation. Pigment Cell Res. 2000;13(Suppl 8):156–162. doi: 10.1034/j.1600-0749.13.s8.28.x. [DOI] [PubMed] [Google Scholar]
- 4.Cui R, et al. Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell. 2007;128(5):853–864. doi: 10.1016/j.cell.2006.12.045. [DOI] [PubMed] [Google Scholar]
- 5.Gerstenblith MR, Goldstein AM, Fargnoli MC, Peris K, Landi MT. Comprehensive evaluation of allele frequency differences of MC1R variants across populations. Hum Mutat. 2007;28(5):495–505. doi: 10.1002/humu.20476. [DOI] [PubMed] [Google Scholar]
- 6.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet. 1995;11(3):328–330. doi: 10.1038/ng1195-328. [DOI] [PubMed] [Google Scholar]
- 7.Beaumont KA, Shekar SN, Cook AL, Duffy DL, Sturm RA. Red hair is the null phenotype of MC1R. Hum Mutat. 2008;29(8):E88–E94. doi: 10.1002/humu.20788. [DOI] [PubMed] [Google Scholar]
- 8.Newton RA, et al. Activation of the cAMP pathway by variant human MC1R alleles expressed in HEK and in melanoma cells. Peptides. 2005;26(10):1818–1824. doi: 10.1016/j.peptides.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 9.Sánchez Más J, et al. Loss-of-function variants of the human melanocortin-1 receptor gene in melanoma cells define structural determinants of receptor function. Eur J Biochem. 2002;269(24):6133–6141. doi: 10.1046/j.1432-1033.2002.03329.x. [DOI] [PubMed] [Google Scholar]
- 10.Sturm RA, et al. The role of melanocortin-1 receptor polymorphism in skin cancer risk phenotypes. Pigment Cell Res. 2003;16(3):266–272. doi: 10.1034/j.1600-0749.2003.00041.x. [DOI] [PubMed] [Google Scholar]
- 11.D’Orazio JA, et al. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443(7109):340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- 12.Valverde P, et al. The Asp84Glu variant of the melanocortin 1 receptor (MC1R) is associated with melanoma. Hum Mol Genet. 1996;5(10):1663–1666. doi: 10.1093/hmg/5.10.1663. [DOI] [PubMed] [Google Scholar]
- 13.Landi MT, et al. MC1R germline variants confer risk for BRAF-mutant melanoma. Science. 2006;313(5786):521–522. doi: 10.1126/science.1127515. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Kraft P, Colditz GA, Wong J, Hunter DJ. Melanocortin 1 receptor variants and skin cancer risk. Int J Cancer. 2006;119(8):1976–1984. doi: 10.1002/ijc.22074. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy C, et al. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol. 2001;117(2):294–300. doi: 10.1046/j.0022-202x.2001.01421.x. [DOI] [PubMed] [Google Scholar]
- 16.Mitra D, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature. 2012;491(7424):449–453. doi: 10.1038/nature11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song X, et al. alpha-MSH activates immediate defense responses to UV-induced oxidative stress in human melanocytes. Pigment Cell Melanoma Res. 2009;22(6):809–818. doi: 10.1111/j.1755-148X.2009.00615.x. [DOI] [PubMed] [Google Scholar]
- 18.Dumaz N, Marais R. Integrating signals between cAMP and the RAS/RAF/MEK/ERK signalling pathways. Based on the anniversary prize of the Gesellschaft für Biochemie und Molekularbiologie Lecture delivered on 5 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 2005;272(14):3491–3504. doi: 10.1111/j.1742-4658.2005.04763.x. [DOI] [PubMed] [Google Scholar]
- 19.Dumaz N, Light Y, Marais R. Cyclic AMP blocks cell growth through Raf-1-dependent and Raf-1-independent mechanisms. Mol Cell Biol. 2002;22(11):3717–3728. doi: 10.1128/MCB.22.11.3717-3728.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donzelli M, Draetta GF. Regulating mammalian checkpoints through Cdc25 inactivation. EMBO Rep. 2003;4(7):671–677. doi: 10.1038/sj.embor.embor887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nilsson I, Hoffmann I. Cell cycle regulation by the Cdc25 phosphatase family. Prog Cell Cycle Res. 2000;4:107–114. doi: 10.1007/978-1-4615-4253-7_10. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt A, Nebreda AR. Inhibition of Xenopus oocyte meiotic maturation by catalytically inactive protein kinase A. Proc Natl Acad Sci USA. 2002;99(7):4361–4366. doi: 10.1073/pnas.022056399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pirino G, Wescott MP, Donovan PJ. Protein kinase A regulates resumption of meiosis by phosphorylation of Cdc25B in mammalian oocytes. Cell Cycle. 2009;8(4):665–670. doi: 10.4161/cc.8.4.7846. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Protein kinase A modulates Cdc25B activity during meiotic resumption of mouse oocytes. Dev Dyn. 2008;237(12):3777–3786. doi: 10.1002/dvdy.21799. [DOI] [PubMed] [Google Scholar]
- 25.Dumaz N, et al. In melanoma, RAS mutations are accompanied by switching signaling from BRAF to CRAF and disrupted cyclic AMP signaling. Cancer Res. 2006;66(19):9483–9491. doi: 10.1158/0008-5472.CAN-05-4227. [DOI] [PubMed] [Google Scholar]
- 26.Dumaz N, Marais R. Protein kinase A blocks Raf-1 activity by stimulating 14-3-3 binding and blocking Raf-1 interaction with Ras. J Biol Chem. 2003;278(32):29819–29823. doi: 10.1074/jbc.C300182200. [DOI] [PubMed] [Google Scholar]
- 27.Marquette A, André J, Bagot M, Bensussan A, Dumaz N. ERK and PDE4 cooperate to induce RAF isoform switching in melanoma. Nat Struct Mol Biol. 2011;18(5):584–591. doi: 10.1038/nsmb.2022. [DOI] [PubMed] [Google Scholar]
- 28.Forrest A, Gabrielli B. Cdc25B activity is regulated by 14-3-3. Oncogene. 2001;20(32):4393–4401. doi: 10.1038/sj.onc.1204574. [DOI] [PubMed] [Google Scholar]
- 29.Buscà R, et al. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. EMBO J. 2000;19(12):2900–2910. doi: 10.1093/emboj/19.12.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Luca M, et al. Alpha melanocyte stimulating hormone (alpha MSH) stimulates normal human melanocyte growth by binding to high-affinity receptors. J Cell Sci. 1993;105(Pt 4):1079–1084. doi: 10.1242/jcs.105.4.1079. [DOI] [PubMed] [Google Scholar]
- 31.Astuti P, et al. MAPK pathway activation delays G2/M progression by destabilizing Cdc25B. J Biol Chem. 2009;284(49):33781–33788. doi: 10.1074/jbc.M109.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.