Abstract
Polyadenylation of pre-mRNAs is critical for efficient nuclear export, stability, and translation of the mature mRNAs, and thus for gene expression. The bulk of pre-mRNAs are processed by canonical nuclear poly(A) polymerase (PAPS). Both vertebrate and higher-plant genomes encode more than one isoform of this enzyme, and these are coexpressed in different tissues. However, in neither case is it known whether the isoforms fulfill different functions or polyadenylate distinct subsets of pre-mRNAs. Here we show that the three canonical nuclear PAPS isoforms in Arabidopsis are functionally specialized owing to their evolutionarily divergent C-terminal domains. A strong loss-of-function mutation in PAPS1 causes a male gametophytic defect, whereas a weak allele leads to reduced leaf growth that results in part from a constitutive pathogen response. By contrast, plants lacking both PAPS2 and PAPS4 function are viable with wild-type leaf growth. Polyadenylation of SMALL AUXIN UP RNA (SAUR) mRNAs depends specifically on PAPS1 function. The resulting reduction in SAUR activity in paps1 mutants contributes to their reduced leaf growth, providing a causal link between polyadenylation of specific pre-mRNAs by a particular PAPS isoform and plant growth. This suggests the existence of an additional layer of regulation in plant and possibly vertebrate gene expression, whereby the relative activities of canonical nuclear PAPS isoforms control de novo synthesized poly(A) tail length and hence expression of specific subsets of mRNAs.
The poly(A) tail at the 3′ end is an essential feature of virtually all eukaryotic mRNAs that influences stability, nuclear export, and translational efficiency of the mRNAs (1, 2). It is synthesized after RNA polymerase II has transcribed past the cleavage and polyadenylation site and associated signal sequences (3, 4). These sequences are bound by several protein complexes, including Cleavage-stimulation Factor (CstF) and Cleavage and Polyadenylation Specificity Factor (CPSF) in animals and their counterparts in yeast and presumably in plants (2, 5). The complexes cleave the nascent pre-mRNA at the prospective polyadenylation site and recruit poly(A) polymerase (PAPS) to add the poly(A) tail.
The poly(A) tail is synthesized by PAPSs, with the bulk of cellular pre-mRNAs being polyadenylated by canonical nuclear PAPSs (cPAPSs) (5, 6) that share substantial sequence identity with human poly(A) polymerase-α (PAPOLA), bovine poly(A) polymerase, or the yeast enzyme Pap1p (7–9). Although the Saccharomyces cerevisiae and Drosophila melanogaster genomes only encode one cPAPS, which is essential for growth (7, 10), three such cPAPSs are found in humans: PAPOLA (PAPα), PAPOLB (PAPβ), and PAPOLG (PAPγ) (11). Of these, PAPOLA is thought to be the main PAPS in somatic cells. PAPOLA and PAPOLG proteins contain a C-terminal regulatory region next to the highly conserved catalytic N-terminal domain and are found either in both nucleus and cytoplasm (PAPOLA) or only in the nucleus (PAPOLG) of cells throughout the human body (9, 11–14). By contrast, PAPOLB lacks the C-terminal region, is exclusively cytoplasmic, and is only found in testis cells, where it is required to extend the poly(A) tail of cytoplasmic mRNAs encoding sperm-related proteins (15); as a consequence, male mice mutant for PAPOLB are sterile.
The Arabidopsis thaliana genome encodes four cPAPS proteins, termed PAPS1 to PAPS4 (16, 17). PAPS3 resembles PAPOLB in lacking an extended C-terminal region, being localized in the cytoplasm and expressed mainly in the male gametophytes (the pollen). By contrast, PAPS1, PAPS2, and PAPS4 all contain an extended C-terminal region, localize exclusively to the nucleus, and are expressed throughout the plant (2, 16–18). All four proteins have nonspecific polyadenylation activity in vitro, suggesting that they represent functional cPAPSs (16, 19). On the basis of the failure to identify homozygous transfer DNA (T-DNA) insertion mutants for any of the three genes, it was concluded that all of them are essential for plant growth and development (17).
Gene expression can be regulated via a number of mechanisms impinging on the mRNA 3′ end. The choice between alternative 3′ end cleavage sites is widely used to regulate gene expression in both animal and plant development, for example via the exclusion or inclusion of microRNA target sites in the resulting 3′ UTRs (20–24). Additionally, modulating the length of the poly(A) tails on mRNAs in the cytoplasm by the opposing actions of cytoplasmic PAPS (e.g., PAPOLB) and deadenylases can be used to control the expression of the encoded proteins (1, 15). However, it is currently unclear whether polyadenylation by nuclear cPAPS can also contribute to the control of specific gene expression. In principle, this could occur in either of two ways. First, pre-mRNAs could be differentially sensitive to variations in the total cPAPS activity provided by one or more functionally interchangeable cPAPS isoforms; such a mechanism may underlie specific developmental phenotypes in weak mutants of D. melanogaster cPAPS (25). Second, some mRNAs may be exclusively or preferentially polyadenylated by one cPAPS in organisms with more than one isoform. Given such target specificity, modulating the balance of activities between the isoforms could then be used to alter the length of the de novo synthesized poly(A) tails, and hence ultimately gene expression, of subsets of mRNAs. Target specificity has at present only been observed for noncanonical PAPS (6, 26), such as Star-PAP, which is required for the cellular response to oxidative stress.
Here we provide evidence for functional specialization and target specificity among A. thaliana nuclear cPAPS isoforms. Mutations affecting different isoforms cause very different phenotypes that depend on the divergent C-terminal domains of the proteins. In particular, reduction of PAPS1 activity disrupts polyadenylation of SMALL AUXIN UP RNA (SAUR) mRNAs and causes leaf growth defects due to reduced SAUR function and a constitutive pathogen response. We propose that this specificity of PAPS isoforms provides an additional level of regulating plant gene expression.
Results and Discussion
paps1 Mutants Show Organ-Specific Effects on Growth.
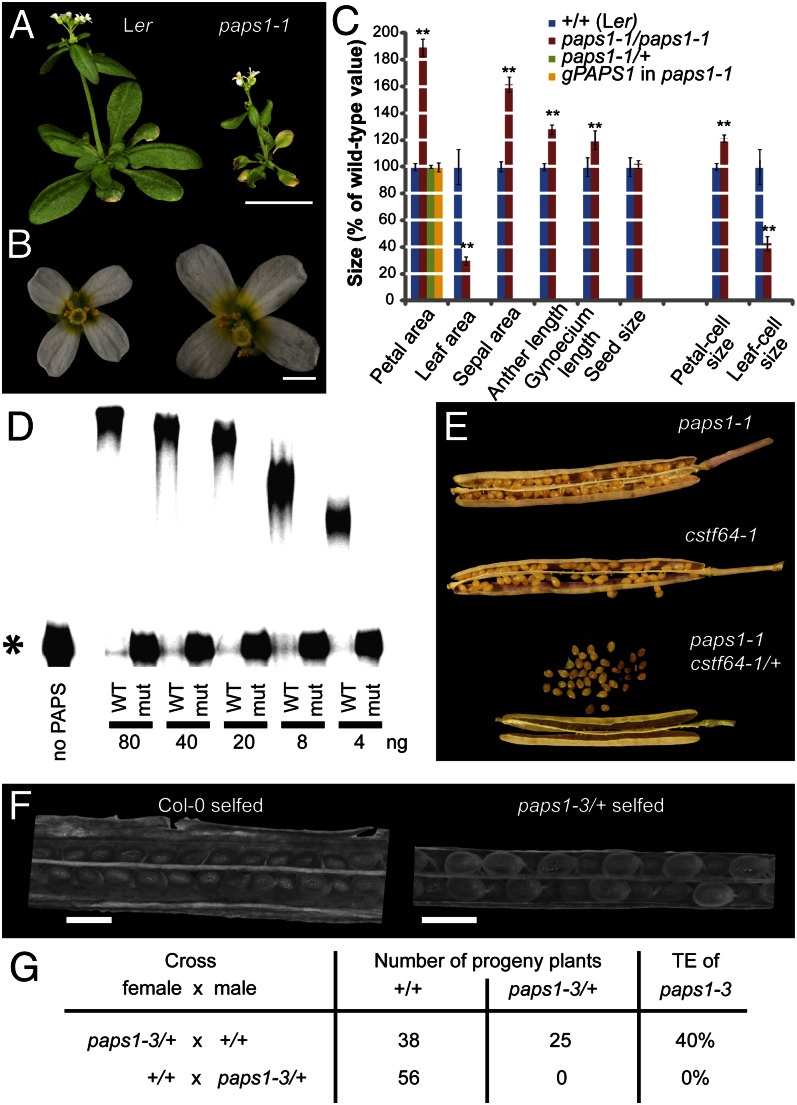
From an ethyl-methanesulfonate (EMS) mutagenesis screen, we identified a unique recessive mutation causing opposite effects on the growth of leaves and flowers, termed paps1-1 (Fig. 1 A–C). Although the size of paps1-1 mutant leaves is reduced to less than one-third of the wild-type value, mutant petals and other floral organs are larger than in wild type, with petals reaching almost twice the wild-type size. At the cellular level, the reduced leaf size is largely due to a defect in cell expansion (Fig. 1C). Conversely, the size of paps1-1 mutant petal cells is only increased by 21%, indicating that the bulk of the difference in petal size is due to a higher number of cells (Fig. 1C). Thus, PAPS1 function is required to allow normal cell expansion in leaves and to limit cell proliferation in petals.
Fig. 1.
Loss of PAPS1 function leads to altered organ growth. (A and B) Whole-plant (A) and flower images (B) of the indicated genotypes. (C) Quantification of organ and cell sizes from the indicated genotypes. Values are mean ± SE from at least 5 (leaves), 20 (petals, sepals, anthers), 7 (gynoecia), or 55 (seeds) organs per genotype, normalized to the wild-type mean. (D) Autoradiograph of in vitro nonspecific polyadenylation assay. The indicated amounts of wild-type PAPS1 protein (WT) or of the mutant form encoded by the paps1-1 allele (mut) were used. Asterisk indicates the unpolyadenylated RNA substrate. (E) Micrographs of opened siliques from the indicated genotypes. Note the aborted seeds produced by paps1-1 cstf64-1/+ plants. (F) Light micrographs of opened siliques from Col-0 (Left) and paps1-3/+ heterozygous plants (Right) after selfing. (G) Transmission efficiency (TE) of the paps1-3 mutant allele through the male and the female gametophyte. The result for the first cross is not significantly different from the expected 50:50 ratio (P = 0.10, χ2 test). (Scale bars, 1 cm in A, 1 mm in B, 500 μm in F.)
The paps1-1 Mutation Reduces the Activity of PAPS1 in Canonical Nuclear Polyadenylation of pre-mRNAs.
To determine the molecular basis of the paps1-1 mutant phenotype, we isolated the affected gene by mapping and sequencing of candidate genes. This identified a C-to-T transition typical for EMS-induced mutations in the At1g17980 gene coding for PAPS1 (Fig. S1A). The mutation causes an amino acid substitution of serine for proline at position 313. The mutated proline lies in a linker peptide between the nucleotidyl-transferase domain and the RNA-binding domain within the N-terminal catalytic region of the protein and is very highly conserved in PAPSs from yeast, plants, and animals (Fig. S1B). Complementation of the paps1-1 mutant with a wild-type genomic copy of the PAPS1 locus restored a wild-type phenotype (Fig. S1C). The paps1-1 allele is temperature sensitive (Fig. S1D); in contrast to growth at 23 °C, seedlings grown at 28 °C showed a very severe phenotype with bleaching and almost complete growth inhibition, indicating that the phenotypes seen at lower temperatures result from only a moderate reduction in PAPS1 activity (see also below).
To determine the effect of the paps1-1 mutation on the protein’s activity, we performed in vitro polyadenylation assays using purified recombinant protein (27). Whereas the wild-type protein showed robust polyadenylation activity, virtually no enzymatic activity was observed when using the mutant form (Fig. 1D and Fig. S1E).
To genetically determine whether PAPS1 is indeed involved in canonical pre-mRNA processing in the nucleus, we combined the paps1-1 allele with a mutant allele of the CSTF64 locus encoding the sole A. thaliana homolog to the Cstf64 subunit of the cleavage-stimulation factor complex (28). We could not recover any double homozygous cstf64-1 paps1-1 mutants, and the siliques from cstf64-1/+; paps1-1/paps1-1 plants contained 25% aborted seeds (Fig. 1E and Fig. S1F), indicating that the double mutant genotype confers embryo lethality. This contrasts with full seed set in either single mutant. Together, this synthetic lethality and the results of the in vitro assay strongly suggest that the paps1-1 mutation affects nuclear polyadenylation of transcripts.
PAPS1 Activity Is Essential for Male Gametophyte Function.
To determine the effects of a complete loss of PAPS1 activity, we studied a presumed null allele with a T-DNA insertion in the fifth intron within the region coding for the N-terminal catalytic protein domain (paps1-3; Fig. S1A). It was not possible to recover plants homozygous for the paps1-3 allele. To determine whether this is due to embryonic lethality or to a gametophytic defect, we analyzed the seeds developing on paps1-3/+ heterozygous plants and performed reciprocal crosses. There was no evidence for either embryo lethality or a female gametophytic defect from analyzing the siliques of paps1-3/+ plants, because we did not detect aborted seeds or unfertilized ovules (Fig. 1F). Consistent with this, the paps1-3 mutant allele was normally transmitted through the female gametophyte (Fig. 1G). By contrast, when applying pollen from paps1-3/+ plants to wild-type stigmas, none of the progeny carried the mutant allele (Fig. 1G), indicating that the paps1-3 mutation causes a male gametophytic defect. Pollen from paps1-1 mutant plants and from paps1-3/+ heterozygotes was morphologically normal and viable (Fig. S1G).
Thus, PAPS1 represents an essential gene for male gametophyte function, and a progressive reduction of remaining PAPS1 activity in the diploid sporophyte causes increasingly more severe phenotypic defects as seen in the paps1-1 allele (see above).
The Three Canonical Nuclear Poly(A) Polymerases Encoded by the A. thaliana Genome Are Functionally Specialized.
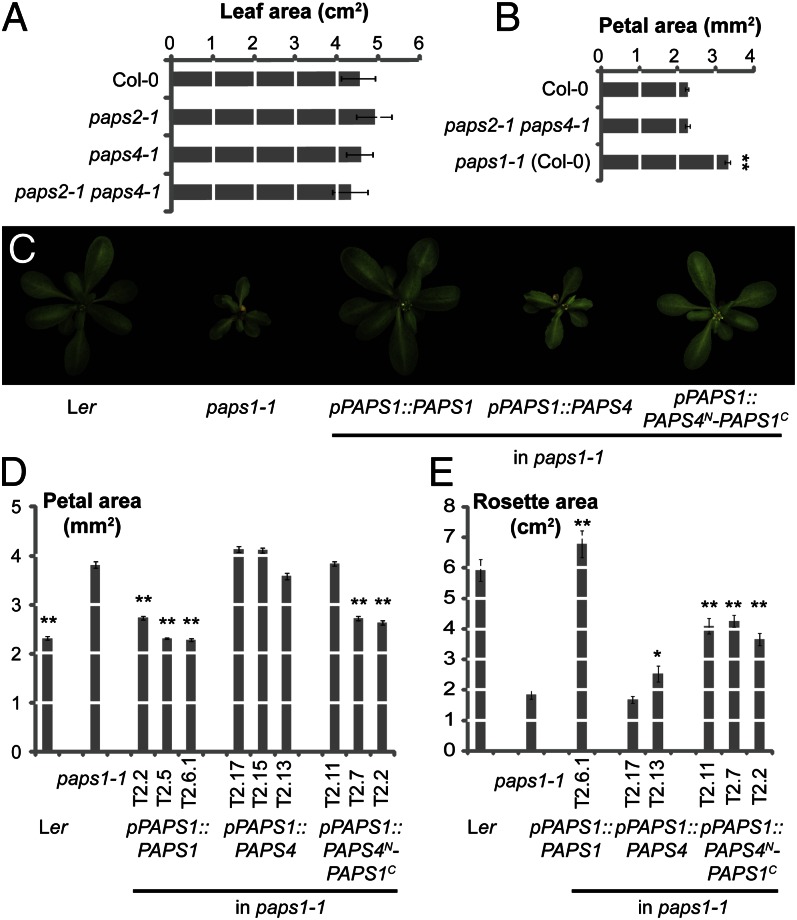
To determine the roles of the other two canonical nuclear PAPSs in A. thaliana, putative null alleles were isolated for PAPS2 and PAPS4. In both cases the eighth exon within the region coding for the catalytic N-terminal domain was disrupted, and no full-length mRNA could be detected from the mutant alleles (Fig. S2 B, D, and E). Both single mutants and the paps2-1 paps4-1 double mutant were viable (Fig. S2B) and showed normal leaf and petal growth (Fig. 2 A and B). To test whether the much more severe phenotypes resulting from loss of PAPS1 function were simply due to PAPS1 being responsible for most polyadenylation in Arabidopsis, we determined bulk poly(A) tail lengths in paps1-1 mutant and wild-type seedlings. There was virtually no change in the distribution of bulk poly(A) tail lengths (Fig. S2 F and G), despite the very severe mutant phenotype under the growth conditions used (Fig. S1D). Together, these results indicate that most transcripts can be redundantly polyadenylated by either PAPS1 or PAPS2/PAPS4 but that a small subset of critical transcripts is exclusively or preferentially targeted by PAPS1.
Fig. 2.
The three nuclear PAPS isoforms in A. thaliana fulfill distinct functions. (A and B) Quantification of leaf (A) and petal size (B) in the indicated genotypes. Values are mean ± SE from at least five leaves and 20 petals per genotype. **Significantly different from wild-type value at P < 0.01 (t test). (C) Top views of plants of the indicated genotypes. (D and E) Quantification of petal (D) and rosette (E) size in the indicated genotypes. Individual bars represent independent transformant lines for the transgenic plants. Values are mean ± SE from at least 15 petals and at least six rosettes per genotype. Asterisks (*,**) indicate significant difference from paps1-1 mutant value at P < 0.05 (*) or P < 0.01 (**) according to t test with Bonferroni correction.
The three proteins PAPS1, PAPS2, and PAPS4 share highly conserved N-terminal catalytic regions, whereas the C-terminal domains are more divergent (Fig. S2A). We therefore asked whether the functional divergence apparent from the different mutant phenotypes was due to differences at the protein level. Introducing the PAPS4 coding region under the control of the pPAPS1 promoter (pPAPS1::PAPS4) into a paps1-1 background did not complement the mutant phenotype (Fig. 2 C–E and Fig. S2C). By contrast, when a chimeric protein consisting of the catalytic domain from PAPS4 and the C-terminal region from PAPS1 was expressed under the control of the pPAPS1 promoter in paps1-1 mutants (pPAPS1::PAPS4N-PAPS1C), it was able to substantially rescue the growth phenotype in leaves and particularly in flowers (Fig. 2 C–E and Fig. S2C). Thus, divergence in the C-terminal domains of the proteins is responsible for functional specialization among the PAPS isoforms in A. thaliana.
Polyadenylation of SAUR mRNAs Is Defective in paps1 Mutants.
To determine the molecular basis for the paps1 mutant phenotypes, we compared transcript abundances in paps1-1 mutant vs. wild-type leaves and flowers, using microarray hybridization. A total of 1,130 and 779 genes were misregulated more than twofold in paps1-1 mutant leaves and flowers compared with wild type, respectively (Tables S1 and S2). Two hundred sixty-one genes were misregulated in both organs, suggesting that despite a substantial overlap in the molecular phenotypes of mutant leaves and floral organs, many transcript changes were specific to one or the other organ type, potentially contributing to the different growth phenotypes.
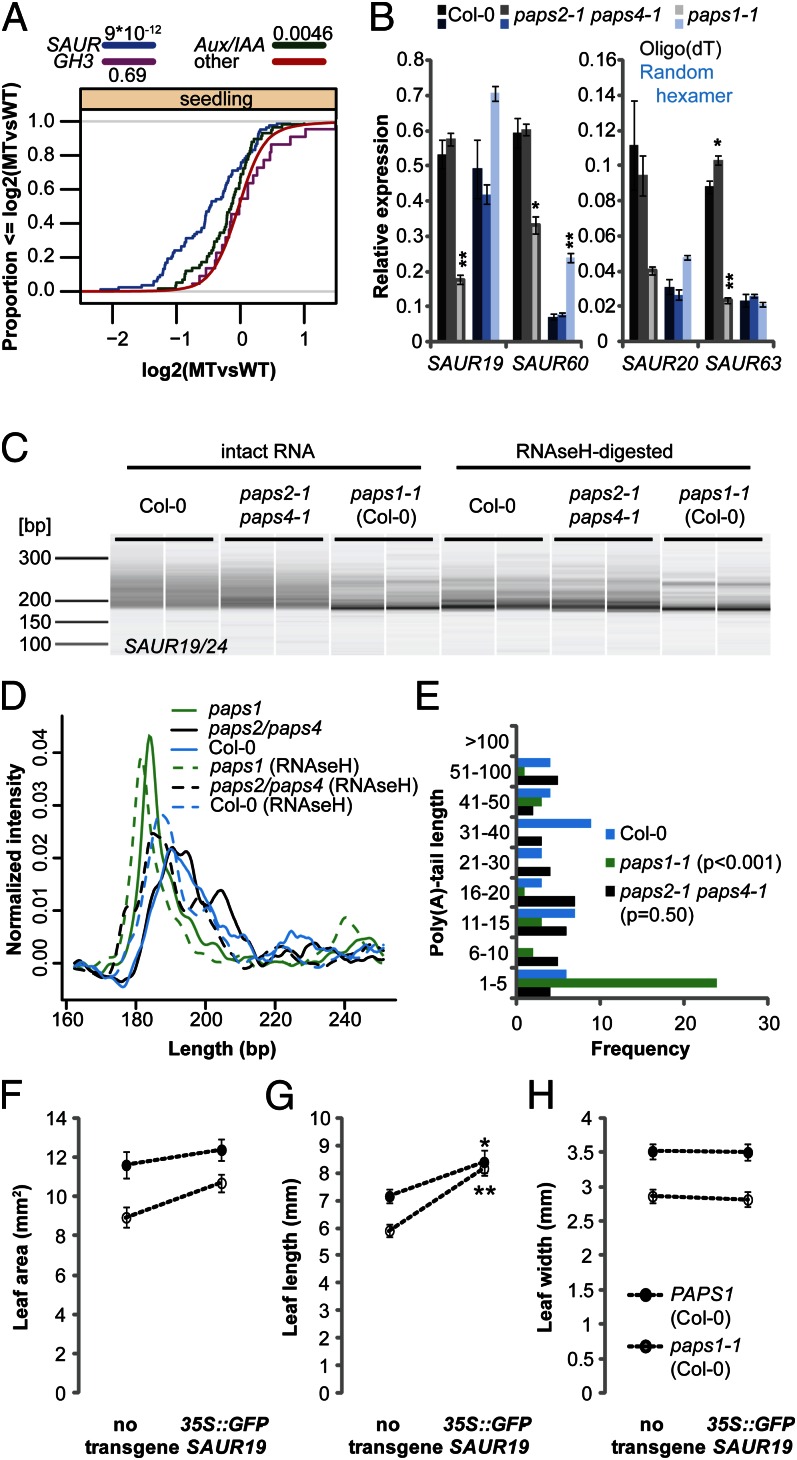
We found a significantly reduced hybridization signal for the family of SAUR mRNAs (29–31) from paps1-1 mutants compared with wild type, particularly in seedlings (Fig. 3A and Fig. S3A; median and average fold-change of 0.72). This was not accompanied by a comparable misregulation of other auxin-responsive genes, such as Aux/IAA or GH3 family members (Fig. 3A and Fig. S3A), indicating that the paps1-1 mutation did not interfere with auxin response as such. Testing individual SAUR transcripts using quantitative RT-PCR (qRT-PCR) with oligo(dT) priming confirmed the reduced signal specifically from paps1-1 mutants but not from paps2-1 paps4-1 mutants (Fig. 3B). However, no such effect was observed when using random hexamers to prime the reverse transcription, with comparable or even higher signals in paps1-1 mutants than in wild type (Fig. 3B). This suggests that the weaker signal on the microarrays and in the oligo(dT)-primed qRT-PCR was due to less efficient oligo(dT)-primed reverse transcription because of a shorter poly(A) tail, not due to reduced abundance of the tested SAUR mRNAs.
Fig. 3.
Defective polyadenylation of SAUR mRNAs in paps1 contributes to reduced leaf growth. (A) Cumulative distribution plot of the expression levels of SAUR, GH3, and Aux/IAA family members in paps1-1 vs. wild-type seedlings. The y axis indicates the fraction of genes with a log2-expression ratio less than or equal to the value on the x axis. Numbers in legend are P values of a Wilcoxon rank-sum test. “other,” all remaining genes on the array. (B) Expression of the indicated SAUR genes in paps1-1 and paps2-1 paps4-1 mutant seedlings compared with wild-type. Values from oligo(dT)-primed cDNA are shown in gray shades, those from random hexamer-primed cDNA in blue shades. Values shown are the means ± SE from three (Col-0 and paps2-3 paps4-3) or two (paps1-1) biological replicates, normalized to the constitutive reference gene PDF2 (AT1G13320). Plants had been kept at 30 °C for 2 h before harvesting. *P < 0.05; **P < 0.01 (Student t test). (C) Bioanalyzer electropherogram of RT-PCR–amplified 3′ ends of SAUR19/24 transcripts from the indicated genotypes. Two biological replicates per genotype are shown. RNA had been left untreated (Left) or poly(A) tails had been digested with RNAseH and oligo(dT) (Right) before reverse transcription. (D) Normalized signal intensities of the PCR products in C. Averages of the two biological replicates per genotype are shown. (E) Length distribution of poly(A) tails as determined by sequencing subcloned individual PCR products from intact RNA in C. P values in legend are from a Wilcoxon rank-sum test. (F–H) Leaf area (F), length (G), and width (H) of wild-type or paps1-1 mutant plants with or without the 35S::GFP-SAUR19 transgene. Asterisks (*,**) indicate significant difference from value in the absence of the transgene at P < 0.05 (*) or P < 0.01 (**) according to t tests with Bonferroni correction.
To determine whether PAPS1 activity was indeed required for polyadenylation of SAUR mRNAs, we compared their poly(A) tail lengths between paps1-1 mutants and wild type using a PCR-based assay to amplify part of the coding sequence/3′ UTR and the entire poly(A) tail (SI Results and Discussion). Before harvesting, seedlings were kept at 30 °C for 2 h to largely abolish the remaining activity of the temperature-sensitive mutant protein (see above). PCR products for SAUR19/24 and SAUR62/63/66/68 mRNAs from paps1-1 mutants were shorter than from wild type or paps2 paps4 double mutants (Fig. 3 C and D, Fig. S3 D and E, and SI Results and Discussion). To confirm that the different lengths of the PCR products indeed reflected a difference in poly(A) tail length and not in the choice of cleavage site, we subcloned and sequenced individual molecules. This indicated that the choice of 3′ end cleavage site was not affected (Fig. S3 B and G) and confirmed the dramatic reduction in the lengths of the poly(A) tails specifically in paps1-1 mutants but not in paps2-1 paps4-1 plants (Fig. 3E and Fig. S3F). The median lengths of poly(A) tails determined from subcloned molecules from wild type, paps2-1 paps4-1, and paps1-1 were 22, 17, and 2 for SAUR19/24 and 19, 20, and 2 for SAUR62/63/66/68, respectively. Measuring the SAUR19/24 poly(A) tail length from nuclear RNA of wild-type and paps1-1 inflorescences showed the same difference as seen for total RNA (Fig. S4 A–C), arguing that the shorter poly(A) tails are indeed due to defective nuclear polyadenylation, rather than faster cytoplasmic deadenylation in the mutant. The most parsimonious explanation of these results is that SAUR transcripts are polyadenylated directly and exclusively by the PAPS1 isoform.
We asked whether the phenotypic rescue of the paps1 leaf-growth defect by the chimeric PAPS4N-PAPS1C protein was mirrored at the molecular level by a rescue of the SAUR mRNA polyadenylation defect. The poly(A) tails on SAUR19/24 mRNAs were longer in paps1-1 plants expressing the chimeric PAPS4N-PAPS1C protein than in plants expressing PAPS4 under the control of the pPAPS1 promoter or in nontransgenic paps1-1 mutants, yet they did not reach the wild-type length (Fig. S4 D–F). This indicates that the divergent C-terminal domains of the PAPS proteins influence the mapping of PAPS isoforms to at least some of their presumed targets, possibly via binding to different forms of 3′ end processing factors (3, 5).
Reduced SAUR Activity Contributes to the Leaf-Growth Defect in paps1 Mutants.
A recent report demonstrated that the activity of the SAUR19-24 subfamily is required for normal cell expansion in hypocotyls and leaves (31). Hypocotyls in paps1-1 are longer than in wild type (Fig. S5A), suggesting that other expansion-promoting effects override a possibly reduced SAUR activity in this organ. To determine whether reduced SAUR19-24 activity contributed to the defect in leaf growth in paps1-1 mutants, we introduced a 35S::GFP-SAUR19 transgene with the 3′ UTR of the nopaline synthase gene (nos terminator) into the mutant background. The transgene promoted growth, especially in the leaf-length direction, in both the wild-type and the paps1-1 mutant background; however, it had a much stronger effect in the latter, both in absolute and in relative terms (leaf length +17% in wild-type, +38% in paps1-1 background; Fig. 3 F–H). Indeed, leaf length was indistinguishable between 35S::GFP-SAUR19 and paps1-1; 35S::GFP-SAUR19 plants (Fig. 3G). This nonadditive effect indicates that the reduced leaf length in paps1-1 mutants is due to lower SAUR activity, because otherwise the GFP-SAUR19 transgene would be expected to have the same effect in both genetic backgrounds.
The above experiment suggests that SAUR19 protein levels are lower in paps1-1 mutant than in wild-type leaves. However, the polyadenylation defect of SAUR mRNAs in paps1 mutants does not seem to destabilize the mRNAs, as indicated by the qRT-PCR experiments on random hexamer-primed cDNA (Fig. 3B), as reported for several previous examples of stable mRNAs with very short poly(A) tails (32–34). Consistent with this, introducing the dst2 mutant (35) into the paps1-1 background to alleviate the inherent instability of SAUR mRNAs did not rescue the paps1-1 phenotype (Fig. S5B). This suggests that in paps1 mutants either nuclear export or translation efficiency of SAUR mRNAs is reduced, either of which could lead to reduced SAUR protein levels.
paps1-1 Mutant Leaves Show a Constitutive Pathogen Response.
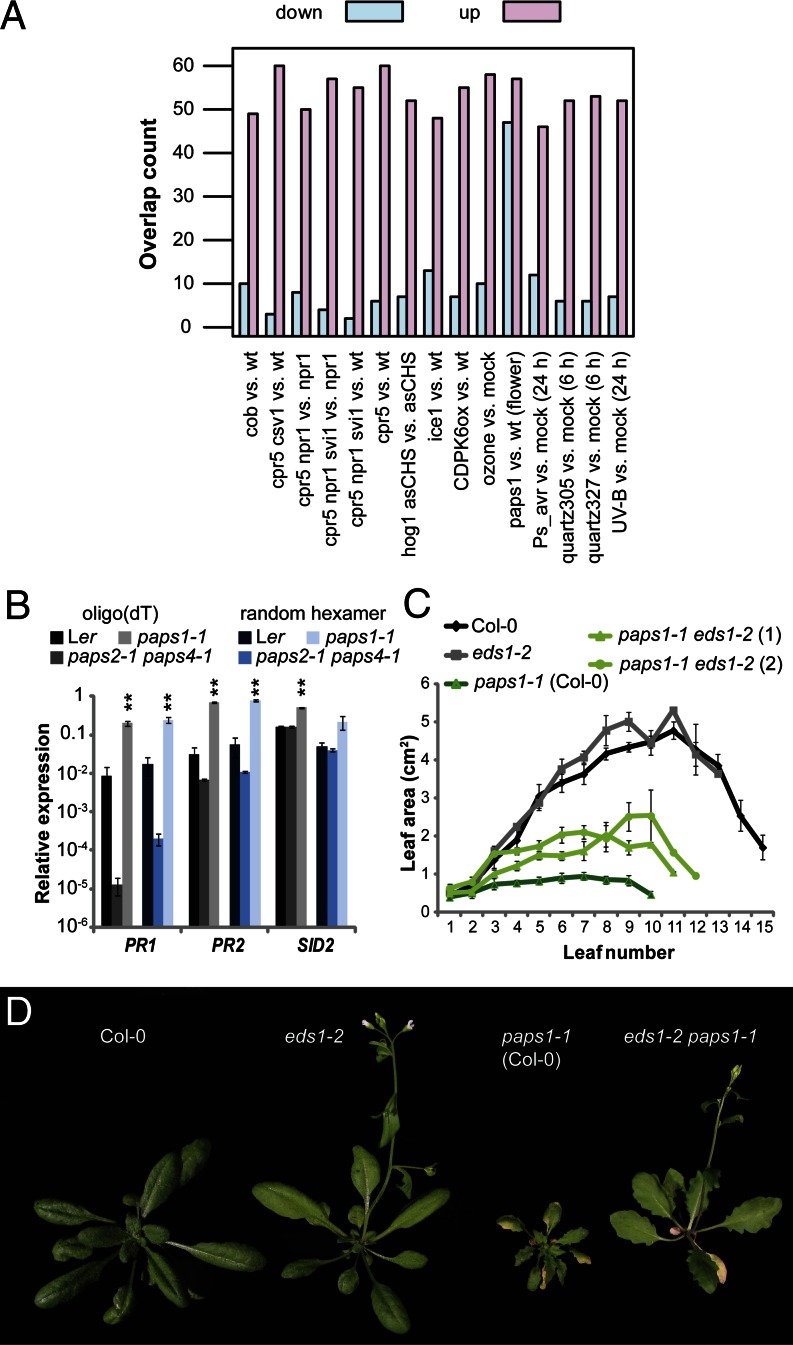
To identify additional biological processes affected by the paps1 mutation, we compared the misregulated genes to more than 600 published A. thaliana microarray studies using MASTA (36). This identified a strong overlap with genes affected in the constitutive expression of pathogenesis-related genes5 (cpr5) mutant and in response to pathogen infection and other stresses (Fig. 4A and Fig. S6A). In particular, the overlap was almost as strong with genes affected in cpr5 vs. wild type as with genes affected in cpr5 npr1 vs. npr1. This suggests that the constitutive pathogen response in paps1-1 is independent of NON-EXPRESSOR OF PR1 (NPR1), a master regulator of the SA-mediated pathogen response (Fig. 4A) (37). This in turn suggests the activation of the ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1)/ PHYTOALEXIN DEFICIENT 4 (PAD4)-dependent pathogen response in paps1-1, which forms part of the cpr5 phenotype (38). The signature of a constitutive pathogen response was also evident at the single-gene level, with several classic marker genes specifically up-regulated in paps1-1 mutant vs. wild-type leaves but not in paps2-1 paps4-1 plants (e.g., PR1, PR2, SID2, and the defensins PDF1.2b, PDF1.2c, and PDF1.4) (Fig. 4B and Table S1).
Fig. 4.
Reduced leaf growth in paps1 is partly due to an EDS1-dependent pathogen response. (A) Overlap of genes misregulated in leaves of paps1-1 mutants vs. wild-type with genes misregulated in the experiments indicated. “Down” and “up” refer to the direction of the expression change in paps1-1 vs. wild type. Table S3 defines the abbreviations used. (B) Expression of the indicated genes in paps1-1 mutant seedlings compared with wild-type, using oligo(dT)-primed (gray shades) or random hexamer-primed cDNA (blue shades). Values shown are the means ± SE from three biological replicates, normalized to the constitutive reference gene PDF2 (AT1G13320). (C) Quantification of leaf area in the indicated genotypes. Values shown are means ± SE from at least three plants per genotype. (D) Whole-plant phenotypes of the indicated genotypes.
Constitutive activation of the pathogen response results in reduced leaf growth due to reduced cell expansion (39). We therefore tested whether an EDS1/PAD4-dependent constitutive pathogen response contributes to the paps1-1 phenotype. Indeed, leaf growth in the eds1-2 paps1-1 and the pad4-1 paps1-1 double mutants was substantially rescued (Fig. 4 C and D and Fig. S6 B and C). However, petal overgrowth was not rescued (Fig. S6D). Thus, an EDS1/PAD4-dependent constitutive pathogen response contributes to the reduced leaf growth but not to the petal overgrowth in paps1 mutants. This indicates that PAPS1, but not PAPS2 or PAPS4, negatively modulates the plant pathogen response. However, none of several pathogen-response associated genes tested (e.g., EDS1, NPR1, PR1, PR2, SID2, SIZ1, WRKY18), some of which are affected in the microarray analysis, showed a robust change in the lengths of their poly(A) tails.
Functional Specialization Among PAPS Isoforms Provides an Additional Level of Gene Regulation.
Our results demonstrate that the three canonical nuclear PAPSs in Arabidopsis fulfill different functions owing to their divergent C-terminal domains. The very different mutant phenotypes, the results of analyzing bulk poly(A)-tail length, and the defects in polyadenylation of SAUR mRNAs in paps1 but not paps2 paps4 mutants strongly suggest that a fraction of transcripts is exclusively or preferentially targeted by PAPS1. As outlined in the Introduction, such target specificity provides an opportunity for regulating gene expression by modulating the balance of activities among the PAPS isoforms. PAPS1 is phosphorylated on several residues in its C-terminal domain [http://phosphat.mpimp-golm.mpg.de (40)], suggesting posttranslational modification as one way of altering PAPS1 activity. Reduced PAPS1 activity causes a constitutive pathogen response via an EDS1/PAD4-dependent mechanism. Thus, it is tempting to speculate that response to pathogen infection may be a scenario in which modulation of PAPS1 activity is used to alter the mRNA polyadenylation status and thus expression of a subset of pathogen-response factors. A genome-wide approach to determine poly(A) tail lengths will be required to identify the pathogen-response genes whose polyadenylation depends on PAPS1 to conclusively test this notion. Finally, we note that because homologs to PAPS1 and PAPS2/PAPS4 are found throughout higher plants in phylogenetically well-supported clades (17), this mode of regulation may function broadly in higher plants.
Materials and Methods
Plant Materials and Growth Conditions.
The paps1-1 mutation was identified in an EMS-mutagenesis screen in the Landsberg erecta (Ler) background and back-crossed three times to Ler before analysis. For comparison with mutants in the Col-0 background, the paps1-1 mutation was introgressed into Col-0 by three rounds of back-crossing. Details of T-DNA insertion lines and other mutants used can be found in SI Materials and Methods.
Plant growth conditions were as described previously (41). All measurements were done with plants grown at 23 °C, unless otherwise stated. For the experiment involving the 35S::GFP-SAUR19 transgene plants were grown on 1/2 MS plates including 1% sucrose at 21 °C (day) and 14 °C (night).
Genotyping Mutant Alleles.
For genotyping the paps1-1 allele, a dCAPS marker (oSV126 and oSV166) was used. The PCR product (210 bp) from the mutated allele is cut by EcoRI. For genotyping of T-DNA insertion alleles, gene-specific primers (called LP and RP primer) that flank the T-DNA insertion site, and a T-DNA right border primer (BP) were used. These are listed in SI Materials and Methods.
Phenotypic Analysis, Measurements of Organ and Cell Sizes.
Dissected organs were scanned, and their size was measured using ImageJ software (http://rsbweb.nih.gov/ij/). Petal-cell size was determined essentially as described previously (42). For determining cell size in leaves, leaves were fixed in FAA solution [10% (wt/vol) formaldehyde, 5% (vol/vol) acetic acid, 50% (vol/vol) ethanol], cleared with chloral hydrate, and observed using differential phase contrast. Further details can be found in SI Materials and Methods.
In Vitro Polyadenylation Assay and Measurement of Bulk Poly(A) Tail Length.
Nonspecific polyadenylation assays were performed essentially as described previously (27), as was the measurement of bulk poly(A) tail lengths (43). Further details can be found in SI Materials and Methods.
Microarray Analysis.
Transcriptomes of paps1-1 mutant and wild-type seedlings and flowers (details in SI Materials and Methods) were compared using the Agilent Arabidopsis 4 × 44K oligo microarray. Two-color microarrays were normalized using the loess method (44). Differentially expressed genes were identified using the R/Bioconducor package Limma (45).
qRT-PCR and Measurements of Poly(A) Tail Length.
Total RNA was prepared by the hot phenol method (46), DNase-digested, and reverse-transcribed using oligo(dT)17 or random-hexamer primers. Expression levels were analyzed using a Roche LightCycler 480. Poly(A) tail length was determined using the Affymetrix Poly(A) Tail-Length Assay Kit. Details and primers used are described in SI Materials and Methods.
Molecular Cloning and Plant Transformation.
The floral dip transformation protocol was carried out as described previously (47). Details of molecular cloning can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Isabel Bäurle, Cyril Zipfel, and members of the Lenhard laboratory for discussion; Timothy Wells, Christiane Schmidt, and Doreen Mäker for excellent plant care; and Jane Parker and Pamela Green for seeds. S.L.V was supported by the Rotation PhD program from the John Innes Centre. Work in the M.L. laboratory was supported by Deutsche Forschungsgemeinschaft Grant Le1412/3-1. D.L. and T.L. were supported by Deutsche Forschungsgemeinschaft Grant La606/13-1 and the EU-INTEREG IV Upper Rhine program. W.M.G. was supported by National Institutes of Health (NIH) Grant GM067203 and National Science Foundation Grant MCB-0817205. N.R. and J.L.M. were supported by NIH Grant GM28983.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE48821).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303967110/-/DCSupplemental.
References
- 1.Eckmann CR, Rammelt C, Wahle E. Control of poly(A) tail length. Wiley Interdiscip Rev RNA. 2011;2(3):348–361. doi: 10.1002/wrna.56. [DOI] [PubMed] [Google Scholar]
- 2.Hunt AG. Messenger RNA 3′ end formation in plants. Curr Top Microbiol Immunol. 2008;326:151–177. doi: 10.1007/978-3-540-76776-3_9. [DOI] [PubMed] [Google Scholar]
- 3.Millevoi S, Vagner S. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res. 2010;38(9):2757–2774. doi: 10.1093/nar/gkp1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proudfoot NJ. Ending the message: Poly(A) signals then and now. Genes Dev. 2011;25(17):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65(7-8):1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmidt MJ, Norbury CJ. Polyadenylation and beyond: Emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip Rev RNA. 2010;1(1):142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- 7.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354(6353):496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 8.Raabe T, Bollum FJ, Manley JL. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353(6341):229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- 9.Thuresson AC, Aström J, Aström A, Grönvik KO, Virtanen A. Multiple forms of poly(A) polymerases in human cells. Proc Natl Acad Sci USA. 1994;91(3):979–983. doi: 10.1073/pnas.91.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juge F, Zaessinger S, Temme C, Wahle E, Simonelig M. Control of poly(A) polymerase level is essential to cytoplasmic polyadenylation and early development in Drosophila. EMBO J. 2002;21(23):6603–6613. doi: 10.1093/emboj/cdf633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13(11):1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topalian SL, et al. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol. 2001;21(16):5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyriakopoulou CB, Nordvarg H, Virtanen A. A novel nuclear human poly(A) polymerase (PAP), PAP gamma. J Biol Chem. 2001;276(36):33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- 14.Zhao W, Manley JL. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16(5):2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kashiwabara S, et al. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science. 2002;298(5600):1999–2002. doi: 10.1126/science.1074632. [DOI] [PubMed] [Google Scholar]
- 16.Addepalli B, Meeks LR, Forbes KP, Hunt AG. Novel alternative splicing of mRNAs encoding poly(A) polymerases in Arabidopsis. Biochim Biophys Acta. 2004;1679(2):117–128. doi: 10.1016/j.bbaexp.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Meeks LR, Addepalli B, Hunt AG. Characterization of genes encoding poly(A) polymerases in plants: Evidence for duplication and functional specialization. PLoS ONE. 2009;4(11):e8082. doi: 10.1371/journal.pone.0008082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hunt AG, et al. Arabidopsis mRNA polyadenylation machinery: Comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics. 2008;9:220. doi: 10.1186/1471-2164-9-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt AG, Meeks LR, Forbes KP, Das Gupta J, Mogen BD. Nuclear and chloroplast poly(A) polymerases from plants share a novel biochemical property. Biochem Biophys Res Commun. 2000;272(1):174–181. doi: 10.1006/bbrc.2000.2755. [DOI] [PubMed] [Google Scholar]
- 20.Di Giammartino DC, Nishida K, Manley JL. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43(6):853–866. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ji Z, Lee JY, Pan Z, Jiang B, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106(17):7028–7033. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherstnev A, et al. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol. 2012;19(8):845–852. doi: 10.1038/nsmb.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu X, et al. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci USA. 2011;108(30):12533–12538. doi: 10.1073/pnas.1019732108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murata T, et al. The hiiragi gene encodes a poly(A) polymerase, which controls the formation of the wing margin in Drosophila melanogaster. Dev Biol. 2001;233(1):137–147. doi: 10.1006/dbio.2001.0205. [DOI] [PubMed] [Google Scholar]
- 26.Mellman DL, et al. A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451(7181):1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 27.Takagaki Y, Ryner LC, Manley JL. Separation and characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52(5):731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 28.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science. 2010;327(5961):94–97. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 29.Chae K, et al. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J. 2012;71(4):684–697. doi: 10.1111/j.1365-313X.2012.05024.x. [DOI] [PubMed] [Google Scholar]
- 30.Hagen G, Guilfoyle T. Auxin-responsive gene expression: Genes, promoters and regulatory factors. Plant Mol Biol. 2002;49(3-4):373–385. [PubMed] [Google Scholar]
- 31.Spartz AK, et al. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 2012;70(6):978–990. doi: 10.1111/j.1365-313X.2012.04946.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijer HA, et al. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35(19):e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu H, Das Gupta J, Schoenberg DR. The poly(A)-limiting element is a conserved cis-acting sequence that regulates poly(A) tail length on nuclear pre-mRNAs. Proc Natl Acad Sci USA. 1999;96(16):8943–8948. doi: 10.1073/pnas.96.16.8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng J, Schoenberg DR. mRNA with a <20-nt poly(A) tail imparted by the poly(A)-limiting element is translated as efficiently in vivo as long poly(A) mRNA. RNA. 2005;11(7):1131–1140. doi: 10.1261/rna.2470905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson MA, Perez-Amador MA, Lidder P, Green PJ. Mutants of Arabidopsis defective in a sequence-specific mRNA degradation pathway. Proc Natl Acad Sci USA. 2000;97(25):13991–13996. doi: 10.1073/pnas.240354097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reina-Pinto JJ, Voisin D, Teodor R, Yephremov A. Probing differentially expressed genes against a microarray database for in silico suppressor/enhancer and inhibitor/activator screens. Plant J. 2010;61(1):166–175. doi: 10.1111/j.1365-313X.2009.04043.x. [DOI] [PubMed] [Google Scholar]
- 37.Dong X. NPR1, all things considered. Curr Opin Plant Biol. 2004;7(5):547–552. doi: 10.1016/j.pbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 38.Clarke JD, Aarts N, Feys BJ, Dong X, Parker JE. Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. 2001;26(4):409–420. doi: 10.1046/j.1365-313x.2001.2641041.x. [DOI] [PubMed] [Google Scholar]
- 39.Bowling SA, et al. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6(12):1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durek P, et al. PhosPhAt: The Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Res. 2010;38(Database issue):D828–D834. doi: 10.1093/nar/gkp810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Disch S, et al. The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr Biol. 2006;16(3):272–279. doi: 10.1016/j.cub.2005.12.026. [DOI] [PubMed] [Google Scholar]
- 42.Horiguchi G, Fujikura U, Ferjani A, Ishikawa N, Tsukaya H. Large-scale histological analysis of leaf mutants using two simple leaf observation methods: Identification of novel genetic pathways governing the size and shape of leaves. Plant J. 2006;48(4):638–644. doi: 10.1111/j.1365-313X.2006.02896.x. [DOI] [PubMed] [Google Scholar]
- 43.Preker PJ, Lingner J, Minvielle-Sebastia L, Keller W. The FIP1 gene encodes a component of a yeast pre-mRNA polyadenylation factor that directly interacts with poly(A) polymerase. Cell. 1995;81(3):379–389. doi: 10.1016/0092-8674(95)90391-7. [DOI] [PubMed] [Google Scholar]
- 44.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31(4):265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 45.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 46.Box MS, Coustham V, Dean C, Mylne JS. Protocol: A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods. 2011;7:7. doi: 10.1186/1746-4811-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.