Significance
The assembly of immunoglobulin and T-cell receptor genes by V(D)J (variable, diversity, joining) recombination must strike a balance between maximum diversification of antigen receptors and favoring gene segments with specialized functions. We quantified the use of V gene segments in the primary T-cell receptor β repertoire, defining the relative contribution of 13 parameters in shaping their recombination efficiencies. Computational analysis of these data provides a unifying model, revealing a minimal set of five parameters that predict Vβ use. This model building approach will help predict how natural alterations of large V clusters impact immune receptor repertoires.
Keywords: lymphocytes, T-cell receptor, gene regulation
Abstract
The primary antigen receptor repertoire is sculpted by the process of V(D)J recombination, which must strike a balance between diversification and favoring gene segments with specialized functions. The precise determinants of how often gene segments are chosen to complete variable region coding exons remain elusive. We quantified Vβ use in the preselection Tcrb repertoire and report relative contributions of 13 distinct features that may shape their recombination efficiencies, including transcription, chromatin environment, spatial proximity to their DβJβ targets, and predicted quality of recombination signal sequences (RSSs). We show that, in contrast to functional Vβ gene segments, all pseudo-Vβ segments are sequestered in transcriptionally silent chromatin, which effectively suppresses wasteful recombination. Importantly, computational analyses provide a unifying model, revealing a minimum set of five parameters that are predictive of Vβ use, dominated by chromatin modifications associated with transcription, but largely independent of precise spatial proximity to DβJβ clusters. This learned model-building strategy may be useful in predicting the relative contributions of epigenetic, spatial, and RSS features in shaping preselection V repertoires at other antigen receptor loci. Ultimately, such models may also predict how designed or naturally occurring alterations of these loci perturb the preselection use of variable gene segments.
Gene activity is regulated at multiple levels to coordinate expression during development. At a most basic level, the collection of cis-acting elements for a genetic locus recruits transcription factors that alter its chromatin environment to either induce or repress gene activity. Emerging studies indicate that the 3D conformation of a locus also plays an important role in the regulation of its composite genes (1). At most genes, many levels of control are integrated to achieve the requisite gene expression state. For example, transcriptional promoters interact with their cognate enhancers over considerable distances in the linear genome to generate “hubs” where the two cis elements are in spatial proximity (1, 2).
All of these regulatory strategies are used to generate functional Ig (Ig) and T-cell receptor (Tcr) genes during lymphocyte development (3). Each antigen receptor (AgR) locus is composed of multiple variable (V), joining (J), and sometimes diversity (D) gene segments that are assembled by the process of V(D)J recombination, creating a potential variable region exon (4). Recombination is mediated by the RAG-1/2 enzymatic complex, which is expressed in all developing lymphocytes and recognizes semiconserved recombination signal sequences (RSSs) flanking all AgR gene segments (5). On selection of two compatible gene segments by RAG-1/2, recombination proceeds via a DNA break/repair mechanism, ultimately fusing the two selected segments (4, 5).
The assembly of AgR genes is strictly regulated despite a common collection of genomic RSS targets and expression of recombinase in all resting (G0/G1) lymphocyte precursors (6). The most obvious level of regulation is lineage specificity. The RAG-1/2 complex assembles Tcr genes in precursor T cells, whereas Ig genes are targeted in precursor B cells. Even within an AgR locus, gene segment recombination is ordered, with D–J rearrangements preceding V–DJ. Numerous studies support a key role for chromatin accessibility in determining the recombination potential of gene segments (7). The primary RAG-1/2 targets in a given cell type are transcriptionally active and DNase hypersensitive, two hallmarks of accessible chromatin. Indeed, RAG-2 binds directly to a histone modification that accompanies transcription [trimethylated histone H3 lysine 4 (H3K4me3)], providing a link between chromatin and recombinase targeting (8, 9). At all AgR loci, activation of (D)J clusters is dependent on communication between at least one distal enhancer and a proximal promoter, which triggers transcription of the unrearranged (D)J segments (10). Recent studies indicate that the high transcriptional activity focuses RAG-1/2 binding at (D)J clusters, forming “recombination centers” into which V gene segments must be brought (11).
Although chromatin accessibility explains most aspects of RAG-1/2 deposition at recombination centers, this feature is not sufficient to ensure rearrangement of the distant V segments. Insertion of a powerful Tcra enhancer (Ea) into Tcrb maintains chromatin accessibility at nearby Vβ gene segments but does not facilitate their recombination at a stage of thymocyte development in which only Tcra genes rearrange (12). Subsequent studies have shown that long-range recombination of V segments requires changes in the 3D structure of an AgR locus, bringing the V cluster into spatial proximity with (D)J recombination centers located up to 3.2 Mb away (13–15). Long-range interactions and locus conformations are determined in large part by CCCTC-binding factor (CTCF) and cohesin, factors that bind numerous sites throughout the mammalian genome forming loops containing the intervening DNA (16). With regard to AgR loci, deletion of CTCF, its binding sites, or essential cohesin subunits disrupt spatial interactions at Igk, Igh, and Tcra, respectively, and perturb V to (D)J recombination (17–20).
In addition to lineage, stage, and allele specificity, it is also likely that the relative use of gene segments is regulated to shape the primary repertoire of V(D)J rearrangements in precursor lymphocyte populations. During subsequent stages of lymphocyte development, V gene segment use is an important component of positive/negative selection and, in some cases, is a primary determinant of functional subsets within a lineage (e.g., TRVB13-2 for iNKT cells) (21). As such, each species may have evolved toward a unique frequency profile for V use at each AgR locus, balancing requirements for receptor diversity, production of functional subsets, and efficacy of given V segments for antigens expressed by common pathogens. The mechanisms that sculpt preselection V repertoires likely incorporate a combination of the chromatin and spatial features described above. However, their relative contributions to the efficiency of long-range V to (D)J recombination at any AgR locus remain unknown.
We now address this basic question in adaptive immunity, beginning with the molecular determinants that shape Vβ use in preselection thymocytes. The Tcrb locus is an attractive starting point for building such models because it contains a manageable set of 35 Vβ segments for molecular analysis; the cis elements controlling recombination also are well defined (Fig. 1A). New experimental data for chromatin profiles, spatial proximity, and transcription, as well as predictions of RSS quality, were incorporated into a computational analysis that weights each of these features in determining Vβ recombination frequencies. Our data and analyses indicate that Tcrb adopts a 3D structure in which the relative proximity of each Vβ gene segment to DβJβ clusters is not a significant determinant in its recombination frequency. Instead, each Vβ gene segment has sufficient spatial access to the DβJβ recombination center, and use is fine-tuned by local Vβ chromatin environments, with a particular emphasis on transcription-dependent histone modifications. Indeed, these chromatin features are absent at nonfunctional Vβ gene segments regardless of their RSS quality or precise proximity to DβJβ clusters. This model-building approach should help unravel the primary determinants of preselection V use at other AgR loci and in predicting how natural alterations of large V clusters may impact immune receptor repertoires.
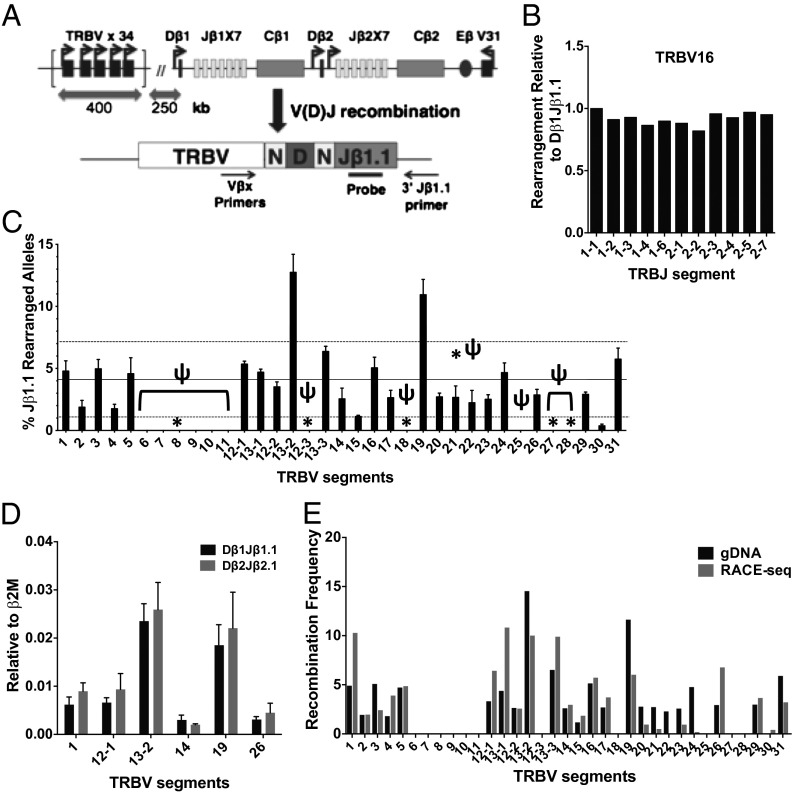
Fig. 1.
Preselection Tcrb V repertoire. (A) Schematic representation of the murine Tcrb locus (Upper) and Taqman assay (Lower) used to quantify VβDβ1Jβ1.1 recombination products. Bold arrows near gene segments denote promoters (Upper). N, N-regions (nontemplated regions of diversification); locations of primers and probes for Taqman assays are shown (Lower). (B) Distribution of V(D)J rearrangements from high-throughput sequencing involving select Vβ segments and each of the 11 functional Jβ segments. The distribution for a given Vβ-Jβ combination is calculated as the number of unique reads for that combination divided by the total number of unique reads for the corresponding Jβ element. Data are represented relative to the distribution of Vβ-Jβ1.1, where percent total Vβ-Jβ1.1 is set to a value of 1. (C) Preselection Vβ repertoire. Taqman real-time PCR quantification of VβDβ1Jβ1.1 rearrangements was performed on gDNA from DN3 thymocytes. Signals from each assay were normalized to values obtained from an assay for the invariant β2M gene. Average levels from three independent DN3 preparations are shown (n = 3, ±SEM). Recombination frequencies are shown as the percent contribution of a given Vβ segment to the total level of Jβ1.1 rearrangement. Pseudogenes are denoted by ψ and gene segments with nonfunctional RSSs are marked with an asterisk. The average Vβ use and SD are denoted by dotted black lines. (D) Taqman real-time PCR assays measuring VβDβ1Jβ1.1 vs. VβDβ2Jβ2.1 rearrangements in DN3 thymocytes were quantified as described in C. (E) Comparison of Vβ use values in DN3 thymocytes using gDNA- vs. mRNA-based methods. Average values from gDNA assay (n = 3) and RNA-5′RACE seq (n = 2) are shown.
Results
Preselection Tcrb Repertoire.
Recent deep sequencing studies of mRNA corresponding to VβDβJβ combinations expressed in peripheral CD4+ T lymphocytes have provided an approximation of the postselection Tcrb repertoire (22). However, our goal is to understand variables that impact the efficiency of long-range Vβ to DβJβ recombination, which shapes the preselection Tcrb repertoire. Accordingly, these analyses must be performed on primary thymocytes before their positive or negative selection, which may alter the Vβ repertoire. Preferably, a DNA-based assay should be used to quantify Vβ use because mRNA expression of VβDβJβ rearrangements may be influenced by promoter strength or message stability. We developed the requisite assay (see below), which was applied to genomic DNA (gDNA) from sorted double negative (DN3) cells (>95% purity; CD4−, CD8−, CD25high, CD44low), a developmental stage in which Vβ to DβJβ recombination occurs at a high frequency, but the vast majority of cells have yet to undergo Tcrb-dependent selection (6). We reasoned that the relative frequency of rearrangements in this cell population involving a particular Vβ segment, regardless of whether the joins are productive or out of frame, accurately reflects its recombination potential.
Initially, we deep sequenced products of a multiplex PCR amplification that incorporates primers for each mouse Vβ and Jβ gene segment, analogous to an approach described previously for analysis of human Tcrb repertoires (23). However, when applied to our DN3 thymocyte samples, a small subset of the mouse Vβ primers exhibit amplification biases in the multiplexing platform, limiting their usefulness for establishing relative Vβ frequencies. In contrast, this approach yields a relative Jβ use similar to that observed in prior studies, suggesting no significant bias in the Jβ primers (Fig. S1A) (22). In keeping with this, we noticed that the collection of VβDβJβ rearrangements for each Jβ segment has a nearly identical Vβ distribution. For example, TRBV16 is used in 8.6% of all rearrangements involving Dβ1Jβ1.1. A nearly identical percentage of Dβ1Jβ1.2 rearrangements, or any other Dβ-Jβ combination, use the TRBV16 gene segment (7.5–8.6%). The Jβ-independent frequency of Vβ use held true for all Vβ gene segments (Fig. 1B; Fig. S1B). Moreover, recent studies have reported similar Vβ use for rearrangements involving either Dβ1 or Dβ2 (22). Thus, an accurate depiction of Vβ use can be established from a simplified approach in which levels of Vβ rearrangements to a single Jβ gene segment are measured quantitatively.
Accordingly, we designed Taqman PCR assays to independently measure rearrangements between Jβ1.1 and each of the 35 Vβ gene segments that undergo V to DJ recombination (Fig. 1A). We also prepared control plasmids containing each of the Vβ-Jβ1.1 combinations to serve as templates for standard curves. Initial experiments verified that all Vβ-Jβ1.1 plasmids amplified with comparable efficiencies (±5%) using Vβ-specific primers with a Jβ1.1 primer/probe combination. Control PCR assays revealed no significant cross-reactivity of Vβ-specific primers with off-target Vβ segments. Standard curves were used to quantify levels of each Vβ-Dβ1Jβ1.1 recombination product in gDNA from sorted DN3 thymocytes. The relative frequencies of Vβ use were consistent in three biological replicates and averaged values are shown in Fig. 1C. Similar Vβ frequencies were observed in assays measuring a subset of Vβ-Dβ2Jβ2.1 rearrangements (Fig. 1D), confirming the Dβ and Jβ independence of Vβ use. Consistent with previous observations, analysis of gDNA from DN-depleted thymocytes revealed only a few modest differences in Vβ use, indicating that the pre- and postselection Vβ repertoires in mouse thymocytes are largely comparable (Fig. S1C) (24). In contrast, deep sequencing of the 5′-RACE library from two DN3 samples yielded a distribution that differed at a subset of Vβ segments compared with our quantitative gDNA-based assay (Fig. 1E). These findings suggest that mRNA levels corresponding to rearrangements involving some Vβ gene segments may not accurately reflect their recombination frequency in preselection thymocytes.
Overall, we observe a >10-fold range in relative Vβ use. Only TRBV13-2 (formerly Vβ8.2) and TRBV19 (formerly Vβ6) are significantly overrepresented in the primary repertoire of Tcrb rearrangements. The preponderance of TRBV13-2 is consistent with analyses using a restricted set of Vβ-specific antibodies from T-cell populations (24). In contrast, rearrangements were undetectable for 11 of the 35 Vβ segments. Five of these 11 “inert” gene segments are predicted to have nonfunctional RSSs (Fig. 1C, asterisks), crippling their recognition by the RAG-1/2 recombinase. Six of the remaining inert gene segments have functional RSSs, but are pseudogene segments due to disruptions in their coding potentials (ψ; Fig. 1C). A lack of VβDβJβ rearrangements involving these six pseudogene segments flanked by functional RSSs indicates that other factors influence their recombination efficiencies (see below). Only two functional Vβs, TRBV15 and TRBV30, were underused compared with the remaining 22 functional segments, which displayed only a modest variability in their use (approximately threefold range). These repertoire data suggest that Tcrb has evolved to normalize use of nearly all functional Vβ segments, perhaps by modulating the three determinants of long-range recombination efficiency: RSS quality, spatial proximity, and chromatin environment.
Spatial Access of Vβ Gene Segments to the DβJβ Recombination Center.
Long-range recombination of V gene segments at all Ig and Tcr loci is facilitated by a contraction process, which places the V cluster into spatial proximity with distal (D)J targets located 0.1–3.2 Mb away in the linear genome (3, 25). Deletion of transcription factors or cis elements that disrupt locus contraction significantly impair V to (D)J recombination, supporting a functional link between these processes (13, 26–28). Additional evidence indicates that V clusters fold into a compact rosette-like structure, which may permit extensive interactions between a recombination center and many or all of its upstream V segments (14). Alternatively, the spatial architecture of V clusters may sculpt the repertoire by positioning a subset of V segments closer to their (D)J targets (efficient rearrangement) while spatially excluding others (inefficient rearrangement). Indeed, emerging studies at Igk suggest that Vκ pseudogene segments may be spatially excluded from interactions with Jκ substrates, perhaps minimizing their recombination potential (29).
To test whether spatial proximity is a key determinant in shaping the preselection Tcrb repertoire, we measured interaction frequencies between restriction fragments spanning each Vβ segment and fragments spanning either of the two DβJβ clusters using chromosome conformation capture (3C) (1). In the linear genome, the distance between these restriction fragments range from 250 to 700 kb (except for TRBV31, which is ∼3 kb downstream of Eβ and rearranges by inversion). 3C assays were performed on cross-linked chromatin from RAG1-deficient thymocytes, a predominantly DN3 cell population in which Tcrb is in an active germ-line conformation. The use of RAG-deficient thymocytes circumvents complications in data analysis that arise from active Tcrb rearrangement. Although we cannot rule out a role for RAG-1 in defining the precise 3D conformation of Tcrb (30), prior studies demonstrate that RAG proteins are dispensable for locus contraction (15).
We measured the cross-linking efficiency of each Vβ-containing HindIII fragment to three downstream vantage points within the Tcrb recombination center. Specifically, we probed Vβ cross-linking to HindIII fragments containing either of its two substrates (Dβ1 or Dβ2), or the transcriptional enhancer Eβ, which generates active chromatin over the DβJβ clusters (10, 31). Regardless of the vantage point, nearly all Vβ gene segments interact more frequently with the DβJβ recombination center in DN thymocytes compared with CD19+ pro-B cells purified from RAG-deficient bone marrow (Fig. 2A; Fig. S2 A and B). These data verify and extend previous analyses showing that Tcrb adopts a T cell–specific conformation, juxtaposing the Vβ cluster with its DβJβ targets (15).
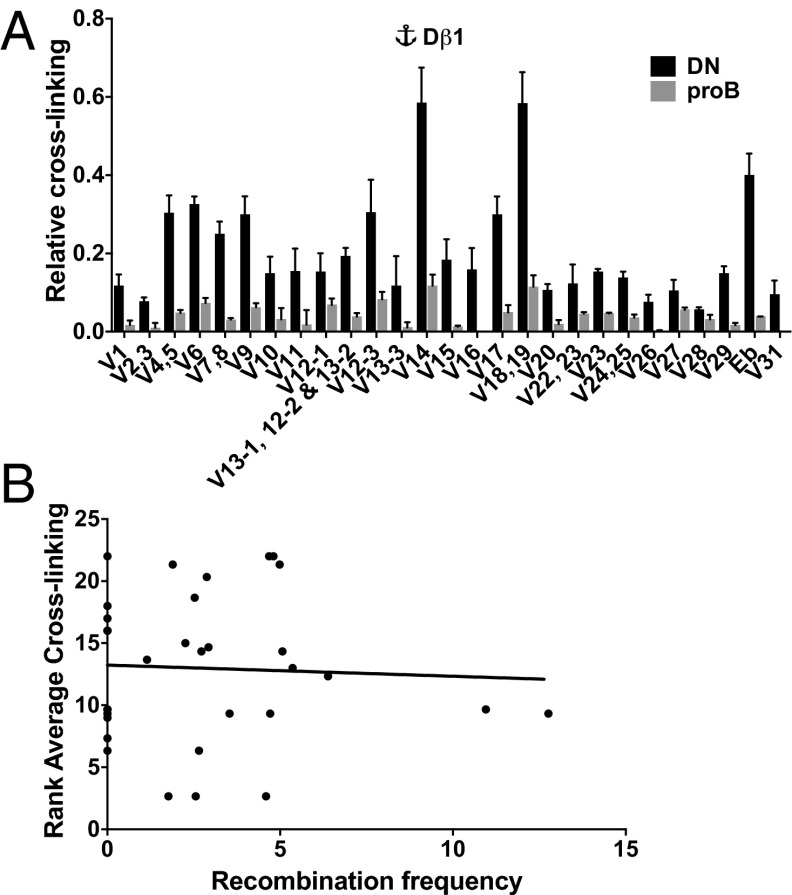
Fig. 2.
Role of Vβ spatial proximity in shaping the Tcrb repertoire. (A) 3C analysis of RAG-deficient thymocytes showing relative cross-linking frequencies between a Dβ1 anchor and HindIII fragments spanning Vβ gene segments. Data are presented as mean ± SEM (n = 3). (B) Spearman correlation of Vβ use and average ranked values for 3C cross-linking frequency from three viewpoints within the recombination center (Dβ1, Dβ2, and Eβ). The Spearman correlation coefficient shows no significance (rs = 0.035, P = 0.85).
Of particular note, interaction levels measured from a given vantage point (e.g., Dβ1) display significant differences across the collection of Vβ segments (Fig. 2A). There were also differences in interactions between specific Vβ segments and two vantage points. For example, the fragments spanning TRBV1 or TRBV18/19 both interact with Dβ1 at a much higher frequency than with Dβ2 (Fig. 2A; Fig. S2A). Conversely, TRBV17 displays a greater interaction with Dβ2 (Fig. 2A; Fig. S2A). Despite these differences, the TRBV1 and TRBV19 segments are used with indistinguishable frequencies in recombination products involving either Dβ1 or Dβ2 (Fig. 1D). In contrast to preliminary findings at Igk (29), a group of pseudogene segments spanning TRBV6–TRBV11 each interact with DβJβ clusters at a relatively high frequency, but these gene segments are absent from the preselection Tcrb repertoire despite having functional RSSs. These findings suggest that relative Vβ use in the preselection Tcrb repertoire cannot be fully explained by differences in their spatial proximity to the DβJβ regions.
To more rigorously investigate the relationship between spatial proximity and long-range recombination, we performed Spearman ranking correlations for 3C and Vβ repertoire data. Because the absolute values of 3C data cannot be quantitatively compared between the three assays, we first ranked cross-linking efficiencies of the Vβ segments within each vantage point (Table S1). No significant correlations between 3C ranking and TRBV rearrangement are observed for any of the three individual viewpoints within the DβJβ recombination center. We also calculated the average ranking for each Vβ segment over the three assays (Dβ1, Dβ2, and Eβ) and compared these values with relative use in VβDβJβ joins (Table S1). As shown in Fig. 2B, there is an absence of significant correlation between Vβ use and its average rank for interactions with the DβJβ recombination center. Consistent with this finding, we also observe no obvious correlation between the recombination frequency of a Vβ segment and its proximity to CTCF binding. We conclude that, although gross locus contraction is important to bring the entire Vβ cluster into spatial proximity with its Dβ substrates, the precise magnitude of each Vβ–Dβ interaction is not a primary determinant of recombination efficiency. Instead, our 3C and repertoire data indicate that once Tcrb is contracted in DN thymocytes, the large Vβ cluster adopts a conformation in which spatial access of Vβ segments to the recombination center is not limiting.
Role of RSS Quality in Determining Vβ Use.
Despite general conservation of the heptamer-spacer-nonamer configuration, RAG-1/2 substrates exhibit substantial variation compared with the consensus RSS sequence: (CACAGTG)–12- or 23-bp spacer–(ACAAAAACC) (32, 33). In vivo replacement or natural variants of RSSs can alter the use of gene segments, including those within the Tcrb recombination center (34–36). In vitro studies using plasmid substrates have defined the effects of positional substitutions within the consensus RSS on recombination efficiency (32, 37, 38). Thus, one component of nonrandom Vβ use is likely the quality of its flanking RSS.
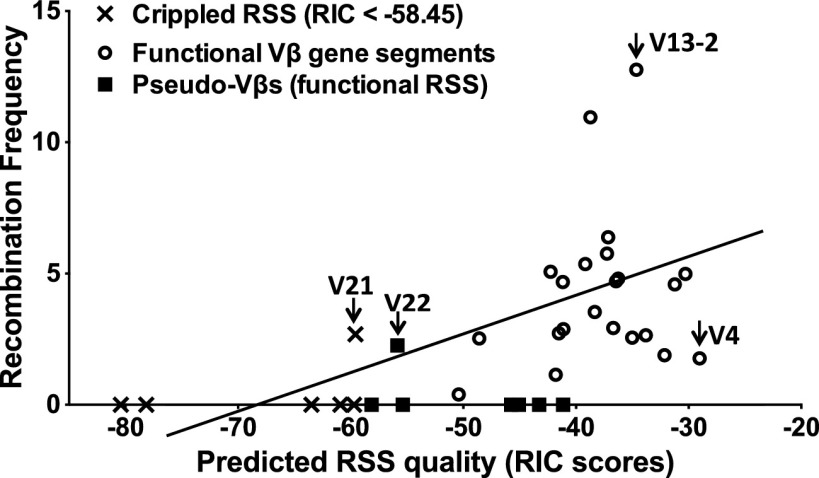
To examine this possibility, we took advantage of an algorithm (www.itb.cnr.it/rss/) that predicts the RSS quality of any given sequence (39). In brief, this algorithm calculates the theoretical recombination potential of an RSS using a statistical model that assigns a score based on the contribution of each nucleotide within the heptamer-spacer-nonamer sequence. The algorithm output is a recombination signal information content (RIC) score, which predicts the quality of an input RSS with a reasonable degree of accuracy based on data from plasmid recombination substrates (40). For Tcrb, 6 of the 35 Vβ gene segments are flanked by nonfunctional RSSs with a RIC score of <−58.5, the threshold defined by Cowell et al. (39), (TRBV8, 12-3, 18, 21, 27, and 28). The remaining 29 Vβ segments have a substantial range in predicted RSS quality, with RIC scores between −29 (TRBV4) and −58.2 (TRBV11). Recombination is undetectable for five of the six Vβ segments flanked by RSSs that score below the functional threshold (Fig. 1C). The exception is TRBV21, which rearranges at a detectable level, but is predicted to have a marginally nonfunctional RSS (RIC score, −58.6) consisting of a consensus heptamer and a 22-bp rather than 23-bp spacer.
The correlation between RIC scores and Vβ use is shown in Fig. 3. Although a positive correlation is apparent, the magnitude of Vβ use diverges significantly from linearity compared with predicted RSS quality. In general, Vβ RSSs with lower quality (RIC scores, −45 to −58) are either inert or rearrange at a level below the average frequency. RSSs with RIC scores >−45 exhibit a broad range of Vβ recombination frequencies, as highlighted by the following examples: (i) TRBV13-2 is the most frequently used segment but shares a nearly identical RIC score with TRBV14, which rearranges at an average frequency; and (ii) six Vβ segments (TRBV7, 15, 16, 20, 24, and 26) have nearly indistinguishable RIC scores (−41 to −42), but one Vβ is recombinationally inert (TRBV7) and the remaining five display an eightfold range in their utilization. We cannot rule out the possible contribution of coding sequences adjacent to each RSS in altering its quality as a RAG-1/2 substrate. Inspection of coding flanks revealed only a small subset with features predicted to attenuate RAG cleavage (e.g., AT or pyrimidine stretches for TRBV12-1, 12–2, 14, 17, and 29) (41–44). However, as shown below, the recombination frequency of these gene segments correlate best with features of associated chromatin. Together, our data indicate that, although predicted RSS qualities contribute to the formation of a preselection Tcrb repertoire, other levels of control clearly impact Vβ use.
Fig. 3.
Correlation between Vβ utilization and predicted RSS quality. The correlation between predicted Vβ RIC23 scores and observed Vβ recombination frequencies (Fig. 1B), yielding a Spearman’s rank correlation coefficient rs = 0.6456, P < 0.0001.
Role of Chromatin Environment in Determining Vβ Recombination Potential.
Chromatin accessibility at gene segments has been studied extensively as a determinant of the tissue- and stage-specific mechanisms controlling V(D)J recombination (6, 7). Germ-line transcription of gene segments leads to the deposition of H3K4me3, a histone modification that is recognized by RAG2 and augments endonuclease function of the RAG complex (8, 9, 45). As such, levels of chromatin accessibility and transcription at each Vβ segment may help determine its use in the preselection Tcrb repertoire.
The emerging approach of “chromatin profiling” uses combinatorial patterns of histone modifications, nucleosome density, and factor binding to assess the epigenetic status of genomic regions (46). To compare epigenetic landscapes at the 35 Vβ segments, we generated chromatin profiling data from RAG1-deficient thymocytes using ChIP assays in combination with Tcrb microarrays (ChIP-chip) or deep sequencing. We also performed formaldehyde-assisted isolation of regulatory elements (FAIRE), which identifies nucleosome-depleted regions in the genome (47). The new ChIP-chip (P300, H3K27ac, H3K4me2), ChIP-seq (H3ac, H3K4me3, and CTCF), and FAIRE-Chip data from RAG-deficient thymocytes were combined with epigenomic data available in public repositories (H3K4me1, RNA Pol II, and H3K9me2) from RAG-deficient thymocytes (48). We used a published methodology to integrate cross-platform data derived from ChIP-chip and Chip-seq (49). In addition to nucleosome depletion (FAIRE), the analyzed features characterize active promoter regions (transcription, RNA Pol II, H3K4me3, and H3ac), active regulatory elements (H3K4me1, H3K27ac, and P300), poised chromatin (H3K4me2), insulators (CTCF), and silent chromatin (H3K9me2).
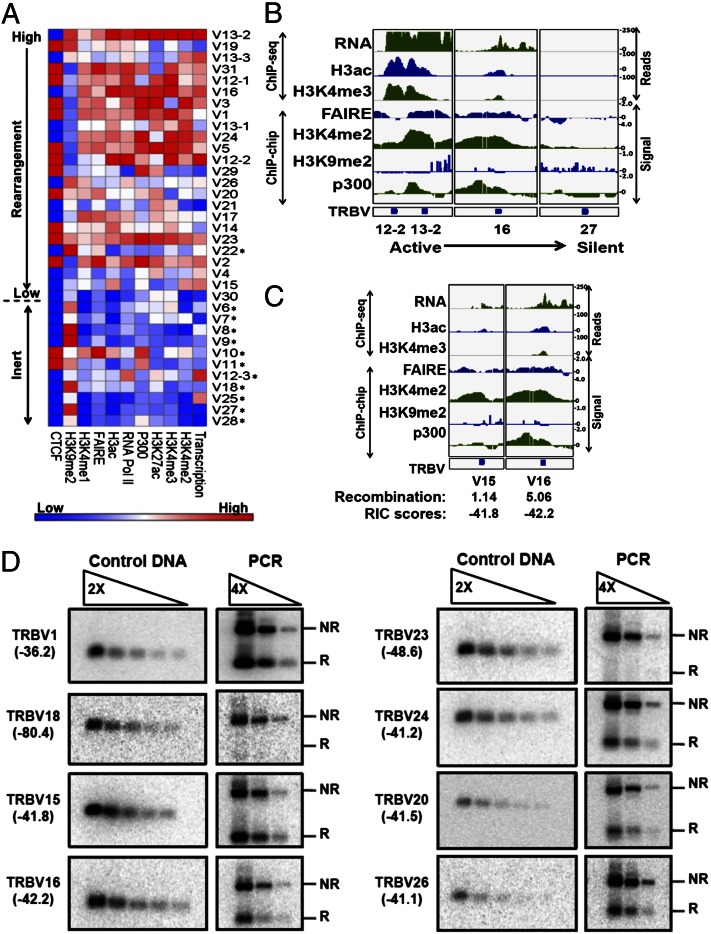
Relative intensities for each feature at the 35 Vβ segments (±1 kb) are represented as a heat map in Fig. 4A. Examples of several features for selected gene segments in chromatin environments ranging from highly active to silent are depicted in Fig. 4B. Overall, most of the Vβ segments that participate in Vβ to DβJβ recombination exhibit higher levels of active chromatin features than the inert Vβ elements (H3K4me, RNA Pol II/transcription, and histone acetylation). In contrast, the repressive H3K9me2 modification was enriched over many of the inert Vβ segments. One region within the Vβ cluster containing the TRBV12-2 and 13-2 gene segments is conspicuously active (Fig. 4B), with high levels of germ-line transcripts and other features associated with open chromatin, including one of the few discernible P300 peaks. As noted above, TRBV13-2 is also the most frequently rearranged gene segment in DN3 thymocytes, suggesting a dominant correlation between open chromatin and long-range recombination efficiency. Consistent with this possibility, many of the pseudogene segments, even those containing functional RSSs, are expressed at a low level and are associated with chromatin that lacks activating histone marks (Fig. 4A, asterisks). In silico analysis of Vβ upstream sequences (−1 kb to leader) for predicted transcription factor binding profiles (TRASFAC/JASPAR databases) revealed no distinguishable differences between functional and pseudo-Vβ gene segments. Promoter activity as measured by luciferase assays in a transfected pre–T-cell line show that all tested upstream Vβ regions from recombinationally active gene segments (11/11) are functional promoters. In contrast, only some of the tested regions upstream of pseudogene segments (4/8) exhibit promoter activity (ψ; Fig. S3), indicating no clear correlation between Vβ utilization and promoter strength. Thus, it appears that the mouse Vβ cluster has evolved multiple strategies to silence chromatin at nonfunctional gene segments.
Fig. 4.
Role of chromatin landscape in Vβ use. (A) Relative intensities of various chromatin features (transcription, RNA Pol II, P300, histone modification signals, and proximal CTCF sites) at the 35 Vβ segments are represented as a heatmap. The log2 values of ChIP-Seq or ChIP-Chip signal intensities at the Vβ segment (±1 kb) for each of the above features were quantified using BEDtools, and the relative intensity for each feature was plotted as a heatmap. CTCF intensities are represented as binary values of 1 or 0 assigned for presence or absence of CTCF within 1 kb of the Vβ segment. Asterisks denote pseudo-V gene segments. (B) Profiles for transcription (RNA), nucleosome depletion (FAIRE), P300, and indicated histone modifications are shown at select Vβ segments. RNA-seq data for transcription, ChIP-seq data for H3ac and H3K4me3, and ChIP-chip data (signal = log2 ratio of ChIP DNA/input DNA) for H3K4me2, P300, and FAIRE are displayed. See Materials and Methods for sources of epigenomic data. (C) Epigenetic profiles at Vβ segments highlighting the influence of chromatin landscapes on gene segment use. (D) An equimolar mixture of the eight indicated Vβ 23-RSS deletion substrates was assayed for rearrangement in conjunction with the 5′Dβ1 12-RSS following transfection into 293T cells with RAG-1/2 expression vectors (40). Rearrangements were detected by PCR using primers shared by all of the substrates (NR, not rearranged; R, Vβ rearranged to Dβ1). RIC scores for each TRBV-RSS are shown in parentheses. Rearrangements for each substrate were detected using probes specific to the given Vβ segment. A semiquantitative measure of rearrangement efficiencies was obtained by comparing twofold dilutions of Vβ plasmid inserts (3 ng–500 ng, Left) with fourfold dilutions of the PCR product (Right). Shown are data from one representative PCR amplification of four independent transfections. Control DNA and PCR products for each Vβ substrate are on the same blot. The TRBV15, 16, 20, 24, and 26 RSSs exhibit similar recombination efficiencies based on this semiquantitative assay (RIC scores all approximately −42), whereas the TRBV18 and 23 RSSs exhibit minimal rearrangement (lower RIC scores) and TRVB1 rearranges most efficiently (best RIC score).
A reasonable concordance was observed between chromatin environments and recombination efficiencies when comparing Vβ segments with equivalent RIC scores. For example, TRBV15 and TRBV16 are predicted to have RSSs of nearly identical qualities but reside in distinct chromatin environments. The elevated levels of transcription and activating histone marks at TRBV16 correspond to an elevated level of recombination (Fig. 4C). In some cases, both the predicted RSS quality and chromatin environment apparently contribute to Vβ use. For example, TRBV23 and TRBV24 are both transcriptionally active and have comparable chromatin features (see heatmap in Fig. 4A); however, the lower predicted RSS quality for TRBV23 (−48.6) compared with TRBV24 (−41.2) correlates with an attenuated level of recombination. We also noted that contributions of chromatin to rearrangement frequencies may derive from different combinations of features. TRBV20 and TRBV26 exhibit nearly identical use (2.7% and 2.9%) and RIC scores (−41.5 and −41.1), but patterns of specific chromatin features at these gene segments differ significantly (see heat map in Fig. 4A). To further validate these comparisons, we performed semiquantitative assays to measure the qualities of eight Vβ-RSSs using plasmid-based substrates (including the six Vβ-RSSs mentioned above). The relative qualities of these RSSs, tested in conjunction with a natural target (5′Dβ1-RSS), are in line with predictions from RIC scores (Fig. 4D), further supporting our conclusions. Together, these profiling studies indicate a strong contribution of chromatin environment to Vβ recombination frequencies but also suggest that individual parameters of chromatin accessibility may affect substrate use in a weighted manner.
Computational Analysis of Vβ Use Determinants.
Our data indicate that predicted RSS qualities and chromatin landscapes likely contribute in a combinatorial manner to the efficiency of long-range Tcrb assembly. To examine these combinatorial relationships, we used classification and regression analyses comparing chromatin features and predicted RSS quality with Vβ use. These analyses were guided by recent computational strategies devised to predict gene expression levels based on patterns of histone modifications (50, 51). We applied one validated approach (50) to study whether chromatin features, predicted RSS quality, and spatial proximity are predictive of the observed Vβ repertoire.
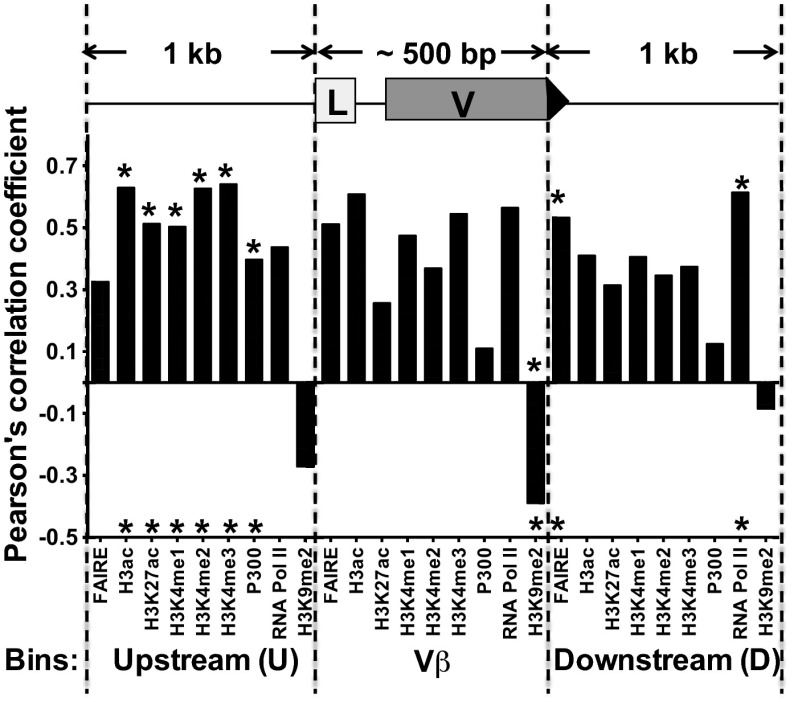
The chosen computational approach takes into account (i) the signal intensity of each chromatin feature, (ii) levels of germ-line transcription, (iii) RIC scores, and (iv) spatial proximity based on the average 3C rank score. With regard to chromatin features, distinct positional profiles are observed for various histone marks. For example, H3K4me3 is enriched over active promoters and progressively wanes along gene bodies. Accordingly, we divided the regions spanning each Vβ segment into three bins: the Vβ segment itself (leader to RSS), its upstream promoter region (1 kb 5′ of leader), and its downstream region (1 kb 3′, including the RSS). For each feature, we computed Pearson correlation coefficients for the three bins vs. Vβ recombination frequencies (Fig. 5). We find the best correlation for a majority of histone modifications in the upstream/promoter bin (H3K4me1, H3K4me2, H3K4me3, P300, H3ac, and H3K27ac). In contrast, repression by H3K9me2 was most correlative in the bin that contains Vβ segments. FAIRE and RNA Pol II signals have very similar predictive abilities over both the Vβ and its downstream bins. These findings are strikingly similar to correlations observed between chromatin features and gene expression (50, 51), further underscoring the relationship between transcriptional activity and Vβ recombination frequencies. A particularly satisfying outcome of this analysis is the correlation between FAIRE signals and the bins flanking RSSs, presumably reflecting a requirement for nucleosome depletion at RAG-1/2 targets (52, 53).
Fig. 5.
Spatial distribution of chromatin features and predictive potential for Vβ use. The regions surrounding each Vβ segment were divided into three bins (see schematic); U, upstream (1 kb); V, Vβ gene body; D, downstream (1 kb). Signal densities for each chromatin feature in the spatial bins were correlated with recombination frequencies, yielding a Pearsons’s correlation coefficient for each bin. The coefficients were used to determine the best bin, which are denoted by asterisks.
Next, we identified features that are most predictive of whether a Vβ segment will rearrange at any frequency or will remain inert. For this and the remaining analyses, we used signal intensities only from bins exhibiting the highest correlation between each chromatin mark and Vβ use (Fig. 5, asterisks). A computational approach called random forest was used (50), which randomly tests combinations of binned features for their predictive abilities to classify gene segments as active or inert (Fig. 6A). This analysis revealed that three features—predicted RSS quality, FAIRE, and RNA Pol II signals—are sufficient to classify the recombination potential of a given Vβ segment with a high level of confidence. The classifications are also evident from linear regression analysis on these three features relative to Vβ recombination frequencies (Fig. 6B; 30/35 segments predicted correctly). When we used the random forest algorithm, but focused only on values for RIC score, FAIRE, and RNA Pol II signals, 32/35 Vβ segments classified correctly as active vs. inert (Materials and Methods). The three exceptions common to both random forest– and linear regression–based classifications are TRBV15, 21, and 22; segments predicted to be inert but exhibiting detectable levels of recombination. These outliers could reflect partial compensation by chromatin features other than the factors determined by our algorithms. Notwithstanding, the most important predictive features of recombinational competency are linked mechanistically to RAG substrate quality (RIC score), substrate accessibility (nucleosome depletion), and RNA Pol II association.
Fig. 6.
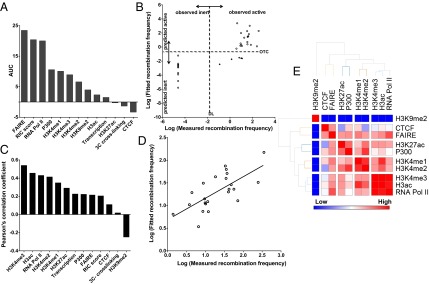
Computational analysis of Vβ use determinants. (A) Features that distinguish rearranging from inert Vβ segments (classifier step; Materials and Methods). Random forest analysis was performed on the shown features to classify Vβ segments. AUC, area under the curve, which represents the relative contribution of each feature to the learned classification scheme. (B) Scatter plot representing the classifier step in the two-step model. Linear regression between observed and fitted frequencies using the three most discriminative features for recombining vs. inert Vβ gene segments (RIC scores, FAIRE signal, and RNA Pol II occupancy). Each symbol represents a Vβ gene segment. Data were generated from the natural logarithm values of recombination frequencies (observed and fitted). The dashed horizontal line represents the optimal threshold for classifying (OTC) rearranging from nonrearranging segments based on the linear combination of the three features. The dashed vertical line represents the detection limit (DL) of Taqman assays used for measuring recombination. Open circles correspond to Vβ segments predicted accurately; black diamonds correspond to outliers. Two of these five exceptions were resolved when the random forest algorithm was applied using the three classification features (RIC score, FAIRE, RNA Pol II). (C) Pearson correlation to rank factors that fine tune Vβ use in the two-step model (regressor step; Materials and Methods). (D) Scatter plot of overall correlation between natural log values of observed and fitted (predicted) frequencies using the five core parameters (H3K4me3, H3K4me2, transcription, P300, and CTCF). Each circle represents one rearranging Vβ segment. The line indicates the best fit between measured and fitted rearrangement frequencies and reflects a strong correlation (Pearson correlation coefficient, 0.69; P = 0.03). (E) Cluster analysis highlights similarities in epigenetic information provided by individual chromatin features.
We next moved beyond black and white classifications to analyze the relative importance of Vβ features in fine-tuning recombination frequencies of the 23 active gene segments. For this purpose, we performed linear regression on the selected bins for each feature vs. frequency values. As shown in Fig. 6C, the features that correlate most significantly with Vβ use are H3K4 methylation, H3Ac, and RNA Pol II occupancy, which normally associate with transcriptionally active regions. The repressive H3K9me2 mark correlates negatively with levels of Vβ recombination. In contrast to its dominant role as a determinant for recombinational competence, RIC scores for the 23 active Vβ gene segments correlate poorly with their relative levels of rearrangement. A similar discordance between recombination frequencies and RSS qualities for a limited set of mouse VH and Vк gene segments has been described previously (54, 55). These findings suggest that chromatin environment, rather than predicted RSS quality, is the dominant feature for fine-tuning Vβ use in long-range recombination.
We next investigated whether various combinations of the 13 features included in this study are predictive of Vβ recombination efficiencies. As a starting point, we examined the predictive capacity of all 13 features using linear regression (Fig. S4A). This analysis yielded a correlation coefficient for best fit of 0.78, which was statistically insignificant (P > 0.05). We next tested whether a subset of these 13 features correlate in a significant manner with observed frequencies of Vβ use. For this purpose, we examined various subsets of features, ranging from a single feature to 12 of the 13 variables in all possible combinations. This combinatorial analysis yielded a set of five features that correlate significantly with Vβ use (Fig. 6D; Pearson correlation coefficient = 0.69, P = 0.03). In descending order of contribution to the fitted model, the identified features were H3K4me3, H3K4me2, transcription, P300, and CTCF. The first four features largely determine the efficiency for most TCRBV segments, whereas the remaining feature, CTCF proximity, improves the fit for several outliers that are poorly predicted by H3K4me3, H3K4me2, transcription, and P300. When further analyzed by clustering, we found that the four chromatin features (H3K4me3, H3K4me2, P300, and CTCF) in this set of five core parameters represent four classes of related marks that share a significant portion of epigenetic information (Fig. 6E). For example, H3K4me3 correlates strongly with H3ac and RNA Pol II occupancy, three features enriched near active promoters, in essence encapsulating the information content of the entire class. The relative contributions of the five core features to the accuracy of fit and the corresponding linear regression formula are provided in Fig. S4B.
Together, the computational analyses derive a two-tiered model for predicting Vβ use in the preselection Tcrb repertoire. First, RIC scores in combination with nucleosome and RNA Pol II densities discriminate active from inert substrates. The recombination frequency of the active Vβ set can be discerned from values for the five core parameters identified by statistical correlations. Moreover, this basal set of five parameters may be useful in future studies to predict the impact on preselection Vβ repertoires of naturally occurring or engineered perturbations at Tcrb.
Discussion
We took an integrative approach to define the molecular determinants of Vβ recombination frequencies, an important component of the preselection Tcrb repertoire. Prior studies have examined the independent effects of RSS quality, 3D architecture, transcription, or chromatin accessibility on recombination of specified gene segments. However, our unified analysis shows how these features impact the efficiency of long-range V to (D)J recombination at an endogenous AgR locus. Using several independent computational approaches, we find that (i) RSS quality and nucleosome density are the major determinants of whether a given Vβ segment will participate in Tcrb gene assembly, (ii) the relative use of a Vβ segment is fine-tuned by its chromatin environment, (iii) the optimal epigenetic landscape for Vβ recombination is a blend of transcriptional activation marks, nucleosome depletion, and a lack of the repressive H3K9me2 mark, and (iv) the precise magnitude of spatial proximity between a Vβ segment and the DβJβ recombination center does not significantly influence its relative utilization. Collectively, we find that a minimum set of five features can be measured to predict the recombination frequency of a competent Vβ segment with a high degree of accuracy.
A critical component of our study was a determination of the preselection Vβ repertoire. The relative use of Vβ segments may have important consequences with regard to AgR-mediated thymic selection, the production of functional T-cell subsets that use specific Vβ segments, or the baseline antigenic profile recognized by emerging T lymphocytes. We used a DNA-based approach to directly quantify rearrangement levels of the 35 Vβ segments in sorted DN3 thymocytes. This approach avoids two caveats of prior repertoire analyses, biases introduced by thymocyte selection or by mRNA expression differences, both of which were observed in our companion assays. We find that only a few functional Vβ segments are either over- or underused in the preselection Tcrb repertoire. One of the overused Vβ segments, TRBV13-2 (formerly Vβ8.2), is enriched in invariant natural killer (iNKT) cells, a subset of lymphocytes that respond to lipid antigens and produce a robust cytokine response. We postulate that the ideal chromatin environment encompassing TRBV13-2 has evolved to augment its rearrangement efficiency, ensuring a sufficient production of iNKT cells, which provide a rapid cellular immune response to numerous foreign antigens. Notwithstanding, rearrangement levels for the vast majority of functional Vβ gene segments (18/22) fall within a threefold range. The relatively limited range of distribution likely reflects a requirement to maximize Tcrb diversity before its pairing with Tcra for subsequent selection by MHC-peptide complexes.
As shown here, the normalization of Vβ use results predominantly from the chromatin environment encompassing each gene segment, with perhaps a minor contribution from its RSS quality. The dominance of chromatin in fine-tuning Vβ use was evident from several outlier gene segments. The TRBV15 and TRBV30 segments are underused compared with all of the other functional Vβ elements, likely because they are poorly transcribed or lack most features of active chromatin. Likewise, nearly all of the pseudogene segments that are flanked by functional RSSs reside in a repressive chromatin environment. For the latter category, we provide evidence that some, but not all, germ-line promoters associated with pseudo-Vβ segments have been incapacitated, despite their retention of potential factor binding sites found in functional Vβ promoters. Another potential mechanism for pseudogene suppression could be their localization to the nuclear periphery or lamina (56). However, the precise underlying mechanisms that sequester these pseudogene segments in repressive chromatin, preventing wasteful recombination, remain to be defined.
With regard to the collection of rearranging Vβ segments, the dominant chromatin features in determining their relative use are associated with active transcription. The strongest correlations exist between recombination efficiencies, histone acetylation (H3ac), H3K4 methylation, nucleosome depletion, and RNA Pol II occupancy. Although a link between this transcriptional epigenetic state and recombination has long been appreciated, its dominant role in sculpting the primary repertoire of antigen receptors is a unique finding of our study. One likely mechanism for this relationship is the affinity of RAG complexes for chromatin bearing the H3K4me3 mark. Prior ChIP-seq studies demonstrate that RAG-1/2 is bound to the DβJβ recombination center in DN thymocytes but is relatively absent from the Vβ cluster (11). This reflects the extremely high levels of H3K4me3 on DβJβ chromatin compared with Vβ segments (∼10-fold difference) (11). Based on our integrative model, we suggest that after Tcrb contracts, prebound RAG-1/2 complexes at the DβJβ recombination center may preferentially target Vβ segments that are most enriched for transcription-associated marks, including H3K4me3. Thus, the strength of each Vβ promoter within its native chromosomal context may be a dominant feature for shaping the preselection Tcrb repertoire.
One important aspect of our study is that the precise magnitude of association between a Vβ segment and DβJβ clusters, as measured by 3C, does not contribute discernibly to its level of use. Clearly, general locus contraction is an important mechanism for bringing V segments into spatial proximity with their distant (D)J substrates (3). However, the spatial architecture adopted by the large Vβ cluster in DN thymocytes must provide sufficient access to all of its composite gene segments by RAG-1/2 bound at the DβJβ recombination center. Recent studies of Igk suggest that most V segments within this locus also may have similar spatial access to their target J segments (29). Given the 10-fold range in cross-linking efficiencies between various Vβ segments and the two DβJβ clusters, we conclude that spatial constraints on long-range Vβ to DβJβ recombination are binary rather than digital, requiring only that target gene segments cross a threshold of spatial proximity. Presumably, this spatial threshold is surpassed via a combination of locus contraction and folding of the Vβ cluster into a more compact structure.
In conclusion, a combination of epigenetic, spatial, transcriptional, and RSS features were used to identify the dominant determinants for sculpting the preselection Vβ repertoire. We concede that a model for Vβ use may not completely apply to all other AgR loci. Indeed, pseudo-Vк segments interact inefficiently with their target Jк cluster, perhaps suppressing their recombination (29). In contrast, pseudo- and functional Vβ segments interact indistinguishably with their DβJβ substrates. Recombination of pseudo-Vβ segments is, instead, suppressed by sequestration into inactive chromatin. This distinction may reflect a more dominant role for spatial constraints at the much larger Igk locus. Notwithstanding, much of the relevant epigenetic and RSS quality data necessary to build predictive models for other AgR loci are available publicly. In most cases, the lacking features are reliable DNA-based analysis of V use and complete sets of 3C data covering V clusters. We suspect that as multiplex PCR approaches improve, eliminating primer bias, comprehensive preselection repertoires for all AgR loci will emerge. Current methods for quantifying spatial proximity on a global scale lack the resolution of focused 3C assays; however, technical improvements and increased sequencing depths may soon overcome these obstacles. The learned model-building strategy used here should be a valuable guide for defining relative contributions of epigenetic, spatial, and RSS features in shaping preselection V repertoires. Ultimately, these models should also be valuable for predicting how designed or naturally occurring alterations of AgR loci perturb the preselection V repertoire. These alterations could range from targeted RSS and promoter substitutions to natural variant AgR alleles that lack portions of the large V clusters, creating “holes” in the immune repertoire. Indeed, a striking parallel exists between the use of several mouse and human Vβ orthologs (33), underscoring the potential utility of our model to predict the effects of human TCRB polymorphisms on primary repertoire formation.
Materials and Methods
Cell Purification and Antibodies.
Thymocytes from C57BL/6 mice (4–6 wk) were depleted of CD4+ and CD8+ cells using magnetic activates cell separation (MACS) (Miltenyi Biotec). The remaining DN cells were stained and sorted for the CD25hi/CD44low DN3 population, yielding a >95% purity. CD19+ bone marrow cells from RAG-deficient mice were purified using MACS in conjunction with CD19 microbeads (Miltenyi Biotec), providing a >90% pure population of pro-B cells. The list of antibodies used is given in SI Materials and Methods.
High-Throughput Sequencing of Tcrb Rearrangement.
gDNA from sorted DN3 cells was amplified by multiplex PCR for Vβ-Dβ-Jβ rearrangements, and the amplicons were deep sequenced by Adaptive Biotechnologies. The gene segment use was analyzed using ImmunoSEQ Analyzer software.
5′ RACE.
Total RNA (0.5 μg) from DN3 thymocytes was converted to cDNA, and 5′ RACE was performed using a Cβ primer (5′-AGCTCCACGTGGTCAGGGAAGAA-3′) following the manufacturer’s protocol (Ambion). The RACE product was blunted, concatemerized, and sonicated to an average size of 175 bp. The sheared fragments were ligated with Illumina adapters and sequenced using an Illumina HiSeq-2000 to provide paired-end reads extending 101 bases. Raw reads were de-multiplexed, and unique FASTA reads were obtained using the FASTX tool kit (http://hannonlab.cshl.edu/fastx_toolkit). For quality control, a portion of the 5′ RACE product was cloned, and individual clones were sequenced. Sequences were analyzed using IMGT High-V quest (www.imgt.org) (57).
Quantitative PCR for VβDβJβ Rearrangements.
We designed a panel of Taqman PCR assays using probes and primers specific for either Jβ1.1 or Jβ2.1 gene segments in combination with a primer specific for each of the 35 Vβ segments. We also generated a collection of plasmids containing each Vβ cloned directly upstream of either Jβ1.1 or Jβ2.1 in an orientation that mimics the corresponding V-D-J rearrangement product. For this purpose, Jβ1.1 or Jβ2.1 segments were amplified by PCR from mouse gDNA and cloned into the NotI/BamHI sites of pBS-KSII. Subsequently, Vβ segments were amplified and cloned upstream of the Jβ region. The specificity of Vβ primers was confirmed by BLAST searches and a panel of PCR assays showing that amplification of control plasmids containing other Vβ segments was detected at <1% compared with the bona fide target. Template plasmids were used to generate standard curves, allowing us to correct for minor differences in PCR efficiency between each of the assays. Total Vβ-DbJb1.1 or Vβ-DbJb2.1 rearrangement product (alleles) was quantified relative to amounts of an unrearranged region within the genome (b2-microglobulin) using the formula E−Ct(V-Jβ)/E−Ct(B2M), where E is the primer efficiency. The list of primers and probes used is given in Table S2.
Chromosome Conformation Capture.
3C assays were performed on 107 RAG1-deficient C57BL/6 DN thymocytes or CD19+ pro-B cells using HindIII as described in Hagège et al. (58). Primers and probes designed for HindIII fragments corresponding to each vantage point in the recombination center (Dβ1, Dβ2, and Eβ) were used in Taqman assays with primers specific for each Vβ gene-containing fragment. Standard curves were generated for these Taqman assays using HindIII-digested bacterial artificial chromosomes (BACs) spanning the entire Tcrb locus, which were then ligated to yield a library of all possible products. Interaction between the nearest neighbor fragments in the ERCC3 gene was set as 1. Cross-linking frequencies were calculated as described in Hagège et al. (58). A list of primers, probe sequences, and BAC clones are provided in Table S3.
ChIP and FAIRE.
ChIP experiments for H3K4me2, H3K27ac, and P300 were performed with chromatin from RAG-deficient thymocytes (C57BL/6) as described previously (59). The ChIP DNA was purified using a Qiagen DNA purification kit and subjected to whole genome amplification (Sigma), labeled, and hybridized to custom Nimblegen microarrays according to the manufacturer’s protocol by Mogene. Total input DNA was used as the hybridization control. A subset of ChIP-Chip data was verified at various locations throughout Tcrb using quantitative PCR (qPCR; data not shown). FAIRE was performed on cross-linked nuclei from RAG-deficient DN thymocytes and purified pro-B cells using published methods (47). Purified FAIRE DNA was used for subsequent analyses by qPCRs or array hybridization. DNA from non–cross-linked cells, processed in parallel, was used as reference samples. Model-based analysis of 2-color arrays (MA2C, version 1.4.1) was used to normalize the microarray data, detect peaks, and generate University of California, Santa Cruz (UCSC) wiggle (WIG) files.
ChIP-seq experiments were performed as above using chromatin from RAG-deficient thymocytes (C57BL/6) for H3ac, H3K4me3, and CTCF. ChIP-seq data for RNA Pol II, H3K4Me1, and ChIP-Chip data for H3K9me2 from RAG-deficient thymocytes were downloaded from www.comline.fr/ciml/ (48). The ChIP-seq raw data were aligned to the mouse reference genome (mm9) using Bowtie 0.12.8. The resulting binary sequence alignment maps (BAM) files were used to generate UCSC wiggle (WIG) files and peaks using model-based analysis of ChIP-seq software (MACS, version 1.4.2). The list of antibodies used in ChIP experiments is given in SI Materials and Methods.
RNA-seq.
Total RNA from RAG-deficient DN thymocytes was extracted using an Ambion Ribopure kit. Ribosomal RNA was removed using Ribo-ZERO (EpiCentre). mRNA was fragmented and reverse-transcribed to yield double-stranded cDNA, which was sequenced on an Illumina HiSeq-2000 using paired-end reads extending 101 bp. Raw data were de-multiplexed and aligned to the mouse reference genome (mm9) using TopHat 1.4.1. Transcript abundances were estimated from the alignment files using Cufflinks.
Luciferase Assays.
The Eβ enhancer was amplified and cloned into the BamHI site of pGL3 (Promega). Each tested upstream Vβ region (300–500 bp) was amplified and cloned into the XhoI/HindIII sites of the Eβ-containing vector. T3 cells (60) were transfected transiently with firefly (4 μg) and Renilla (40 ng) luciferase plasmids using electroporation. After 24 h, the transfected cells were assayed for firefly and Renilla activities. A list of primers is provided in Table S4.
V(D)J Recombination Substrates.
A Dβ1-Jβ1.1 rearrangement that includes the 5′ Dβ1-RSS was amplified from thymus DNA and cloned into pCDNA3.1. Each recombination substrate includes the specified Vβ-RSS together with its upstream and downstream flanking sequences (80 and 130 bp, respectively), which were cloned 5′ to the DβJβ1.1 join (deletion substrates). An inert yellow fluorescent protein (YFP) coding sequence was inserted as a stuffer between the Vβ and Dβ1-Jβ1.1 elements. A list of Vβ-specific primers is provided in Table S5.
Recombination Substrate Assays.
Human embryonic kidney 293T cells were transfected with an equimolar mixture of eight recombination substrates (TRBV1, 15, 16, 18, 20, 23, 24, and 26), pEBB-RAG1, and pEBB-RAG2, using Trans-IT 293 (Mirus) (40). Plasmid substrates were recovered 48 h posttransfection and digested with NotI to minimize unrearranged PCR products and DpnI to cut untransfected substrates (40). The digested DNA mixture was amplified with primers that are common to all substrates—one that recognizes plasmid sequence upstream of the Vβs and one specific for Jβ1.1 (dsT7-CAAGCTGGCTAGCGTTTAAAC and J1.1TR-CTCGAATATGGACACGGAGGACATGC). PCR was performed for 30 cycles on serial fourfold dilutions of recovered substrates. The products were separated on 1% agarose gels, transferred to Zetaprobe (BioRad), and probed with labeled Vβ-specific oligonucleotides.
Computational Analysis.
Regression analysis was performed following a two-step procedure that is a simplified version of the protocol described previously (50).
Step 1.
For each of the chromatin features analyzed, the region spanning Vβ segments was divided into three bins: jth Vj segment itself, 1 kb immediately upstream (Uj), and 1 kb immediately downstream of the V segment (Dj). The signal intensity of each bin (3 bins × 35 Vβs, 105 total bins) was measured from the UCSC WIG files containing either read counts (ChIP-seq) or MA2C scores (ChIP-chip) using BEDtools. The signal intensities were then converted to the natural logarithm of their values. To eliminate any ln(0) values in the computational analyses, a pseudocount of 1 was added to the read counts. Pearson’s correlation coefficients were then used to define which of the three bins (Vj, Uj, Dj) correlate best with V recombination frequencies. The bin for each feature with the highest correlation coefficient was used in further analyses. Recombination frequencies fj for Vj regions (expressed in percent of overall use) were transformed into their natural logarithm values [ln(fj + 0.01), where 0.01 is an added pseudocount]. The Vβ gene segments were then classified as rearranging or nonrearranging, and random forest classification was used to determine which of the features distinguish best between rearranging and inert Vβ gene segments (R package; RandomForest).
Step 2.
Linear regression analysis was performed for 13 variables using data corresponding to only the subset of 23 rearranging Vβ segments (nonzero recombination frequency) using R package (leaps) to identify the most important regressors for recombination levels. The analysis was further refined to determine a reduced set of variables that attains statistical significance (Tables S6–S8 and Dataset S1).
Supplementary Material
Acknowledgments
We thank Drs. Barry Sleckman, Baeck-Seung Lee, and David Schatz for valuable comments and reagents. We thank the Genome Technology Access Center in the Department of Genetics at Washington University School of Medicine in St. Louis for help with genomic analyses. The Center is partially supported by National Cancer Institute Cancer Center Support Grant P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant UL1RR024992 from the National Center for Research Resources, a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical Research. This research was supported by NIH Grants AI 079732, AI 081224 and CA 156690 (to E.M.O.) and AI 082918 (to A.J.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE49234 and GSE48817).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304048110/-/DCSupplemental.
References
- 1.Dekker J. Gene regulation in the third dimension. Science. 2008;319(5871):1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shih HY, et al. Tcra gene recombination is supported by a Tcra enhancer- and CTCF-dependent chromatin hub. Proc Natl Acad Sci USA. 2012;109(50):E3493–E3502. doi: 10.1073/pnas.1214131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bossen C, Mansson R, Murre C. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol. 2012;30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 4.Bassing CH, Swat W, Alt FW. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 2002;109(Suppl):S45–S55. doi: 10.1016/s0092-8674(02)00675-x. [DOI] [PubMed] [Google Scholar]
- 5.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11(4):251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 6.Cobb RM, Oestreich KJ, Osipovich OA, Oltz EM. Accessibility control of V(D)J recombination. Adv Immunol. 2006;91:45–109. doi: 10.1016/S0065-2776(06)91002-5. [DOI] [PubMed] [Google Scholar]
- 7.Feeney AJ. Genetic and epigenetic control of V gene rearrangement frequency. Adv Exp Med Biol. 2009;650:73–81. doi: 10.1007/978-1-4419-0296-2_6. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27(4):561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews AG, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450(7172):1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oestreich KJ, et al. Regulation of TCRbeta gene assembly by a promoter/enhancer holocomplex. Immunity. 2006;24(4):381–391. doi: 10.1016/j.immuni.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 11.Ji Y, et al. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141(3):419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson A, Kondilis HD, Khor B, Sleckman BP, Krangel MS. Regulation of T cell receptor beta allelic exclusion at a level beyond accessibility. Nat Immunol. 2005;6(2):189–197. doi: 10.1038/ni1157. [DOI] [PubMed] [Google Scholar]
- 13.Guo C, et al. Two forms of loops generate the chromatin conformation of the immunoglobulin heavy-chain gene locus. Cell. 2011;147(2):332–343. doi: 10.1016/j.cell.2011.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jhunjhunwala S, et al. The 3D structure of the immunoglobulin heavy-chain locus: Implications for long-range genomic interactions. Cell. 2008;133(2):265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skok JA, et al. Reversible contraction by looping of the Tcra and Tcrb loci in rearranging thymocytes. Nat Immunol. 2007;8(4):378–387. doi: 10.1038/ni1448. [DOI] [PubMed] [Google Scholar]
- 16.Rubio ED, et al. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci USA. 2008;105(24):8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seitan VC, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476(7361):467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribeiro de Almeida C, et al. The DNA-binding protein CTCF limits proximal Vκ recombination and restricts κ enhancer interactions to the immunoglobulin κ light chain locus. Immunity. 2011;35(4):501–513. doi: 10.1016/j.immuni.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Guo C, et al. CTCF-binding elements mediate control of V(D)J recombination. Nature. 2011;477(7365):424–430. doi: 10.1038/nature10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang Y, Zhou X, Hewitt SL, Skok JA, Garrard WT. A multifunctional element in the mouse Igκ locus that specifies repertoire and Ig loci subnuclear location. J Immunol. 2011;186(9):5356–5366. doi: 10.4049/jimmunol.1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godfrey DI, Hammond KJ, Poulton LD, Smyth MJ, Baxter AG. NKT cells: Facts, functions and fallacies. Immunol Today. 2000;21(11):573–583. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 22.Ndifon W, et al. Chromatin conformation governs T-cell receptor Jβ gene segment usage. Proc Natl Acad Sci USA. 2012;109(39):15865–15870. doi: 10.1073/pnas.1203916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robins HS, et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood. 2009;114(19):4099–4107. doi: 10.1182/blood-2009-04-217604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson A, Maréchal C, MacDonald HR. Biased V beta usage in immature thymocytes is independent of DJ beta proximity and pT alpha pairing. J Immunol. 2001;166(1):51–57. doi: 10.4049/jimmunol.166.1.51. [DOI] [PubMed] [Google Scholar]
- 25.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296(5565):158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 26.Fuxa M, et al. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18(4):411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynaud D, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9(8):927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu H, et al. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21(10):1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin YC, et al. Global changes in the nuclear positioning of genes and intra- and interdomain genomic interactions that orchestrate B cell fate. Nat Immunol. 2012;13(12):1196–1204. doi: 10.1038/ni.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaumeil J, et al. Higher-order looping and nuclear organization of Tcra facilitate targeted rag cleavage and regulated rearrangement in recombination centers. Cell Rep. 2013;3(2):359–370. doi: 10.1016/j.celrep.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spicuglia S, et al. TCRalpha enhancer activation occurs via a conformational change of a pre-assembled nucleo-protein complex. EMBO J. 2000;19(9):2034–2045. doi: 10.1093/emboj/19.9.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: A functional definition of the joining signals. Genes Dev. 1989;3(7):1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 33.Livák F. Evolutionarily conserved pattern of gene segment usage within the mammalian TCRbeta locus. Immunogenetics. 2003;55(5):307–314. doi: 10.1007/s00251-003-0577-6. [DOI] [PubMed] [Google Scholar]
- 34.Posnett DN, et al. Level of human TCRBV3S1 (V beta 3) expression correlates with allelic polymorphism in the spacer region of the recombination signal sequence. J Exp Med. 1994;179(5):1707–1711. doi: 10.1084/jem.179.5.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu C, et al. Dramatically increased rearrangement and peripheral representation of Vbeta14 driven by the 3’Dbeta1 recombination signal sequence. Immunity. 2003;18(1):75–85. doi: 10.1016/s1074-7613(02)00515-0. [DOI] [PubMed] [Google Scholar]
- 36.Nadel B, et al. Decreased frequency of rearrangement due to the synergistic effect of nucleotide changes in the heptamer and nonamer of the recombination signal sequence of the V kappa gene A2b, which is associated with increased susceptibility of Navajos to Haemophilus influenzae type b disease. J Immunol. 1998;161(11):6068–6073. [PubMed] [Google Scholar]
- 37.Feeney AJ, Tang A, Ogwaro KM. B-cell repertoire formation: Role of the recombination signal sequence in non-random V segment utilization. Immunol Rev. 2000;175:59–69. [PubMed] [Google Scholar]
- 38.Jung D, et al. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 2003;18(1):65–74. doi: 10.1016/s1074-7613(02)00507-1. [DOI] [PubMed] [Google Scholar]
- 39.Cowell LG, Davila M, Yang K, Kepler TB, Kelsoe G. Prospective estimation of recombination signal efficiency and identification of functional cryptic signals in the genome by statistical modeling. J Exp Med. 2003;197(2):207–220. doi: 10.1084/jem.20020250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee AI, et al. A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol. 2003;1(1):E1. doi: 10.1371/journal.pbio.0000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cuomo CA, Mundy CL, Oettinger MA. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol. 1996;16(10):5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerstein RM, Lieber MR. Coding end sequence can markedly affect the initiation of V(D)J recombination. Genes Dev. 1993;7(7B):1459–1469. doi: 10.1101/gad.7.7b.1459. [DOI] [PubMed] [Google Scholar]
- 43.Olaru A, Patterson DN, Villey I, Livák F. DNA-Rag protein interactions in the control of selective D gene utilization in the TCR beta locus. J Immunol. 2003;171(7):3605–3611. doi: 10.4049/jimmunol.171.7.3605. [DOI] [PubMed] [Google Scholar]
- 44.Yu K, Lieber MR. Mechanistic basis for coding end sequence effects in the initiation of V(D)J recombination. Mol Cell Biol. 1999;19(12):8094–8102. doi: 10.1128/mcb.19.12.8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: Implications for translocations. Mol Cell. 2009;34(5):535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (Formaldehyde Assisted Isolation of Regulatory Elements) Methods. 2009;48(3):233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pekowska A, et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 2011;30(20):4198–4210. doi: 10.1038/emboj.2011.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, et al. MM-ChIP enables integrative analysis of cross-platform and between-laboratory ChIP-chip or ChIP-seq data. Genome Biol. 2011;12(2):R11. doi: 10.1186/gb-2011-12-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dong X, et al. Modeling gene expression using chromatin features in various cellular contexts. Genome Biol. 2012;13(9):R53. doi: 10.1186/gb-2012-13-9-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karlić R, Chung HR, Lasserre J, Vlahovicek K, Vingron M. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci USA. 2010;107(7):2926–2931. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon J, Morshead KB, Guyon JR, Kingston RE, Oettinger MA. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol Cell. 2000;6(5):1037–1048. doi: 10.1016/s1097-2765(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 53.Osipovich O, et al. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat Immunol. 2007;8(8):809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- 54.Williams GS, et al. Unequal VH gene rearrangement frequency within the large VH7183 gene family is not due to recombination signal sequence variation, and mapping of the genes shows a bias of rearrangement based on chromosomal location. J Immunol. 2001;167(1):257–263. doi: 10.4049/jimmunol.167.1.257. [DOI] [PubMed] [Google Scholar]
- 55.Aoki-Ota M, Torkamani A, Ota T, Schork N, Nemazee D. Skewed primary Igκ repertoire and V-J joining in C57BL/6 mice: Implications for recombination accessibility and receptor editing. J Immunol. 2012;188(5):2305–2315. doi: 10.4049/jimmunol.1103484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452(7184):243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- 57.Lefranc MP, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37(Database issue):D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hagège H, et al. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2(7):1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 59.Degner SC, et al. CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc Natl Acad Sci USA. 2011;108(23):9566–9571. doi: 10.1073/pnas.1019391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferrier P, et al. Separate elements control DJ and VDJ rearrangement in a transgenic recombination substrate. EMBO J. 1990;9(1):117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.