Abstract
Complement component C1, the complex that initiates the classical pathway of complement activation, is a 790-kDa assembly formed from the target-recognition subcomponent C1q and the modular proteases C1r and C1s. The proteases are elongated tetramers that become more compact when they bind to the collagen-like domains of C1q. Here, we describe a series of structures that reveal how the subcomponents associate to form C1. A complex between C1s and a collagen-like peptide containing the C1r/C1s-binding motif of C1q shows that the collagen binds to a shallow groove via a critical lysine side chain that contacts Ca2+-coordinating residues. The data explain the Ca2+-dependent binding mechanism, which is conserved in C1r and also in mannan-binding lectin-associated serine proteases, the serine proteases of the lectin pathway activation complexes. In an accompanying structure, C1s forms a compact ring-shaped tetramer featuring a unique head-to-tail interaction at its center that replicates the likely arrangement of C1r/C1s polypeptides in the C1 complex. Additional structures reveal how C1s polypeptides are positioned to enable activation by C1r and interaction with the substrate C4 inside the cage-like assembly formed by the collagenous stems of C1q. Together with previously determined structures of C1r fragments, the results reported here provide a structural basis for understanding the early steps of complement activation via the classical pathway.
Keywords: structural biology, innate immunity
The innate immune system targets invading microbes by recognizing pathogen-associated molecular patterns. One such innate defense is complement, which triggers lysis and opsonization of invading cells and stimulates inflammatory and adaptive immune responses (1). In the classical pathway, a large multicomponent assembly (the C1 complex) binds to immune complexes, protein modulators (e.g., C-reactive protein), and polyanionic structures on pathogens to initiate complement activation. It is composed of a recognition subcomponent, C1q (460 kDa), and two serine protease subcomponents, C1r (90 kDa) and C1s (80 kDa) in a 1:2:2 ratio, with an overall molecular mass of ∼790 kDa. C1q has a bouquet-like structure assembled from six collagenous stems, each with a C-terminal globular head. Binding to pathogens induces a conformational change that drives activation of the zymogen proteases in stepwise fashion: C1r first autoactivates, then activates C1s (2). C1s subsequently cleaves substrates C4 and C4b-bound C2, to form the C3 convertase (C4b2a), the downstream component of the reaction cascade.
C1r and C1s are modular proteases each with two N-terminal complement C1r/C1s, Uegf and bone morphogenetic protein-1 (CUB) domains, separated by an epidermal growth factor (EGF)-like domain, followed by two complement control modules (CCP) and a C-terminal serine protease (SP) domain (3). In the absence of C1q, they form an elongated S-shaped heterotetramer with two central C1r polypeptides, linked via their catalytic domains (CCP1-CCP2-SP), each flanked by a C1s chain (4). A clue to the likely mode of binding between C1r and C1s is provided by the crystal structure of the C1s CUB1-EGF fragment (5) and structures of two related proteases of the lectin pathway of complement: the CUB1-EGF-CUB2 domains of mannan-binding lectin-associated serine protease (MASP)-2 (6) and MASP-1/-3 (7), in which polypeptides are arranged as antiparallel X-shaped dimers. Residues at the interface are conserved in C1r, suggesting that C1r and C1s bind to each other in the same way.
Solution studies and electron microscopy show that the C1r-C1s tetramer folds up to form a relatively compact structure when it binds to C1q (8, 9); however, relatively little is known about the conformational changes that enable binding or how the resulting complex permits activation. Calcium plays a central role in the assembly and is necessary for binding of C1r to C1s and of the protease tetramer to C1q (10). Recent work has identified separate binding sites for C1q in the CUB1 and CUB2 domains of C1r and in CUB1 of C1s, providing a total of six sites on a C1r-C1s tetramer, one for each collagenous stem (11, 12). In this study, we have determined the structures of a series of fragments of C1s, including C1s bound to a collagen-like peptide from C1q and a tetramer that reveals how the proteases pack together at the center of the complex. Together, these structures show how subcomponents form the large C1 complex and explain important steps in the activation process.
Results
Structure Determination of the C1s–C1q Collagen Complex.
C1q is assembled from three different polypeptide chains (designated A, B, and C; encoded by three closely related and clustered C1q genes) that combine to form six heterotrimeric subunits (13). Despite the relatively low degree of conservation among higher vertebrates, the C-terminal half of the collagenous domain contains a hexapeptide motif in all species: Hyp-Gly-Lys-Xaa-Gly-Pro/Tyr/Asn (where Hyp is hydroxyproline), which mediates binding to C1r and C1s (14). For cocrystallization, we designed a short collagen-like peptide containing the sequence Hyp-Gly-Lys-Leu-Gly-Pro at the center. We have shown that C1r and C1s recognize this sequence and that the lysine residue at the center of the motif is essential for binding (12).
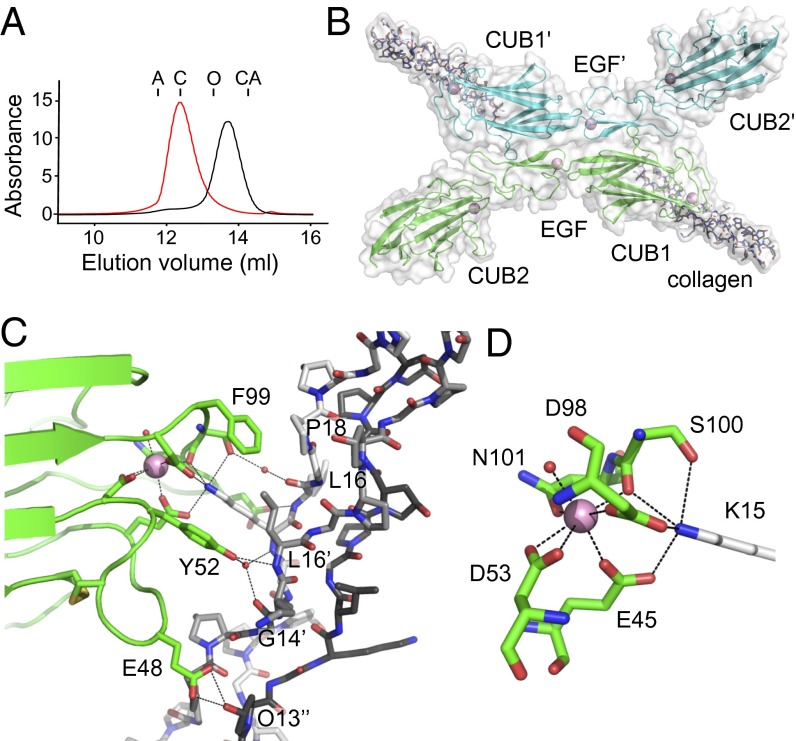
The CUB1-EGF-CUB2 region of C1s was produced in Chinese hamster ovary cells, and gel filtration revealed that it is a Ca2+-dependent dimer (Fig. 1A). Crystals were grown from a mixture of C1s and peptide in the presence of Ca2+ to enable binding. The asymmetric unit of the crystal contained a single complex, with the collagen bound to the CUB1 domain of C1s. The CUB1-EGF-CUB2 domains of C1s are arranged linearly with three Ca2+: one in each CUB domain and one within the EGF-like domain, near the CUB1-EGF junction (Fig. S1). Monomers associate to form an X-shaped dimer (Fig. 1B) through lateral association of the CUB1 and EGF-like domains about the crystallographic symmetry axis. The interface features a number of hydrophobic side chains with a total buried surface of 1,923 Å2 (5). There are no additional contacts from CUB2.
Fig. 1.
Structure of C1s–C1q collagen complex. (A) Gel filtration of the CUB1-EGF-CUB2 fragment of C1s (35 kDa) in Ca2+ (red) and EDTA (black; each at 2 mM). Samples were separated on a Superdex 200 column equilibrated in 25 mM Tris⋅HCl at pH 7.4, containing 150 mM NaCl. Elution positions of aldolase (A; 158 kDa), conalbumin (C; 75 kDa), ovalbumin (O; 43 kDa), and carbonic anhydrase (CA; 29 kDa) are shown. (B) Structure of the C1s X-shaped dimer (green and blue) bound to the collagen-like peptide of C1q. (C) Close-up of the C1s/C1q collagen interface showing the residues that participate in the interaction. The leading, middle, and trailing strands are in black, gray, and white, respectively. (D) The Ca2+-binding site of the CUB1 domain of C1s showing the six coordinating ligands arranged in a tetragonal bipyramid and the interaction with Lys15 of the C1q collagen-like peptide. Waters are shown as red spheres and Ca2+ are in pink, throughout.
The C1s–C1q Collagen Interface.
The three collagen-like peptides form a right-handed helix with a characteristic one residue stagger between adjacent strands. Each chain binds to C1s, with the leading, middle, and trailing strands accounting for 8, 47, and 45% respectively, of the buried surface of the collagen helix of 418 Å2 (of a total of 836 Å2 including both surfaces; Table 1). Key to the interaction is Lys15 of the trailing strand, which contacts the carboxylate groups of Glu45 and Asp98 and the hydroxyl and carbonyl groups of Ser100, three of the five residues that coordinate the Ca2+ of CUB1 (Fig. 1 C and D). Additional contacts include hydrogen bonds between the hydroxyl group of Tyr52 of C1s and the backbone amide of Leu16 of the middle chain and water-mediated hydrogen bonds to the carbonyl of Gly14 and the backbone amide of Lys15 of the of the middle and trailing chains, respectively. In addition, a hydrogen bond connects Glu48 and the hydroxyl group of Hyp13 of the leading strand, and a water-mediated hydrogen bond links the hydroxyl group of Ser100 to the carbonyl group of Leu15 of the trailing chain. The remaining contacts are hydrophobic and include Phe99 stacking against Pro18 of the trailing strand and Tyr52 against the β-carbon of Leu16 of the middle strand.
Table 1.
Surface area of the collagen peptide of C1q buried (in Å2) upon complex formation with C1s
| Collagen residue | Leading | Middle | Trailing |
| Gly11 | 0 | 0.6 | 0 |
| Pro12 | 0 | 39.5 | 0 |
| Hyp13 | 32.6 | 60.9 | 0 |
| Gly14 | 0 | 4.9 | 1.7 |
| Lys15 | 0 | 4.3 | 136.1 |
| Leu16 | 0 | 70.7 | 23.7 |
| Gly17 | 0 | 0 | 2.8 |
| Pro18 | 0 | 0 | 29.2 |
| Hyp19 | 0 | 0 | 11.2 |
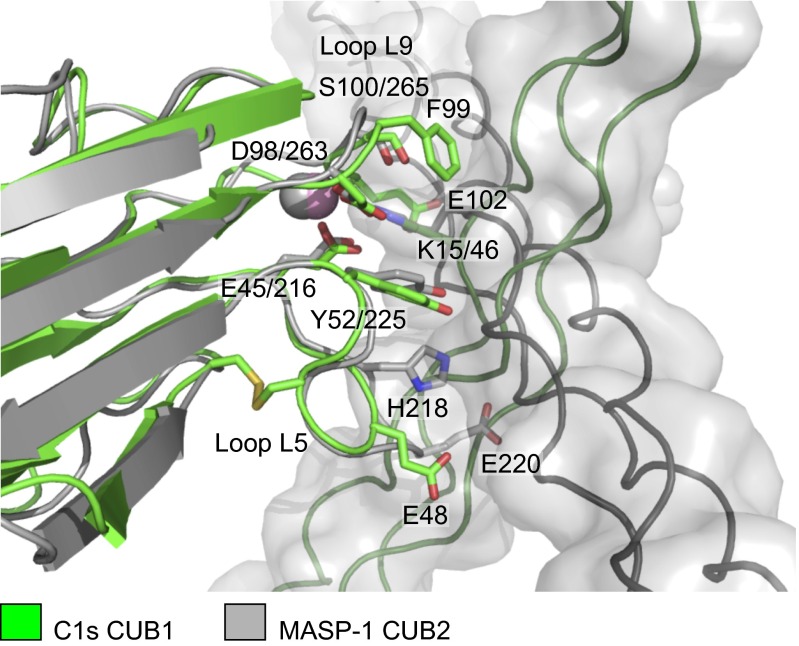
The structure is compatible with previous mutagenesis data that highlight the importance of the lysine residue in the collagen and Glu45 and Asp98 in C1s, and explains the Ca2+ dependence of binding (11, 12). It shares a number of similarities with the complex between the CUB2 domain of MASP-1 bound to a collagen-like peptide of MBL of the lectin pathway (15). In particular, both structures feature a lysine residue from the collagen binding to Ca2+-coordinating residues in the CUB domain. Remarkably, however, the orientation of the collagen helix differs by ∼90° in the two structures, highlighting the versatility of the binding arrangement, in which differences in the size and composition of two loops (L5 and L9) of the CUB dictate the orientation of the collagen helix (Fig. 2). Binding residues are conserved in the CUB1 domains of C1r, MASP-1/-3, and MASP-2, indicating that the interaction mechanism is conserved in initiating complexes of the lectin and classical pathways (Fig. S2).
Fig. 2.
Comparison of CUB1–collagen and CUB2–collagen complexes of the classical and lectin pathways. Overlay of the C1s–C1q collagen structure (green) with the MASP-1/-3-MBL collagen structure (gray; PDB ID code: 3POB) (15). Key elements of the complexes superimpose including the lysine residue of the collagen, the Ca2+ and Ca2+-coordinating residues in the CUB domains together with a tyrosine residue, which contacts the collagen in each structure. However, the orientation of the collagen helices differs by ∼90°. Glu48 of loop L5 and Glu102 in loop L9 of C1s sandwich the collagen helix creating the shallow binding groove. Glu102 and other residues in loop L9 prevent binding in the alternative (horizontal) orientation through steric clashes. Loop L5 is longer in MASP-1 than in C1s and contributes toward binding via His218, which forms hydrogen bonds to the collagen, and blocks binding in the alternative (vertical) orientation (e.g., through Glu220, which would clash with the collagen chains).
Notably, only one of the lysine residues in the collagen forms contacts with C1s, yet the binding motif is present in all three chains of human C1q: A chain, OGK59VGY; B chain, OGK61VGP; and C chain, OGK58NGP. Recent mutagenesis data show that LysB61 and LysC58 are essential for binding, whereas LysA59 is less important, so it may not bind to C1r or C1s (16). From the structure, it is not possible to determine whether the B or C chains of C1q contribute the lysine side chain, because the register of the collagenous domain of C1q is not known (i.e., which of the A, B, and C chains is leading, middle, and trailing). In addition, a valine from the B chain or an asparagine from the C chain at position 16 of the collagen could be accommodated into the binding groove of C1s (Fig. 1C). Interestingly, in the MASP-1 CUB2-collagen structure, the key lysine projects from the leading strand of the collagen rather than the trailing strand. By analogy, the four CUB1 domains (two each from C1r and C1s) probably contact different lysine residues to the two CUB2 domains of C1r when the C1r-C1s tetramer binds to C1q, explaining why both LysB61 and LysC58 are essential for C1 complex formation.
Structure of the C1s Tetramer.
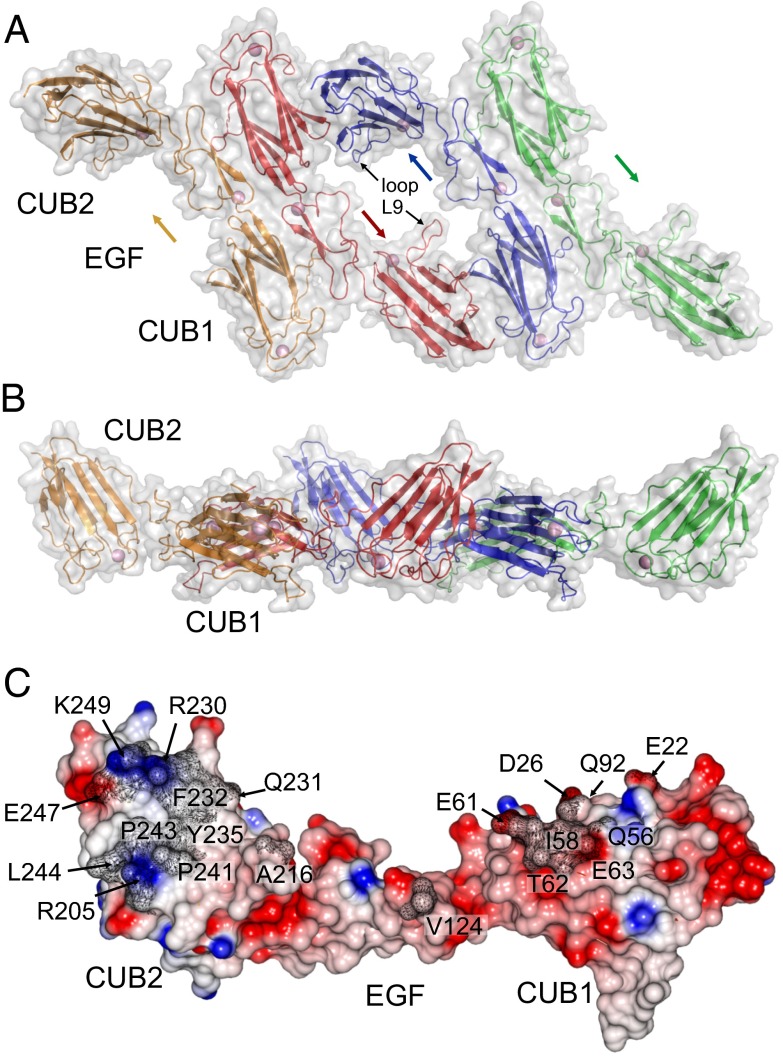
The yield of recombinant C1r was very low (both full-length and the CUB1-EGF-CUB2 fragment), and we were not able to generate sufficient material for cocrystallization with C1s. Nevertheless, in the absence of collagen peptide, C1s crystallized to form a tetramer, which mimics the likely organization of C1r and C1s polypeptides in the C1 complex (Fig. 3 A and B). The tetramer is a flat ring with an outer diameter of ∼120 Å. Two CUB2 domains from the outermost polypeptides project from opposite sides of the ring to give a maximum separation of ∼190 Å (between the two C termini). The structure is assembled from two X-shaped dimers linked via a connecting interface at the center. This interaction, in turn, is made possible by the unusual position of CUB2 in C1s, which is rotated by ∼55° (Fig. S1C) compared with human MASP-1 and rat MASP-2, enabling it to pack against the CUB1 domain of its partner. Comparison with the collagen-bound structure indicates that there are no major changes to C1s upon collagen binding. The unusual orientation of CUB2 is maintained through a network of interactions with the EGF-like domain (Fig. S1D).
Fig. 3.
Structure of the C1s tetramer. Top (A) and side (B) views of the C1s tetramer in which two X-shaped dimers are connected via a unique central interface. Arrows indicate the direction of the C1s chains. (C) Side view of the central C1s–C1s interface showing the electrostatic potential. Residues buried by the interaction are labeled and shaded in gray.
The central C1s–C1s interface encompasses 1,148 Å2 of buried surface, with a small hydrophobic core at the center, involving Ile58 of CUB1 and Phe232, Tyr235, and Pro241 of CUB2, surrounded by polar and charged residues (Fig. 3C). Hydrogen bonds link the side chain of Thr62 with carbonyl oxygen of Pro24, the side chain of Asp61 with the amide of Gly233, and carbonyl oxygen of Asp61 with the hydroxyl of Tyr235. Reciprocal contacts are made via loop L9 of CUB2 (Fig. 3A), which extends across a central gap in the tetramer to contact a loop in the EGF-like domain. In addition, there is overall charge complementarity, with the binding regions of CUB1 and CUB2 having net negative and positive charges, respectively. A C1s–C1s interaction at the center of the C1 complex has been predicted in previous molecular dynamics and modeling studies (11, 12, 17) but has not been observed directly. In the absence of C1q, the interaction must be weak because only C1s dimers are detected in solution (corresponding to the X-shaped dimers), but is stabilized when C1r-C1s tetramers bind to C1q, explaining how the elongated proteases form the compact structures observed in electron micrographs of the C1 complex (9).
Structural Organization of C1s.
Previous attempts to crystallize full-length C1s were unsuccessful. Structures of the catalytic domains of zymogen C1s (CCP1-CCP2-SP; PDB ID code: 4J1Y) (18) and of CCP2-SP of the activated enzyme have been determined (PDB ID code: 1ELV) (19), but the way in which the N- and C-terminal domains are linked together is not known. To address this question, we characterized two fragments encompassing the CUB2-CCP1 junction: CUB2-CCP1 and CUB2-CCP1-CCP2. These fragments were produced in Escherichia coli and refolded from inclusion bodies. Both fragments were crystallized and structures were refined to 2.0 Å and 2.9 Å, respectively (Table S1). The three domains are arranged linearly, with the CUB1-CCP1 junction stabilized through an extensive interface involving hydrophobic interactions, hydrogen bonds, and a salt bridge (Fig. S3B). The position of the CUB2 and CCP1 domains is similar in both structures with a rmsd of 1.6 Å over 183 C-α atoms (Fig. S3C). The CCP1-CCP2 interface also has a hydrophobic component, but the CCP2 domain is rotated by ∼30° in comparison with the structure of zymogen C1s (Fig. S3C) (18), suggesting some flexibility at the CCP1-CCP2 junction.
Combining all of the structures of C1s reveals an extended L-shaped architecture with the base and stem formed from the CUB1-EGF-CUB2 and catalytic domains, respectively (Fig. S3D). The domain interfaces are all well-defined and overlapping structures superimpose closely suggesting little flexibility overall; the exception being the CCP1-CCP2 junction, which is likely to permit some movement, as discussed above. The configuration of the zymogen and active forms of C1s is also similar. Although activation leads to changes within the SP domain itself, its relationship with the CCP2 domain remains the same (rmsd of 1.9 Å for 289 C-α atoms over the two domains) (19). To verify that the assembled structure reflects the solution configuration of C1s, sedimentation coefficients were calculated from the atomic coordinates of the monomer and the (X-shaped) dimer. Calculated values of 4.2 and 5.7 S are in excellent agreement with experimental values of 4.3 and 5.6 S (20).
Discussion
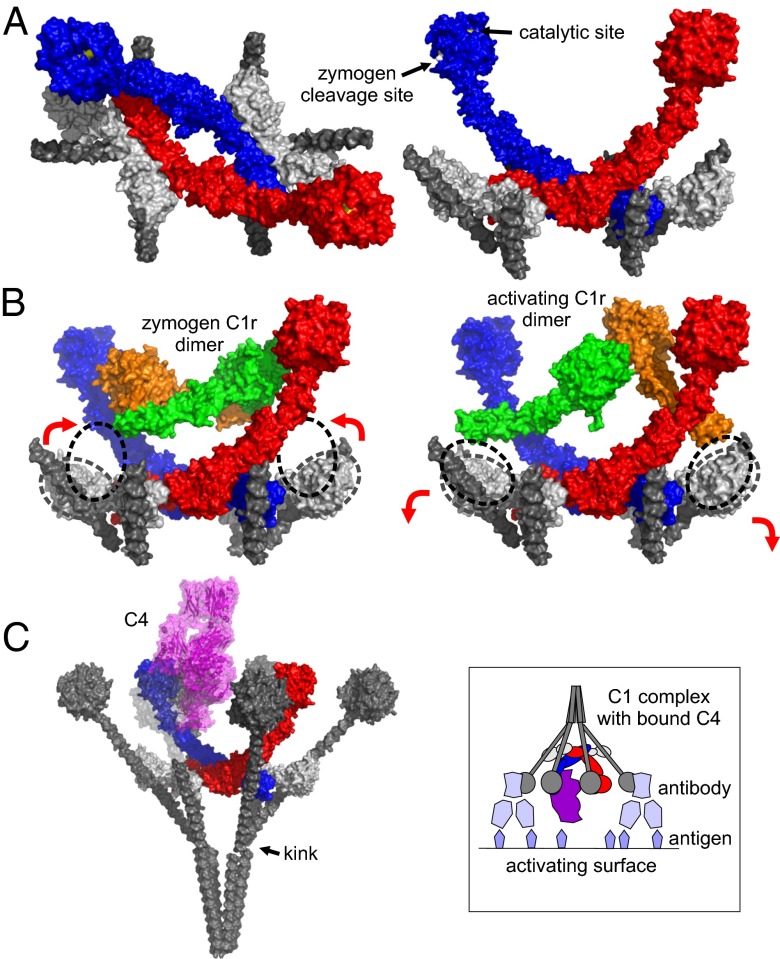
Combining the structures described here with existing structures of the catalytic domains of C1r reveals a comprehensive picture of the C1 complex and the changes that must occur during activation. The center is formed from the CUB1-EGF-CUB2 domains of the proteases that together form a ring-shaped tetramer, with C1s polypeptides innermost. The six collagenous stems of C1q are arranged around the outside of the ring (Fig. 4A). Upon extension to their full length, they converge close to an interruption in the collagen-like domain, often called the C1q kink (Fig. 4C). The structure is thus entirely compatible with the bouquet-like architecture of C1q observed by electron microscopy (21).
Fig. 4.
Structural organization of the C1 complex and the steps leading to activation and substrate recognition. (A) Superposition of the C1s tetramer, the C1s–collagen complex and the catalytic domains of C1s reveals the arrangement of subcomponents at the center of the C1 complex. C1s polypeptides are show in red and blue, the collagen is in gray, and the putative C1r CUB1-EGF-CUB2 fragments are in white. The MASP-1 CUB2-MBL collagen complex (PDB ID code: 3POB) (15) was overlaid onto the CUB2 domains of C1r to position the additional two collagen helices. (B) The catalytic domains of zymogen C1r (PDB ID code: 1GPZ) (18) and active C1r in a putative enzyme-product conformation (PDB ID code: 2QYO) (31) fit between the arms of C1s and can be connected by rotation of the CUB2 domains of C1r. The positions of the C1r CUB2 domains together with the resting position (based on the structure of the C1s tetramer) are indicated by the dotted lines. (C) Superposition of the catalytic domains of C1s with the corresponding domains of MASP-2 bound to C4 (PDB ID code: 4FXG) (24) shows that C4 can fit inside the cage-like assembly formed by the collagenous stems of C1q. The stems have been extended to their full length, and the globular domains of C1q (PDB ID code: 1PK6) (32) are positioned on top. The upper part of the front left stem and the globular domain is partially transparent so that the position of C4 (purple) can be seen. (Inset) Schematic representation of the C1 complex with C4 attached bound to antibody molecules on an activating surface.
Surprisingly, C1s is located entirely within the cage-like structure formed by the collective collagenous stems of C1q (even allowing for flexibility at the CCP1-CCP2 junction of C1s). The two central C1s polypeptides curve upwards to form a U-shape with the protease domains at the tips of the U, and the catalytic sites point inwards toward the center of the complex (Fig. 4A). Their central position would ensure that they are protected from other serum proteases before activation, which could potentially trigger adventitious activation. No structure is available for the N-terminal domains of C1r. However, it has the same modular structure as C1s (with ∼40% identity) and C1q binding residues are conserved (Fig. S2), so the overall architecture and mode of binding to the collagenous stems is likely to be the same. Indeed, in the absence of C1r, two C1s dimers bind to C1q, which although nonfunctional, because C1r is required to activate C1s, maintains the overall stoichiometry of the complex (22).
The position of the subcomponents in the fully assembled complex suggests a simple mechanism for steps leading to protease activation. The catalytic domains of C1r fit between the arms of C1s, without any steric clashes (Fig. 4B). In the preactivated complex, these domains form a head-to-tail dimer that can be connected to the N-terminal fragments by rotation of the CUB2 domains of C1r (Fig. 4B) (19). Upon target recognition, splaying apart of the two C1q stems bound to the CUB2 domain of C1r would cause the SP domains to separate; repositioning of the CUB2 domains back to their resting positions would align the SP domains so that one protease can catalyze the cleavage of its partner (and vice versa). Once activated, each C1r arm can access the zymogen cleavage site of C1s, located on the back of the SP domain (Fig. 4A). C1r is much more flexible than C1s (23), and flexibility about the CUB2 domain in particular is likely to permit the necessary conformational changes that trigger activation of the C1 complex.
Recognition of C4 involves an extended binding site encompassing both CCP modules and the SP domain itself (24). Overlaying the structure of the homolog MASP-2 bound to C4, shows that C4 fits inside the cage-like structure where catalysis could subsequently occur (Fig. 4C). Although C4 can still pass between the stems of C1q, the enclosed environment would help to ensure that its product, C4b, attaches to the activating surface close enough to allow subsequent proteolysis of C2 once bound to C4b, thus facilitating coordinated activation of the reaction cascade.
Materials and Methods
Protein Production and Crystallization.
The collagen peptide Ac(GPO)4-GKL-(GPO)4NH2 was made by Generon Ltd. The CUB1-EGF-CUB2 fragment of C1s incorporating a C-terminal hexahistidine tag was produced in Chinese hamster ovary cells and purified by affinity chromatography on a nickel-Sepharose affinity column as described (12). Protein was purified further by gel filtration on a Superdex 200 16/60 column (GE Healthcare) in 20 mM Tris at pH 7.5, 50 mM NaCl, and 2 mM CaCl2 and was concentrated by filtration using a 10-kDa molecular mass cutoff membrane (Amicon) before crystallization.
The CUB2-CCP1 and CUB2-CCP1-CCP2 fragments of C1s were produced in E. coli and refolded from inclusion bodies. Briefly, the cDNA were amplified by PCR and cloned into the NcoI and EcoR1 restriction sites of expression vector pET28a. Cells were grown in Power Prime broth (Molecular Dimensions), induced during midlog phase with IPTG (1 mM), and harvested after growth at 37 °C for an additional 16 h. Inclusion bodies were isolated and protein refolded as described (23). Proteins were purified initially by ion exchange chromatography on a 10-mL Q-Sepharose column, using a 0.05–1 M gradient of NaCl in 20 mM Tris⋅HCl at pH 8.0; followed by gel filtration on a Superdex 75 16/60 column (GE Healthcare) in 20 mM Tris at pH 7.5 containing 50 mM NaCl and 2 mM CaCl2.
All crystals were grown by using the sitting-drop vapor diffusion method by mixing equal volumes (1.2 + 1.2 μL) of protein and reservoir solution. Crystals of the C1s–collagen complex were grown from a mixture of C1s (3.5–5 mg/mL; 0.1–0.15 μM) containing trimeric peptide (0.5 μM) mixed with 20% (vol/vol) PEG 3350, 50 mM Tris⋅HCl at pH 8.5, and 100 mM NaBr. Similar conditions were used to crystallize C1s alone, except that the concentration of NaBr was 200 mM. The CUB2-CCP1 (5 mg/mL) was crystallized in 20% (vol/vol) PEG 10000 in 100 mM Tris⋅acetate buffer at pH 8.0, containing 200 mM KH2PO4 and the CUB2-CCP1-CCP2 fragments in 24–30% (vol/vol) PEG 8000 containing 100 mM Bis-Tris propane⋅HCL at pH 6.3 with 200 mM potassium citrate. All crystals were transferred to reservoir solution containing 20% (vol/vol) glycerol before storage in liquid nitrogen and were maintained at 100 K during data collection.
Diffraction data were collected at Diamond Light Source and were processed with iMosflm. Phases were determined by molecular replacement with Phaser (25) using the MASP-2 CUB1-EGF-CUB2 (6) and Gly-Pro-Hyp collagen peptide (26) structures as search models. Models were optimized by using cycles of manual refinement with Coot and refinement in Refmac5 (27), part of the CCP4 software suite (28), and in Phenix (29).
Calculation of Predicted Sedimentation Coefficients.
The predicted sedimentation coefficients of the assembled C1s monomer and dimer were calculated from the atomic coordinates, using the program HYDROPRO (30).
Supplementary Material
Acknowledgments
We thank Diamond Light Source (DLS) for access to beamlines I04 and I04-1, the UK Midlands Block Allocation Group mx310, and DLS beamline scientists for their help with data collection. Funding for this work was provided by Medical Research Council UK Grant G1000191/1 (to R.W., P.C.E.M. and W.J.S.) and Wellcome Trust Grant 087848 (to A.R.G.). P.G. is supported by the Hungarian Scientific Research Fund Grants NK77978 and NK100834.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4LMF, 4LOR, 4LOT and 4LOS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311113110/-/DCSupplemental.
References
- 1.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: A key system for immune surveillance and homeostasis. Nat Immunol. 2010;11(9):785–797. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wallis R, Mitchell DA, Schmid R, Schwaeble WJ, Keeble AH. Paths reunited: Initiation of the classical and lectin pathways of complement activation. Immunobiology. 2010;215(1):1–11. doi: 10.1016/j.imbio.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forneris F, Wu J, Gros P. The modular serine proteases of the complement cascade. Curr Opin Struct Biol. 2012;22(3):333–341. doi: 10.1016/j.sbi.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Tschopp J, Villiger W, Fuchs H, Kilchherr E, Engel J. Assembly of subcomponents C1r and C1s of first component of complement: Electron microscopic and ultracentrifugal studies. Proc Natl Acad Sci USA. 1980;77(12):7014–7018. doi: 10.1073/pnas.77.12.7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregory LA, Thielens NM, Arlaud GJ, Fontecilla-Camps JC, Gaboriaud C. X-ray structure of the Ca2+-binding interaction domain of C1s. Insights into the assembly of the C1 complex of complement. J Biol Chem. 2003;278(34):32157–32164. doi: 10.1074/jbc.M305175200. [DOI] [PubMed] [Google Scholar]
- 6.Feinberg H, et al. Crystal structure of the CUB1-EGF-CUB2 region of mannose-binding protein associated serine protease-2. EMBO J. 2003;22(10):2348–2359. doi: 10.1093/emboj/cdg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teillet F, et al. Crystal structure of the CUB1-EGF-CUB2 domain of human MASP-1/3 and identification of its interaction sites with mannan-binding lectin and ficolins. J Biol Chem. 2008;283(37):25715–25724. doi: 10.1074/jbc.M803551200. [DOI] [PubMed] [Google Scholar]
- 8.Perkins SJ. Molecular modelling of human complement subcomponent C1q and its complex with C1r2C1s2 derived from neutron-scattering curves and hydrodynamic properties. Biochem J. 1985;228(1):13–26. doi: 10.1042/bj2280013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strang CJ, Siegel RC, Phillips ML, Poon PH, Schumaker VN. Ultrastructure of the first component of human complement: Electron microscopy of the crosslinked complex. Proc Natl Acad Sci USA. 1982;79(2):586–590. doi: 10.1073/pnas.79.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villiers CL, Arlaud GJ, Painter RH, Colomb MG. Calcium binding properties of the C1 subcomponents C1q, C1r and C1s. FEBS Lett. 1980;117(1):289–294. doi: 10.1016/0014-5793(80)80964-1. [DOI] [PubMed] [Google Scholar]
- 11.Bally I, et al. Identification of the C1q-binding sites of human C1r and C1s: A refined three-dimensional model of the C1 complex of complement. J Biol Chem. 2009;284(29):19340–19348. doi: 10.1074/jbc.M109.004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips AE, et al. Analogous interactions in initiating complexes of the classical and lectin pathways of complement. J Immunol. 2009;182(12):7708–7717. doi: 10.4049/jimmunol.0900666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reid KB. Complete amino acid sequences of the three collagen-like regions present in subcomponent C1q of the first component of human complement. Biochem J. 1979;179(2):367–371. doi: 10.1042/bj1790367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallis R, et al. Localization of the serine protease-binding sites in the collagen-like domain of mannose-binding protein: Indirect effects of naturally occurring mutations on protease binding and activation. J Biol Chem. 2004;279(14):14065–14073. doi: 10.1074/jbc.M400171200. [DOI] [PubMed] [Google Scholar]
- 15.Gingras AR, et al. Structural basis of mannan-binding lectin recognition by its associated serine protease MASP-1: Implications for complement activation. Structure. 2011;19(11):1635–1643. doi: 10.1016/j.str.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Bally I, et al. Expression of recombinant human complement C1q allows identification of the C1r/C1s-binding sites. Proc Natl Acad Sci USA. 2013;110(21):8650–8655. doi: 10.1073/pnas.1304894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beveridge AJ, Wallis R, Samani NJ. A molecular dynamics study of C1r and C1s dimers: Implications for the structure of the C1 complex. Proteins. 2012;80(8):1987–1997. doi: 10.1002/prot.24087. [DOI] [PubMed] [Google Scholar]
- 18.Perry AJ, et al. A molecular switch governs the interaction between human complement protease C1s and its substrate, complement C4. J Biol Chem. 2013;288(22):15821–15829. doi: 10.1074/jbc.M113.464545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaboriaud C, Rossi V, Bally I, Arlaud GJ, Fontecilla-Camps JC. Crystal structure of the catalytic domain of human complement c1s: A serine protease with a handle. EMBO J. 2000;19(8):1755–1765. doi: 10.1093/emboj/19.8.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arlaud GJ, Reboul A, Meyer CM, Colomb MG. Purification of proenzymic and activated human C1s free ofC1r. Effect of calcium and ionic strength on activated C1s. Biochim Biophys Acta. 1977;485(1):215–225. doi: 10.1016/0005-2744(77)90208-x. [DOI] [PubMed] [Google Scholar]
- 21.Brodsky-Doyle B, Leonard KR, Reid KB. Circular-dichroism and electron-microscopy studies of human subcomponent C1q before and after limited proteolysis by pepsin. Biochem J. 1976;159(2):279–286. doi: 10.1042/bj1590279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon PH, Schumaker VN. Measurement of macromolecular interactions between complement subcomponents C1q, C1r, C1s, and immunoglobulin IgM by sedimentation analysis using the analytical ultracentrifuge. J Biol Chem. 1991;266(9):5723–5727. [PubMed] [Google Scholar]
- 23.Major B, et al. Calcium-dependent conformational flexibility of a CUB domain controls activation of the complement serine protease C1r. J Biol Chem. 2010;285(16):11863–11869. doi: 10.1074/jbc.M109.098541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kidmose RT, et al. Structural basis for activation of the complement system by component C4 cleavage. Proc Natl Acad Sci USA. 2012;109(38):15425–15430. doi: 10.1073/pnas.1208031109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bella J, Eaton M, Brodsky B, Berman HM. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266(5182):75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, Number 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 29.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.García De La Torre J, Huertas ML, Carrasco B. Calculation of hydrodynamic properties of globular proteins from their atomic-level structure. Biophys J. 2000;78(2):719–730. doi: 10.1016/S0006-3495(00)76630-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kardos J, et al. Revisiting the mechanism of the autoactivation of the complement protease C1r in the C1 complex: Structure of the active catalytic region of C1r. Mol Immunol. 2008;45(6):1752–1760. doi: 10.1016/j.molimm.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 32.Gaboriaud C, et al. The crystal structure of the globular head of complement protein C1q provides a basis for its versatile recognition properties. J Biol Chem. 2003;278(47):46974–46982. doi: 10.1074/jbc.M307764200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.