Abstract
Insulin-degrading enzyme (IDE) selectively degrades the monomer of amyloidogenic peptides and contributes to clearance of amyloid β (Aβ). Thus, IDE retards the progression of Alzheimer’s disease. IDE possesses an enclosed catalytic chamber that engulfs and degrades its peptide substrates; however, the molecular mechanism of IDE function, including substrate access to the chamber and recognition, remains elusive. Here, we captured a unique IDE conformation by using a synthetic antibody fragment as a crystallization chaperone. An unexpected displacement of a door subdomain creates an ∼18-Å opening to the chamber. This swinging-door mechanism permits the entry of short peptides into the catalytic chamber and disrupts the catalytic site within IDE door subdomain. Given the propensity of amyloidogenic peptides to convert into β-strands for their polymerization into amyloid fibrils, they also use such β-strands to stabilize the disrupted catalytic site resided at IDE door subdomain for their degradation by IDE. Thus, action of the swinging door allows IDE to recognize amyloidogenicity by substrate-induced stabilization of the IDE catalytic cleft. Small angle X-ray scattering (SAXS) analysis revealed that IDE exists as a mixture of closed and open states. These open states, which are distinct from the swinging door state, permit entry of larger substrates (e.g., Aβ, insulin) to the chamber and are preferred in solution. Mutational studies confirmed the critical roles of the door subdomain and hinge loop joining the N- and C-terminal halves of IDE for catalysis. Together, our data provide insights into the conformational changes of IDE that govern the selective destruction of amyloidogenic peptides.
Keywords: M16 metalloprotease, X-ray crystallography, substrate recognition
Proteins in living organisms face acute and chronic challenges to their integrity, which necessitate proteostatic processes to protect their functions (1). Protein–protease networks play a key role in proteostasis by ensuring proper protein function through protein turnovers (2). Amyloidogenic peptides, such as amyloid β (Aβ) and amylin, present a major challenge to proteostasis, because they can form toxic aggregates that impair diverse physiological functions and contribute to human diseases (3, 4). Insulin-degrading enzyme (IDE), a Zn2+-metalloprotease, prefers to degrade amyloidogenic peptides to prevent the formation of amyloid fibrils (3). Exemplary substrates of IDE are insulin and Aβ, which are critical for the development of type 2 diabetes mellitus (DM2) and Alzheimer’s disease (AD), respectively. Genetic analyses strongly support functional roles of IDE in the clearance of insulin and Aβ (2, 3). In humans, several single nucleotide polymorphisms at the IDE locus on human chromosome 10q are associated with DM2 and late-onset AD (5, 6).
Structural analyses have provided significant insights to substrate recognition and catalysis by IDE. IDE has two ∼50-kDa αβαβα N-terminal (IDE-N) and C-terminal (IDE-C) halves, which are linked by a short hinge loop and come together to form an enclosed catalytic chamber (Fig. S1) (3, 7). IDE uses the size, shape, and charge distribution of its chamber (∼13,000 Å3) to selectively engulf structurally diverse peptides, such as insulin, Aβ, tumor growth factor-α, macrophage inflammatory protein-1α (MIP-1α), natriuretic peptides, and amylin (7–11). IDE substrates are presumably unraveled inside the catalytic chamber and then stochastically cleaved in regions that have a high propensity to convert into a β-strand for the formation of intermolecular cross-β sheets, the fundamental structural element of amyloid fibrils (7–9).
IDE belongs to the M16 metalloprotease family, which contributes to diverse biological functions; for example, the clearance of amyloidogenic peptides (12), the processing and clearance of mitochondrial signal peptides by mitochondrial processing peptidase (MPP) and presequence peptidase (PreP) (13, 14), and the degradation of hemoglobin as the food source by falcilysin in the malaria parasite (15). Crystal structures are available for all three subfamilies: M16A (e.g., IDE and pitrilysin), M16B (e.g., MPP and cytochrome bc1 complex core), and M16C (e.g., PreP and falcilysin) (Fig. S1) (7, 13, 14, 16–19). All M16 proteases contain two homologous ∼50-kDa domains, one of which contains a conserved HXXEH zinc ion-binding motif. However, the linker between these two domains is distinct among the three subfamilies. They are joined by a short hinge loop in M16A and by a helical hairpin in M16C, whereas no linker is present in M16B.
Despite significant progress, many key questions regarding structures and functional roles of IDE conformational states throughout the entire catalytic cycle remain unresolved. Crucially, the molecular basis for the recognition of amyloidogenic peptides by IDE remains elusive. On the basis of the available structures of M16 proteases, the displacement of IDE-N and IDE-C likely mediates the open–closed conformational switch required for the entrance of diverse substrates and the exit of products. However, the crystal structures of IDE have been determined only for the closed state, and its conformational states in solution have not been assessed. Furthermore, the role that the linker between IDE-N and IDE-C plays in catalysis, if any, remains undetermined, because the active enzyme can be reconstituted by mixing separated IDE-N and IDE-C (20). Here we combine structural, biophysical, and biochemical approaches to address these questions.
Results and Discussion
Swinging Door State of IDE Revealed in the IDE-Fab(IDE) Complex.
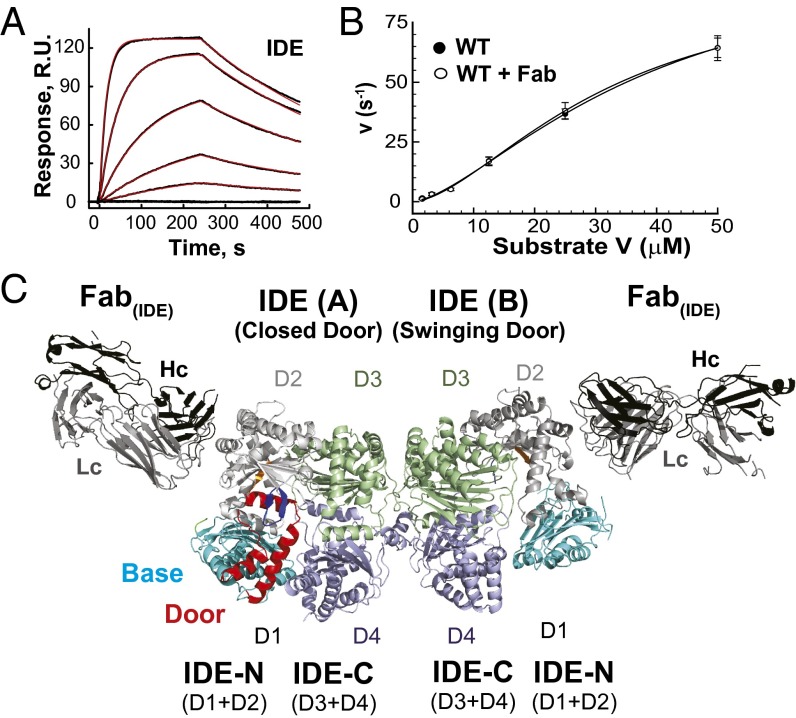
Crystal structures of human and rat IDE have only revealed the IDE dimer that has the fully enclosed catalytic chamber (7, 21). We suspect that the inability to capture another conformational state of IDE might be due to extensive intermolecular contacts conducive to crystal formation that preferentially stabilizes the closed state (Fig. S2B). Thus, we aimed to alter surface characteristics of IDE by using an antibody fragment as a crystallization chaperone (22). To do so, we biotinylated IDE in Escherichia coli, immobilized IDE on streptavidin-coated plates, used it to probe a phage-display library, and identified a synthetic antibody in the form of an antigen-binding fragment (Fab). This crystallization chaperone system has effectively facilitated crystallization of membrane proteins and RNA that are otherwise difficult to crystallize (23–25). The final synthetic antibody, termed Fab(IDE), bound human IDE with a KD value of 4 nM, as determined by surface plasmon resonance (Fig. 1A). It did not affect the steady-state kinetics of IDE when measured with substrate V, a short fluorogenic substrate that mimics bradykinin (BK) (Fig. 1B). We purified the IDE-Fab(IDE) complex to homogeneity (Fig. S2A), which was then crystallized. We then solved this structure at 3.4-Å resolution with satisfactory data statistics (Fig. 1C, Fig. S2 D–F, and Table S1).
Fig. 1.
Functional properties of Fab(IDE) and structure of IDE-Fab(IDE) complex. (A) Sensorgram traces for the interaction of Fab(IDE) to immobilized IDE. Black lines show experimental data, and red lines display the best fit of the global 1:1 Langmuir model, which resulted in association [ka = 2.18 ± 0.02 (×105) M−1s−1] and dissociation (kd = 5.87 ± 0.05 (×10−3) s−1] rate constants and the derived equilibrium constant (KD = 3.72 ± 0.03 nM). (B) Fab(IDE) does not affect IDE catalytic activity. Catalytic activity of IDE in the absence and presence of the equal molar Fab(IDE) was measured at varying concentrations of Substrate V. Data points represent mean ± SD (n = 3). A sigmoidal curve (Hill equation with Hill coefficient = ∼2) was fit to each data set. (C) Ribbon representation of IDE-Fab(IDE) structure [Protein Data Bank (PDB) ID 4IOF]. The base, door, P-loop, G-loop, H-loop, D2, D3, and D4 of IDE are colored cyan, red, green, orange, blue, light gray, light green, and light blue, respectively, and such color scheme is used throughout the article. Heavy and light chains (Hc and Lc) are colored black and gray, respectively.
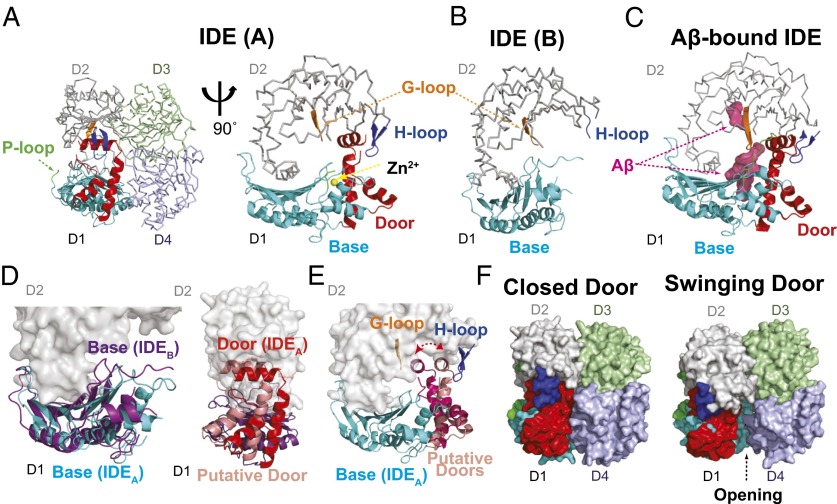
The asymmetric unit of the IDE-Fab(IDE) complex contains an IDE dimer and two Fab(IDE) (Fig. 1C). Of the two chains, A and B, of the IDE dimer, chain B represents a unique “swinging door state,” revealing key regions and residues involved in the dynamics, and therefore the catalysis of IDE that we seek to test. As defined previously, IDE-N and IDE-C are each made up of two homologous domains, D1/D2 and D3/D4, respectively (Fig. 1C) (7). IDE chain A is similar to IDE closed state, described previously (rmsd = 0.8 Å; Fig. S3), except for three disordered segments in IDE-N (amino acids 209–215, 287–297, and 366–369) and a small rigid body movement of IDE D1 away from D4 [0.7-Å displacement from the center of mass (COM) of D1 and D4]. In contrast, IDE chain B is distinct from IDE chain A and the putative open state depicted by the structure of pitrilysin (Fig. 2 A and B and Fig. S1). We found more disordered regions in IDE-N (amino acids 171–236, 284–298, 367–368, 456, and 492–496) in chain B than chain A, whereas IDE-C in chain B is nearly identical to the IDE closed state (Fig. S3). The contacts between the two chains within the IDE dimer are similar to that observed previously (7). D1 in both chains and D2 in chain B within IDE-N possess higher average thermal B factors than D3 and D4 in IDE-C (60–77 Å2 vs. 42–49 Å2) (Fig. S3). This likely represents natural thermal motions of IDE because only IDE-C is involved in IDE dimerization.
Fig. 2.
Comparison of two IDE chains in IDE-Fab(IDE) complex. (A) IDE A chain (closed door state). IDE D1 and IDE D2–4 are shown as ribbon and line representations, respectively. (A–C) Comparison among the subdomains and loops in chain A and chain B of IDE in the IDE-Fab(IDE) complex (PDB ID 4IOF) and those in Aβ-bound IDE (PDB ID 2G47). (D) Superimposition of swinging door base (purple) and closed base (cyan) and of closed door (red) and putative door in the swinging door state (pink), shown to indicate the proposed swinging movement between states. (E) Two putative positions of door subdomain to depict the potential swinging door motion. (F) Surface representation of the IDE dimer in closed- and swinging-door states; putative door subdomain in the swinging door state, described in D, is placed to show the 11- to 18-Å opening of the IDE catalytic chamber in the swinging door state.
Fab(IDE) mediates extensive intermolecular contacts in crystal packing, resulting in fewer interactions among IDE dimers (Fig. S2C). Fab(IDE) bound IDE D2 via extensive van der Waal interactions and an extensive network of hydrogen bonds (Fig. S4A). The “elbow” angle between the variable and constant domains was significantly different between the two Fab(IDE) in the asymmetric unit, corresponding to a 12-Å rigid body displacement of the COM of the constant domains (Fig. S4B) (26). We speculate that the Fab–Fab contacts and the flexibility between the variable and constant domains of Fab(IDE) have enabled IDE to crystallize in two distinct conformations within a single crystal. This further demonstrates that Fab serves as an effective crystallization chaperone to reveal conformational states that are otherwise difficult to crystallize (22).
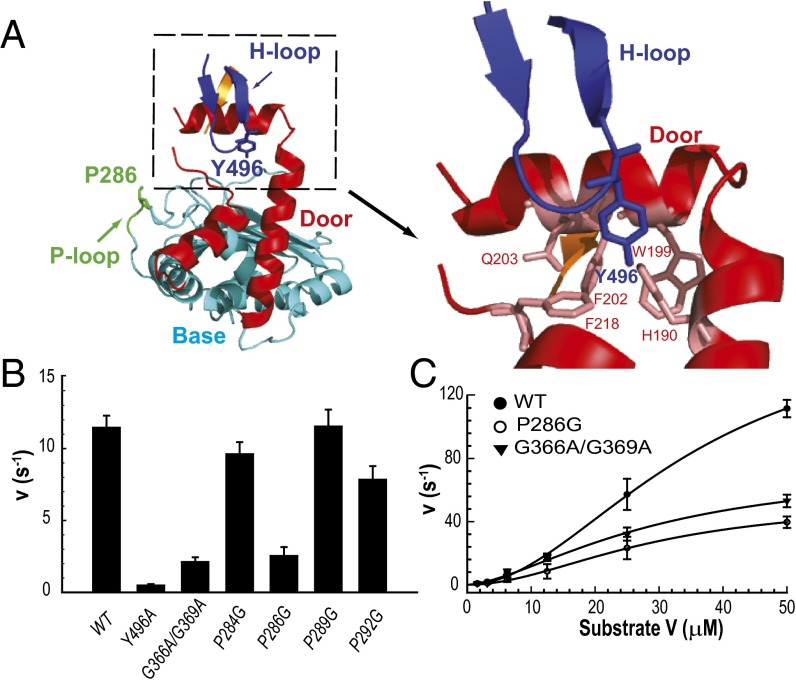
A comparison of chains A and B of the IDE dimer reveals regions exhibiting dramatic differences. There are two subdomains of D1, which we will subsequently refer to as the door (amino acids 170–241) and base (amino acids 47–170 and 242–281), and three loops, which we term the proline-rich (P) loop (amino acids 284–298), glycine-rich (G) loop (amino acids 361–369), and hydrophobic residues at the tip (H) loop (amino acids 490–500) (Figs. 1C and 2 A and B and Fig. S5). All three loops are evolutionarily conserved among vertebrate and insect IDEs (Fig. S6). In chain B, both the door subdomain and P loop are invisible (Fig. 2B and Fig. S5). Importantly, the door subdomain includes E189, which, together with H108 and H112 within the conserved HXXEH motif, binds the catalytic zinc (Fig. 2A), and key hydrophobic residues (W199 and F202) in the catalytic cleft for substrate recognition (Fig. 2 A and C). The comparison also reveals that the base subdomain undergoes a rigid body pendulum motion to move toward the G loop and away from IDE D4 and H loop (3.3-Å COM displacement and 5.4° relative to D2 COM; Fig. 2D and Fig. S7). The door subdomain is connected to the base subdomain, which is linked by the P loop to IDE D2. Thus, the P loop likely drives the motion of base subdomain to move door subdomain away from the H loop so that the hydrophobic contacts between the hydrophobic pocket formed by F202 and W199 of the door subdomain with Y496 in the H loop would then be lost. Consequently, the IDE door subdomain would undergo a rigid-body swing motion, resulting in multiple conformational states to make the door subdomain not visible in chain B (Fig. 2E). Such motion creates an opening of IDE catalytic chamber (Fig. 2F). We thus define chain B of IDE as the “swinging door” state and chain A as the “closed door” state.
Roles of IDE Swinging Door in Substrate Recognition and Rate of Catalysis.
We used mutational studies to examine the role of the swinging door in the catalysis of IDE. As described above, three loops surrounding the door and base subdomains exhibit substantial conformational differences between chain A and B, and thus we predicted that mutations in these loops should affect the kinetic parameters of IDE. The P loop is rich in proline (4 of 15 residues) and connects IDE D1 and D2 (Fig. 2A). It is fully disordered in the IDE swinging door state and partially disordered in the closed state. The restricted dihedral angles of the proline residues could confine how the base subdomain leads the motion of the door subdomain. Indeed, a P loop mutation at residue P286 to glycine (P286G), but not at P284, P289, and P292, resulted in a significant reduction of IDE catalysis by predominantly affecting Vmax without altering the apparent Km or Hill coefficient (∼2) (Fig. 3 B and C and Fig. S8). The H loop contains a highly conserved hydrophobic residue, Y496, which makes substantial contact with the door subdomain in chain A (Fig. 3A) and could act to lock the door subdomain in a catalytically competent state. Consistent with this view, the Y496A mutation dramatically impaired the enzymatic activity of IDE (Fig. 3B and Fig. S8). The G loop is rich in glycine (four of nine residues) and is adjacent to the door subdomain, opposite the H loop. The inherent structural flexibility of glycine could accommodate the swing motion of the door subdomain in the swinging door state of IDE (Fig. 2B). Glycine to alanine mutations within the G loop, which should restrict the freedom of the dihedral angle of residues G366 and G369, also profoundly reduced the catalytic rate of IDE, primarily by reducing Vmax (Fig. 3B and Fig. S8). (IDE G361A/G362A also has reduced enzymatic activity. However, these mutations also dramatically reduce IDE production, and purified IDE G361A/G362A protein has noticeable contamination of IDE fragments, which complicates the interpretation.) Together, these mutation studies support the importance of the three loops and consequently the swinging door mechanism in IDE function.
Fig. 3.
Characterization of IDE loop mutants. (A) Detailed interactions of select residues within the P-loop and H-loop with other subdomains located in IDE D1 at the closed state. (B) Relative activities of IDE (WT) and IDE mutants within the H-loop (Y496A), G-loop (G366A/G369A), and P-loop (P284G, P286G, P289G, P292G) at an enzyme concentration of 6.25 nM. Activity was followed by measuring fluorescence resulting from the cleavage of 0.5 μM Substrate V at 37 °C. (C) Comparison of the kinetics of IDE (WT) with P-loop (P286G) or G-loop (G366A/G369A) mutations at varying concentrations of Substrate V, as indicated. A sigmoidal curve was fit to each respective data set, with Vmax (170, 53, 76 s−1), Km (35, 29, 30 μM), and the Hill coefficient (2.0, 1.9, 1.6) calculated for WT, P286G, and G366A/G369A, respectively, with mean ± SD shown (n = 3).
The swinging door motion offers a possible mechanism for the preferential degradation of amyloidogenic peptides by IDE. The salient feature of amyloidogenic peptides (e.g., Aβ) is their high propensity to convert to a β-stranded structure, self-assemble into an intermolecular cross β-sheet, and thus form insoluble amyloid fibrils (4). Our previous structures of the IDE dimer in complex with Aβ and amylin reveal that regions of amyloidogenic peptides that form a β-strand in amyloids docks onto β6 within the IDE door subdomain to form an intermolecular, antiparallel β-sheet (Fig. 2C) (7, 8). Because the door subdomain contains residues crucial for binding catalytic zinc and forming the catalytic cleft, the swing motion prevents IDE from hydrolyzing its substrates (Fig. 2 A and E). We hypothesize that substrates within the catalytic chamber can only be degraded when the IDE door subdomain is locked in place. The high propensity of amyloidogenic peptides to adopt a β-strand would thus allow these peptides to form the intermolecular β-sheet with β6 and thus stabilize the door subdomain. This mechanism leads to the recognition of the key feature of amyloidogenic peptides.
Conformational States of IDE in Solution.
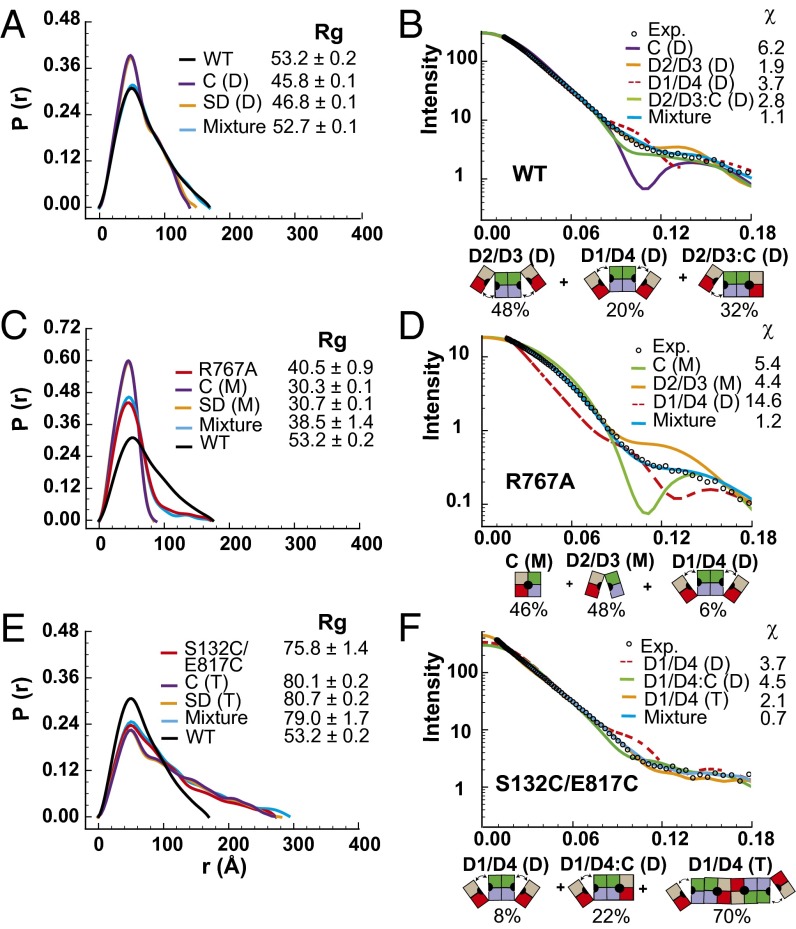
Because the ∼11- to 18-Å opening in the swinging door state will not allow IDE to capture large substrates such as insulin, other conformational state(s) with larger opening of the catalytic cavity must exist in the catalytic cycle of IDE (Fig. S5). Thus, we probed the global shape of IDE using small angle X-ray scattering (SAXS) to assess the presence of such conformation(s) (Fig. 4). Because the KD for IDE dimerization is ∼10 nM, WT IDE should exist mostly as a dimer under our conditions for SAXS (9 μM) (9). We found that the radius of gyration (Rg) of WT IDE was 53 Å with Dmax = 175 Å, noticeably larger than those predicted using the closed or swinging door state of the IDE dimer (∼46 Å) (Fig. 4A). We also observed high χ values (5.4–6.4), and the lack of the intensity dip along the q range predicted for a population of dimers exclusively in the closed conformation (Fig. 4B and Fig. S9). Thus, WT IDE in solution did not exist predominantly in the closed state. We also analyzed two IDE mutants, R767A and S132C/E817C. Because the R767A mutation disrupts the dimerization of IDE, IDE R767A had an Rg value (40 Å) smaller than that of WT IDE, but a comparable Dmax value. This agrees with biochemical data that IDE R767A exists mostly as a monomer, but some dimers can still form (Fig. 4C) (10). IDE S132C/E817C introduces a disulfide bond to lock IDE D1 and D4 together, which only becomes active under reducing conditions (7). The Rg value for IDE S132C/E817C was much larger (76 Å) than that of WT IDE (Fig. 4E). It is known that insulin can induce IDE dimers to form a tetramer by facilitating a substrate-induced switch to the closed state (9). Thus, the ability of S132C/E817C to preferentially stay in the closed state may promote tetramerization and thereby explain the increase in Rg value. Thus, we also found that SAXS data of neither R767A nor S132C/E817C fit the models of the closed states (Fig. 4 C–F and Fig. S9).
Fig. 4.
SAXS analysis of IDE. Pair distribution functions and scattering curves of WT IDE (A and B) and IDE R767A (C and D), and IDE S132C/E817C (E and F). Curve fitting is based on atomic models using the program CRYSOL (single model) or OLIGOMER (mixture). A protein concentration of 1 mg/mL was used for the calculation of all scattering profiles. D2/D3, D2/D3 pivot; D1/D4, D1/D4 pivot; C, closed state; M, monomer; D, dimer; T, tetramer. The diagrams and ratios shown below the scattering profiles represent the distribution of mixture that could best fit the SAXS data.
We then used our SAXS data to assess the conformational states of IDE in solution by modeling the rigid body motion between IDE-N and IDE-C on the basis of structures of the M16 family (Fig. S1). We found that a D2/D3 pivot model fit the SAXS data of WT IDE well (Fig. 4B), which had the same degree of opening as depicted in an IDE homolog, E. coli pitrilysin (pdb = 1q2l; χ = 1.9). Models with small variations in translation or rotation for the pivoting between IDE D2 and D3 could still fit the data with similar χ values (Fig. S9). Such D2/D3 pivot models would allow IDE to capture larger substrates such as insulin and Aβ (Fig. S5). We also tested models with various degrees of pivoting between the IDE D1 and D4 that must exist in the disulfide bond-locked IDE S132C/E817C (D1/D4 pivot) and incremental increases of separation between IDE-N and IDE-C via a parallel-spring motion (Fig. S9). However, only a few models generated a reasonable fit (χ = 2.5 for best model of parallel spring; χ = 3.7 for D1/D4 pivot), and none fit better than that of D2/D3 pivot.
We then tested whether the use of multiple models to account for multiple conformation states in solution would allow a better fit. Although no improvement could be made with any combination of two states, the mixture of three IDE dimeric states (D2/D3 pivot, D1/D4 pivot, and a mixed D2/D3 pivot and the closed state in which the door subdomain is either closed or in the swinging state) significantly improved the fit to the data of WT IDE (χ = 1.1; Fig. 4 A and B). We use the closed/swinging door state here because the swinging and closed door states generate nearly indistinguishable SAXS profiles (Fig. 4 A, C, and F). We did not find good fit with any single state to SAXS data of R767A or S132C/E817C. However, the mixture of largely D2/D3 pivot and closed/swinging door monomeric states could also fit the data of IDE R767A well (χ = 1.2; Fig. 4 C and D). The mixture of IDE dimers and tetramers that are in the D1/D4 pivot and closed/swinging door states also generated a good fit for IDE S132C/E817C (χ = 0.7; Fig. 4 E and F). These data suggest that IDE in solution adopts multiple conformations, including the closed/swinging door, D2/D3 pivot, and D1/D4 pivot states. This is consistent with the dynamic nature of proteins in general and conformational diversity revealed by structures of M16B bacterial protease (16, 27) (Fig. S1). Because many possible states may exist and SAXS data have limited resolution, the precise states and dynamics of various conformations of IDE in solution await future investigation.
Together, our studies reveal that IDE exists as a mixture of closed and open conformations in solution and has at least two types of motions: the swinging door in IDE D1 and pivoting between IDE-N and IDE-C (Fig. 5A). We speculate that the swinging door motion can occur in both closed and open states of IDE. The swinging door motion could explain why IDE has a higher Vmax for shorter peptides (e.g., BK) than for larger peptides (e.g., insulin and Aβ) (Fig. 5A). The rate of insulin degradation by IDE is ∼10 s−1; however, the estimated rate of BK degradation is at least 20-fold higher (3, 9, 28). The door swing motion creates an ∼11- to 18-Å opening (Fig. 2F), which is only large enough for short or unfolded peptides like BK to enter the catalytic chamber, whereas larger peptides such as insulin are excluded. Larger substrates would only be able to enter the catalytic chamber of IDE by a mechanism distinct from the opening created by the swinging door, likely by the pivoting between IDE-N and IDE-C, as described below. Thus, conformational transitions of IDE from the closed to open states for substrate capture followed by substrate unraveling inside the closed catalytic chamber are required for IDE to degrade larger peptides. In contrast, the smaller conformational transition between the swinging and closed door states should be sufficient for IDE to capture and degrade smaller peptides, which should occur more frequently.
Fig. 5.
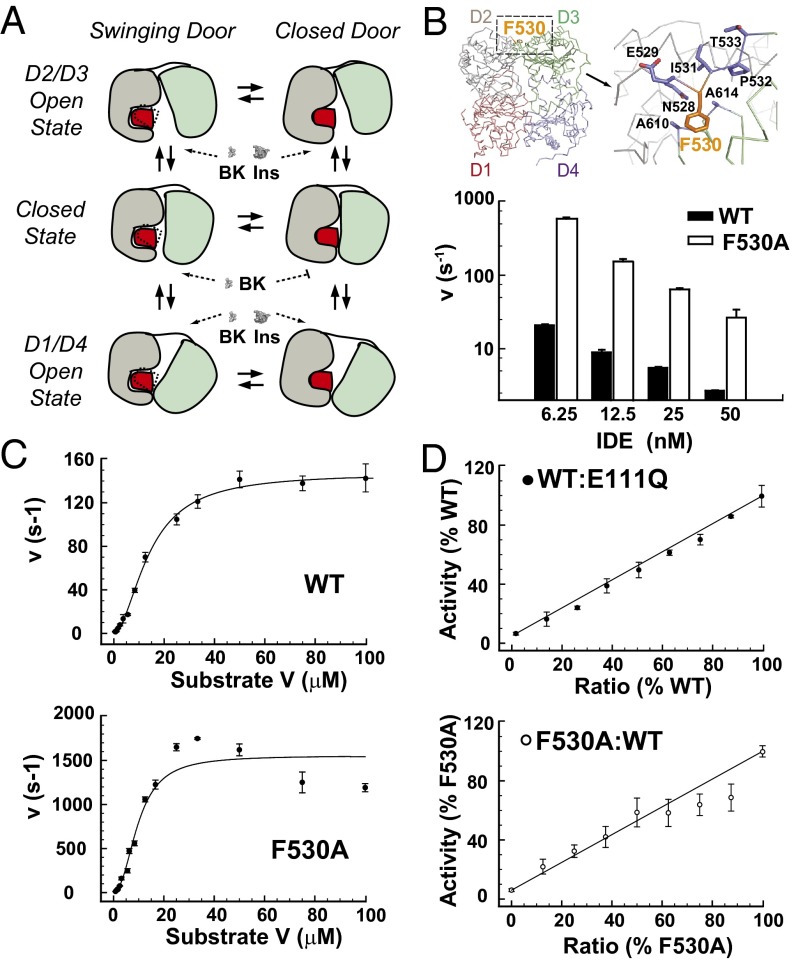
Characterizations of IDE F530A. (A) Model of motions of IDE monomer. Ins, insulin. (B) Detailed interaction of residue F530 in the linker region between IDE-N and IDE-C. Activity was followed by measuring fluorescence resulting from the cleavage of 0.5 μM Substrate V at 37 °C using varied concentrations of protein (6.25–50 nM), with lower enzyme concentrations yielding the greatest change in relative activity. (C) Comparison of kinetic rate of WT to the F530A (6.25 nM) linker mutation at varying concentrations of Substrate V, as indicated. A sigmoidal curve was fit to the WT data set with Vmax, Km, and Hill coefficient (150 s−1, 14 μM, 1.8) calculated, respectively, with a high goodness of fit (R2 = 0.99). Because of the biphasic kinetic nature of the linker region mutation, a hyperbolic curve was fit to F530A protein with two distinct Vmax (1,500, 15 s−1) and Km (10, 0.8 μM) values and a Hill coefficient of 2.3 (R2 = 0.97). (D) WT and F530A IDE (25 nM) were mixed together and with a catalytically inactive mutant, E111Q, as a control at various ratios. Means ± SD represent at least three experiments.
Role of the Hinge Region Between IDE-N and IDE-C in Catalysis.
Catalytic activity and substrate selectivity of IDE is allosterically regulated by its oligomerization (dimer/tetramer), binding to substrates, ATP, and cellular partners (3, 9, 21, 28–31). For example, the mixed oligomer of WT and mutant IDE can exhibit enzyme kinetics distinctly different from the homogeneous counterparts, suggesting cross-talk between subunits (31). ATP binds the catalytic chamber of IDE-C to selectively accelerate the degradation of the short peptides (e.g., substrate V) (21, 30). Nestin, an intermediate filament protein, can enhance the degradation of substrate V by IDE while suppressing degradation of insulin (29). IDE-N and IDE-C made separately can come together for catalysis without a physical linker (20). This raises the question whether the hinge loop between IDE D2 and D3 contributes to IDE catalysis. We postulated that the conformational switches of IDE monomer shown in Fig. 5A can occur within the oligomerized IDE and the hinge loop is critical for the proper pivoting motion between IDE-N and IDE-C to regulate the allostery of IDE.
To examine the role of linker region between IDE-N and IDE-C, we performed alanine-scanning mutagenesis on the hydrophobic residues within the range of residues 497–560. We found that mutation of the partially buried F530 residue to alanine (F530A) rendered IDE hyperactive, with up to a 20-fold enhancement in degrading substrate V (Fig. 5B). We then examined whether the hinge loop mutation could alter allosteric regulation of IDE. Similar to the WT enzyme, kinetic analysis using substrate V as a model substrate revealed that F530A has a Hill coefficient of ∼2 (Fig. 5C). However, the F530A mutant presented a biphasic kinetic curve best fit by two distinct Km and Vmax values (Fig. 5C), suggesting that the F530A mutation confers distinct kinetic parameters to each monomer within an IDE dimer. If so, combining F530A and WT IDE would create a mixed dimer with a catalytic rate that deviates from the prediction based on the line drawn between the activities of WT IDE and F530A (Fig. 5D). Indeed, we observed noticeable and progressively lower rates than predicted when the ratio of F530A to total IDE went from 62% to 88% (Fig. 5D). As a control, the mixing of WT IDE with the catalytically inactive mutant E111Q did not result in deviated rates of catalysis (Fig. 5D). Hence, our data suggest that the hinge loop could indeed modulate the communication between active sites, and hence the allostery of IDE.
In summary, the swinging door motion uncovered by this study significantly advances our mechanistic understanding of IDE function. The motion permits IDE to recognize the key feature of amyloidogenic peptides, which tend to convert to β-strands for the formation of intermolecular cross-β sheet; such β-strands stabilize the door subdomain, leading to their degradation by IDE. Furthermore, the swinging door motion permits the entrance of only short peptides, thus explaining the higher kcat of IDE for shorter peptides (e.g., BK) compared with those for longer peptides (e.g., insulin and Aβ). Furthermore, each chain within an IDE dimer can adopt distinct conformations [e.g., the swinging door and closed states in our IDE-Fab(IDE) complex]. Such conformational diversity and structural dynamics of IDE fit well with the intricate allostery of IDE. Our finding of the hyperactive mutation at the hinge loop also suggests a new way to boost IDE catalysis. Because IDE is a key protease in the destruction of amyloidogenic peptides (12) and novel biologically relevant substrates of IDE, such as MIP-1α and calcitonin gene-related peptide, continue to be discovered (11, 32), future studies will address how conformational dynamics are linked to the catalytic cycle of IDE and how to control such processes. Such information can also provide avenues to design IDE-based therapies for modulating proteostasis in humans.
Methods
Construction, Expression, and Purification of Human IDE and IDE Mutants.
Human IDE mutants were generated by site-directed mutagenesis, and proteins were expressed in E. coli Rosetta (DE3) as previously described (9). His-tagged, biotinylated IDE was expressed in E. coli BL21 (DE3) that carried two plasmids, one for IDE with an N-terminal His-tag and a C-terminal AviTag (GLNDIFEAQKIEWHE), and the other for E. coli BirA, a biotin ligase that transfers Biotin to AviTag. IDE proteins were purified using Ni-NTA, source-Q, and Superdex 200 columns.
Identification, Purification, and Characterization of an IDE-Binding Synthetic Antibody, Fab(IDE).
We isolated an IDE-binding synthetic antibody as the antigen-binding fragment (Fab) from a phage-display library as previously described, using 100, 100, 50, and 20 nM biotinylated IDE for sorting in rounds 1, 2, 3, and 4, respectively (33). Fab was expressed in E. coli strain 55244 and purified using a HiTrap protein G HP column, as previously described (25). Surface plasmon resonance measurements were carried out at 20 °C on a Biacore 3000 by immobilizing His-tagged IDE onto a Ni-NTA chip and then injecting 1.2–100 nM of Fab at a flow rate of 30 μL/min (25, 34).
Crystallization and Structure Determination of IDE-Fab(IDE) Complex.
IDE-Fab(IDE) complex was formed by mixing equimolar Fab(IDE) and IDE, purified by an S200 column, and crystallized in 0.1 M sodium cacodylate (pH 6.5), 0.2 M MgCl2, and 10% (vol/vol) PEG-3000 at 18 °C by hanging drop vapor diffusion. For data collection, crystals were equilibrated in reservoir buffer with 30% (vol/vol) glycerol and flash frozen in liquid nitrogen. Diffraction data were collected at 100K on the 19-ID beamline at Argonne National Laboratory. Data sets were processed using HKL2000 and the CCP4 suite, and the structure of IDE-Fab(IDE) complex was determined by molecular replacement, using the cysteine-free IDE-E111Q structure (3CWW) and Fab fragment in Fab-bound KcsA (3PJS) as search models. Model building and refinement were performed by using REFMAC, PHENIX, and COOT without NCS restraints of IDE or Fab(IDE). The final model (pdb = 4IOF) has Rwork = 23% and Rfree = 28%.
Enzymatic Assays.
Enzymatic activity of IDE was measured by monitoring cleavage of a fluorogenic BK-mimetic substrate of IDE, substrate V (7-methoxycoumarin-4-yl-acetyl-RPPGF-SAFK-2,4-dinitrophenyl; R&D Systems), on a Tecan Safire microplate reader using an excitation wavelength of 327 nm and emission wavelength of 395 nm (7, 30). Reactions were carried out at 37 °C, using 100 µL of indicated concentrations of substrate V in 50 mM potassium phosphate buffer (pH 7.3) with addition of 1 µL of IDE protein (6–25 nM) to initiate the reaction. Degradation of substrate V was assessed by monitoring fluorescence increase for 20 min at 10-s intervals, with nine reads per well per time point. To calculate enzymatic activity, background subtraction and linear regression fitting were used to find initial velocity, whereas specific activity (s−1) was determined by comparing the maximal fluorescence converted from the known quantity of substrate V by IDE.
SAXS Data.
SAXS data were collected at the 18-ID (BioCAT) beamline using the Mar 165 CCD detector at room temperature (23 °C) and an incident X-ray wavelength of 1.033 Å. All data processing was performed using IGOR Pro with macros written by the BioCAT staff and further analyzed with GNOM (35). CRYSOL was used to calculate solution scattering of models and to fit these models to experimental scattering data (36), whereas OLIGOMER was used to estimate distribution of each conformational state (37).
Supplementary Material
Acknowledgments
We thank Qing Guo for his technical assistance, the staff of the Advanced Photon Source Structural Biology Center and Biophysics Collaborative Access Team in assisting data collection, and Tobin Sosnick for his helpful comments on SAXS data analysis. This work was supported by National Institutes of Health Grants GM81539 (to W.-J.T.) and GM072688 (to S.K.), and an American Health Assistance Foundation grant (S.K.). Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Basic Energy Sciences, under Contract W-31-109-ENG-38.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4IOF).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304575110/-/DCSupplemental.
References
- 1.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 2.De Strooper B. Proteases and proteolysis in Alzheimer disease: A multifactorial view on the disease process. Physiol Rev. 2010;90(2):465–494. doi: 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- 3.Malito E, Hulse RE, Tang WJ. Amyloid β-degrading cryptidases: Insulin degrading enzyme, presequence peptidase, and neprilysin. Cell Mol Life Sci. 2008;65(16):2574–2585. doi: 10.1007/s00018-008-8112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 5.Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: The AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- 6.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 7.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443(7113):870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo Q, Manolopoulou M, Bian Y, Schilling AB, Tang WJ. Molecular basis for the recognition and cleavages of IGF-II, TGF-alpha, and amylin by human insulin-degrading enzyme. J Mol Biol. 2010;395(2):430–443. doi: 10.1016/j.jmb.2009.10.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manolopoulou M, Guo Q, Malito E, Schilling AB, Tang WJ. Molecular basis of catalytic chamber-assisted unfolding and cleavage of human insulin by human insulin-degrading enzyme. J Biol Chem. 2009;284(21):14177–14188. doi: 10.1074/jbc.M900068200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ralat LA, et al. Insulin-degrading enzyme modulates the natriuretic peptide-mediated signaling response. J Biol Chem. 2011;286(6):4670–4679. doi: 10.1074/jbc.M110.173252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren M, et al. Polymerization of MIP-1 chemokine (CCL3 and CCL4) and clearance of MIP-1 by insulin-degrading enzyme. EMBO J. 2010;29(23):3952–3966. doi: 10.1038/emboj.2010.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurochkin IV. Insulin-degrading enzyme: Embarking on amyloid destruction. Trends Biochem Sci. 2001;26(7):421–425. doi: 10.1016/s0968-0004(01)01876-x. [DOI] [PubMed] [Google Scholar]
- 13.Johnson KA, et al. The closed structure of presequence protease PreP forms a unique 10,000 Angstroms3 chamber for proteolysis. EMBO J. 2006;25(9):1977–1986. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor AB, et al. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure. 2001;9(7):615–625. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- 15.Eggleson KK, Duffin KL, Goldberg DE. Identification and characterization of falcilysin, a metallopeptidase involved in hemoglobin catabolism within the malaria parasite Plasmodium falciparum. J Biol Chem. 1999;274(45):32411–32417. doi: 10.1074/jbc.274.45.32411. [DOI] [PubMed] [Google Scholar]
- 16.Aleshin AE, et al. Crystal and solution structures of a prokaryotic M16B peptidase: An open and shut case. Structure. 2009;17(11):1465–1475. doi: 10.1016/j.str.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maruyama Y, Chuma A, Mikami B, Hashimoto W, Murata K. Heterosubunit composition and crystal structures of a novel bacterial M16B metallopeptidase. J Mol Biol. 2011;407(1):180–192. doi: 10.1016/j.jmb.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 18.Ohtsuka J, Ichihara Y, Ebihara A, Nagata K, Tanokura M. Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase. Proteins. 2009;75(3):774–780. doi: 10.1002/prot.22365. [DOI] [PubMed] [Google Scholar]
- 19.Xia DD, et al. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277(5322):60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li P, Kuo WL, Yousef M, Rosner MR, Tang WJ. The C-terminal domain of human insulin degrading enzyme is required for dimerization and substrate recognition. Biochem Biophys Res Commun. 2006;343(4):1032–1037. doi: 10.1016/j.bbrc.2006.03.083. [DOI] [PubMed] [Google Scholar]
- 21.Noinaj N, et al. Anion activation site of insulin-degrading enzyme. J Biol Chem. 2012;287(1):48–57. doi: 10.1074/jbc.M111.264614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koide S. Engineering of recombinant crystallization chaperones. Curr Opin Struct Biol. 2009;19(4):449–457. doi: 10.1016/j.sbi.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koldobskaya Y, et al. A portable RNA sequence whose recognition by a synthetic antibody facilitates structural determination. Nat Struct Mol Biol. 2011;18(1):100–106. doi: 10.1038/nsmb.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uysal S, et al. Crystal structure of full-length KcsA in its closed conformation. Proc Natl Acad Sci USA. 2009;106(16):6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang X, et al. Identification of a tetratricopeptide repeat-like domain in the nicastrin subunit of γ-secretase using synthetic antibodies. Proc Natl Acad Sci USA. 2012;109(22):8534–8539. doi: 10.1073/pnas.1202691109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilson IA, Stanfield RL. Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol. 1994;4(6):857–867. doi: 10.1016/0959-440x(94)90267-4. [DOI] [PubMed] [Google Scholar]
- 27.Henzler-Wildman K, Kern D. Dynamic personalities of proteins. Nature. 2007;450(7172):964–972. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 28.Malito E, et al. Molecular bases for the recognition of short peptide substrates and cysteine-directed modifications of human insulin-degrading enzyme. Biochemistry. 2008;47(48):12822–12834. doi: 10.1021/bi801192h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chou YH, Kuo WL, Rosner MR, Tang WJ, Goldman RD. Structural changes in intermediate filament networks alter the activity of insulin-degrading enzyme. FASEB J. 2009;23(11):3734–3742. doi: 10.1096/fj.09-137455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Im H, et al. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem. 2007;282(35):25453–25463. doi: 10.1074/jbc.M701590200. [DOI] [PubMed] [Google Scholar]
- 31.Song ES, Rodgers DW, Hersh LB. Mixed dimers of insulin-degrading enzyme reveal a cis activation mechanism. J Biol Chem. 2011;286(16):13852–13858. doi: 10.1074/jbc.M110.191668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YG, Lone AM, Nolte WM, Saghatelian A. Peptidomics approach to elucidate the proteolytic regulation of bioactive peptides. Proc Natl Acad Sci USA. 2012;109(22):8523–8527. doi: 10.1073/pnas.1203195109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller KR, et al. T cell receptor-like recognition of tumor in vivo by synthetic antibody fragment. PLoS ONE. 2012;7(8):e43746. doi: 10.1371/journal.pone.0043746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koide A, Wojcik J, Gilbreth RN, Hoey RJ, Koide S. Teaching an old scaffold new tricks: Monobodies constructed using alternative surfaces of the FN3 scaffold. J Mol Biol. 2012;415(2):393–405. doi: 10.1016/j.jmb.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Svergun DI. Mathematical methods in small-angle scattering data analysis. J Appl Cryst. 1991;24(5):485–492. [Google Scholar]
- 36.Svergun D, Barberato C, Koch M. CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28(6):768–773. [Google Scholar]
- 37.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36(5):1277–1282. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.