Abstract
Rubber oxygenase A (RoxA) is one of only two known enzymes able to catalyze the oxidative cleavage of latex for biodegradation. RoxA acts as a processive dioxygenase to yield the predominant product 12-oxo-4,8-dimethyl-trideca-4,8-diene-1-al (ODTD), a tri-isoprene unit. Here we present a structural analysis of RoxA from Xanthomonas sp. strain 35Y at a resolution of 1.8 Å. The enzyme is a 75-kDa diheme c-type cytochrome with an unusually low degree of secondary structure. Analysis of the heme group arrangement and peptide chain topology of RoxA confirmed a distant kinship with diheme peroxidases of the CcpA family, but the proteins are functionally distinct, and the extracellular RoxA has evolved to have twice the molecular mass by successively accumulating extensions of peripheral loops. RoxA incorporates both oxygen atoms of its cosubstrate dioxygen into the rubber cleavage product ODTD, and we show that RoxA is isolated with O2 stably bound to the active site heme iron. Activation and cleavage of O2 require binding of polyisoprene, and thus the substrate needs to use hydrophobic access channels to reach the deeply buried active site of RoxA. The location and nature of these channels support a processive mechanism of latex cleavage.
Keywords: natural rubber, isoprenoids, protein crystallography, c-type cytochromes, dioxygenases
Natural rubber, or coutchouc, is a hydrocarbon biopolymer with a long history of use in a wide range of industrial and technical applications. The most significant source of natural rubber is the sap of the rubber tree, Hevea brasiliensis, which has been commercially cultivated for more than a century. Different types of rubber are also produced by other plants and fungi, and although synthetic rubbers have become available, the natural product still constitutes the most important raw material for the latex industry.
The latex milk of H. brasiliensis contains 25–35% of a high molecular weight polyisoprene with some 100–10,000 cis-1,4-isoprene units per molecule. Hevea polyisoprenes are presumably terminated by dimethylallyl groups at both ends and by two trans-conjugated isoprene units next to one unit of the dimethylallyl end (1–4). Latex, used by the plant to seal injuries of the bark and to kill invading insects by capturing them in coagulated rubber plugs, is a resilient and stable polymer, and the mere fact that it does not accumulate in nature is evidence of the existence of pathways for its biodegradation.
Despite the economic importance of rubber and the sheer amounts produced and used in industry, knowledge of the mechanistic basis of latex biodegradation is limited. Different rubber-degrading microorganisms have been described, some of which have been isolated and characterized (5–8). Two of these microorganisms, Streptomyces sp. K30 (9) and Xanthomonas sp. 35Y (7), form characteristic clearing zones around the colonies when grown on latex agar. The latex-degrading enzymes have been identified in both cases. They do not demonstrate recognizable sequence homologies, and thus likely represent two convergent solutions to the same biochemical problem. The latex-clearing protein (Lcp) (9, 10) from Streptomyces sp. K30 is a 43-kDa polypeptide without a known cofactor or prosthetic group, whereas the rubber oxygenase RoxA from Xanthomonas sp. 35Y is a 75-kDa c-type cytochrome with two heme groups (11, 12). In the latter, the porphyrins are covalently attached via thioether bonds to the cysteine residues of heme attachment motifs of sequence CXXCH. RoxA has been isolated and its spectroscopic properties have been assessed by UV/vis and electron paramagnetic resonance (EPR) spectroscopy (13).
Formation of a clearing zone depends on secretion of RoxA into the medium, where it catalyzes the oxidative cleavage of the polyisoprene molecule, notably with the tri-isoprene 12-oxo-4,8-dimethyl-trideca-4,8-diene-1-al (ODTD) as the predominant cleavage product (Fig. 1B) (14). This reaction is slow and lacks specificity, with minor amounts of other products with between two and six isoprene units detected as well (11). All such products contained carbonyl functions, as aldehydes or ketones, at the termini, and 18O-labeling studies verified that both oxygen atoms incorporated into the polymer during cleavage are derived from O2 rather than from water, making RoxA a functional dioxygenase (14). The predominant formation of a triisoprenoid product points toward an internal mechanism for measuring chain length (a so-called “molecular ruler”). Thus, the reactivity of RoxA is most likely processive, that is, the polyisoprene chain is degraded linearly from one end.
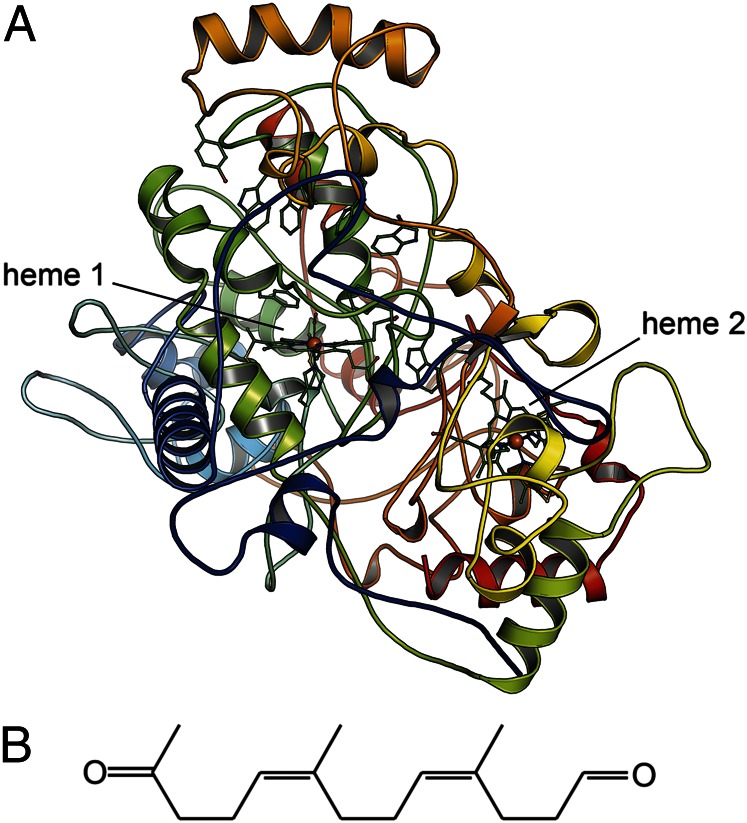
Fig. 1.
Reaction product and structure of Xanthomonas sp. RoxA. (A) The monomeric RoxA in cartoon representation, colored from blue at the N terminus to red at the C terminus. The two heme groups and functionally relevant residues are rendered as sticks. (B) The rubber dioxygenase RoxA cleaves linear poly-cis-1,4-isoprene into trimeric units of ODTD.
To obtain insight into the functionality of RoxA, we crystallized the protein (15) and determined its 3D structure to a resolution of 1.8 Å, using multiwavelength anomalous dispersion methods for solving the crystallographic phase problem. RoxA is a unique type of dioxygenase with an unusual fold and a series of obvious adaptations to achieve its unique functionality.
Results
Xanthomonas RoxA Is a Globular Enzyme with an Unusually Low Amount of Secondary Structure.
Crystals of RoxA belong to the monoclinic space group P21, with two monomers per asymmetric unit, comprising residues 15–662, that are identical within the limits of experimental error (rmsd for all atom positions, 0.14 Å). With the exception of a small, two-stranded β-sheet composed of residues 420–422 and 500–502, the secondary structure is α-helical, but the proportion of secondary structures with respect to all of the loop regions within the protein is unusually small. More than two-thirds (67.3%) of the RoxA polypeptides are organized as loops, whereas only 31.6% form helices (Fig. 1A). As detailed below, numerous loop regions form extended patches with no or little secondary structure elements that cover the protein surface, and thus discerning the internal domain structure of RoxA is not straightforward. The presence of two disulfide bridges (C32–C88 and C574–C586) within these surface loops is uncommon in c-type cytochromes. The disulfide bonds are important for RoxA activity, because chemical reduction almost completely inhibits the enzyme (11).
The high sensitivity of RoxA activity to ionic and nonionic detergents (11) may be explained by the low resilience of these surface loops. The two heme groups are buried within the protein matrix and spaced at an Fe–Fe distance of 21.4 Å, which by itself precludes efficient direct electron transfer. Heme group 1 is bound to the polypeptide chain via thioether bonds to the Sγ atoms of residues C191 and C194 within the first heme-binding motif. H195 serves as a proximal axial ligand to the heme iron, whereas the distal axial position is not occupied by a protein ligand. The second heme is attached to the C-terminal heme-binding motif, with C390 and C393 forming thioether bonds and H394 serving as a proximal axial ligand. This heme group features the bis-histidinyl coordination typically seen in low-potential c-type hemes involved in electron transfer processes, with H641 as a distal axial ligand.
Midpoint redox potentials for the two heme groups are –65 mV and –130 to –160 mV, respectively (13). These values are in stark contrast to the potential difference of 650 mV observed for CcpA peroxidases (16), but lie within the range seen for MauG (–159 to –244 mV) (17). Such similar potential values lead to efficient electronic coupling of the centers, hindering unambiguous assignment of the values to individual hemes.
RoxA Is Evolutionarily Derived from CcpA Peroxidases.
Although our database searches did not reveal any obvious homologs of RoxA, signature motifs of another class of diheme c-type cytochromes, the CcpA/MauG family, have been reported previously (12). CcpA peroxidases are dimeric hydroperoxide reductases with an intricate activation mechanism involving a structural rearrangment of three loop regions on reduction of one heme group, making the substrate binding site at the distal axial position of the other heme group accessible for H2O2 binding (16). The monomeric MauG, although structurally similar to CcpA, is not a metabolic enzyme, but rather a maturation factor for methylamine dehydrogenase (MADH), where it is involved in the formation of the cofactor tryptophan tryptophylquinone (TTQ) (18, 19). Neither enzyme acts as a dioxygenase, and both enzymes are only approximately half the size of RoxA.
Structural superposition of RoxA with a representative diheme peroxidase, such as MacA (20) or CcpA (21) from Geobacter sulfurreducens, showed that despite the absence of recognizable sequence similarities, the folding cores of both proteins aligned very closely. Differences between RoxA (Fig. 2A) and G. sulfurreducens CcpA (Fig. 2B) manifested in all connecting loop regions, leading to drastically altered morphology of the two proteins, with significant consequences in terms of their functional properties. The most obvious of these consequences were seen in three long insertions in loop regions of RoxA that topologically corresponded to the loops that undergo conformational changes on reductive activation of CcpA peroxidases. In RoxA, we observed no rearrangement of these loops in a dithionite-reduced crystal structure obtained from recombinant protein. The loops form an elevated, tower-like structure above the active site heme group and thus are ideally placed for a key role in interaction with the insoluble substrate.
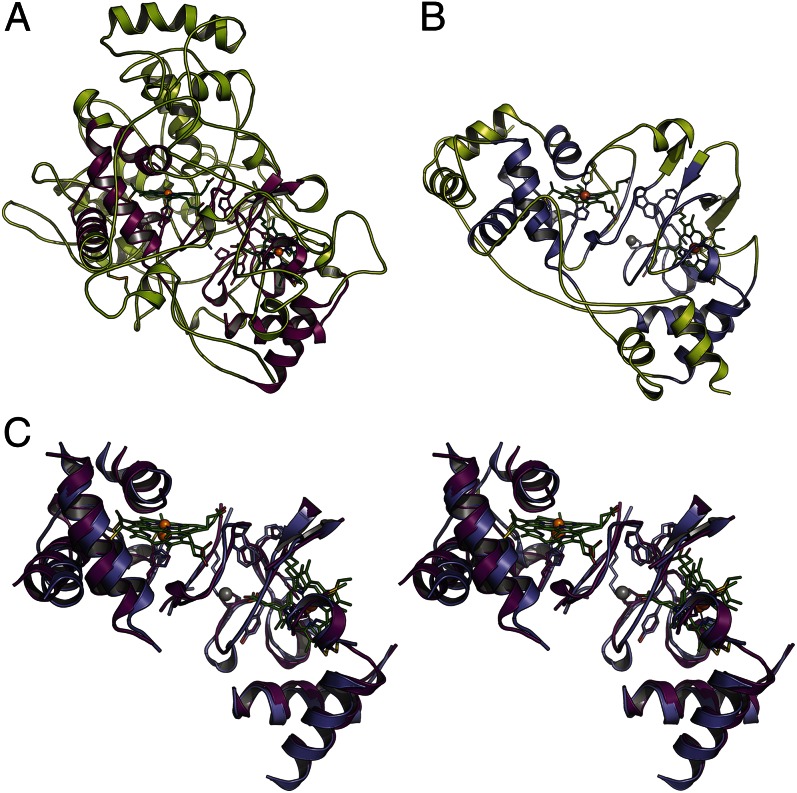
Fig. 2.
RoxA and CcpA peroxidases share a common ancestry. (A) In RoxA, the conserved folding core shared with CcpA peroxidases forms only the central part of the molecule, whereas nearly the entire protein surface is decorated with extended-loop regions. (B) In the peroxidase CcpA from G. sulfurreducens, this core composes most of the protein structure. (C) Stereo representation of the conserved core in RoxA (violet) and CcpA (purple), showing that disconnected peptide fragments almost completely retain their spatial orientation.
The core regions of RoxA and CcpA were aligned with an overall rmsd of 1.2 Å for all atoms (Fig. 2C). This conserved region included stretches of elongated protein backbone and characteristic residues, such as W302, which is located at this very position in all CcpA and MauG orthologs characterized to date, where it provides an electron conduit bridging the relatively large interheme distance (16, 22). The rigidity of this central part of the protein obviously provides a platform for the highly divergent evolution of all external loops. In RoxA, these loop regions cover almost the entire protein surface, and whereas the two aforementioned disulfide bridges help stabilize the overall fold of the enzyme, the progressive, evolutionary process of loop extension has likely precluded the formation of ordered secondary structures. As a result, the extracellular enzyme RoxA is indeed optimized for stability, despite its remarkable absence of helical regions and pleated sheets.
Active Site of RoxA Is Located at the Distal Axial Position of a Pentacoordinate Heme Group.
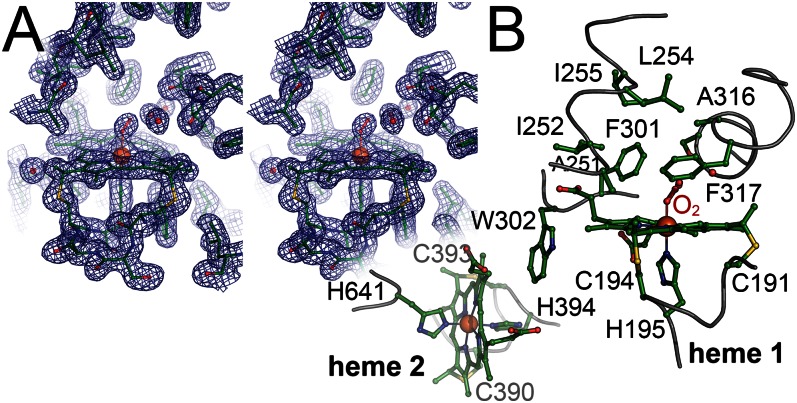
Heme group 1 of RoxA has residue H195 as a proximal axial ligand to the heme iron, and the accessible distal axial position is a suggested candidate for the active site that also corresponds to the hydrogen peroxide-binding site in CcpA peroxidases and to the O2-binding site in MauG. Heme group 1 exhibits a pronounced saddling distortion from planarity (Fig. 3A). An extended internal cavity surrounds the putative active site, lined by the hydrophobic residues F317, A251, I252, F301, L254, I255, and A316 (Fig. 3B). This provides sufficient space to accommodate three water molecules that are visible as well-defined electron density maxima (Fig. 3A and Fig. S1). The water molecules form hydrogen bonds with the backbone carbonyl of Glu314 and the backbone amide of Phe317 (W1); the backbone carbonyl of Ser313, W3, and the distal axial ligand at heme 2 (W2); and the β-hydroxy group of Ser313, the Nε2 atom of Gln300, and W2 (W3). At the same time, no amino acid side chains of RoxA are sufficiently close to the distal axial coordination position to interact with a bound ligand (Fig. 3B). The closest amino acid (5 Å) to the distal heme iron, Phe317, was previously shown to be important for RoxA activity (23).
Fig. 3.
Heme group environment in RoxA. (A) Stereo representation of the distal pocket at heme group 1 with a bound O2 molecule. The displayed 2Fo – Fc electron density map is contoured at the 1 σ level. (B) Amino acid residues at and around the two heme groups. Three regions of the protein form a spacious, hydrophobic cavity above the distal side of heme 1. W302, a residue also conserved in CcpA peroxidases, bridges the hemes. Heme group 1 is linked to the protein via C191 and C194, with H195 as a proximal axial ligand, and heme group 2 is attached through C390 and C393, with H394 as a proximal axial ligand and H641 as a distal axial ligand.
Structural and Spectroscopic Data Indicate Bound O2 at the Active Site Heme.
Electron density maps consistently indicated the presence of a small molecule at the distal axial position (Fig. 3). Owing to its free coordination site and partially occupied 3d shell, the heme iron in heme group 1 is well suited for binding the diradical O2. Reversible O2 binding to heme is known from hemoglobins and their relatives, and the bound molecule can be reductively activated to yield an aggressive oxidant, such as the Fe(IV)=O species observed in cytochrome P450. Identifying a small-molecule ligand coordinating a metal site at moderate resolution is not straightforward, however. Because dioxygen is a cosubstrate of the polyisoprene cleavage reaction, RoxA as isolated may be present in an oxygenated form. This capacity is relevant for the enzymatic mechanisms, and we have compiled evidence from different sources to support this interpretation.
Monoatomic and diatomic ligands are difficult to distinguish in electron density maps, particularly in the case of unsaturated compounds, such as CO or O2, with short bond lengths. Nonetheless, the available high-quality maps clearly showed an elongated feature of Fo – Fc difference electron density (Fig. 4A). Refinement of a water molecule at this position resulted in a remaining positive maximum on the Fo – Fc map, with no such feature observed for the water molecules present in the active site cavity (Fig. 4B). In contrast, modeling a bound O2 molecule at a typical angle perfectly matched the observed electron density maximum (Fig. 4C). Independent evidence of the presence of oxygenated RoxA was obtained from electron excitation spectroscopy.
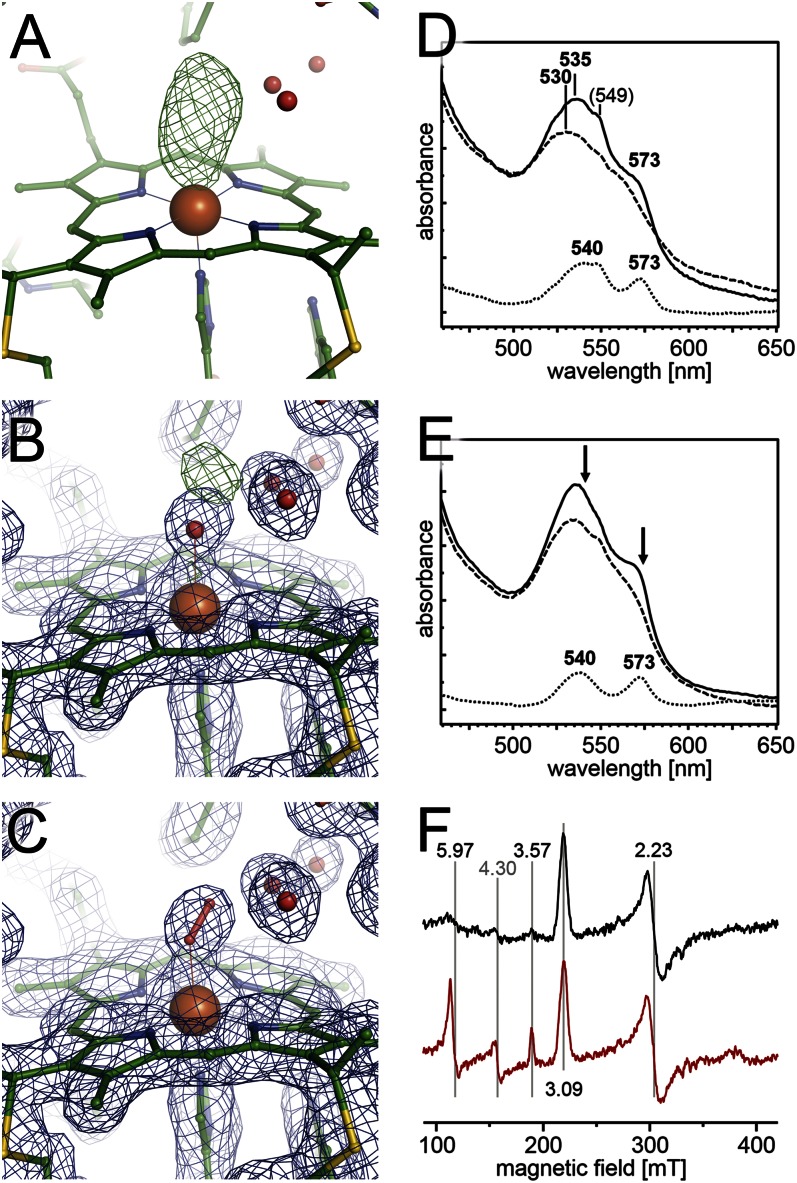
Fig. 4.
RoxA binds O2 at the active site heme group. (A) An Fo – Fc difference electron density map contoured at the +3.5 σ level shows an elongated feature at the distal axial position of heme 1. (B) When modeled with a water molecule at a distance of 2.1 Å, a positive difference density feature remains. (C) An O2 ligand at a distance of 1.9 Å fully satisfies the observed density maximum. (D) RoxA as isolated shows distinct UV/vis features (solid line) that yield to a normal Fe(III) spectrum on oxidation with K3[Fe(CN)6] (dashed line). The difference spectrum (dotted line) shows peaks at 540 and 573 nm. (E) A similar effect is observed when protein as isolated (solid line) is treated with pyrogallol to remove bound O2 (dashed line). The same maxima as in D are observed in a difference spectrum (dotted line). Arrows indicate loss of absorption on removal of dioxygen. (F) EPR spectra. Comparison of RoxA as isolated (black) with enzyme that was dithionite-reduced and subsequently reoxidized with K3[Fe(CN6)] (red) as in D, and gives rise to additional features at g = 3.57 and g = 5.97, representing a low/high-spin equilibrium at an Fe(III) heme that was previously EPR-silent.
A previous study found an increase in the α-band at 549 nm after removal of molecular oxygen (N2 atmosphere) or under low gas pressure conditions (vacuum), reflecting partial reduction of one of the heme groups (13). The effect is characteristic of dioxygen-binding proteins, such as globins, under low oxygen pressure, when the (reversible) removal of O2 leaves the heme iron in the reduced state. The presence of an O2 ligand in the resting state of RoxA is further supported by changes in the Q-band region that slowly appeared on oxidation with ferricyanide (Fig. 4D), as well as on the addition of pyrogallol, which is not an oxidizing agent but nonetheless consumes O2 or O2– (Fig. 4E). Remarkably, similar effects were found when chemically (dithionite) reduced RoxA was anaerobically reoxidized with ferricyanide. The reoxidation resulted in decreased Q-bands, as documented by a drop in the maximum from 535 nm to approximately 530 nm and loss of the shoulder at 573 nm. The difference spectra of RoxA as isolated minus O2-depleted (Fig. 4 D and E, dotted lines) display two maxima at 539–540 nm and 572–573 nm. This spectrum is strongly reminiscent of an oxygenated heme spectrum as seen in hemoglobin (24, 25) and myoglobin (26, 27).
EPR analysis of RoxA isolated from latex culture showed two well-separated sets of signals indicative of two ferric centers, one of which was considerably weaker in intensity than the other and was subject to variations from batch to batch (13). This finding suggests that one heme center is only partially present in a paramagnetic Fe(III) state. With the assumption of O2 ligation, this can be explained by a π back-bonding effect of the ligand that conveys a diamagnetic (EPR-silent) Fe(II) character to the heme iron (28). Moreover, in preparations of recombinant RoxA expressed over 2 d in the absence of rubber latex, signals assigned to heme 1 were absent or appeared only at low intensity, and high-spin signals were more intense in dithionite-reduced RoxA after reoxidation (13).
To investigate these findings, we reduced RoxA with dithionite under exclusion of dioxygen (N2 atmosphere) and reoxidized with ferricyanide. The resulting EPR spectrum showed additional high-spin signals at g = 5.97 and low-spin signals at g = 3.57 (Fig. 4F, red trace), likely originating from heme 1. Signals originating from heme 2 that also retained bis-histidinyl coordination on reduction with dithionite reappeared with identical intensity after reoxidation at g = 3.09 and g = 2.23 (Fig. 4F).
These findings suggest that the N-terminal heme center is present in a reduced state in a considerable fraction of a given RoxA preparation. Given that a ferrous state can be readily distinguished by UV/vis spectroscopy, reduction of heme 1 can be excluded. Rather, the dioxygen coordination to heme 1 can be described as an Fe2+‒ O2 <=> Fe3+‒ O2‒ equilibrium, matching the diamagnetic ferric-superoxide state (24, 29–31) caused by partial π back-bonding (28). Such an oxygenated heme is EPR-silent, but gives rise to a characteristic UV/vis spectrum distinct from that of a reduced heme group (Fig. 4E). In summary, our results support that dioxygen, and not a water molecule, represents the distal ligand of heme 1.
Polyisoprene Can Access the Active Site via Hydrophobic Tunnels.
A processive mechanism of RoxA requires that the linear isoprene polymer be threaded into the enzyme to pass the active site and exit, cleaved into ODTD units, along a distinct pathway. In CcpA peroxidases, reductive activation of the enzyme leads to major conformational changes in three loop regions that yield access to the active site (16, 20). A dithionite-reduced structure of RoxA, solved at a resolution of 2.6 Å, provided only limited information on structural details, such as the positions of water molecules and the nature of the active site ligand, but reduction clearly did not lead to rearrangements that exposed the active site. In this respect, RoxA is distinct from most CcpA peroxidases but similar to MauG, in which no conformational changes were observed.
The three loop regions that open up the active site on reduction of CcpA peroxidases are present in RoxA and compose residues 250–284 (loop 1), 300–320 (loop 2), and 422–499 (loop 3) (Figs. 5 and S2). Although appearing structurally rigid, they form an extended, dome-like structure on the distal side of heme group 1, making them the part of RoxA that the substrate chain must cross to access the reactive center. No open channel leading from the buried active site of RoxA to the protein surface was visible in the structure, but this was not unexpected, given the fact that a channel should exclude water while allowing the substrate to pass. Such properties can be realized by flexible, apolar, or aromatic amino acid side chains that form a hydrophobic barrier but can move to accommodate the entry of the substrate polymer (“hydrophobic brushes”), as have been described in protein crystal structures.
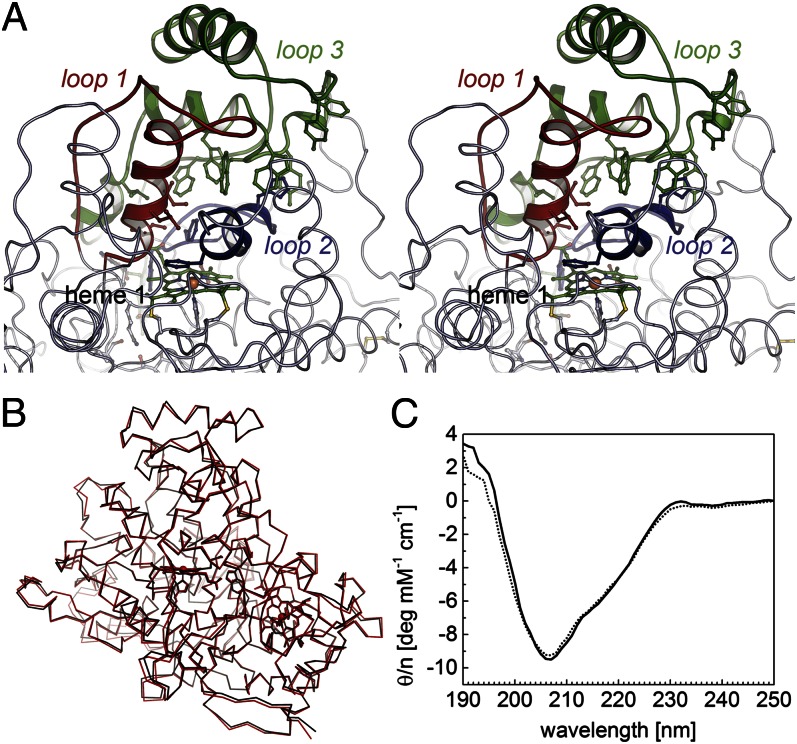
Fig. 5.
Architecture of the regulatory loop regions and possible active site access. (A) The topological equivalents of loop regions 1 (red), 2 (blue), and 3 (green) that rearrange on reductive activation of Ccp peroxidases are extended in RoxA and form a dome-like structure above the distal face of heme group 1. Note the chain of five tryptophans and one tyrosine residues in loop 3, as well as H312 and Y462, which can form H bonds with the terminus of the substrate chain and thus may act as the molecular ruler required to generate the trimeric product ODTD. (B) Superposition of an oxidized (black) and a dithionite-reduced (red) structure of RoxA with no major loop rearrangements. (C) CD spectra of oxidized (solid line) and dithionite-reduced (dotted line) RoxA showing that the conformation of the enzyme is independent of its redox state (see also Fig. S2).
Loop regions 1, 2, and 3 of RoxA provide hydrophobic amino acid side chains that face the distal active site cavity (Fig. 5A). In loop 1, these are shorter-chain residues, such as I252, L254, and I255, whereas the notable residue in loop 2 is F317, which was recently identified as relevant for enzymatic activity (23). A remarkable feature was observed in loop 3, in which a series of aromatic residues (mostly tryptophan) line the interface with the other two loop regions. Starting with the residues closest to the active site, these are W495, W445, W450, W449, and Y462, the latter of which lies at the protein surface. In addition, W449 and Y452 sandwich residue H312 belonging to loop region 2 through hydrophobic stacking interactions. Within the protein matrix, the rotational flexibility of the bulky tryptophan side chains could possibly create a transient hydrophobic pathway for the polyisoprene substrate of RoxA.
Discussion
The extracellular diheme cytochrome c RoxA is excreted by Xanthomonas to functionalize and cleave the inert latex biopolymer poly(cis-1,4-isoprene) into the more soluble ODTD units. These units are sufficiently small to be taken up for further use inside the cell through an as-yet unidentified outer membrane protein. Only five sequences for roxA orthologs discovered since 2009 are available in the databases, indicating either that RoxA is rare in nature or that the subset of available (meta)genomes underrepresents latex-degraders. Using molecular dioxygen as a cosubstrate, catalysis by RoxA proceeds slowly, likely because the threading of the substrate chain into the enzyme is rather inefficient.
Despite virtually negligible homologies on the level of the primary structure, RoxA is evidently evolutionarily related to both CcpA peroxidases (Fig. 2) and the TTQ biosynthesis protein MauG (22). The three systems are mechanistically distinct, although all rely on a structurally conserved core with the characteristic arrangement of hemes and bridging tryptophan (Fig. 2C). CcpA peroxidases store one electron on their high-potential heme group before granting access to the active site at the low-potential heme group through a conformational change. This allows a concerted two-electron transfer from both hemes to the substrate hydrogen peroxide, yielding the product water and a Fe(IV)=O oxoferryl adduct at the active site that is sufficiently stable to persist until another electron is delivered by a redox partner (16).
In MauG, in contrast, a total of six electrons must be transferred within a protein complex to complete the biosynthesis of TTQ, the catalytic cofactor of the enzyme MADH. To this end, MauG consecutively activates three molecules of O2 in a process involving a unique bis-Fe(IV) intermediate, with an Fe(IV)=O oxoferryl species at the pentacoordinate high-spin heme and an unusual His-Tyr–ligated ferryl iron at the low-spin heme group (22, 32). Together, the cofactors in this state are functionally equivalent to compound I of monoheme peroxidases, such as cytochrome P450, a Fe(IV)=O porphyrin cation radical (33). MADH, the actual substrate of MauG, does not bind close to the high-spin heme group; instead, electrons are transferred through the hemes and two tryptophan residues to the enzyme precursor, creating a spatial separation of 40 Å between the site of O2 activation and reduction of the protein cofactor (22).
Evidently, neither of these mechanisms applies to the rubber dioxygenase RoxA, in which both oxygen atoms of an O2 molecule are incorporated into the substrate and no net redox reaction occurs. A mechanism for this reaction has been suggested previously (14) (Fig. S3). Our analysis shows that in its resting state, as isolated, RoxA binds O2 at heme group 1, giving rise to spectroscopic properties reminiscient of globins not observed in MauG (18). In the oxygenated state, the Q-bands of globins usually occur at around 540 nm and 575 nm (28). For the heme dioxygenases indolamine-2,3-dioxygenase and tryptophan-2,3-dioxygenase, oxygen adducts were identified by maxima at 542 nm and 576 nm (34) and 543 nm and 576 nm (35, 36), respectively. However, compared with the stable oxygen bound to globins, these oxygenated compounds are short-lived and seen only after a preceding chemical reduction, given that heme dioxygenases are isolated in the fully oxidized state (37). In contrast, RoxA is isolated with an O2-ligated heme. Considering that the optical spectrum and activity of isolated RoxA do not change significantly after storage for days to weeks on ice, the oxygenated state of RoxA must be very stable indeed. This property is unusual for a heme dioxygenase and, to the best of our knowledge, has not been described previously.
Stable O2 binding readies the enzyme for the complicated step of proper threading in the substrate isoprene polymer. We suggest that the unusual aggregation of bulky aromatic side chains in the vicinity of the distal side of heme 1 creates transient hydrophobic channels. The preferred generation of the trimer product ODTD implies that RoxA has a mechanism for determining the length of the substrate chain from its end to the next cleavage site. This molecular ruler requires the presence of two substrate channels leading from the protein surface to the active site and back to the surface.
In an extended conformation, the length of an ODTD molecule is ∼14 Å; thus, we searched for functional groups close to the protein surface capable of interacting with the carbonyl function at the end of the polyisoprene chain. Two of the aromatic residues in loops 2 and 3, H312 and Y462, fulfill this requirement. Both H312 and Y462 are ideally positioned to form hydrogen bonds that arrest the substrate chain, so that the bond connecting the third and fourth monomer is in close proximity to the O2 ligand at heme 1. After oxidative cleavage, the resulting ODTD product dissociates, and RoxA slides along the isoprene chain until the terminus is again arrested by hydrogen bonds. In this model, processivity and the existence of two substrate channels are necessary prerequisites for the observed formation of ODTD, and the uniform substrate distribution then allows a specific uptake system in the outer membrane of Xanthomonas to optimally exploit rubber as a carbon source for growth. The solubility of ODTD in water is low, and despite extensive trials, we were not able to obtain complexes of RoxA with substrate analogs or the product. Further mutagenesis studies are needed to verify the role of possible hydrogen bond-forming residues that act as molecular rulers for measuring the substrate.
Materials and Methods
Isolation of Native and Recombinant RoxA.
The extracellular enzyme was obtained either from native, rubber-degrading cultures of Xanthomonas sp. strain 35Y (11) or by recombinant production in a homologous expression system (38) following a modified protocol (13). All steps were carried out at 295 K. In brief, the supernatant of a Xanthomonas sp. liquid culture was concentrated by cross-flow filtration and loaded onto a Q-Sepharose Fast Flow column (Q-FF 50/11) preequilibrated with 20 mM Tris⋅HCl buffer (pH 8.5) at a flow rate of 3 mL·min–1. RoxA bound to the column and was eluted in a concentration step at 50 mM NaCl. RoxA-containing fractions were pooled and concentrated by ultrafiltration (30-kDa molecular weight cutoff), then subjected to a second chromatographic step on hydroxyapatite. The column (CHT 16/10) was equilibrated with potassium phosphate buffer (10 mM; pH 6.8), and the pooled RoxA was loaded after buffer exchange to 10 mM potassium phosphate (pH 6.8) on a HiPrep 26/10 desalting column. The column was developed with a linear gradient of 10–200 mM potassium phosphate (pH 6.8) and RoxA eluted at ∼40 mM. Fractions were pooled and concentrated by ultrafiltration to 5 mg·mL–1. The addition of 300 mM NaCl on ice was required to prevent precipitation of RoxA at higher protein concentrations. Purity was analyzed by SDS/PAGE and by determination of the quotient of absorption at 406/280 nm, which was 1.35 for pure RoxA (13). Purified RoxA was flash-frozen and stored in liquid nitrogen.
Crystallization and Data Collection.
Crystals of RoxA were obtained by the sitting drop vapor diffusion method as described previously (15). A four-wavelength anomalous dispersion experiment was carried out at the K-edge of iron to optimally exploit the phasing power of the two metal ions in the heme groups of RoxA (15). In addition, a high-resolution dataset was obtained from crystals of oxidized RoxA that yielded diffraction data to resolutions better than 1.8 Å. Crystals of native RoxA belong to the monoclinic space group P21, with two monomers per asymmetric unit. Diffraction data were indexed and integrated with XDS (39) and scaled with SCALA (40) (Table 1).
Table 1.
Data collection and refinement statistics
| Statistic | Value |
| Data collection | |
| Space group | P21 |
| Wavelength,Å | 0.9184 |
| Cell dimensions | |
| a, b, c, Å | 72.4, 97.1, 101.1 |
| α, β, γ,° | 90.0, 98.4, 90.0 |
| Resolution, Å | 50.0–1.80 (1.85–1.80)* |
| Rmerge | 0.106 (0.586) |
| Rp.i.m. | 0.051 (0.292) |
| I/σI | 10.4 (2.0) |
| Completeness, % | 98.6 (97.0) |
| Multiplicity | 5.1 (4.8) |
| Refinement | |
| Resolution, Å | 35.8–1.8 (1.84–1.80) |
| No. of reflections | 119,356 (6,310) |
| Rwork/ Rfree | 0.168/0.220 (0.306/0.350) |
| No. of atoms | |
| Protein | 10,200 |
| Ligand/ion | 172 |
| Water | 1,762 |
| B-factors | |
| Protein | 21.9 |
| Ligand/ion | 16.8 |
| Water | 33.2 |
| rmsd | |
| Bond lengths, Å | 0.019 |
| Bond angles, ° | 2.721 |
Values in parentheses represent the highest-resolution shell.
Structure Solution and Refinement.
The four Fe sites were located with SHELXD (41). SHARP (42) was used for site refinement and phase calculations (15). The quality of the resulting electron density map was sufficient for model building using Coot (43), and a nearly complete model was built manually before crystallographic refinement was carried out with REFMAC (44). Medium noncrystallographic symmetry restraints were retained between the two monomers of the oxidized crystal form and the four monomers of the reduced form. In the oxidized model, 1,762 water molecules and 2 molecules of Hepes originating from the crystallization buffer were modeled, and the structure was refined to final Rcryst/Rfree values of 0.168/0.220 at 1.8-Å resolution (Table 1). The structure was validated using PROCHECK (45), which yielded three residues in disallowed regions of the Ramachandran plot. All three of these residues had very well-defined electron densities, with L565 and F516 in close proximity to heme 2 and F301 directly in the active site cavity at the distal side of heme 1. Molecular images were created with PyMOL version 1.5r4 (Schrödinger).
Electron Paramagnetic Resonance Spectroscopy.
EPR spectra were recorded on a Bruker Elexsys 500 continuous-wave spectrometer equipped with an Oxford Cryosystems helium cryostat in X-band (9.43 GHz) under conditions described previously (13).
PDB ID Code.
Coordinates and structure factors for RoxA have been deposited with the Protein Data Bank at http://www.pdb.org (PDB ID code 4B2N).
Supplementary Material
Acknowledgments
We thank the staff at beamline X06DA at the Swiss Light Source for their excellent assistance with diffraction data collection. We also thank Reinhard Braaz for assistance with the isolation of RoxA, Eva-Maria Burger and Thomas Spatzal for help with electron paramagnetic resonance data collection, and Peter Kroneck for stimulating discussions. This work was supported by the Deutsche Forschungsgemeinschaft (Grants Ei-520/1, Ei-520/5 and Je-152/13).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4B2N).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305560110/-/DCSupplemental.
References
- 1.Takahashi Y, Kumano T. Crystal structure of natural rubber. Macromolecules. 2004;37:4860–4864. [Google Scholar]
- 2.Nyburg SC. A statistical structure for crystalline rubber. Acta Crystallogr. 1954;7:385. [Google Scholar]
- 3.Bunn CW. Molecular structure and rubberlike elasticy, II: The stereochemistry of chain polymers. Proc R Soc Lond A Math Phys Sci. 1942;180:40. [Google Scholar]
- 4.Tanaka Y, Sakdapipanich JT. Chemical structure and occurence of natural polyisoprenes. In: Koyama T, Steinbüchel A, editors. Biopolymers: Polyisoprenoids. Weinheim, Germany: Wiley-VCH; 2001. pp. 1–25. [Google Scholar]
- 5.Jendrossek D, Tomasi G, Kroppenstedt RM. Bacterial degradation of natural rubber: A privilege of actinomycetes? FEMS Microbiol Lett. 1997;150(2):179–188. doi: 10.1016/s0378-1097(97)00072-4. [DOI] [PubMed] [Google Scholar]
- 6.Linos A, et al. Biodegradation of cis-1,4-polyisoprene rubbers by distinct actinomycetes: Microbial strategies and detailed surface analysis. Appl Environ Microbiol. 2000;66(4):1639–1645. doi: 10.1128/aem.66.4.1639-1645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchii A, Takeda K. Rubber-degrading enzyme from a bacterial culture. Appl Environ Microbiol. 1990;56(1):269–274. doi: 10.1128/aem.56.1.269-274.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yikmis M, Steinbüchel A. Historical and recent achievements in the field of microbial degradation of natural and synthetic rubber. Appl Environ Microbiol. 2012;78(13):4543–4551. doi: 10.1128/AEM.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose K, Tenberge KB, Steinbüchel A. Identification and characterization of genes from Streptomyces sp. strain K30 responsible for clear zone formation on natural rubber latex and poly(cis-1,4-isoprene) rubber degradation. Biomacromolecules. 2005;6(1):180–188. doi: 10.1021/bm0496110. [DOI] [PubMed] [Google Scholar]
- 10.Yikmis M, Steinbüchel A. Importance of the latex-clearing protein (Lcp) for poly(cis-1,4-isoprene) rubber cleavage in Streptomyces sp. K30. Microbiologyopen. 2012;1(1):13–24. doi: 10.1002/mbo3.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braaz R, Fischer P, Jendrossek D. Novel type of heme-dependent oxygenase catalyzes oxidative cleavage of rubber (poly-cis-1,4-isoprene) Appl Environ Microbiol. 2004;70(12):7388–7395. doi: 10.1128/AEM.70.12.7388-7395.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jendrossek D, Reinhardt S. Sequence analysis of a gene product synthesized by Xanthomonas sp. during growth on natural rubber latex. FEMS Microbiol Lett. 2003;224(1):61–65. doi: 10.1016/S0378-1097(03)00424-5. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt G, Seiffert G, Kroneck PMH, Braaz R, Jendrossek D. Spectroscopic properties of rubber oxygenase RoxA from Xanthomonas sp., a new type of dihaem dioxygenase. Microbiology. 2010;156(Pt 8):2537–2548. doi: 10.1099/mic.0.038992-0. [DOI] [PubMed] [Google Scholar]
- 14.Braaz R, Armbruster W, Jendrossek D. Heme-dependent rubber oxygenase RoxA of Xanthomonas sp. cleaves the carbon backbone of poly(cis-1,4-Isoprene) by a dioxygenase mechanism. Appl Environ Microbiol. 2005;71(5):2473–2478. doi: 10.1128/AEM.71.5.2473-2478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M, Braaz R, Jendrossek D, Einsle O. Crystallization of the extracellular rubber oxygenase RoxA from Xanthomonas sp. strain 35Y. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2008;64(Pt 2):123–125. doi: 10.1107/S1744309108001206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pettigrew GW, Echalier A, Pauleta SR. Structure and mechanism in the bacterial dihaem cytochrome c peroxidases. J Inorg Biochem. 2006;100(4):551–567. doi: 10.1016/j.jinorgbio.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Li XH, Feng ML, Wang YT, Tachikawa H, Davidson VL. Evidence for redox cooperativity between c-type hemes of MauG which is likely coupled to oxygen activation during tryptophan tryptophylquinone biosynthesis. Biochemistry. 2006;45(3):821–828. doi: 10.1021/bi052000n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, et al. MauG, a novel diheme protein required for tryptophan tryptophylquinone biogenesis. Biochemistry. 2003;42(24):7318–7325. doi: 10.1021/bi034243q. [DOI] [PubMed] [Google Scholar]
- 19.Li XH, Jones LH, Pearson AR, Wilmot CM, Davidson VL. Mechanistic possibilities in MauG-dependent tryptophan tryptophylquinone biosynthesis. Biochemistry. 2006;45(44):13276–13283. doi: 10.1021/bi061497d. [DOI] [PubMed] [Google Scholar]
- 20.Seidel J, et al. MacA is a second cytochrome c peroxidase of Geobacter sulfurreducens. Biochemistry. 2012;51(13):2747–2756. doi: 10.1021/bi300249u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann M, Seidel J, Einsle O. CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase. J Mol Biol. 2009;393(4):951–965. doi: 10.1016/j.jmb.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Jensen LMR, Sanishvili R, Davidson VL, Wilmot CM. In crystallo posttranslational modification within a MauG/pre-methylamine dehydrogenase complex. Science. 2010;327(5971):1392–1394. doi: 10.1126/science.1182492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birke J, Hambsch N, Schmitt G, Altenbuchner J, Jendrossek D. Phe317 is essential for rubber oxygenase RoxA activity. Appl Environ Microbiol. 2012;78(22):7876–7883. doi: 10.1128/AEM.02385-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittenberg JB, Wittenberg BA, Peisach J, Blumberg WE. On the state of the iron and the nature of the ligand in oxyhemoglobin. Proc Natl Acad Sci USA. 1970;67(4):1846–1853. doi: 10.1073/pnas.67.4.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zijlstra WG, Buursma A. Spectrophotometry of hemoglobin: Absorption spectra of bovine oxyhemoglobin, deoxyhemoglobin, carboxyhemoglobin, and methemoglobin. Comp Biochem Physiol B. 1997;118:743–749. [Google Scholar]
- 26.Bowen WJ. The absorption spectra and extinction coefficients of myoglobin. J Biol Chem. 1949;179(1):235–245. [PubMed] [Google Scholar]
- 27.Yamazaki I, Yokota KN, Shikama K. Preparation of crystalline oxymyoglobin from horse heart. J Biol Chem. 1964;239:4151–4153. [PubMed] [Google Scholar]
- 28.Momenteau M, Reed CA. Synthetic heme dioxygen complexes. Chem Rev. 1994;94:659–698. [Google Scholar]
- 29.Jensen KP, Ryde U. How O2 binds to heme: reasons for rapid binding and spin inversion. J Biol Chem. 2004;279(15):14561–14569. doi: 10.1074/jbc.M314007200. [DOI] [PubMed] [Google Scholar]
- 30.Pauling L, Coryell CD. The magnetic properties and structure of hemoglobin, oxyhemoglobin and carbonmonoxyhemoglobin. Proc Natl Acad Sci USA. 1936;22(4):210–216. doi: 10.1073/pnas.22.4.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss JJ. Nature of iron-oxygen bond in oxyhaemoglobin. Nature. 1964;202:83–84. doi: 10.1038/202083b0. [DOI] [PubMed] [Google Scholar]
- 32.Abu Tarboush N, et al. Functional importance of tyrosine 294 and the catalytic selectivity for the bis-Fe(IV) state of MauG revealed by replacement of this axial heme ligand with histidine. Biochemistry. 2010;49(45):9783–9791. doi: 10.1021/bi101254p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang YT, et al. MauG-dependent in vitro biosynthesis of tryptophan tryptophylquinone in methylamine dehydrogenase. J Am Chem Soc. 2005;127(23):8258–8259. doi: 10.1021/ja051734k. [DOI] [PubMed] [Google Scholar]
- 34.Hirata F, Ohnishi T, Hayaishi O. Indoleamine 2,3-dioxygenase: Characterization and properties of enzyme–O2 complex. J Biol Chem. 1977;252(13):4637–4642. [PubMed] [Google Scholar]
- 35.Capece L, et al. The first step of the dioxygenation reaction carried out by tryptophan dioxygenase and indoleamine 2,3-dioxygenase as revealed by quantum mechanical/molecular mechanical studies. J Biol Inorg Chem. 2010;15(6):811–823. doi: 10.1007/s00775-010-0646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yeh SR, et al. (2010) Comparative studies of human indoleamine 2,3-dioxygenase and tryptophan dioxygenase. XXII International Conference on Raman Spectroscopy (John Wiley & Sons Ltd, New York), p. 283.
- 37.Batabyal D, Yeh SR. Substrate–protein interaction in human tryptophan dioxygenase: The critical role of H76. J Am Chem Soc. 2009;131(9):3260–3270. doi: 10.1021/ja807969a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hambsch N, Schmitt G, Jendrossek D. Development of a homologous expression system for rubber oxygenase RoxA from Xanthomonas sp. J Appl Microbiol. 2010;109(3):1067–1075. doi: 10.1111/j.1365-2672.2010.04732.x. [DOI] [PubMed] [Google Scholar]
- 39.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62(Pt 1):72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 41.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64(Pt 1):112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 42.de la Fortelle E, Irwin JJ, Bricogne G. SHARP: A maximum-likelihood heavy-atom parameter refinement and phasing program for the MIR and MAD methods. Crystallographic Computing. 1997;7:1–9. [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. Procheck: A program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.