Significance
How cancer cells become invasive is key to understanding malignancy. Perturbations in O-glycosylation are strongly correlated with invasiveness. Here we report that tumor cells display relocation of O-glycosylation initiating glycosyltransferases from the Golgi apparatus to the endoplasmic reticulum (ER). ER-located O-glycosylation stimulates cell migration and invasiveness, which depend on cell surface O-glycoproteins. Inhibition of the glycosyltransferases in the ER reduces tissue invasion and metastasis formation in mice. Our study suggests that control of glycosylation via the subcellular localization of glycosyltransferases is a critical mechanism driving invasiveness in tumor cells.
Keywords: invasion, Src, COPI, Thomsen-Friedenreich antigen, C1GalT
Abstract
Invasiveness underlies cancer aggressiveness and is a hallmark of malignancy. Most malignant tumors have elevated levels of Tn, an O-GalNAc glycan. Mechanisms underlying Tn up-regulation and its effects remain unclear. Here we show that Golgi-to-endoplasmic reticulum relocation of polypeptide N-acetylgalactosamine-transferases (GalNAc-Ts) drives high Tn levels in cancer cell lines and in 70% of malignant breast tumors. This process stimulates cell adhesion to the extracellular matrix, as well as migration and invasiveness. The GalNAc-Ts lectin domain, mediating high-density glycosylation, is critical for these effects. Interfering with the lectin domain function inhibited carcinoma cell migration in vitro and metastatic potential in mice. We also show that stimulation of cell migration is dependent on Tn-bearing proteins present in lamellipodia of migrating cells. Our findings suggest that relocation of GalNAc-Ts to the endoplasmic reticulum frequently occurs upon cancerous transformation to enhance tumor cell migration and invasiveness through modification of cell surface proteins.
Invasiveness is an important hallmark that differentiates malignant from benign tumors (1). Invasion of surrounding tissues often underlies the lethality of tumors, either at the primary site, as for glioblastomas, or through the formation of metastases, as for most solid tumors (2, 3). Whether the mechanisms underlying invasiveness share a common basis in different tumors remains an open question; albeit, the embryonic process of epithelio-mesenchymal transition has been proposed to be a driver (3).
The induction of invasiveness is likely to involve changes both at the cell surface and in the levels of secreted factors. Therefore, both surface proteins and their attached glycans—the complex carbohydrates linked to most cell surface and secreted proteins—are likely to regulate cancer cell invasiveness. Indeed, changes in N-glycans have been proposed to have an important impact on cancer progression (4, 5).
Tn is a small O-glycan present at high levels in most primary and metastatic carcinomas that correlates with metastatic potential and poor prognosis (6, 7). Normal tissues, on the other hand, express much lower levels of Tn (8). Currently, the mechanisms underlying the increase in Tn levels as well as how Tn contributes to invasiveness remain unclear. The minimal Tn epitope is N-acetyl-d-galactosamine, which is α-linked to Ser or Thr residues (O-GalNAc) and can be recognized by lectins such as Helix pomatia lectin (HPL) (9) and Vicia villosa B4 lectin (VVL) (10).
Tn is formed by specific glycosyltransferases called UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyl-transferases (GalNAc-Ts), which comprise a family of ∼20 enzymes that initiate O-GalNAc glycosylation (hereafter referred to as O-glycosylation for simplicity) by catalyzing the addition of GalNAc onto polypeptides (11). GalNAc-Ts can modify a variety of cell surface and secreted proteins (12, 13), and are unique among glycosyltransferases for their ricin-like lectin domain (11, 14). This domain binds O-GalNAc and promotes secondary GalNAc addition on neighboring positions in a peptidic sequence (15, 16). Cancer cell glycoproteins display O-GalNAc at high density (17), suggesting that the lectin domain could be important for Tn expression in tumors.
Normally, GalNAc-Ts are localized in the Golgi apparatus (18) and Tn is only detected in the cis-portion of the Golgi (19). Subsequently, other glycosyltransferases rapidly elongate Tn into other, more complex O-glycans. The first step is usually carried out by C1GalT, which adds galactose to O-GalNAc and converts Tn into the T antigen, alias core 1 O-glycan, which is itself further elongated by other enzymes (20–22). Loss of C1GalT or its essential private chaperone C1GALT1C1 (Cosmc) leads to the strong up-regulation in Tn expression in patients with Tn syndrome, a rare hematological disorder (23–25). By extension, it has been proposed that high Tn tumors have lost C1GalT activity (26). This interpretation has led to hypotheses that Tn must be abundant at the surface of cancer cells and is an aberrant structure that could be targeted by the immune system or by exogenous antibodies. Although this theory has led to efforts to develop anti-Tn immunotherapy and vaccines (27, 28), the loss of C1GalT activity has only been demonstrated in a limited set of cell lines and tumors (21, 26, 29).
Recently, we reported that activation of the invasion-promoting kinases EGF receptor (EGFR), platelet-derived growth factor receptor (PDGF-R) and Src induce Golgi-to-endoplasmic reticulum (ER) trafficking of GalNAc-Ts specifically through activation of the COPI coatomer transport machinery (30). Here we report that relocation of GalNAc-Ts to the ER markedly increases Tn staining and is a common phenomenon in human malignant breast tumors. We demonstrate that relocation of GalNAc-Ts promotes cell adhesion, cell motility, and invasiveness; it also induces the accumulation of Tn-bearing proteins in lamellipodia at the cell surface. Using a Tn-specific soluble lectin, we show that inhibition of these surface Tn-bearing proteins reduces cancer cell motility in vitro. In addition, compartment-specific inhibition of GalNAc-Ts through the expression of an ER-targeted lectin reduces Tn levels as well as cancer cell motility in vitro and invasiveness in vivo.
Overall, our results link a membrane trafficking process to the regulation of glycosylation and in turn the control of cellular invasive behavior.
Results
ER Localization of GalNAc-T2 Is Sufficient to Drive High Tn Expression.
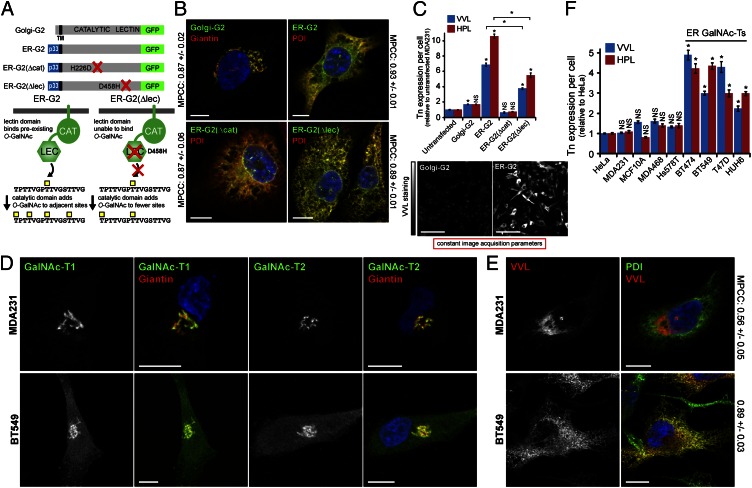
We previously determined that Golgi-to-ER relocation of GalNAc-Ts upon growth factor treatment enhances O-GalNAc glycosylation (30). To dissect the effects of an ER localization of GalNAc-Ts, we constructed a chimeric version of human GalNAc-T2 that constitutively localized to the ER (ER-G2) (18) (Fig. 1A). ER-G2 was stably expressed in MDA-MB-231 (hereafter called MDA231) breast cancer cells. Cells were also generated to express wild-type GalNAc-T2 (Golgi-G2), an inactive ER chimera with a point mutation (H226D) in the catalytic domain [ER-G2(Δcat)], or an ER chimera with an inactivating point mutation (D458H) in the lectin domain [ER-G2(Δlec)] (Fig. 1A). All constructs were homogeneously expressed and at comparable levels, except Golgi-G2, for which the expression was elevated relative to its ER counterparts (Fig. S1A). As expected, Golgi-G2 colocalized with the Golgi-marker Giantin, whereas ER-targeted chimeras colocalized with the ER-marker protein disulphide isomerase (PDI). Colocalization was supported by high mean Pearson correlation coefficients (MPCC) between stainings (Fig. 1B).
Fig. 1.
ER-localized GalNAc-Ts drive high Tn expression. (A) Schematic of GalNAc-T2-GFP constructs used in this study. (B) Golgi-G2 and ER-targeted chimeras colocalize with Golgi (Giantin) and ER (PDI) markers, respectively. (Scale bars, 10 µm.) (C) Mean Tn expression ± SEM in model GalNAc-T2 cells. *P < 0.01 relative to untransfected. NS, not significant. (Scale bars, 100 µm.) (D) GalNAc-T1 and -T2 staining in cells with low (MDA231) or high (BT549) Tn levels. Golgi stained with Giantin. (Scale bar, 10 µm.) (E) Costaining Tn with VVL and ER with anti-PDI antibody. (Scale bars, 10 µm.) (F) Mean of Tn expression ± SEM in cancer cells. *P < 0.01 relative to HeLa cells. Nuclei stained using Hoechst.
Automated microscopic quantification of VVL fluorescence in hundreds of cells acquired under constant parameters revealed that although Golgi-G2 increased Tn expression by twofold, ER-G2 expression caused a 7- to 10-fold increase in Tn. This effect was strictly dependent on a functional catalytic domain as ER-G2(Δcat) did not yield any increase (Fig. 1C). ER-G2(Δlec) induced about three- to fivefold Tn increase, indicating that about 50% of the signal is depending on a functional lectin domain (Fig. 1C). Overall, these results indicate that localization of GalNAc-Ts in the ER leads to high levels of Tn staining.
Several Cancer Cell Lines Display Constitutive ER-Localized GalNAc-Ts.
We next investigated the subcellular localization of GalNAc-Ts in nine human cancer cell lines by staining for the ubiquitously expressed GalNAc-T1 and -T2 (11). These enzymes were localized in the Giantin-labeled Golgi in HeLa, MDA231, MCF10A, and MDA-MB-468 cells but detected outside the Golgi in BT474, BT549, T47D, and HUH6 cells (Fig. 1D and Fig. S1B). In agreement with GalNAc-Ts staining, Tn staining was primarily restricted to the Golgi in the first group (Fig. 1E and Fig. S1C), whearas, in the second group, there was extensive colocalization with PDI (Fig. 1E and Fig. S1C). This trend was confirmed by quantifying the VVL-PDI MPCC, showing 0.56 (± 0.05) in MDA231 versus 0.89 (± 0.03) in BT549 cells (Fig. 1E). For comparison, the MPCC values of poorly colocalized PDI and Giantin were 0.25 (± 0.02) in MDA231 and 0.32 (± 0.02) in BT549 (see Table S1 for all MPCC values).
Tn levels were next quantified as outlined above and found to be higher in cells containing ER-localized GalNAc-Ts (up to fivefold) than in cells with mostly Golgi-localized GalNAc-Ts (Fig. 1F). The presence of GalNAc-Ts in the ER suggests that the growth factor-inducible relocation process is constitutively activated in some cancer cell lines. GalNAc-T relocation is dependent on the Src kinase, the vesicle forming small GTPase Arf1 and the COPI coat (30). Overnight treatment of BT549 cells with 10 µM PP2, a potent Src inhibitor, led to the relocation of Tn staining within the GM130-labeled Golgi apparatus, indicating that a fraction of ER-localized GalNAc-Ts had translocated to the Golgi (Fig. S2A). Similarly, inhibiting COPI vesicle biogenesis in BT549 cells through the over-expression of a GTP-locked Arf1 isoform, Arf1(Q71L)-GFP, also shifted Tn staining from the ER to the Golgi (Fig. S2B).
We next tested whether MDA231, MCF10A, and MDA468 cells could be induced to relocate their primarily Golgi-localized GalNAc-Ts to the ER. Treatment with 100 ng/mL EGF for 2 h promoted ER redistribution of GalNAc-T1 and led to enzymatic activity in the ER, as judged by colocalization of Tn with PDI (Fig. S2 C–E). Increased Tn levels in these EGF-treated cells was also observed, albeit to a lesser extent (<twofold) relative to cancer cell lines with constitutive ER-localized GalNAc-Ts (up to fivefold) (Fig. 1F and Fig. S2F).
Taken together, these results suggest that high Tn levels in some cancer cells arise from endogenous stimulation of GalNAc-Ts transport from Golgi to ER.
ER-Localized GalNAc-Ts and C1GalT Deficiency Result in Distinct Glycophenotypes.
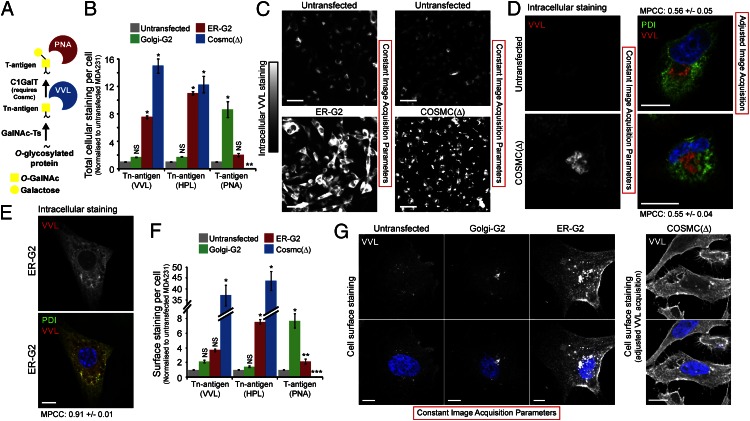
Tn expression has been proposed to result from a reduction in C1GalT or COSMC activity (Fig. 2A) (26). To compare the loss of this enzyme with the ER localization of GalNAc-Ts, MDA231 cells were genetically depleted of the C1GalT chaperone Cosmc [Cosmc(Δ)] using a zinc-finger nuclease (13). Automated microscopic quantification confirmed that T antigen expression revealed with peanut agglutinin (PNA lectin) staining was abolished in Cosmc(Δ) cells. Total cellular Tn staining (on cells fixed and permeabilized with a mild detergent) was strongly up-regulated upon Cosmc depletion (12- to 15-times) and was comparable to Tn expression in ER-G2 cells (Fig. 2B). Despite similar increases in Tn levels, the Tn staining patterns in these two model systems were distinct. Tn was localized primarily at the cell surface and Golgi in Cosmc(Δ) cells (Fig. 2 C and D) and, by contrast, mostly in the ER in ER-G2 cells (Fig. 2 C and E). This change is reflected by low and high VVL-PDI MPCC values in Cosmc(Δ) and ER-G2 cells, respectively (Table S1).
Fig. 2.
Loss of C1GalT activity and ER localization of GalNAc-Ts result in distinct glyco-phenotypes. (A) Schematic of O-GalNAc glycosylation pathway and lectin reactivity. (B and C) Mean expression ± SEM of T and Tn antigens in genetically modified MDA231. *P < 0.01 (ANOVA) and **P < 0.01 (t test) relative to untransfected cells. (D) Costaining VVL and PDI in Cosmc(Δ) and untransfected MDA231 cells. Weaker VVL staining in untransfected cells required adjusted acquisition. (Scale bars, 10 µm.) (E) Costaining VVL and PDI in ER-GalNAc-T2 cells. (Scale bar, 10 µm.) (F and G) Mean cell surface expression ± SEM of T and Tn antigens in genetically modified MDA231. (Scale bars, 10 µm.) *P < 0.01, **P < 0.05 (ANOVA), and ***P < 0.01 (t test) relative to untransfected cells. Nuclei stained using Hoechst.
Overexpression of Golgi-localized GalNAc-T2 in Golgi-G2 cells led to a large eightfold increase in T antigen expression, suggesting that Tn added in the Golgi is efficiently elongated by C1GalT. In contrast, T antigen levels increased by only twofold in ER-G2 cells, suggesting that only a fraction of GalNAc added in the ER is modified by Golgi-localized C1GalT (Fig. 2B).
Next, we stained cell surface Tn using VVL and HPL on nonpermeabilized cells. No significant perinuclear Golgi-like pattern could be detected in these experiments, confirming the specificity of the surface labeling. As expected, Cosmc(Δ) cells displayed a dramatic increase in surface Tn (36- to 44-times), consistent with a block of GalNAc elongation. ER-G2 cells also showed an increase in surface Tn expression but significantly less than for Cosmc(Δ) cells (four- to sevenfold), whereas Golgi-G2 cells showed no significant increase (Fig. 2 F and G). Thus, although Cosmc(Δ) and ER-G2 cells showed comparable total increases in total Tn, surface Tn is approximately 10-times higher in Cosmc(Δ) cells (Fig. 2 B and F). This finding suggests that only a small fraction of Tn-bearing proteins reach the surface unmodified in cells containing ER-localized GalNAc-T2. Why this Tn is not modified during transit to the cell surface is not clear at this stage.
Thus, ER-localized GalNAc-Ts and C1GalT deficiency result in distinct O-glycosylation phenotypes despite both increasing total cellular Tn levels.
ER-Localized GalNAc-Ts Drive High Tn Expression in Human Breast Carcinomas.
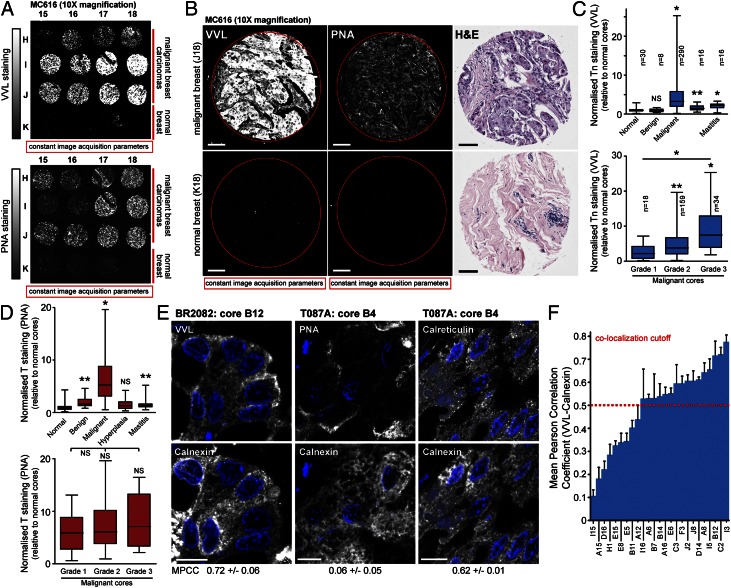
To investigate the mechanistic basis for high Tn levels in tumors, we first screened two human breast cancer tissue microarrays (BR2082, MC616) containing 360 histological cores encompassing malignant and benign tumors as well as hyperplasia, mastitis and normal tissue. After VVL staining, imaging was performed at 10× magnification under constant acquisition parameters (Fig. 3 A and B) and the fluorescence per pixel was quantified and normalized to mean fluorescence of cores from normal breast tissue (Fig. S3A and Table S2). This analysis revealed that Tn staining in malignant breast tumor cores was 4.7-fold higher than that in normal tissue or benign tumor cores (Fig. 3C). Cores labeled for hyperplasia and mastitis had significantly smaller (1.6- and 2-fold, respectively) increases in Tn levels (Fig. 3C). Separating malignant cores into histological grades highlighted that Tn staining increased with the grade or severity of the tumor (Fig. 3C). Increases in Tn levels for grade 2 (4.6-fold) and grade 3 (8.7-fold) cores was comparable with that observed in ER-G2 cells and cancer cell lines displaying ER-localized GalNAc-Ts (Figs. 1 C and F, and 3C). Similar results were observed by separating malignant cores in the MC616 array into histological stages (Fig. S3B). Fig. S4 shows a more detailed comparison of VVL and H&E staining in tumor cores from the MC616 array.
Fig. 3.
Human breast carcinomas with high Tn expression contain ER-localized GalNAc-Ts activity. (A and B) Tn (VVL) and T (PNA) expression in microarrays of human breast tumor biopsies. H&E staining is shown in B. (Scale bars, 100 µm.) (C and D) Quantification of Tn (C) and T (D) staining in human breast cancer biopsies. *P < 0.01 and **P < 0.05 relative to normal tissue cores (Mann–Whitney). NS, not significant. Histological grade confirmed by pathologist. Grade 1: well differentiated; Grade 2: moderately differentiated; Grade 3: poorly differentiated. (E) Costaining with VVL or PNA and ER-marker Calnexin in human breast tumor biopsies. Costaining of Calnexin and ER-protein Calreticulin for comparison. (Scale bar, 10 µm.) (F) Quantification of VVL-Calnexin colocalization in 25 randomly acquired malignant high Tn breast cancer cores from the BR2082 array. Nuclei stained using Hoechst.
To study the subcellular pattern of VVL staining, we next imaged the tissue arrays at 100× magnification. This process revealed that ∼70% of high Tn (>threefold) malignant cores displayed an ER pattern, as determined by eye (Fig. 3E). To quantify this phenomenon, we measured MPCC values between VVL and the ER marker Calnexin in costained tumor cores. Baselines for this study were obtained with a smaller breast cancer tissue microarray (T087A) with different marker combinations. Costaining for Calreticulin and Calnexin, both abundant ER-localized chaperones, displayed an average MPCC of 0.64 (± 0.01) (Fig. 3E and Fig. S3C) (31). In contrast, costaining for PNA and Calnexin exhibited an average MPCC of 0.17 (± 0.02), consistent with PNA staining for T antigen localized in the Golgi and at the cell surface (Fig. 3E and Fig. S3 C and E) (32).
Two cores (B3, B4) in the T087A array contained high Tn levels (>threefold) relative to that in normal tissue cores and in both cores, the stainings for VVL and Calnexin colocalized (Fig. S3D). VVL-Calnexin MPCC values in these two cores (B3: 0.62 ± 0.05 and B4: 0.63 ± 0.01) were significantly higher than that for cores with low Tn levels (mean of 0.35 ± 0.03 for all samples) (Fig. S3 C and E). This finding suggests that a high level of Tn is correlated with an ER pattern and derives from an ER localization of GalNAc-Ts, whereas at low levels Tn staining is not localized in the ER. From this analysis and the control stainings we concluded that an MPCC ≥ 0.5 was an indication of positive Calnexin/VVL colocalization (Fig. S3D).
Next, VVL-Calnexin MPCC values were measured for 25 randomly chosen high Tn (>threefold) malignant cores from the BR2082 microarray (Fig. 3 E and F). This finding revealed that 16 (64%) cores had MPCC values above 0.5, indicating that Tn staining was significantly localized in the ER. Several cores below this cutoff also displayed a clear ER Tn staining pattern by eye; however, additional VVL staining at the cell surface resulted in a lower VVL-Calnexin MPCC score. Thus, high Tn levels are highly correlated with an ER localization of staining in breast carcinomas.
Loss of C1GalT Activity Is a Rare Event in Breast Carcinomas.
In search for evidence of C1GalT deficiency in breast cancer, T antigen levels (which depend on C1GalT) were quantified using PNA staining in human breast cancer arrays (BR2082, MC616, and T087A), as outlined previously (Fig. 3 A and B). This finding revealed that all high Tn breast carcinoma cores also contained T antigen levels (Fig. 3D, and Fig. S3 A and D), indicating that they contain functional C1GalT. In fact, both T and Tn antigens in malignant cores were markedly increased relative to that in normal tissue cores (Fig. 3 C and D).
We also searched gene-expression datasets from two recently published studies on breast cancer based on a total of 65 invasive ductal carcinoma and 223 normal breast tissue (33, 34) (data publicly available at the European Bioinformatics Institute Expression Atlas). Neither C1GalT nor COSMC expression appeared significantly decreased in most of these breast tumor samples.
Expression levels of Tn and T antigens do not correlate within the tissue cores analyzed here (Fig. S3 A and D). This finding suggests that ER relocation of GalNAc-Ts is not driving T antigen expression. It is possible that variable elongation and in particular sialylation of the T antigen masks an underlying correlation. To test for this possibility, tissue arrays were treated with neuraminidase to remove sialic acid residues before staining with PNA and VVL (Fig. S5). However, this treatment resulted in a more limited increase in PNA staining between normal and malignant tissues, indicating that sialylation is not masking pools of T antigen. Neuraminidase treatment also did not increase the correlation between PNA and VVL staining (Fig. S5A). Finally, T antigen levels, with or without neuraminidase treatment, did not correlate with grade as much as Tn levels (Fig. S5C), further suggesting that Tn and T antigen expression are driven by different biological processes.
In conclusion, high Tn expression in breast carcinoma appears driven by the ER-localization of GalNAc-Ts and not C1GalT deficiency.
ER-GalNAc-T2 Stimulates Cell Adhesion and Motility.
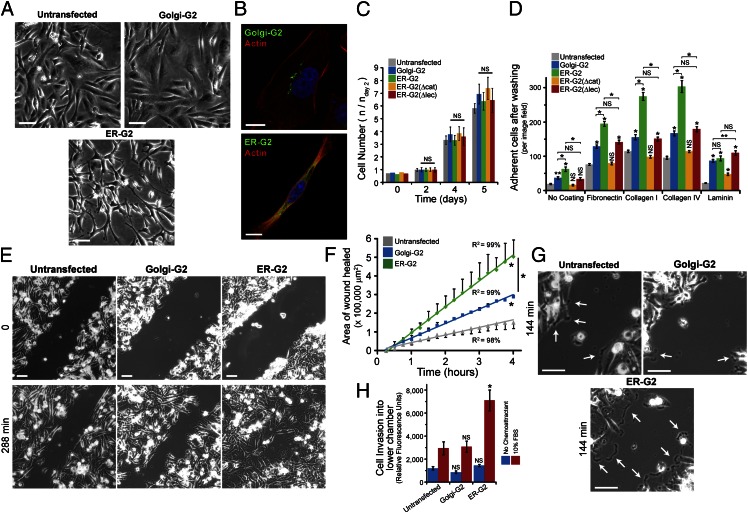
During culturing, it became apparent that ER-G2 cells were more spindle-shaped relative to the polygonal untransfected MDA231 and Golgi-G2–expressing cells (Fig. 4A), as illustrated by actin cytoskeleton staining (Fig. 4B). In contrast, none of the GalNAc-T2-GFP constructs significantly altered MDA231 growth in vitro (Fig. 4C). This morphological alteration suggested changes in cellular adhesion. Cell adhesiveness to uncoated versus fibronectin- (20 µg/mL), laminin- (10 µg/mL), collagen-I– (10 µg/mL), and collagen-IV–coated (5 µg/mL) plastic surfaces was quantified after passaging with 2 mM EDTA to preserve surface proteins. Here we also used ER-G2(Δcat)– and ER-G2(Δlec)–expressing cells, which express ER-localized GalNac-T2 with, respectively, an inactivating point mutation in the catalytic domain and an inactivating point mutation in the lectin domain. ER-G2(Δcat) cells showed no difference in cell adhesiveness across the four coatings compared with the uncoated control. ER-G2 cells showed significantly increased adherence to fibronectin and collagen compared with Golgi-G2 cells. ER-G2(Δlec) cells showed no change in adhesiveness compared with Golgi-G2 cells (Fig. 4D). Thus, ER-GalNAc-T2 increases binding to fibronectin and collagen in a lectin-domain dependent fashion.
Fig. 4.
ER-targeted GalNAc-T2 enhances cell adhesion and migration. (A) Phase-contrast images of untransfected and transgenic GalNAc-T2 cells. (Scale bars, 100 µm.) (B) Actin staining in Golgi-G2 and ER-G2 cells. (Scale bars, 10 µm.) (C) Mean growth rate ± SEM of model GalNAc-T2 cells. (D) Mean adhesive strength ± SEM of model GalNAc-T2 cells. *P < 0.01, **P < 0.05 relative to untransfected or stated samples. (E) Migration assay using scratch wound of cellular monolayer. (Scale bars, 100 µm.) (F) Mean of wound closure ± SEM (n = 2 experiments). *P < 0.01 relative to untransfected cells or stated samples. (G) Lamellipodia (arrows) in migrating ER-G2 compared with untransfected and Golgi-G2 cells. (Scale bars, 100 μm.) (H) Transwell migration assay in a Boyden chamber (n = 3 experiments). *P < 0.01 relative to untransfected cells. Nuclei stained using Hoechst.
Adhesion to fibronectin promotes the migratory properties of cancer cells (35). In a scratch wound-healing assay on fibronectin-coated plastic, ER-G2 cells migrated about four-times faster into the denuded area than untransfected cells, and twice as fast as Golgi-G2 cells (Fig. 4 E and F). The rate of migration was consistently higher during the 4 h of the experiment (Fig. 4F), indicating that enhanced O-glycosylation does not promote a faster response to the scratch but rather sustains faster cell migration throughout. Consistently, examination of the time-lapse photography revealed that lamellipodia in migrating ER-G2 cells were larger and more stable than those in untransfected and Golgi-G2 cells (Fig. 4G and Movies S1–S3). Taken together, these observations suggest that enhanced cell adhesion promotes cell protrusions that lead to faster cell movement.
Increased cell motility is sometimes correlated with enhanced invasiveness through the extracellular matrix (ECM) (36). The invasive potential of Golgi-G2 and ER-G2 cells was compared using a Boyden chamber assay with a barrier made of Matrigel (complex ECM mixture comprising purified basement membrane proteins from the Engelbreth Holm-Swarm tumor) and 10% (vol/vol) FBS as a chemotactic attractant. Over a 24-h course, about three-times more ER-G2 cells were able to go through the ECM barrier than untransfected cells, whereas Golgi-G2 cells did not show any significant increase in invasion (Fig. 4H).
Tn-Bearing Proteins Are Located in Lamellipodia and Act to Regulate Adhesion and Motility.
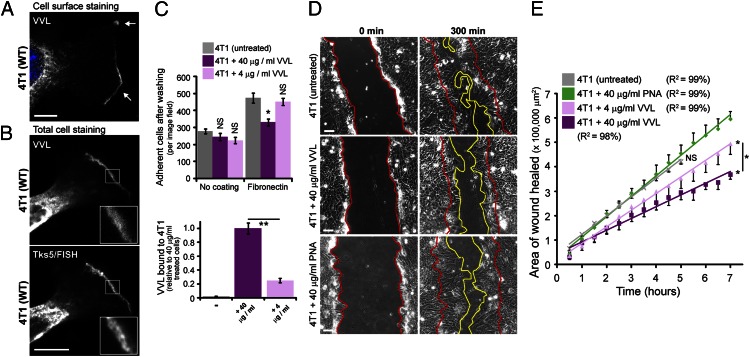
The marked effect of ER-localized O-GalNAc glycosylation on cell motility and invasiveness suggests the involvement of cell-surface glycoproteins. Interestingly, surface staining of 4T1, Hs578T, and HUH6 cancer cells using VVL revealed that they displayed Tn-bearing proteins in cell protrusions such as in lamellipodia stained with the actin regulator Tsk5/FISH (Fig. 5 A and B, and Fig. S6 A and B). Mouse mammary 4T1 tumor cells were selected for the rest of the study because, similar to human Hs578T and HUH6, they displayed evidence of significant O-glycosylation in the ER and are highly metastatic in mice (37). Tn-positive protrusions in 4T1 cells also stained positively for actin, paxillin, and the kinases FAK and Src, suggesting that they were actively protruding (Fig. 5B and Fig. S6C).
Fig. 5.
Surface Tn-modified proteins regulate the adhesiveness and motility of 4T1 carcinoma cells containing ER-localized GalNAc-Ts. (A) Cell surface Tn staining (VVL) in 4T1 cells. (Scale bar, 10 µm.) (B) Costaining Tn (VVL) with the lamellipodia marker Tks5/FISH in 4T1 cells. (Scale bar, 10 µm.) (C) Adhesion of 4T1 cells to fibronectin after VVL preincubation for 20 min and quantification of VVL bound to cells after 20-min incubation. *P < 0.01 relative to untreated cells. (D) Wound healing of 4T1 cells on fibronectin after VVL preincubation for 20 min. (Scale bars, 100 µm.) (E) Mean of wound closure ± SEM (n = 3 experiments). *P < 0.01 relative to PNA treated cells or stated samples. (Scale bars, 10 µm.) Nuclei stained using Hoechst.
We next tested whether interfering with surface Tn-bearing proteins using soluble VVL could interfere with the adhesive properties of 4T1 cells. Strikingly, preincubation with a high VVL concentration (40 µg/mL) for 20 min at room temperature caused an ∼30% reduction in the number of adherent 4T1 cells after washing (Fig. 5C). This effect was observed on fibronectin-coated surfaces but not on uncoated plastic, and was also proportional to VVL concentration and to surface Tn masking, as evaluated by the amount of bound VVL (Fig. 5C). During scratch wound-healing assays on fibronectin, VVL preincubation was inhibitory in a concentration-dependent manner and, at the highest concentration (40 µg/mL), caused a twofold reduction in the rate of 4T1 cell migration (Fig. 5 D and E and Movies S4 and S5). By comparison, preincubation with PNA (40 µg/mL) did not inhibit 4T1 cell adhesion to fibronectin (Fig. S6D) nor did it affect wound healing on fibronectin (Fig. 5 D and E and Movies S6 and S7). Furthermore, preincubation of HeLa cells (lacking endogenous ER-GalNAc-Ts and containing very low surface Tn levels) with either VVL or PNA (40 µg/mL) did not inhibit their migration on fibronectin-coated surfaces in a scratch wound-healing experiment (Fig. S7 A and B and Movies S8–S10).
In sum, surface Tn-bearing proteins are important for the enhanced fibronectin adhesiveness and motility of cells with ER-localized O-GalNAc glycosylation.
Inhibiting O-Glycosylation in the ER Lowers Carcinoma Cell Adhesion.
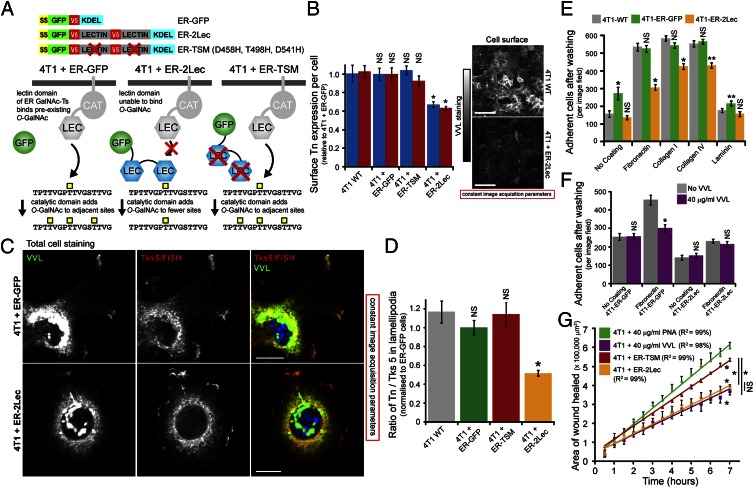
We next sought to stably inhibit O-glycosylation in the ER using a genetic approach. Because the lectin domain of ER-localized GalNAc-T2 is important for elevated surface Tn expression (Fig. 1C), we reasoned that an isolated lectin domain might be able to compete with GalNAc-Ts localized in the ER. We fused an ER-targeting sequence from human growth hormone (residues 1–33) with GFP, two lectin domains from GalNAc-T2 in tandem, and a KDEL sequence; ER-targeting and KDEL sequences ensure inhibitor targeting and retention in the ER, respectively. Controls lacking either the inhibitory lectin domains (ER-GFP) or the functional O-GalNAc sugar binding sites (D458H, T498H, D541H) in the lectin domains (ER-triple sugar mutant or ER-TSM) were also generated (Fig. 6A).
Fig. 6.
Inhibiting ER-localized GalNAc-Ts lowers the adhesiveness and motility of 4T1 carcinoma cells in vitro. (A) Schematic of makeup and effect of the ER-specific GalNAc-T inhibitor. (B) Mean surface Tn expression ± SEM in model 4T1 cells. *P < 0.01 relative to 4T1-WT. (Scale bars, 10 µm.) (C) Costaining Tn (VVL) with the lamellipodia marker Tks5/FISH in model 4T1 cells. (Scale bars, 10 µm.) (D) Quantification of VVL/Tks5 staining ratios from lamellipodia in model 4T1 cells (n = 10 images per sample). *P < 0.01 relative to untreated cells. (E) Mean adhesive strength ± SEM of model 4T1 cells. *P < 0.01, **P < 0.05 relative to 4T1-WT. (F) Mean adhesive strength ± SEM after VVL preincubation for 20 min. *P < 0.01 relative to untreated cells. (G) Mean of wound closure ± SEM of 4T1-TSM and 4T1-ER-2Lec (n = 2 experiments). PNA- and VVL-treated experimental samples from Fig. 5E are shown for comparison. *P < 0.01 relative to PNA-treated cells or stated samples. Nuclei stained using Hoechst.
ER-GFP, ER-TSM, and ER-2Lec were stably and homogeneously expressed at similar levels in 4T1 cells and colocalized with the ER-marker Calreticulin but not the Golgi-marker Giantin, as confirmed by high and low MPCC values, respectively (Fig. S8 A and B, and Table S1). Proliferative growth was comparable between these cell lines in vitro (Fig. S8C). In contrast, ER-2Lec expression reduced intracellular and surface Tn levels by ∼40% (Fig. 6B). Neither ER-GFP nor ER-TSM expression affected cellular Tn levels in 4T1 cells, confirming that the inhibitory effect was dependent on the presence of lectin domains that can bind O-GalNAc (Fig. 6B). Inhibition of glycosylation by ER-2Lec, however, was not complete, as residual Tn staining in the ER was apparent. Nevertheless, the inhibitor was ER-specific as its expression in HeLa cells (with mostly Golgi-localized GalNAc-Ts) did not significantly affect Tn levels (nor cell proliferation) (Fig. S8 D–F).
Notably, ER-2Lec expression in 4T1 cells specifically reduced peripheral Tn staining localized in Tks5/FISH-positive lamellipodia (Fig. 6C). The Tn/Tks5 staining ratio in the lamellipodia of ER-2Lec-expressing cells was reduced by about 50% compared with those of wild-type, ER-GFP, and ER-TSM cells (Fig. 6D).
ER-2Lec expression in 4T1 cells reduced adhesion to fibronectin by about 40% and to collagen-I and -IV by about 30% each, but ER-GFP expression had no significant effect (Fig. 6E). Comparatively, ER-2Lec expression did not affect HeLa cell adhesion (Fig. S8G) or migration on fibronectin-coated surfaces in a scratch wound-healing experiment (Fig. S7 C and D and Movies S8 and S11), demonstrating the specificity of the inhibitor. Furthermore, combining soluble lectin treatment and the expression of ER-2Lec in 4T1 cells did not yield an additive effect, suggesting that both approaches ultimately affect similar glycoproteins (Fig. 6F). Consistently, live 4T1-ER-2Lec cells in these adhesion experiments bound about 80% less VVL than that seen for 4T1-ER-GFP cells, indicating that they contain lower amounts of Tn-bearing proteins at the cell surface (Fig. S8H). Finally, ER-2Lec expression inhibited the migratory potential of 4T1 cells in a scratch wound-healing assay to a similar extent as that observed following incubation with 40 µg/mL VVL; ER-TSM expression, on the other hand, had little effect (Fig. 6G and Movies S12 and S13).
Taken together, these results suggest that ER-2Lec inhibits ER-localized O-GalNAc glycosylation of proteins that mediate cellular adhesion and migration.
ER-Localized O-Glycosylation Promotes Metastasis in Vivo.
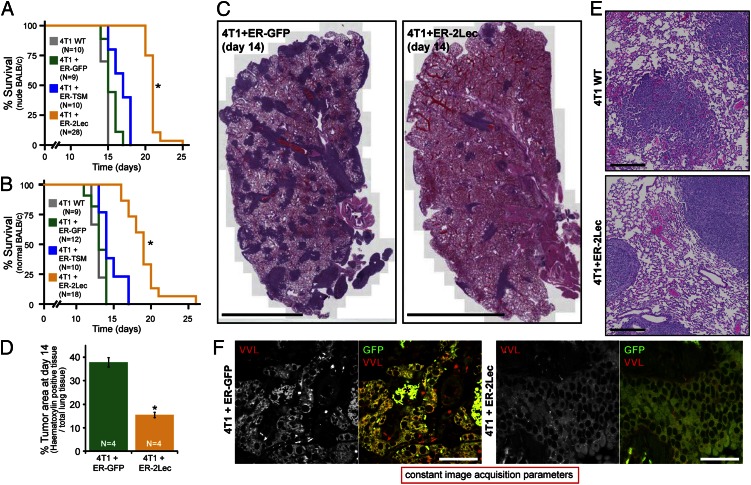
The metastatic potential of the 4T1 cells with or without ER-2Lec inhibitor was evaluated by monitoring nude and nonnude BALB/c mice survival rates after tail-vein injection. Nude mice injected with 4T1-ER-2Lec cells survived longer (median, 21 d) than mice injected with wild-type 4T1 (15 d), 4T1-ER-GFP cells (15 d), and 4T1-ER-TSM cells (17 d) (Fig. 7A). Similar results were obtained after tail-vein injection in nonnude BALB/c mice (Fig. 7B). Next, mice injected with 4T1 cells expressing either the ER-GFP or ER-2Lec constructs were killed at day 14 and the histological appearance of their lungs examined after H&E staining. The amount of metastatic lung nodules revealed by dark purple staining was markedly decreased when 4T1 cells expressed the ER-2Lec construct (Fig. 7C). The quantification using image analysis (Fig. S9A) revealed an average of 60% reduction in the tumor load (Fig. 7D).
Fig. 7.
Inhibiting ER-localized GalNAc-Ts lowers the invasiveness of 4T1 carcinoma cells in vivo. (A and B) Kaplan–Meier survival curves for nude (A) and nonnude (B) BALB/c mice injected with model 4T1 cells. *P < 0.0001 relative to 4T1-WT, 4T1-ER-GFP and 4T1-TSM (Log rank). For statistical comparison of wild-type and ER-2Lec curves: χ2 = 41.4 (nude) and 32.1 (nonnude); hazard ratio = 5.8 (nude) and 5.6 (nonnude). (C) H&E staining reveals metastatic nodules (dark purple) in the lung parenchyma of mice injected with model 4T1 cells. (Scale bars, 2 mm.) (D) Mean percentage of tumor area over total lung surface ± SEM in mice (n = 4) injected with transgenic 4T1 cells. *P < 0.01 relative to 4T1-WT cells. (E) Zoom-in illustrating the morphological differences between nodules. (Scale bars, 200 µm.) (F) Tn staining (VVL) of the lung parenchyma of mice injected with 4T1-ER-GFP and 4T1-ER-2Lec cells. 4T1 cells are marked by GFP fluorescence. (Scale bars, 50 µm.) An asterisk represents erythrocytes.
At higher magnification, the 4T1-ER-2Lec nodules had more defined margins and less trabeculae structures of detaching cells than wild-type and 4T1-ER-GFP nodules, suggesting that inhibited cells invade into lung parenchyma less efficiently (Fig. 7E). A longer incubation time (up to 21 d) for the injected mice resulted in larger 4T1-ER-2Lec nodules, with the well-defined edges becoming even more apparent (Fig. S9B). VVL histological staining indicated that 4T1-ER-2Lec nodules had lower Tn expression relative to 4T1-ER-GFP nodules, highlighting that ER-2Lec remains functional in the tumor context (Fig. 7F).
In sum, ER-localized O-GalNAc glycosylation promotes efficient lung colonization, probably because it promotes extravasation and lung parenchyma invasion through its stimulatory effect on cell adhesion, cell motility and invasiveness.
Discussion
Our study proposes a mechanism to explain a glyco-phenotype observed in most malignant tumors that was first described 30 y ago: the increase in Tn levels (8). Although variations in glycan levels or glyco-phenotypes are usually explained in terms of enzyme expression (38), we find that Tn levels are strongly affected by GalNAc-T subcellular localization and therefore regulated at least in part by signaling and membrane trafficking processes. As GalNAc-T translocation to the ER is highly specific and depends on the COPI coat (30), our result suggests that, in malignant tumor cells, COPI retrograde transport of GalNAc-Ts is markedly up-regulated.
It has been previously shown that loss or inactivation of the main O-GalNAc modifying enzyme, C1GalT, drives Tn expression in a limited set of cancer cell lines and human cancer patients (26). In our study, we find no sign of C1GalT activity deficiency in hundreds of human breast carcinoma samples. In contrast, evidence of ER-localized GalNAc-Ts was found in 70% of high Tn breast carcinomas studied here, implying that the subcellular relocation of GalNAc-Ts is the driving mechanism for Tn levels in most breast tumors.
A rather surprising finding in our study is that most Tn in cancer cell lines and tumors is localized in the ER, and only a small fraction is present at the cell surface; this is in contrast to the effects observed upon C1GalT loss. To date, one of the few preclinical validations of Tn-targeting therapies used C1GalT deficient Jurkat cells as a xenograft cancer model (29, 39). The differences in the amount of cell surface Tn between cells displaying ER O-glycosylation versus C1GalT deficiency suggest it might be important to re-evaluate therapies targeting the Tn antigen. Indeed, these differences may perhaps partly explain the difficulties encountered in human breast cancer clinical trials for anti-Tn therapies (28, 40).
In the cancer cell lines we examined, high Tn levels appear to derive from a perturbation in intracellular signaling, leading to enhanced GalNAc-T retrograde traffic and O-glycosylation initiation in the ER. This finding suggests that ER O-glycosylation is not specific to cancer but rather a physiological phenomenon hyper-activated in tumor cells. Src-dependent signaling cascades known to stimulate GalNAc-T relocation, for example downstream of EGFR and PDGF-R, are frequently hyperactivated in cancer cells (41, 42) and Src activity itself has been shown to increase with tumor grade (43). Recently, we showed that several kinases and phosphatases act as negative regulators of GalNAc-T ER relocation, suggesting loss or reduction in their activity could also contribute to elevated Tn expression in tumor cells (44).
This tight signaling control of the subcellular location of GalNAc-Ts is made intelligible by the pronounced effects of O-GalNAc glycosylation initiation in the ER on cell behavior in vitro and in vivo. These effects likely stem from modification of multiple substrates. Indeed, we recently showed that several ER-resident proteins are O-GalNAc glycosylated (13). Furthermore, ER O-glycosylation could modulate protease processing of cell surface and secreted proteins through modification of cleavage sites targeted by proprotein convertases or disintegrin and metalloproteases (ADAM) (45, 46). Based on Tn-specific lectin staining, it appears that at least one protein affected by ER O-glycosylation reaches the cell surface.
It is possible that for this protein, some O-GalNAc residues are not extended upon trafficking through the Golgi because they are located in a GalNAc cluster, which is suspected to inhibit C1GalT activity (47). Indeed, we find here that the lectin domain of ER-localized GalNAc-Ts plays a critical role in enhancing cell adhesion and migration and the function of the lectin domain is presumably to promote GalNAc clusters formation (15).
The lamellipodial localization of the unknown Tn-bearing glycoproteins argues for a relatively direct role in promoting adhesion and cell motility, but obviously identification will be essential to establish underlying molecular mechanisms promoting cell migration and ECM degradation.
Our study reveals that controlling the subcellular localization of the glycosyltransferases, GalNAc-T, is an important element of control for signaling molecules, such as Src. The modifications catalyzed by these glycosylation enzymes could regulate a wide range of secreted, cell surface- and ER-located factors in a correlated manner, thus possibly facilitating the coordination of a cellular invasion program.
Materials and Methods
Cloning.
The human GALNT2 (NM-004481) pDONR221 clone was described previously (30). GALNT2 mutants were generated by PCR. ER-2Lec, ER-TSM, and ER-GFP were constructed by PCR before cloning into pDONR221. pFB-lenti-CGFP-DEST is a destination vector constructed by replacement of a V5 tag in pLenti6.3-DEST (Invitrogen) with emGFP. pFB-lenti-ssHGH-NGFP-DEST is a destination vector constructed by addition of a N-terminal emGFP tag (fused to an ER signal sequence from human growth hormone) to pLenti6.3-DEST. GALNT2 constructs (lacking 3′ STOP codons) were cloned into pFB-lenti-CGFP-DEST. ER-TSM, ER-2Lec and ER-GFP (with 3′ STOP codons) were cloned into pFB-lenti-ssHGH-NGFP-DEST. Human ARF1 (NM-001024228)-GFP expression clones were described previously (30). All construct sequences were verified before use.
Cell Culture.
HEK293T, MDA231, MDA468, Hs578T, MCF10A, BT474, BT549, and T47D cells were a gift from W. Hong (Institute of Molecular and Cell Biology, Singapore). HeLa cells were a gift from V. Malhotra (Centre for Genomic Regulation, Barcelona, Spain). 4T1 cells were a gift from Q. Zeng (Institute of Molecular and Cell Biology, Singapore). HUH6 cells were a gift from F. Saltel (Institut National de la Santé et de la Recherche Médicale, Bordeaux, France). Genetic ablation of Cosmc in MDA231 cells [Cosmc(Δ)] was performed using a strategy detailed previously (12). All cells were grown at 37 °C in a 10% CO2 incubator. HeLa, MDA231 [wild-type, GalNAc-T2-GFP expressing and Cosmc(Δ)], MDA468, Hs589T, HUH6, and T47D were grown in DMEM with 10% FBS. HEK293T cells were grown in DMEM with 15% FBS. BT474, BT549, and 4T1 (wild-type and lectin-GFP-expressing) cells were grown in RPMI with 10% FBS, 1× nonessential amino acids, 1× sodium pyruvate, and 1× sodium glutamate. MCF10A cells were growth in a 1:1 mixture of DMEM:HamsF12 with 10% FBS, 20 ng/mL EGF, 0.5 µg/mL hydrocortisone and 5 µg/mL insulin.
Lentiviral Particle Production and Cell Infection.
Lentiviral particles were generated by transient transfection of HEK293T cells with a mixture of pLenti, plasmid lentiviral packaging (PLP1), PLP2, and vesicular stomatitis virus-G plasmids using a calcium phosphate method (Invitrogen). The next day, the media was replaced, and two batches of media were harvested at 48-h intervals. These were combined, filtered with a 0.45-µm mesh and centrifuged at 38,700 × g for 3 h. Sedimented lentiviral particles were resuspended in 0.22-µm-filtered HBSS supplemented with 20% sucrose. Cells were infected with virus for 48 h and rapidly expanded. Cells infected with GFP virus were FACs sorted to enrich for expressing cells. Enriched cells were rapidly grown to large numbers and stored in liquid nitrogen. All experiments were performed on cells passaged fewer than 10 times after thawing.
Western Blot Analysis.
MDA-MB-231 cells stably expressing GALNT2 constructs were seeded into 10-cm dishes and grown to confluence (24–48 h). Procedures for cell harvesting and processing for Western blot are outlined as described previously (30).
Adhesion, Invasion, Wound Healing, and Growth Assays.
Wild-type MDA231 cells and those expressing GALNT2 constructs or wild-type 4T1 cells and those expressing ER-GFP, ER-TSM and ER-2Lec were grown to 70–80% confluence before passaging with trypsin (growth and wound healing assays) or 2 mM EDTA (adhesion and invasion assays) to obtain well-separated cell suspensions. Coulter (Beckman Coulter) or Countess (Invitrogen) systems were used to accurately determine initial seeding densities. For growth assays, 7,500–15,000 cells were seeded into each well of a 24-well tissue culture plate (Nunc). Three independent wells were seeded per sample per timepoint. Final cell number was measured using a Coulter system. For adhesion assays, 10,000 (HeLa) or 20,000 (4T1, MDA231) cells were seeded into 96-well black imaging plates (BD Falcon) precoated for 2 h with ECM proteins. In experiments using VVL or PNA preincubation, suspended cells were added to various concentrations of lectin before shaking gently (30 rpm) for 20 min at room temperature and eventual seeding into the coated imaging plates. After 30-min incubation at 37 °C and 10% CO2, wells were washed twice with 0.1% BSA/DMEM (aspirated up/down five times per wash) before staining with Hoescht for 1 h. Cells were imaged using an ImageXpressMICRO wide-field microscope with a 10× Plan Fluor objective and computationally counted (Molecular Devices). For wound-healing assays, cells were seeded onto fibronectin-coated (as above) 35-mm plastic tissue-culture dishes (Ibidi) and grown to confluence (16–24 h). A wound was generated using micropipette tip before washing to remove cell debris. In scratch experiments under VVL (4 or 40 µg/mL) or PNA (40 µg/mL) treatment, the soluble lectin was added 20 min before wound generation. Live phase contrast imaging was performed at 37 °C using a Zeiss Axiovert microscope (model 200M) (Oberkochen) equipped with a CCD camera (AxioCam HRc) with a 20× objective (LD Plan-NEOFLUAR; 20×; N.A. 0.4). Frames were acquired at 3- to 5-min intervals. Areas of wound invasion were calculated using ImageJ (National Institutes of Health). Boyden chamber migration assays were performed as per the manufacturer’s instructions using a cell invasion assay kit (ECM554; Millipore). Cells were incubated for 24 h at 37 °C and 10% CO2 before quantification of invaded cell number.
In Vivo Metastases Assay.
All animal protocols used in this study were approved by Institute of Molecular and Cell Biology Animal Safety and Use Committee. Cells (500,000) were injected in the tail vein of 8- to 12-wk-old nude or nonnude BALB/c mice. Mice were closely monitored and necropsied once moribund or at desired time points. Tissues were fixed in 10% neutral buffered formalin overnight and embedded in paraffin. Sections (5-μm-thick) were stained with a standard protocol for H&E staining. Slides were scanned at 20× using a Leica SCN400 slide scanner (Leica Microsystems). Images were exported to Slidepath Digital Image Hub (Leica Microsystems) for viewing. Images were analyzed using Measure Stained Area algorithms of Slidepath TissueIA software. Data were collated using Microsoft Excel. Scanning and image analysis was performed by the Advanced Molecular Pathology Laboratory, Institute of Molecular and Cell Biology, Singapore.
Immunofluorescence Imaging.
Details are provided in SI Materials and Methods.
Statistical Analysis.
Details are provided in SI Materials and Methods.
Human Tumor Microarray Imaging.
Details are provided in SI Materials and Methods.
Human Tumor Gene Expression Datasets Analysis.
Expression data for both C1GALT and C1GALT1C1 (alias COSMC) were assessed in the datasets E-GEOD-10780 and E-TABM-276, accessible at the European Bioinformatics Institute Expression Atlas (www.ebi.ac.uk/gxa).
Supplementary Material
Acknowledgments
We thank Pernille Roth and Steve Cohen for critical reading and discussion; Jerrold Ward, Keith Rogers, and Susan Rogers at the Institute of Molecular and Cell Biology Histopathology core; and Biomax for the use of images of H&E staining of the breast carcinoma cores (images also at www.biomax.us). This work was supported by the Agency for Science, Technology and Research, Singapore and the Danish Research Foundation (DNRF107).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305269110/-/DCSupplemental.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Teodorczyk M, Martin-Villalba A. Sensing invasion: Cell surface receptors driving spreading of glioblastoma. J Cell Physiol. 2010;222(1):1–10. doi: 10.1002/jcp.21901. [DOI] [PubMed] [Google Scholar]
- 3.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau KS, Dennis JW. N-Glycans in cancer progression. Glycobiology. 2008;18(10):750–760. doi: 10.1093/glycob/cwn071. [DOI] [PubMed] [Google Scholar]
- 5.Ungar D. Golgi linked protein glycosylation and associated diseases. Semin Cell Dev Bioly. 2009;20(7):762–769. doi: 10.1016/j.semcdb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Laack E, et al. Lectin histochemistry of resected adenocarcinoma of the lung: Helix pomatia agglutinin binding is an independent prognostic factor. Am J Pathol. 2002;160(3):1001–1008. doi: 10.1016/S0002-9440(10)64921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konno A, Hoshino Y, Terashima S, Motoki R, Kawaguchi T. Carbohydrate expression profile of colorectal cancer cells is relevant to metastatic pattern and prognosis. Clin Exp Metastasis. 2002;19(1):61–70. doi: 10.1023/a:1013879702702. [DOI] [PubMed] [Google Scholar]
- 8.Springer GF. T and Tn, general carcinoma autoantigens. Science (New York, NY) 1984;224(4654):1198–1206. doi: 10.1126/science.6729450. [DOI] [PubMed] [Google Scholar]
- 9.Newman RA, Uhlenbruck GG. Investigation into the occurrence and structure of lectin receptors on human and bovine erythrocytes milk fat globule and lymphocyte plasma-membrane glycoproteins. Eur J Biochem. 1977;76(1):149–155. doi: 10.1111/j.1432-1033.1977.tb11580.x. [DOI] [PubMed] [Google Scholar]
- 10.Tollefsen SE, Kornfeld R. The B4 lectin from Vicia villosa seeds interacts with N-acetylgalactosamine residues alpha-linked to serine or threonine residues in cell surface glycoproteins. J Biol Chem. 1983;258(8):5172–5176. [PubMed] [Google Scholar]
- 11.Bennett EP, et al. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology. 2012;22(6):736–756. doi: 10.1093/glycob/cwr182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steentoft C, et al. Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat Methods. 2011;8(11):977–982. doi: 10.1038/nmeth.1731. [DOI] [PubMed] [Google Scholar]
- 13.Steentoft C, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ten Hagen KG, Fritz TA, Tabak LA. All in the family: The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13(1):1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 15.Wandall HH, et al. The lectin domains of polypeptide GalNAc-transferases exhibit carbohydrate-binding specificity for GalNAc: Lectin binding to GalNAc-glycopeptide substrates is required for high density GalNAc-O-glycosylation. Glycobiology. 2007;17(4):374–387. doi: 10.1093/glycob/cwl082. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen JW, et al. Lectin domains of polypeptide GalNAc transferases exhibit glycopeptide binding specificity. J Biol Chem. 2011;286(37):32684–32696. doi: 10.1074/jbc.M111.273722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue M, et al. High density O-glycosylation of the MUC2 tandem repeat unit by N-acetylgalactosaminyltransferase-3 in colonic adenocarcinoma extracts. Cancer Res. 2001;61(3):950–956. [PubMed] [Google Scholar]
- 18.Röttger S, et al. Localization of three human polypeptide GalNAc-transferases in HeLa cells suggests initiation of O-linked glycosylation throughout the Golgi apparatus. J Cell Sci. 1998;111(Pt 1):45–60. doi: 10.1242/jcs.111.1.45. [DOI] [PubMed] [Google Scholar]
- 19.Pavelka M, Ellinger A. Localization of binding sites for concanavalin A, Ricinus communis I and Helix pomatia lectin in the Golgi apparatus of rat small intestinal absorptive cells. J Histochem Cytochem. 1985;33(9):905–914. doi: 10.1177/33.9.4020101. [DOI] [PubMed] [Google Scholar]
- 20.Ju T, Brewer K, D’Souza A, Cummings RD, Canfield WM. Cloning and expression of human core 1 beta1,3-galactosyltransferase. J Biol Chem. 2002;277(1):178–186. doi: 10.1074/jbc.M109060200. [DOI] [PubMed] [Google Scholar]
- 21.Ju T, Cummings RD. A unique molecular chaperone Cosmc required for activity of the mammalian core 1 beta 3-galactosyltransferase. Proc Natl Acad Sci USA. 2002;99(26):16613–16618. doi: 10.1073/pnas.262438199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stanley P (2011) Golgi glycosylation. Cold Spring Harb Perspect Biol 3(4):pii a005199. [DOI] [PMC free article] [PubMed]
- 23.Thurnher M, Clausen H, Fierz W, Lanzavecchia A, Berger EG. T cell clones with normal or defective O-galactosylation from a patient with permanent mixed-field polyagglutinability. Eur J Immunol. 1992;22(7):1835–1842. doi: 10.1002/eji.1830220724. [DOI] [PubMed] [Google Scholar]
- 24.Ju T, Cummings RD. Protein glycosylation: Chaperone mutation in Tn syndrome. Nature. 2005;437(7063):1252. doi: 10.1038/4371252a. [DOI] [PubMed] [Google Scholar]
- 25.Crew VK, et al. New mutations in C1GALT1C1 in individuals with Tn positive phenotype. Br J Haematol. 2008;142(4):657–667. doi: 10.1111/j.1365-2141.2008.07215.x. [DOI] [PubMed] [Google Scholar]
- 26.Ju T, Otto VI, Cummings RD. The Tn antigen-structural simplicity and biological complexity. Angew Chem Int Ed Engl. 2011;50(8):1770–1791. doi: 10.1002/anie.201002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo-Man R, et al. A fully synthetic therapeutic vaccine candidate targeting carcinoma-associated Tn carbohydrate antigen induces tumor-specific antibodies in nonhuman primates. Cancer Res. 2004;64(14):4987–4994. doi: 10.1158/0008-5472.CAN-04-0252. [DOI] [PubMed] [Google Scholar]
- 28.Miles D, et al. Theratope® Study Group Phase III multicenter clinical trial of the sialyl-TN (STn)-keyhole limpet hemocyanin (KLH) vaccine for metastatic breast cancer. Oncologist. 2011;16(8):1092–1100. doi: 10.1634/theoncologist.2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ju T, et al. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008;68(6):1636–1646. doi: 10.1158/0008-5472.CAN-07-2345. [DOI] [PubMed] [Google Scholar]
- 30.Gill DJ, Chia J, Senewiratne J, Bard F. Regulation of O-glycosylation through Golgi-to-ER relocation of initiation enzymes. J Cell Biol. 2010;189(5):843–858. doi: 10.1083/jcb.201003055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.High S, Lecomte FJ, Russell SJ, Abell BM, Oliver JD. Glycoprotein folding in the endoplasmic reticulum: A tale of three chaperones? FEBS Lett. 2000;476(1–2):38–41. doi: 10.1016/s0014-5793(00)01666-5. [DOI] [PubMed] [Google Scholar]
- 32.Yuan M, et al. Comparison of T-antigen expression in normal, premalignant, and malignant human colonic tissue using lectin and antibody immunohistochemistry. Cancer Res. 1986;46(9):4841–4847. [PubMed] [Google Scholar]
- 33.Cheng AS, et al. Epithelial progeny of estrogen-exposed breast progenitor cells display a cancer-like methylome. Cancer Res. 2008;68(6):1786–1796. doi: 10.1158/0008-5472.CAN-07-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen DT, et al. Proliferative genes dominate malignancy-risk gene signature in histologically-normal breast tissue. Breast Cancer Res Treat. 2010;119(2):335–346. doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Humphries MJ, Olden K, Yamada KM. A synthetic peptide from fibronectin inhibits experimental metastasis of murine melanoma cells. Science. 1986;233(4762):467–470. doi: 10.1126/science.3726541. [DOI] [PubMed] [Google Scholar]
- 36.Hood JD, Cheresh DA. Role of integrins in cell invasion and migration. Nat Rev Cancer. 2002;2(2):91–100. doi: 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- 37.Lelekakis M, et al. A novel orthotopic model of breast cancer metastasis to bone. Clinical & experimental metastasis. 1999;17(2):163–170. doi: 10.1023/a:1006689719505. [DOI] [PubMed] [Google Scholar]
- 38.Lauc G, Rudan I, Campbell H, Rudd PM. Complex genetic regulation of protein glycosylation. Mol Biosyst. 2010;6(2):329–335. doi: 10.1039/b910377e. [DOI] [PubMed] [Google Scholar]
- 39.Welinder C, Baldetorp B, Borrebaeck C, Fredlund BM, Jansson B. A new murine IgG1 anti-Tn monoclonal antibody with in vivo anti-tumor activity. Glycobiology. 2011;21(8):1097–1107. doi: 10.1093/glycob/cwr048. [DOI] [PubMed] [Google Scholar]
- 40.Heimburg-Molinaro J, et al. Cancer vaccines and carbohydrate epitopes. Vaccine. 2011;29(48):8802–8826. doi: 10.1016/j.vaccine.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Normanno N, et al. Epidermal growth factor receptor (EGFR) signaling in cancer. Gene. 2006;366(1):2–16. doi: 10.1016/j.gene.2005.10.018. [DOI] [PubMed] [Google Scholar]
- 42.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22(10):1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guarino M. Src signaling in cancer invasion. J Cell Physiol. 2010;223(1):14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 44.Chia J, et al. RNAi screening reveals a large signaling network controlling the Golgi apparatus in human cells. Mol Syst Biol. 2012;8:629. doi: 10.1038/msb.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schjoldager KT, et al. A systematic study of site-specific GalNAc-type O-glycosylation modulating proprotein convertase processing. J Biol Chem. 2011;286(46):40122–40132. doi: 10.1074/jbc.M111.287912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schjoldager KT, Clausen H. Site-specific protein O-glycosylation modulates proprotein processing–Deciphering specific functions of the large polypeptide GalNAc-transferase gene family. Biochim Biophys Acta. 2012;1820(12):2079–2094. doi: 10.1016/j.bbagen.2012.09.014. [DOI] [PubMed] [Google Scholar]
- 47.Gerken TA. Kinetic modeling confirms the biosynthesis of mucin core 1 (beta-Gal(1-3) alpha-GalNAc-O-Ser/Thr) O-glycan structures are modulated by neighboring glycosylation effects. Biochemistry. 2004;43(14):4137–4142. doi: 10.1021/bi036306a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.