Abstract
Small molecule iron-chelators, siderophores, are very important in facilitating the acquisition of Fe(III), an essential element for pathogenic bacteria. Many Gram-negative outer-membrane transporters and Gram-positive lipoprotein siderophore-binding proteins have been characterized, and the binding ability of outer-membrane transporters and siderophore-binding proteins for Fe-siderophores has been determined. However, there is little information regarding the binding ability of these proteins for apo-siderophores, the iron-free chelators. Here we report that Bacillus cereus YxeB facilitates iron-exchange from Fe-siderophore to apo-siderophore bound to the protein, the first Gram-positive siderophore-shuttle system. YxeB binds ferrioxamine B (FO, Fe-siderophore)/desferrioxamine B (DFO, apo-siderophore) in vitro. Disc-diffusion assays and growth assays using the yxeB mutant reveal that YxeB is responsible for importing the FO. Cr-DFO (a FO analog) is bound by YxeB in vitro and B. cereus imports or binds Cr-DFO in vivo. In vivo uptake assays using Cr-DFO and FO and growth assays using DFO and Cr-DFO show that B. cereus selectively imports and uses FO when DFO is present. Moreover, in vitro competition assays using Cr-DFO and FO clearly demonstrate that YxeB binds only FO, not Cr-DFO, when DFO is bound to the protein. Iron-exchange from FO to DFO bound to YxeB must occur when DFO is initially bound by YxeB. Because the metal exchange rate is generally first order in replacement ligand concentration, protein binding of the apo-siderophore acts to dramatically enhance the iron exchange rate, a key component of the Gram-positive siderophore-shuttle mechanism.
Iron is terrestrially abundant but biologically scarce because of the low aqueous solubility of Fe(III). However, most organisms depend upon iron as a cofactor for essential processes, including oxygen binding, electron transfer, and catalysis (1). The supply and demand for iron has caused animals, plants, and microorganisms to develop multicomponent systems that increase the availability of iron and transport it to cells. Bacteria secrete small-molecules called siderophores that bind Fe(III) with high affinity and solubilize it (2). Then siderophore-specific transporter systems import the Fe-siderophore across the cell membranes to the cytoplasm (2).
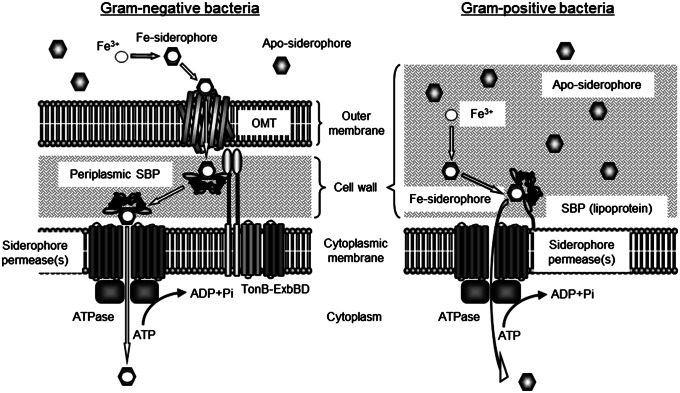
Fe-siderophore transport systems in Gram-positive and Gram-negative bacteria differ (Fig. 1). Gram-negative bacteria use outer-membrane transporters (OMTs), such as Escherichia coli FecA (ferric citrate transporter) (3) and FhuA (ferrichrome transporter) (4), to recognize and bind extracellular Fe-siderophores. Binding a Fe-siderophore signals the TonB-ExbBD system to move the substrate across the outer membrane from the OMT to a periplasmic siderophore-binding protein (periplasmic SBP) (5). The SBP then delivers the Fe-siderophore to the appropriate siderophore-permease(s)-ATPase system to be transported through the inner membrane to the cytoplasm (2). Much less is known about the iron transport of Gram-positive bacteria, even though many are among the most dangerous human pathogens. These bacteria do not have siderophore-binding OMTs. Instead, lipoprotein SBPs anchored to the cell membrane bind extracellular Fe-siderophores to be imported by a siderophore-permease(s)-ATPase system (6). The lipoprotein SBP-permease(s)-ATPase system in Gram-positive bacteria is similar to the periplasmic SBP-permease(s)-ATPase system in Gram-negative bacteria (Fig. 1).
Fig. 1.
Siderophore uptake machineries in Gram-negative bacteria (Left) and Gram-positive bacteria (Right). Gram-negative bacteria possess an OMT of Fe-siderophores. After an OMT recognizes a siderophore, the Fe-siderophore is transferred to a periplasmic SBP using the TonB-ExbBD system (5). The Fe-siderophore bound to the SBP is then imported using the appropriate siderophore-permeases and ATPase. In Gram-positive bacteria, a lipoprotein SBP anchored to the membrane binds a siderophore, and the Fe-siderophore is imported using its siderophore-permeases and ATPase.
Some Gram-negative OMTs and Gram-positive lipoprotein SBPs can bind not only Fe-siderophores but also apo-siderophores. Of Gram-negative OMTs, FhuA of E. coli binds desferrichrome (DFch) and FptA of Pseudomonas aeruginosa binds apo-pyochelin strongly enough that they copurify (7, 8). Of Gram-positive SBPs, YclQ of Bacillus subtilis binds apo-petrobactin (PB) with a dissociation constant (Kd) of 35 nM, whereas it binds Fe-PB with a Kd of 113 nM, indicating that the SBP more strongly binds apo-PB (9). Additionally, FeuA, FpuA, and FatB of Bacillus cereus bind apo-siderophores. The Kds of FeuA for apo-enterobactin (Ent) and Fe-Ent are 36 and 12 nM, respectively. The Kds of FpuA for apo-PB and Fe-PB are 23 and 175 nM, respectively, and the Kds of FatB for apo-PB and Fe-PB are 77 and 127 nM, respectively (10). This finding suggests that several Gram-negative OMTs and Gram-positive SBPs bind apo-siderophores, especially under iron-limited conditions.
It was puzzling to us why Gram-negative OMTs and Gram-positive SBPs involved in iron-transport would bind apo-siderophores; an apo-siderophore is structurally and electronically different from a Fe-siderophore, and usually receptor proteins precisely recognize a specific substrate. For example, the Pseudomonas putida receptor OprB binds glucose but not the related molecules glucuronic acid or maltose (11). In B. subtilis, the membrane protein CitM transports Mg(II)-citrate but not Ca(II)-citrate, and CitH transports Ca(II)-citrate but not Mg(II)-citrate (12, 13). Thus, receptor proteins precisely recognize substrates; hence, the binding of apo-siderophores by OMTs and SBPs should serve a function.
What is the function served by bacterial OMTs and SBPs binding apo-siderophores? One possibility is that apo-siderophores bound to an OMT or SBP can catch Fe(III) from the extracellular milieu and enable iron transport, even if the bacteria do not have a Fe(III) transporter system like B. subtilis YwbLMN (6). Another possibility is that a “siderophore-shuttle” system (14) uses the apo-siderophores to efficiently import Fe(III). The siderophore-shuttle mechanism demonstrated by Stintzi et al. begins with an OMT bound to an apo-siderophore (14) because the concentration of apo-siderophore at the cell surface, where the apo-siderophores are secreted, will be higher than the concentration of Fe-siderophore (7, 8). An Fe-siderophore is then bound by the same OMT, and the increased local concentration near the binding pocket facilitates iron exchange from the Fe-siderophore to the apo-siderophore. This step is the salient feature of the siderophore shuttle; Fe(III) is kinetically labile so as to enable iron exchange (15), although the nonfacilitated iron exchange rate between Fe- and apo-hydroxamate siderophores is extremely slow, with a half-life of nearly 10 d (16). The newly formed Fe-siderophore continues into the cell and the former Fe-siderophore remains in the OMT as an apo-siderophore, ready to participate in the next siderophore-shuttle (14).
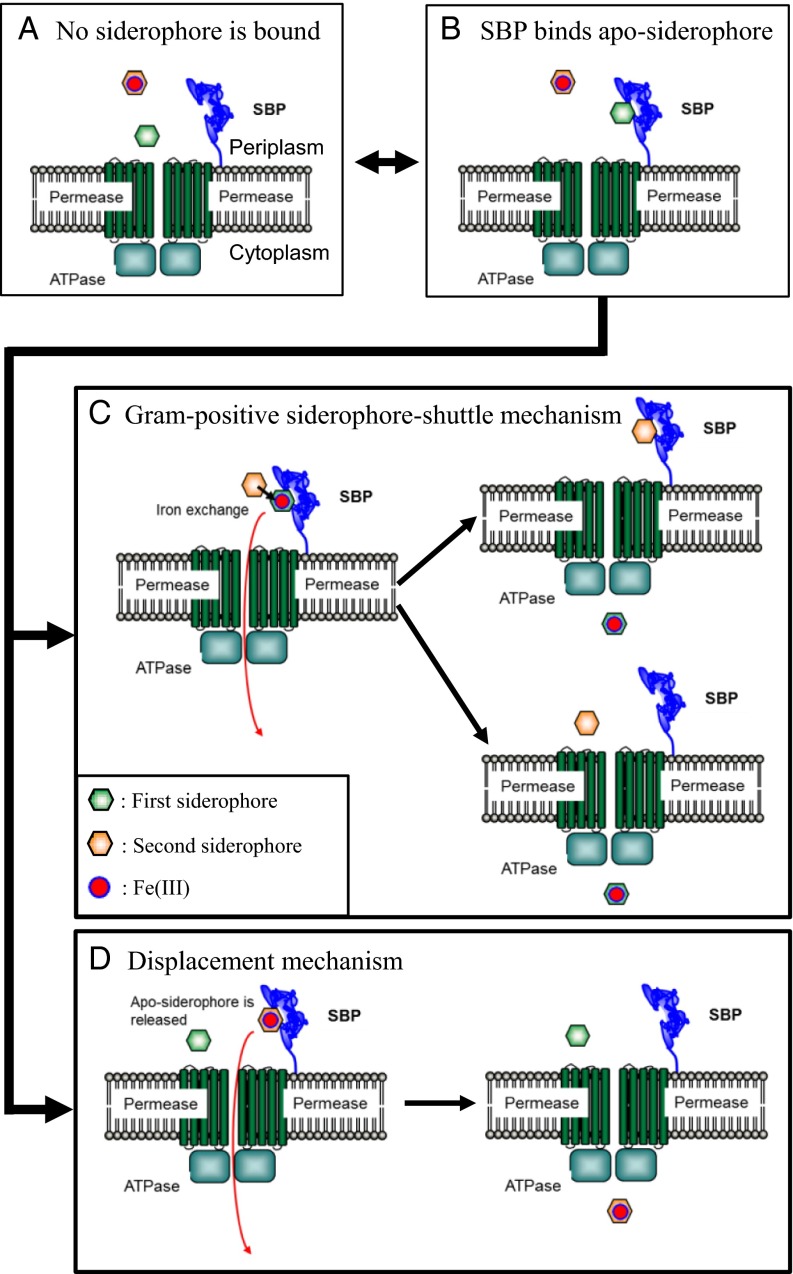
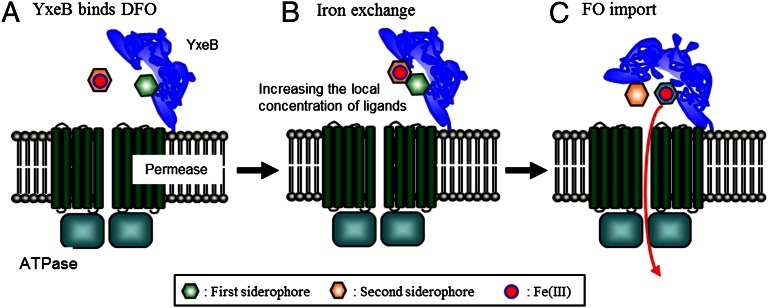
The siderophore-shuttle in Gram-negative bacteria depends on the ability of OMTs to bind apo-siderophores. For the Gram-positive siderophore shuttle mechanism, a lipoprotein SBP binds an apo-siderophore (Fig. 2). Then a Fe-siderophore interacts with the SBP near the apo-siderophore. The increased local concentration complex facilitates iron exchange to the apo-siderophore, and the new Fe-siderophore is passed through the permeases to the cytoplasm (Fig. 2C). The alternative uptake mechanism, when an apo-siderophore is initially bound to the SBP, we will call the “displacement mechanism” (Fig. 2D). In this mechanism, the Fe-siderophore displaces the apo-siderophore from the SBP. No iron exchange takes place, and the original Fe-siderophore passes through the permeases to the cytoplasm. Iron exchange is the distinguishing feature of the two mechanisms.
Fig. 2.
Possible Fe-siderophore uptake systems in Gram-positive bacteria. (A) No siderophore is bound to the SBP. In the situation when Fe-siderophore is bound to the SBP, the siderophore is imported in cytoplasm. (B) On the other hand, when apo-siderophore is bound to the SBP, two Fe-siderophore uptake mechanisms, the Gram-positive siderophore-shuttle mechanism (C) or the displacement mechanism (D), are possible. (C) In the Gram-positive siderophore-shuttle mechanism, iron is transferred from an Fe-siderophore to the apo-siderophore:SBP complex and the new Fe-siderophore is then imported into the cytoplasm. It is not clear whether the resulting apo-siderophore remains bound to the SBP or not after iron-exchange has occurred. (D) In the displacement mechanism, the apo-siderophore is released from the SBP, and the Fe-siderophore occupies the binding pocket. No iron exchange occurs, and the original Fe-siderophore is transported to the cytoplasm.
B. cereus ATCC 14579 uses a lipoprotein SBP called YxeB to bind and import FO (ferrioxamine B) and Fch (10). These two siderophores deliver iron through YxeB, even though B. cereus does not produce the corresponding apo-siderophores DFO (desferrioxamine B) and DFch (17). We report that YxeB uses a Gram-positive siderophore-shuttle mechanism to transport Fe-siderophores when apo-siderophore is present.
Results
YxeB Binds DFO, FO, DFch, and Fch.
Previously, Zawadzka et al. demonstrated that YxeB (BC_0383) binds Fe-siderophores, FO and Fch (10). However, it was unknown if the protein also binds apo-siderophores, DFO or DFch (Fig. S1A). To begin our study of YxeB, the yxeB gene in B. cereus ATCC 14579 was sequenced. Sequencing revealed two different nucleotides in the gene compared with the sequence in the National Center for Biotechnology Information (NCBI). One nucleotide, G555, in the database (the number is with respect to the first nucleotide of the yxeB translational start codon) is incorrect, and the correct nucleotide is A555. The other nucleotide has two variations, TT425A and TC425A, in the laboratory stock. The yxeB genes with TT425A and TC425A encode YxeB-L142 (residue 142 is Leu) and YxeB-S142 (residue 142 is Ser), respectively. Both YxeB-L142 and YxeB-S142 were used in the following fluorescence-quenching assays to measure the binding affinity for several substrates.
The quenching assays of YxeB-L142-6×His show that the protein fluorescence was quenched by FO and Fch (Fig. S1 B and C). The data were fit to a one-to-one binding model using Hyperquad (18) to determine Kds. The Kds for FO and Fch were 38.8 nM and 43.0 nM, respectively (Table 1). Significantly, the protein fluorescence increased upon addition of DFO or DFch, the same as previously reported by Zawadzka et al. for YxeB-V5(epitope tag)-6×His (10). Thus, it is possible that the increasing fluorescence of the YxeB-L142 protein is caused by substrate binding. To confirm this theory, nano–ESI-MS (electrospray ionization-mass spectrometry) analysis of the protein and DFch or Fch complexes was performed. The data show that the protein formed complexes with DFch and Fch (Fig. S2 and Table S1). Additionally, the YxeB-L142 protein mixed with DFO or FO was purified and then analyzed by reverse-phase (RP)-HPLC showing that the protein had bound DFO and FO (Fig. S3 B and F). Thus, it is clear that the increasing fluorescence of the protein is because of siderophore binding.
Table 1.
Dissociation constants (Kd) of YxeB-L142-6×His and YxeB-S142-6×His
A fluorescence-quenching assay of the YxeB-S142 protein was also performed. The fluorescence was quenched by DFO, FO, DFch, and Fch (Fig. S1 D and E) and the calculated Kds for the substrates by Hyperquad (18) were very similar (Table 1). Thus, YxeB-S142 has similar affinity for both the apo and ferric forms of siderophores.
YxeB Is the Sole FO/Fch-Binding Protein.
B. cereus ATCC 14579 produces only PB and BB (bacillibactin), yet it possesses at least 10 genes encoding siderophore binding proteins (17) (B. cereus ATCC 14579 gene annotation in the NCBI genome database). It has been demonstrated that the gene products YxeB, YfiY, FeuA, and FpuA/FatB are a DFO/Fch, schizokinen, Ent/BB, and PB (FpuA and FatB)-binding proteins, respectively (10). FctC was recently identified as a triferric tricitrate-binding protein (19). The FO/Fch-binding ability of the other less characterized siderophore-binding proteins of B. cereus, BC_2208, BC_4363, BC_4416, and BC_5380, was assessed, and none of these proteins bind FO or Fch (Fig. S4). Thus, YxeB is the only DFO/Fch-binding protein in vitro.
To confirm that YxeB is the only DFO/Fch-binding protein in vivo, the yxeB markerless mutant was constructed (Materials and Methods). Because yxeB and the downstream genes, BC_0382 and BC_0381, make an operon, only yxeB is disrupted in the constructed strain, TC111 (yxeB−), while preserving the downstream genes. Fig. S5 A and B show the growth assay of TC111, TC129 (YxeB-L142), and TC128 (YxeB-S142) strains. This assay uses iron-limited minimum medium, but DFO can chelate Fe(III) from the medium even though the iron concentration is very low. The growth of TC129 and TC128 with DFO was better than the growth without DFO, indicating that both the strains can import and use FO. On the other hand, the growth of TC111 with DFO was not better than the growth without DFO, showing that the TC111 strain cannot use FO. This result also shows that PB and BB produced by B. cereus during the experiment do not affect growth in these conditions.
To further assess whether TC111 can use FO and Fch or not, a disc-diffusion assay was performed. In this experiment the cells grow around a disc containing FO or Fch if the substrates can be used. As shown in Fig. S5C, the wild-type strains, TC129 and TC128, grew in halos around the discs containing FO and Fch. However, TC111 did not grow around the discs with FO and Fch, although the strain grew around a disc containing BB (the positive control substrate), indicating that it cannot use FO and Fch. Therefore, the in vivo growth assay and disc-diffusion assay strongly suggest that yxeB is the sole FO/Fch-binding protein. The other SBPs including YfiY, FeuA, FpuA, FatB, and FctC are not associated with FO/Fch uptake. Moreover, Fe(III) coordinated with FO is not transferred to BB and PB produced by B. cereus under the iron-limited condition because the yxeB mutant, TC111, did not grow well.
Cr-DFO Is a FO Analog.
Because Cr(III) is kinetically inert and will not exchange from one siderophore to another on the experimental timescale (15, 20), Cr-DFO was synthesized to probe the presence of metal exchange in the siderophore transport mechanism of YxeB. The affinity of YxeB for Cr-DFO was measured using the fluorescence-quenching assay. The YxeB-L142 protein was quenched by Cr-DFO, and the quenching data were fit by Hyperquad (18) to determine the Kd (Fig. S1B). The Kd for Cr-DFO was similar to the Kd for FO (Kd for Cr-DFO, 69.0 nM; Kd for FO, 38.8 nM) (Table 1). The YxeB-S142 protein was also quenched by both substrates (Fig. S1D). The Kds for Cr-DFO and FO were, respectively, 98.9 nM and 29.1 nM (Table 1). Thus, Cr-DFO can be used as a FO analog, especially with YxeB-L142, as a metal-exchange probe.
To see if B. cereus ATCC 14579 can import Cr-DFO like FO, the amount of chromium derived from Cr-DFO in whole cells was measured using inductively coupled plasma (ICP) elemental analysis. The wild-type strains, TC129 and TC128, could uptake Cr-DFO, whereas the yxeB mutant, TC111 could not (Fig. S6A). Thus, YxeB can import the kinetically inert Cr-DFO.
Cr-DFO/DFO Growth Assay Shows That Cr-DFO Does Not Inhibit B. cereus Growth When DFO Is Present.
Because Cr-DFO is an exchange-inert FO analog (Figs. S1 and S6A), Cr-DFO was used as a FO competitor in growth assays. The optimal concentration of DFO for the growth assay with wild-type TC129 was determined. When the concentration of DFO in the culture was less than 10 nM, the growth was delayed compared with the growth at 10-nM or higher concentration of DFO, indicating that 10 nM DFO is the minimum amount for normal growth (Fig. S6B). Thus, the Cr-DFO growth assays were performed with 10 nM DFO. Including DFO in the medium creates an initial state with YxeB bound to apo-siderophore.
When DFO is not included, merely 10 nM Cr-DFO inhibits growth (Fig. S6 C and D). If B. cereus can import Cr-DFO by the displacement mechanism even when DFO is included in the culture, severe growth delay should be observed (the theory is shown in Fig. S7A). The growth of TC129 and TC128 was not delayed greatly even when 500 nM Cr-DFO, 50-times the DFO concentration, was added to the culture (Fig. S6 C and D). Cr-DFO is not an effective competitor for YxeB in the presence of DFO, which is evidence against the displacement mechanism.
In Vivo Cr-DFO/FO Uptake Assay Shows That YxeB Selectively Imports FO When DFO Is Present.
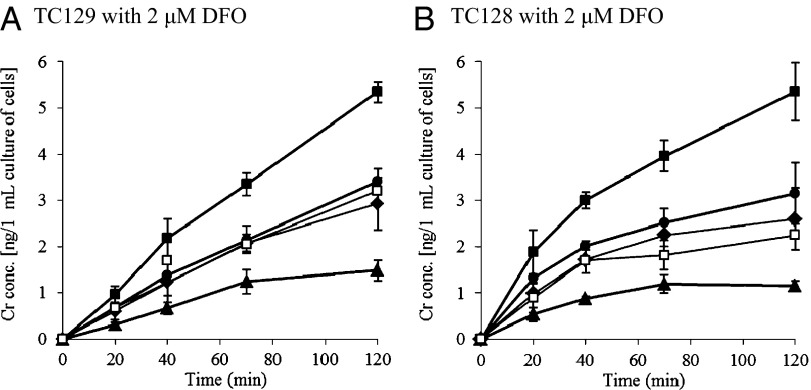
To observe if YxeB selects for FO over Cr-DFO when DFO is present in the culture, in vivo Cr-DFO/FO uptake assays were also performed (the theory is shown in Fig. S7B). In the experiment B. cereus produces BB and PB; however, these siderophores do not affect the experiment because YxeB is the only FO/Cr-DFO-binding protein (Figs. S4 and S5). When 2 μM Cr-DFO was added to cultures of TC129 or TC128 containing 2 μM DFO, 5 ng of chromium per 1 mL of cell culture was imported (closed squares in Fig. 3). When 2 μM FO was added along with 2 μM Cr-DFO to the cultures of TC129 and TC128 containing 2 μM DFO, the amount of imported chromium was drastically reduced to 1 ng of chromium per 1 mL of cell culture (triangles in Fig. 3). Significantly, even with 10-times lower FO concentration than Cr-DFO (0.2 μM FO, 1:10 FO:Cr-DFO) was added to the culture containing 2 μM DFO, the imported chromium was only 2.5 ng in 1 mL of cell culture (circles in Fig. 3), which does not approach the maximum 5 ng of chromium imported per 1 mL of cell culture in the presence of 2 μM DFO (see squares in Fig. 3). Thus, it is clear that the Cr-DFO uptake is disproportionately inhibited by the addition of FO when DFO is present in the culture. In other words, YxeB selectively imports FO over Cr-DFO, the exchange-inert analog, when DFO is present. Such a large selectivity would not be observed for the displacement mechanism and proves that metal exchange is critical to the uptake mechanism.
Fig. 3.
Cr-DFO/FO uptake assay in vivo. The cells of TC129 (A) or TC128 (B) were incubated in iron-limited minimum medium. After 2 μM DFO had been added and the cells had been incubated for 15 min, Cr-DFO (1 or 2 μM) and purified FO (0, 0.2, 0.5, or 2 μM) were added to the culture. After 0-, 20-, 40-, 70-, and 120-min incubation, the cells were harvested and Cr amounts in the whole cells were measured by ICP, as described in Materials and Methods. The optical density at 600 nm of the cultures after 0- or 120-min incubation was 1.2–1.4 (TC129 after 0-min incubation), 1.2–1.3 (TC128 after 0-min incubation), 1.8–2.0 (TC129 after 120-min incubation), and 1.8–1.9 (TC128 after 120-min incubation). The Cr-DFO and FO concentrations used with 2 μM DFO are as follows; 2 μM Cr-DFO (■), 2 μM Cr-DFO and 2 μM FO (1:1) (▲), 2 μM Cr-DFO and 0.5 μM FO (4:1) (♦), 2 μM Cr-DFO and 0.2 μM FO (10:1) (●), 1 μM Cr-DFO (□). Data are the average of two independent experiments. Bars indicate the SEs.
In Vitro Cr-DFO/FO Competition Assay Demonstrates That YxeB Selectively Binds FO When DFO Is Present.
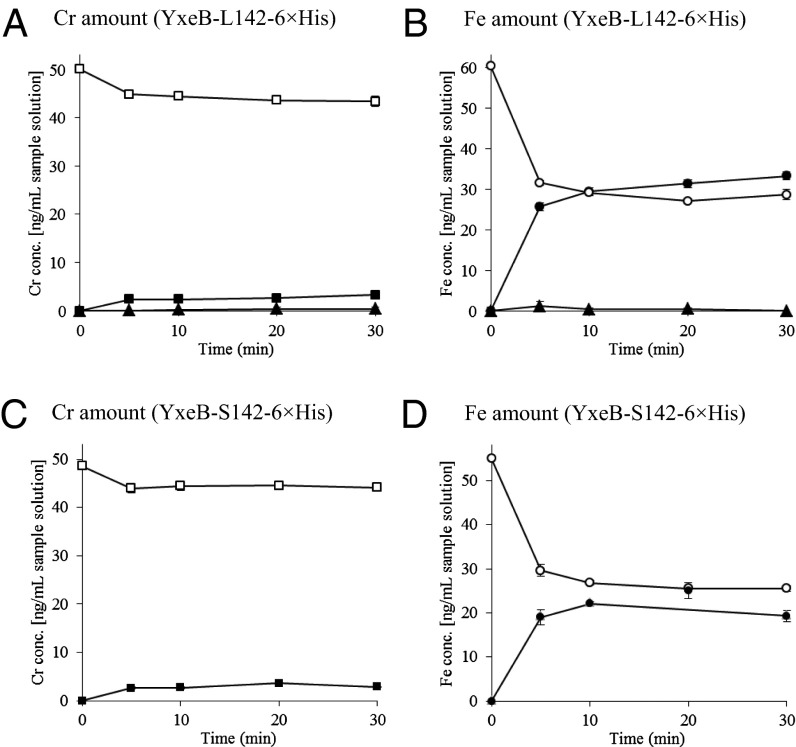
It is possible that the substrate selectivity by YxeB occurs because B. cereus selectively imports FO when DFO is present (Fig. 3). To confirm this possibility, YxeB protein (YxeB-L142-6×His or YxeB-S142-6×His), DFO, and Ni-agarose beads were mixed. A mixture of Cr-DFO/FO was added to the sample, the beads were pelleted, and the chromium and iron levels in the pellet versus supernatant were measured using ICP. As a negative control, Ni-agarose beads without protein did not bind iron or chromium (triangles in Fig. 4 A and B) and the complexes of the YxeB-L142 and YxeB-S142 proteins contained iron from FO as the major substrate (closed circles in Fig. 4 B and D, respectively) but very little chromium from Cr-DFO. Thus, the YxeB protein selectively binds FO over Cr-DFO when DFO is initially bound to the protein, and the bound FO was generated by metal exchange.
Fig. 4.
In vitro Cr-DFO/FO competition assay using YxeB-L142-6×His (A and B) and YxeB-S142-6×His (C and D). After DFO had been bound to the YxeB proteins, Cr-DFO and FO were added to the samples and the bound or unbound amounts of Cr-DFO and FO to YxeB were then measured by ICP, as described in Materials and Methods. (A and C) Amounts of bound (■) and unbound (□) chromium derived from Cr-DFO. (B and D) Amounts of bound (●) and unbound (○) iron derived from FO. The amounts of chromium and iron bound to Ni-agarose beads without YxeB are shown in closed triangles (▲, control experiment). Data represent the average of three independent experiments. Bars are SEs.
Discussion
YxeB Possesses a Gram-Positive Siderophore-Shuttle System.
Growth assays of TC128 (YxeB-S142) and TC129 (YxeB-L142) using Cr-DFO show that addition of only a small amount of Cr-DFO (10 nM) delayed the growth of both strains (Fig. S6 C and D). However, the addition of DFO (10 nM) recovered the growth of both strains even though excess Cr-DFO (500 nM) was in the culture (Fig. S6 C and D). Because the Kds of the YxeB-L142 and YxeB-S142 proteins for FO are similar to the Kds of the proteins for Cr-DFO (Table 1), the addition of Cr-DFO should delay the growth if the strains can import both of the substrates equally by the displacement mechanism (Fig. S7A). The in vivo Cr-DFO/FO uptake assay demonstrates that addition of FO with DFO strongly inhibited the Cr-DFO uptake even though 10-times less FO (0.2 μM) than Cr-DFO (2 μM) was added to the culture (Fig. 3). Because YxeB is the only SBP responsible for FO and Cr-DFO binding and uptake (Figs. S4 and S5), these results show that YxeB selectively imports FO over Cr-DFO. The key difference between FO and Cr-DFO is that Fe(III) exchanges between siderophores but Cr(III) does not. The selectivity for FO shows that metal-exchange is important and that YxeB participates in a Gram-positive siderophore-shuttle mechanism when DFO is present. However, this finding does not eliminate the possibility that YxeB uses a displacement mechanism in vivo because Cr-DFO uptake was strongly inhibited but not completely eliminated by addition of 2 μM FO in the presence of DFO (triangles in Fig. 3). This possibility was eliminated by the next study.
The in vitro Cr-DFO/FO competition assay shows that YxeB binds FO and not Cr-DFO when initially loaded with DFO (Fig. 4), even when the Kd of the YxeB-S142 protein for Cr-DFO is nearly the same as the Kd for FO (Table 1). In the competition assay, 1 μM the YxeB-S142 protein and a 20-fold excess of DFO (20 μM) were used, and it is calculated from the Kd that more than 99% of the protein is bound to DFO. Because only 1 μM FO and 1 μM Cr-DFO were added to the assay solution and YxeB is saturated with DFO, the Gram-positive siderophore-shuttle mechanism (Fig. 2C) or displacement mechanism (Fig. 2D) is responsible for any metal bound to the protein. YxeB-S142 almost exclusively binds iron from FO in the assay (Fig. 4D), and the metal-exchange selectivity indicates that the protein uses the shuttle mechanism instead of the displacement mechanism. Moreover, YxeB-L142 also uses the shuttle mechanism because the protein binds DFO and FO (Fig. S3 B and F) like YxeB-S142 (Fig. S3 C and G), and the in vitro Cr-DFO/FO competition assay shows that the protein binds FO but not Cr-DFO (Fig. 4 A and B). Therefore, both variants of YxeB predominately use the Gram-positive siderophore-shuttle mechanism over the displacement mechanism.
Siderophore Recognition by YxeB.
Fluorescence intensity of the YxeB-L142 protein was increased by the addition of DFO or DFch although the protein fluorescence was quenched by addition of FO and Fch (Fig. S1 B and C). Nano–ESI-MS analysis of the protein:DFch and protein:Fch complexes (Fig. S2) and RP-HPLC analysis of the protein:DFO and protein:FO (Fig. S3) show that the protein binds all of the substrates. Moreover, the shapes of the fluorescence-quenching curves of the YxeB-S142 protein for DFO and DFch were different from the shapes for FO and Fch, although the calculated Kds for their substrates are not different (Table 1 and Fig. S1 D and E). The difference in protein fluorescence points to a difference in protein conformation dependent on whether the bound substrate is an apo- or Fe-siderophore. Because the yxeB gene (BC_0383) makes an operon with putative permease genes, BC_0382 and BC_0381 (10), the conformation change of YxeB may allow the permeases to distinguish between Fe- and apo-siderophores and import only Fe-siderophores.
Comparison Between the Siderophore-Shuttle Systems in Gram-Positive and -Negative Bacteria.
Previously, Stintzi et al. demonstrated that an OMT in Aeromonas hydrophila is a siderophore-shuttle protein (14). From the siderophore shuttle in Gram-negative bacteria, Fe(III) exchange seems to occur in the siderophore-binding pocket surrounded by a β-barrel and extracellular loops (14). In contrast, SBPs in Gram-positive bacteria have the siderophore binding pocket at the surface of the protein, based on the structures of the B. subtilis FeuA:Fe-enterobactin complex (21) and Staphylococcus aureus HtsA:Fe-staphyloferrin A complex (22). Thus, the mechanisms of iron exchange between apo-siderophore and Fe-siderophore in Gram-negative OMT and Gram-positive SBP may differ. Here we show that YxeB has a Gram-positive siderophore-shuttle mechanism.
The E. coli periplasmic SBP FhuD binds FO, gallichrome (an Fch analog), and Fe-coprogen, and the substrate-binding pocket is large to recognize and fit the different substrates (23, 24). Because the YxeB protein (YxeB-L142 and YxeB-S142) binds DFO/FO and DFch/Fch and Gram-negative periplasmic SBPs and Gram-positive lipoprotein SBPs are similar (Fig. 1), it is possible that the YxeB protein also has a large substrate-binding pocket to exchange iron from FO to DFO bound to the protein. The complex analysis of the YxeB-L142 protein and DFch/Fch by ESI-MS suggests it is possible that the protein binds two molecules of DFch and Fch (Fig. S2). Thus, the protein would facilitate iron exchange, which is first order in both Fe- and apo-siderophore, by increasing the local concentration of the apo-siderophore (15).
Significantly, FO and Fch-binding proteins are widely conserved in Gram-positive bacteria although many bacteria do not produce DFO or DFch. B. subtilis possesses a FO-binding protein, YxeB, and a Fch-binding protein, FhuD (6). S. aureus FhuD1 and FhuD2 (25) and Listeria monocytogenes FhuD (26) bind FO and Fch. Streptococcus pnuemoniae also possesses a FO/Fch-binding protein, FhuD (27). In Gram-negative bacteria not only A. hydrophila (14) but also E. coli (28) and Salmonella typhimurium enterica (29) have OMTs for FO/Fch import. Possibly all these FO/Fch-binding proteins use a Gram-positive or -negative siderophore-shuttle mechanism if they can also bind DFO/DFch.
Some SBPs have been cocrystallized with Fe-siderophores. It is known that several SBPs in Gram-positive bacteria and periplasmic SBPs in Gram-negative bacteria, such as B. subtilis FeuA, S. aureus HtsA, and Vibrio cholerae ViuP recognize oxygen atoms that coordinate with iron, and the siderophores seem to nearly fill the binding pocket (21, 22, 30). It is possible that apo-siderophores also fill the binding pocket. However, there are few structural complexes of apo-siderophores in siderophore-binding proteins (including Gram-negative OMTs), including a complex of E. coli FecA and dicitrate (3), although several SBPs and OMTs can bind both Fe-siderophores and apo-siderophores. We hope to characterize an apo-siderophore:SBP or apo-siderophore:OMT complex structure to understand why and how SBPs and OMTs bind apo-siderophores. From this study we conclude that increasing the local ligand concentration in the apo-siderophore:SBP (OMT) complex facilitates iron exchange, and hence apo-siderophore binding plays an important role in iron uptake (Fig. 5) in siderophore-shuttle systems of both Gram-negative and Gram-positive bacteria.
Fig. 5.
Model of the Gram-positive siderophore-shuttle mechanism of YxeB. (A) YxeB binds DFO because the local concentration of apo-siderophore at the cell surface, where the apo-siderophores are secreted, will be higher than the concentration of Fe-siderophore. (B) Iron-exchange from FO to DFO bound to YxeB occurs at the siderophore binding pocket near the surface of the protein. The binding of the apo-siderophore to YxeB acts to dramatically enhance the iron exchange rate by increasing the local ligand concentration. (C) FO is imported in the cytoplasm because of the conformation change of YxeB.
Materials and Methods
See SI Materials and Methods for detailed discussions of all methods described, and Table S2 for a list of strains and plasmids used in this study.
Construction of B. cereus YxeB-L142-6×His and YxeB-S142-6×His Overexpression Plasmids, pCTF97 and pCTF98.
Plasmids for overexpression of YxeB-L142-6×His and YxeB-S142-6×His were created using pET101/D-TOPO vector (Life Technologies). The plasmids contain yxeB-L142 or yxeB-S142, except for the predicted lipoprotein signal sequence.
Construction of Integration Plasmids, pCTF59 in B. cereus Chromosome and pCTF65 for Creating a yxeB Markerless Mutant.
pCTF59 was created for construction of an integration plasmid in the B. cereus chromosome, and pCTF65 was created from pCTF59.
Construction of B. cereus yxeB (BC_0383) Mutant.
The yxeB gene makes an operon with BC_0382 and BC_0381. Thus, a markerless deletion method described by Szurmant et al. (31) was referred to for creating the mutant to express the downstream genes, BC_0382 and BC_0381.
Purification of YxeB-L142-6×His and YxeB-S142-6×His.
YxeB-L142-6×His and YxeB-S142-6×His were purified from E. coli BL21(DE3)(pCTF97 and pCTF98, respectively) cells.
Synthesis and Purification of Cr-DFO and FO.
The detailed method for preparing Cr-DFO and FO is shown in SI Materials and Methods.
Fluorescence-Quenching Experiment.
Fluorescence quenching experiment of YxeB-L142-6×His and YxeB-S142-6×His for DFO, FO, DFch, and Fch was performed as described previously (10, 19). The dissociation constants were calculated fitting the fluorescence-quenching data with a one-site binding model by Hyperquad (18), which uses nonlinear least-squares regression analysis. The method used for the fluorescence-quenching experiments of BC_2208, BC_4363, BC_4416, and BC_5380 for FO, Fch, and Fe-BB is shown in SI Materials and Methods.
Nano–ESI-MS.
The complexes of DFch or Fch and YxeB-L142-6×His in solution were analyzed by nano–ESI-MS in positive mode (Q-TOF Premier, Waters), described previously (19).
YxeB-L142-6×His and YxeB-S142-6×His Binding Assay for DFO and FO Using RP-HPLC.
The complexes of YxeB-L142-6×His or YxeB-S142-6×His and DFO or FO were analyzed by RP-HPLC.
Disc-Diffusion Assay.
Disc diffusion assay was performed as described by Zawadzka et al. (9). The growth of B. cereus TC128 and TC129 (wild-type), and TC111 (yxeB−) strains were assessed whether the cells make a halo or not around a disc containing DFO, DFch, or apo-BB.
Growth Assay Using DFO and Cr-DFO.
B. cereus TC129, TC128, and yxeB markerless strain (TC111) were incubated in iron-limited minimum medium containing 250 µM 2.2′-dipyridyl and several amounts of DFO or Cr-DFO.
Measurement of Cr-DFO Import in Cells.
Cr-DFO import in B. cereus TC129, TC128 and yxeB markerless mutant were measured by ICP using an Optima 7000 DV (PerkinElmer).
In Vivo Cr-DFO/FO Uptake Assay.
B. cereus TC129 and TC128 strains were incubated in iron-limited minimum medium at 37 °C. After 2 μM DFO (final concentration) had been added to the culture, 1 or 2 μM Cr-DFO and 0, 0.2, 0.5, or 2 μM purified FO were added to the culture and the culture was harvested after 0-, 20-, 40-, 70-, and 120-min incubation, followed by centrifuging the samples. The pellets were used for measuring the imported Cr amounts.
Competition Assay Using FO and Cr-DFO in Vitro.
One micromolar YxeB-L142-6×His or YxeB-S142-6×His (final concentration), 20 μM DFO (final concentration), and 100 μL solution of Ni Sepharose 6 Fast Flow agarose beads (Sigma-Aldrich) were mixed in TBS buffer (pH 7.4) and the mixture was gently shaken for 2 h at room temperature. One micromolar FO and 1 μM Cr-DFO (final concentration) were added to the sample, and the sample was gently shaken. After 0-, 5-, 10-, 20-, and 30-min shaking, the sample was collected and centrifuged. The amounts of chromium and iron in the supernatant and pellet were measured by ICP using an Optima 7000 DV (PerkinElmer).
Supplementary Material
Acknowledgments
We thank Dr. Anna M. Zawadzka for nano-electrospray ionization-mass spectrometry analytical advice; Dr. Jide Xu for setting up reverse-phase HPLC; Dr. Elena Kreimer for helping with inductively coupled plasma analysis; and Prof. James A. Hoch and Prof. Marta Perogo for the kind gift of the pBKJ223 plasmid. This work is supported by National Institutes of Health Grants AI11744 (to K.N.R.) and 1S10RR022393-01 for the acquisition of the Q-TOF mass spectrometer.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.J.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304235110/-/DCSupplemental.
References
- 1.Byers BR, Arceneaux JEL. In: Metal Ions in Biological Systems. Sigel A, Sigel H, editors. Vol 35. New York: Marcel Dekker; 1998. pp. 37–66. [Google Scholar]
- 2.Braun V, Hantke K, Köster W. In: Metal Ions in Biological Systems. Sigel A, Sigel H, editors. Vol 35. New York: Marcel Dekker; 1998. pp. 67–145. [PubMed] [Google Scholar]
- 3.Yue WW, Grizot S, Buchanan SK. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J Mol Biol. 2003;332(2):353–368. doi: 10.1016/s0022-2836(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 4.Eisenhauer HA, Shames S, Pawelek PD, Coulton JW. Siderophore transport through Escherichia coli outer membrane receptor FhuA with disulfide-tethered cork and barrel domains. J Biol Chem. 2005;280(34):30574–30580. doi: 10.1074/jbc.M506708200. [DOI] [PubMed] [Google Scholar]
- 5.Koebnik R. TonB-dependent trans-envelope signalling: The exception or the rule? Trends Microbiol. 2005;13(8):343–347. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 6.Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188(10):3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoegy F, et al. Binding of iron-free siderophore, a common feature of siderophore outer membrane transporters of Escherichia coli and Pseudomonas aeruginosa. J Biol Chem. 2005;280(21):20222–20230. doi: 10.1074/jbc.M500776200. [DOI] [PubMed] [Google Scholar]
- 8.Schalk IJ, et al. Copurification of the FpvA ferric pyoverdin receptor of Pseudomonas aeruginosa with its iron-free ligand: Implications for siderophore-mediated iron transport. Biochemistry. 1999;38(29):9357–9365. doi: 10.1021/bi990421x. [DOI] [PubMed] [Google Scholar]
- 9.Zawadzka AM, et al. Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore. Proc Natl Acad Sci USA. 2009;106(51):21854–21859. doi: 10.1073/pnas.0904793106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zawadzka AM, Abergel RJ, Nichiporuk R, Andersen UN, Raymond KN. Siderophore-mediated iron acquisition systems in Bacillus cereus: Identification of receptors for anthrax virulence-associated petrobactin. Biochemistry. 2009;48(16):3645–3657. doi: 10.1021/bi8018674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Berg B. Structural basis for outer membrane sugar uptake in pseudomonads. J Biol Chem. 2012;287(49):41044–41052. doi: 10.1074/jbc.M112.408518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boorsma A, van der Rest ME, Lolkema JS, Konings WN. Secondary transporters for citrate and the Mg2+-citrate complex in Bacillus subtilis are homologous proteins. J Bacteriol. 1996;178(21):6216–6222. doi: 10.1128/jb.178.21.6216-6222.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krom BP, Warner JB, Konings WN, Lolkema JS. Complementary metal ion specificity of the metal-citrate transporters CitM and CitH of Bacillus subtilis. J Bacteriol. 2000;182(22):6374–6381. doi: 10.1128/jb.182.22.6374-6381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stintzi A, Barnes C, Xu J, Raymond KN. Microbial iron transport via a siderophore shuttle: A membrane ion transport paradigm. Proc Natl Acad Sci USA. 2000;97(20):10691–10696. doi: 10.1073/pnas.200318797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotton FA, Wilkinson G. Advanced Inorganic Chemistry: A Comprehensive Text. New York: John Wiley and Sons; 1988. pp. 1283–1334. [Google Scholar]
- 16.Tufano TP, Raymond KN. Coordination chemistry of microbial iron transport compounds. 21. Kinetics and mechanism of iron exchange in hydroxamate siderophore complexes. J Am Chem Soc. 1981;103(22):6617–6624. [Google Scholar]
- 17.Hotta K, Kim CY, Fox DT, Koppisch AT. Siderophore-mediated iron acquisition in Bacillus anthracis and related strains. Microbiology. 2010;156(Pt 7):1918–1925. doi: 10.1099/mic.0.039404-0. [DOI] [PubMed] [Google Scholar]
- 18.Gans P, Sabatini A, Vacca A. Investigation of equilibria in solution. Determination of equilibrium constants with the HYPERQUAD suite of programs. Talanta. 1996;43(10):1739–1753. doi: 10.1016/0039-9140(96)01958-3. [DOI] [PubMed] [Google Scholar]
- 19.Fukushima T, et al. Bacillus cereus iron uptake protein fishes out an unstable ferric citrate trimer. Proc Natl Acad Sci USA. 2012;109(42):16829–16834. doi: 10.1073/pnas.1210131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leong J, Raymond KN. Coordination isomers of biological iron transport compounds, IV, Giometrical isomers of chromic desferriferrioxamine B. J Am Chem Soc. 1975;97(2):293–296. doi: 10.1021/ja00835a011. [DOI] [PubMed] [Google Scholar]
- 21.Peuckert F, et al. The siderophore binding protein FeuA shows limited promiscuity toward exogenous triscatecholates. Chem Biol. 2011;18(7):907–919. doi: 10.1016/j.chembiol.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 22.Grigg JC, Cooper JD, Cheung J, Heinrichs DE, Murphy ME. The Staphylococcus aureus siderophore receptor HtsA undergoes localized conformational changes to enclose staphyloferrin A in an arginine-rich binding pocket. J Biol Chem. 2010;285(15):11162–11171. doi: 10.1074/jbc.M109.097865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke TE, Ku SY, Dougan DR, Vogel HJ, Tari LW. The structure of the ferric siderophore binding protein FhuD complexed with gallichrome. Nat Struct Biol. 2000;7(4):287–291. doi: 10.1038/74048. [DOI] [PubMed] [Google Scholar]
- 24.Clarke TE, Braun V, Winkelmann G, Tari LW, Vogel HJ. X-ray crystallographic structures of the Escherichia coli periplasmic protein FhuD bound to hydroxamate-type siderophores and the antibiotic albomycin. J Biol Chem. 2002;277(16):13966–13972. doi: 10.1074/jbc.M109385200. [DOI] [PubMed] [Google Scholar]
- 25.Sebulsky MT, Heinrichs DE. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J Bacteriol. 2001;183(17):4994–5000. doi: 10.1128/JB.183.17.4994-5000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao Q, et al. Sortase independent and dependent systems for acquisition of haem and haemoglobin in Listeria monocytogenes. Mol Microbiol. 2011;80(6):1581–1597. doi: 10.1111/j.1365-2958.2011.07667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pramanik A, Braun V. Albomycin uptake via a ferric hydroxamate transport system of Streptococcus pneumoniae R6. J Bacteriol. 2006;188(11):3878–3886. doi: 10.1128/JB.00205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sauer M, Hantke K, Braun V. Sequence of the fhuE outer-membrane receptor gene of Escherichia coli K12 and properties of mutants. Mol Microbiol. 1990;4(3):427–437. doi: 10.1111/j.1365-2958.1990.tb00609.x. [DOI] [PubMed] [Google Scholar]
- 29.Kingsley RA, et al. Ferrioxamine-mediated Iron(III) utilization by Salmonella enterica. Appl Environ Microbiol. 1999;65(4):1610–1618. doi: 10.1128/aem.65.4.1610-1618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li N, et al. Unique iron coordination in iron-chelating molecule vibriobactin helps Vibrio cholerae evade mammalian siderocalin-mediated immune response. J Biol Chem. 2012;287(12):8912–8919. doi: 10.1074/jbc.M111.316034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szurmant H, Fukushima T, Hoch JA. The essential YycFG two-component system of Bacillus subtilis. Methods Enzymol. 2007;422:396–417. doi: 10.1016/S0076-6879(06)22020-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.