Abstract
The major capsid protein Vp54 from the prototype chlorovirus Paramecium bursaria chlorella virus 1 (PBCV-1) contains four Asn-linked glycans. The structure of the four N-linked oligosaccharides and the type of substitution at each glycosylation site was determined by chemical, spectroscopic, and spectrometric analyses. Vp54 glycosylation is unusual in many ways, including: (i) unlike most viruses, PBCV-1 encodes most, if not all, of the machinery to glycosylate its major capsid protein; (ii) the glycans are attached to the protein by a β-glucose linkage; (iii) the Asn-linked glycans are not located in a typical N-X-(T/S) consensus site; and (iv) the process probably occurs in the cytoplasm. The four glycoforms share a common core structure, and the differences are related to the nonstoichiometric presence of two monosaccharides. The most abundant glycoform consists of nine neutral monosaccharide residues, organized in a highly branched fashion. Among the most distinctive features of the glycoforms are (i) a dimethylated rhamnose as the capping residue of the main chain, (ii) a hyperbranched fucose unit, and (iii) two rhamnose residues with opposite absolute configurations. These glycoforms differ from what has been reported so far in the three domains of life. Considering that chloroviruses and other members of the family Phycodnaviridae may have a long evolutionary history, we suggest that the chlorovirus glycosylation pathway is ancient, possibly existing before the development of the endoplasmic reticulum and Golgi pathway, and involves still unexplored mechanisms.
Keywords: virus-encoded glycosylation, glucose/asparagine linkage, cytoplasmic glycosylation, glycopeptide, glycobiology
Many viruses, such as rhabdoviruses, herpesviruses, poxviruses, and paramyxoviruses, have structural proteins that are glycosylated. All viruses studied to date use host-encoded glycosyltransferases and glycosidases located in the endoplasmic reticulum (ER) and Golgi apparatus to add and remove sugar residues from their glycoproteins cotranslationally or shortly after translation of the protein. The virus glycoproteins are subsequently transported to host membranes, and progeny viruses then bud through these virus-specific target membranes, which is often the final step in the assembly of infectious virions (1–3). Thus, virus glycoproteins are host-specific because they are glycosylated by the same mechanism as host glycoproteins. Consequently, the only way to alter the glycan structure of virus glycoproteins is to grow the virus in a different host or have a mutation in the protein that alters its glycosylation site.
In contrast, viruses in the genus Chlorovirus (family Phycodnaviridae) differ from this paradigm in that they encode most, if not all, of the machinery to glycosylate their major capsid protein (4). Furthermore, the process probably occurs in the cytoplasm of the infected cell. Chloroviruses are large (190 nm in diameter) icosahedral, plaque-forming, dsDNA-containing viruses with an internal lipid membrane; they have genomes of 290 to 370 kb that contain ∼400 protein-encoding genes (5–7). The prototype chlorovirus, Paramecium bursaria chlorella virus 1 (PBCV-1), infects Chlorella variabilis (formerly known as Chlorella NC64A) that is normally a symbiont in the protozoan P. bursaria. The 330 kb PBCV-1 genome encodes 416 protein-encoding genes of 40 codons or larger (8). The PBCV-1 major capsid protein Vp54 comprises ∼40% of total virus protein. The primary translation product of its gene, a430l, has a predicted weight of 48,165 Da, but the protein undergoes additional posttranslational modifications so that the mature product has a molecular weight of approximately 54 kDa (9).
Previous studies established that glycosylation of Vp54 is unusual: the protein lacks N-acetylglucosamine and N-acetylgalactosamine, typical of Asn-linked (N-linked) and many Ser/Thr-linked (O-linked) glycoproteins (10). Instead, Vp54 glycans were reported to contain seven neutral monosaccharides, glucose, fucose, galactose, mannose, xylose, rhamnose, and arabinose (10). Crystallization of Vp54 at 2 Å resolution revealed four N-linked and two O-linked glycans (11). Comparison of the molecular mass of Vp54 with the predicted molecular weight from the amino acid sequence (48,165 Da) indicates that the protein contains ∼35 sugar moieties, of which ∼20 were detected, but not identified, in the crystal density map. At a minimum, single sugars were attached to 57Ser and 387Ser and branched-chain sugar moieties were attached to 280Asn, 302Asn, 399Asn, and 406Asn. Surprisingly, none of these asparagines reside in a N-X-(T/S) sequence, where X is anything but a Pro, commonly recognized by eukaryotic and prokaryotic enzymes involved in N-glycosylation. Instead, 302Asn, 399Asn, and 406Asn occur in the amino acid sequence (A/G)-N-T-X-T and 280Asn occurs in an A-N-I-P-G sequence. These results explain why previous tests for N-glycosylation in Vp54 were negative (12).
These findings suggest that the structure of the Vp54 glycans differ from typical glycans and prompted us to investigate their structures. The current manuscript describes the unusual structure of the PBCV-1 N-linked oligosaccharides.
Results
Isolation of the Two Largest Vp54 Glycans.
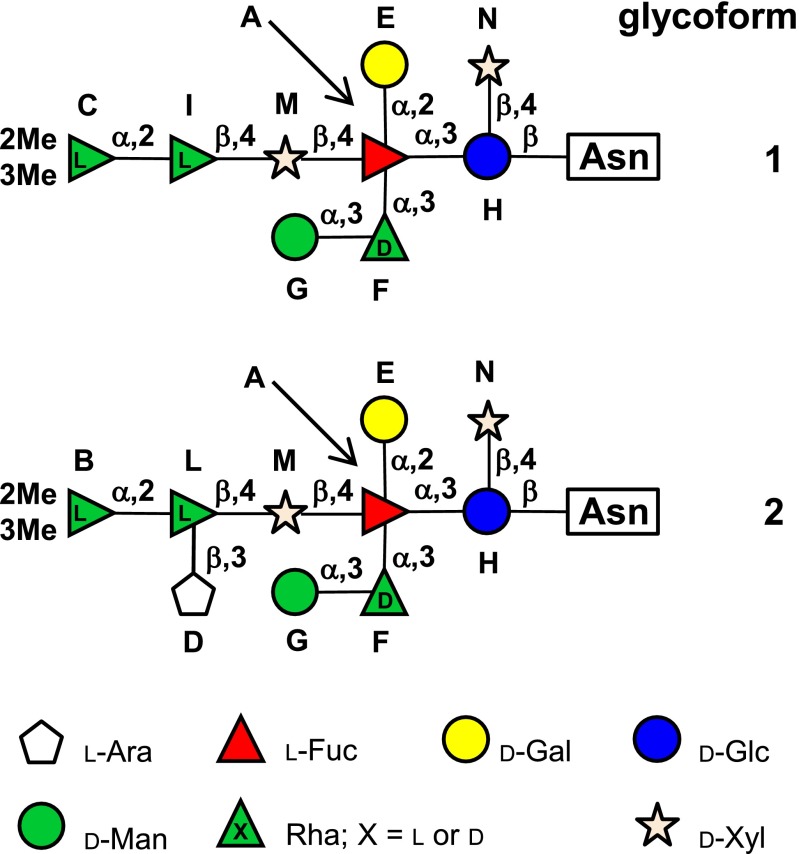
Vp54 was extensively digested with proteinase K to minimize the length and the heterogeneity of the peptide moiety of the glycopeptides. The crude mixture was chromatographed on size-exclusion columns and four fractions containing glycopeptides were isolated. An 1H NMR spectrum of these samples showed the same pattern of the anomeric proton signals (5.7–4.3 ppm; Fig. S1), but differed in the number of signals around 4.5 ppm and for those occurring in the high and low field regions of the spectrum. This information suggested that the samples differed in the peptide portion containing the oligosaccharide, and also explained their different chromatographic retention volumes. With regard to the anomeric profile, not all of the protons occurred with the same intensity, as typically occurs when more glycoforms are present. Therefore, each fraction was investigated by MALDI MS at the same time the full NMR analysis was carried out on fraction 4. These approaches led to the full elucidation of the glycoforms attached to Vp54 (Fig. 1).
Fig. 1.
Structures of glycoforms of the major capsid protein Vp54 from chlorovirus PBCV-1. Residues are labeled with the letter used during the NMR assignment. Glycoform 3 (or 4) differs from 1 (or 2) by lacking a mannose unit.
NMR Characterization of the Two Most Abundant Vp54 Glycoforms.
The complete assignment of all resonances of the glycopeptide mixture in fraction 4 was performed by a combination of homo- and heteronuclear 2D NMR experiments. The heteronuclear single quantum correlation (HSQC) spectrum (Fig. S2; structures and labels in Fig. 1) contained 12 anomeric proton and one ring proton signals (range, 5.7–4.4 ppm), a crowded carbinolic region (4.4–3.0 ppm), two O-Me (3.47 and 3.45 ppm), and, eventually, four methyl signals at approximately 1.3 ppm, typical of 6-deoxy residues (Table S1).
The anomeric proton of H at 5.00 ppm was identified as an N-linked residue by virtue of its correlated carbon chemical shift at 80.5 ppm and therefore located at the reducing end of the oligosaccharide (13). In support of this conclusion, the H anomeric proton in the heteronuclear multiple bond correlation (HMBC) spectrum correlated with a carbonyl at 174.4 ppm, namely Cγ of the Asn residue. The gluco stereochemistry of H determined the efficient magnetization transfer in the total correlation spectroscopy (TOCSY) spectrum (Fig. S3A). Accordingly, the H anomeric signal displayed correlations up to H-6s and, integrating TOCSY information with those from the correlation spectroscopy (COSY) spectrum, the chemical shifts of all of the ring protons were assigned. Carbon chemical shift values of H were then determined analyzing HSQC (Fig. S2), HMBC (Fig. S4A), and HSQC-TOCSY (Fig. S4B) spectra. As a result, residue H was identified as a glucose β-configured at the anomeric center (3JH1H2, 9.1 Hz) and glycosylated at O-4; this last information was deduced by the low field displacement of the corresponding carbon (74.9 ppm) with respect to the standard values (70.6 ppm) (14). In agreement with this information, the HMBC spectrum correlated H-1 of N with C-4 of H (Fig. S4A) and contained an additional long-range correlation connecting H-3 of H to the anomeric proton of residue A. These results indicated that H was glycosylated at O-4 from N and at O-3 from A.
The same spectroscopic approach was applied to the two substituents of H, and in turn to all of the other residues. In some cases, TOCSY connectivity did not propagate along all of the protons of the sugar ring because of the stereochemistry of the residue; therefore, information from the HMBC or the transverse rotating-frame overhauser effect spectroscopy (TROESY) spectra were used to complete assignment of the residue (a detailed discussion is provided in SI Results). N was identified as a terminal β-xylose, whereas A was a fully substituted α-fucose carrying a terminal α-galactose (E) at O-2, an α-rhamnose (F) at O-3, and a β-xylose (M) at O-4 (Fig. 1). The α-rhamnose (F) was substituted at O-3 with a terminal α-mannose (G), whereas the xylose (M) had a β-rhamnose (I) at O-4. The β-rhamnose (I) also had a terminal α-rhamnose (C) at O-2 that was methylated at O-2 and O-3. With the addition of residue C, the structure of the first glycoform (glycoform 1; Fig. 1) was completed.
Spectroscopic investigation of the left minor signals started with L, which exhibited the same pattern found for I (SI Results). Therefore, it was identified as a β-rhamnose linked at O-4 of M. The carbon chemical shift analysis of L revealed that it was glycosylated at O-2 and O-3, and TROESY spectrum (Fig. S3B) showed that O-3 was substituted with D, whereas O-2 was connected to B, a terminal α-rhamnose methylated at O-2 and O-3. The anomeric proton of D (Fig. S3A) correlated with five other protons in the TOCSY spectrum, and their assignment, together with the information from the HSQC spectrum (Table S1), identified this residue as a pentose monosaccharide in the furanose form because of the deshielded value of its C-4 (83.0 ppm). Comparison of D carbon chemical shifts with those of reference compounds (12) identified this unit as a terminal β-arabinofuranose residue. Therefore, the identification of residues L, B, and D allowed a second N-linked oligosaccharide to be constructed (glycoform 2; Fig. 1), which differed from the major glycoform 1 by the presence of an additional residue.
Analysis of other minor signals in the spectra led to the identification of a threonine residue, whose occurrence was confirmed through MS analysis performed on fractions 3 and 4. Finally, the low intensity of the few remaining signals prevented any further spectroscopical assignment. These additional signals indicated that minute amounts of other glycoforms were present; information regarding these species was obtained by GC-MS and MALDI.
Chemical Composition of the Vp54 Glycans.
The determination of the monosaccharide composition and linkage analysis was performed according to standard procedures (15) using fractions 3 and 4. Accordingly, residues identified in the NMR analyses together with their linkage patterns were confirmed. Determination of the absolute configuration of all residues was performed by analyzing the fully acetylated R-(-)-2-octylglycosides (15), which revealed the occurrence of l-arabinose, d-xylose, l-fucose, d-mannose, d-galactose, d-glucose, d- and l-rhamnose, and the 2,3-di-OMe-rhamnose derivative (units B and C; Fig. 1). For this latter residue, it was necessary to synthesize an ad hoc standard, the 4-O-Ac-2,3-di-OMe-rhamnose 2-(-)-octylglycoside. The compound was synthesized by methylating the O-antigen polysaccharide from Salmonella enterica ssp. enterica (16), which contains a 4-substituted l-rhamnose, the proper precursor of the target compound (Fig. S5).
As for the occurrence of d- and l-rhamnose residues, we surmised that the residues I (or L) and F of the two glycoforms possessed the opposite stereochemistry; accordingly, their configuration was inferred applying the same approach used to deduce the C configuration and using the polysaccharide from Kaistella flava (17) as a precursor for the standards (Fig. S6). Therefore, I possessed the l configuration, whereas F was the opposite d. All this information completed the structural data necessary to fully describe the two major glycoforms present in PBCV-1 virus Vp54 capsid protein (Fig. 1). This approach also revealed a small percentage of nonsubstituted d-rhamnose F, as later confirmed by MALDI MS.
Determination of the N-Glycosylation Sites and the Site-Specific Glycan Structure Heterogeneity.
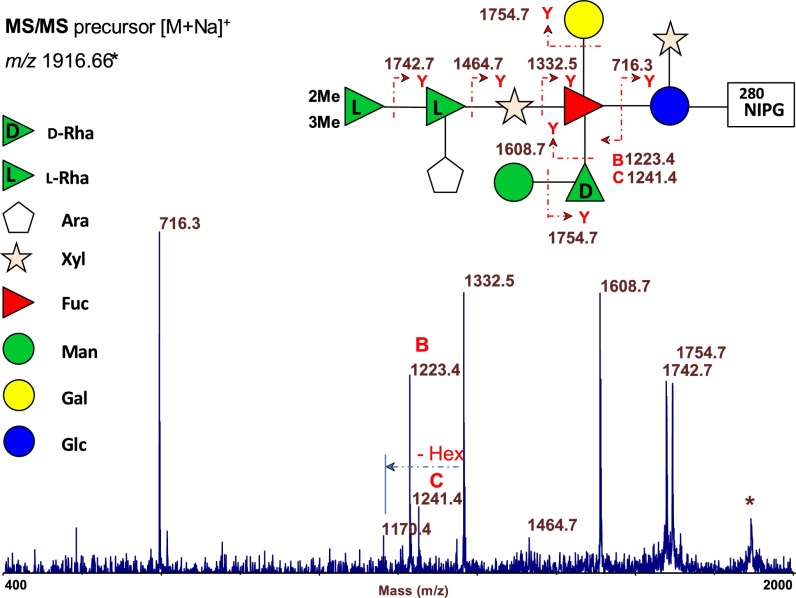
To identify the specific N-glycosylation sites, all Bio-Gel P10 fractions were analyzed by MALDI MS. The spectrum of fraction 2 (Fig. S7) showed two major peaks at m/z 1,916.66 and 1,932.64, and two less intense peaks at m/z 1,784.60 and 1,800.59 (Δm/z 132). The two main ions were ascribed to the Na+ (m/z 1,916.64) and K+ (m/z 1,932.64) ion adducts of the same glycopeptide composed from the 280NIPG amino acidic sequence linked to glycoform 2. The pair of minor peaks at Δm/z 132 (i.e., the 132 mass shift corresponding to a pentose residue) were assigned to the analogous glycopeptide ions composed of the same 280NIPG sequence plus glycoform 1. This interpretation was confirmed by the MS/MS fragmentation spectrum of the precursor ion at m/z 1,916.66 by using MALDI TOF/TOF MS/MS (Fig. 2; nomenclature in ref. 18).
Fig. 2.
MALDI TOF/TOF MS/MS spectrum of the precursor ion at m/z 1,916.66 of glycoform 2, isolated from Vp54 protein after proteinase K digestion and Bio-Gel P10 purification. Structure of the glycopeptide (located at 280NIPG) is drawn together with the fragmentation observed in the spectrum. Ion at m/z 1,170.4 results from dual fragmentation.
MALDI spectra of fractions 3 and 4 comprised glycopeptides consistent with glycoform 1 and glycoform 2 linked to a single Asn or to a NT dipeptide. All these species could be located at glycosylation sites 302Asn, 399Asn, or 406Asn; thus, they do not identify the specific glycosylation site.
Conversely, MALDI spectra of fraction 1 showed two major peaks, at m/z 1,848.58 and 1,980.62, whose assignments, based on the molecular masses and MS/MS spectra (MS/MS of 1,848.58 in Fig. S8A), were consistent with the glycoforms 1 and 2 (both present as Na+ ions) linked at the 399NTET amino acid sequence. The MS spectrum also contained a pair of less intense peaks at m/z 1,686.51 and m/z 1,818.55 that differed by a hexose monosaccharide (Δm/z 162) from the two main ions. Taking into consideration the compositional data, these peaks were assigned to two different glycoforms, named 3 and 4, linked at the same tetrapeptide, with the same structure of glycoforms 1 and 2, but lacking the mannose residue G.
To obtain meaningful glycopeptides with amino acid lengths suitable for site identification by MS, we digested Vp54 with a more selective protease. Trypsin was expected to produce two glycopeptides, the first containing 280Asn and 302Asn and the second containing 399Asn and 406Asn, with estimated molecular masses greater than m/z 6,300; these sizes are too large to acquire useful MS and MS/MS spectra. We then tried thermolysin because of its predicted specificity. Reverse-phase HPLC of the glycoprotein digestion with thermolysin yielded 25 fractions, mostly consisting of complex mixtures of peptides and glycopeptides; they were all analyzed by MALDI MS. Glycopeptides were identified by their specific spacing patterns (Δm/z 132 or 162) between the glycoforms, and by their MALDI TOF/TOF MS/MS fragmentation patterns. This process led to the identity of several glycopeptides composed of glycoforms 1 to 4 linked to 302Asn, 399Asn, or 406Asn. Fig. S8 B–D shows the MS/MS spectra of the main glycopeptide ions, containing glycoform 1 as the glycan moiety. In summary, the combined digestion procedure revealed the identity of the oligosaccharide(s) present at a specific glycosylation site together with their proportion. Glycoform 2 is the prominent species at 280Asn whereas glycoform 1 predominates over the other three glycoforms at the other glycosylation sites, even though most of the glycosylation heterogeneity occurs at 399Asn and 302Asn (Table 1).
Table 1.
Proportion and type of oligosaccharides found at each glycosylation site in PBCV-1 major capsid protein Vp54
| Position | Glycoform, % | |||
| 1 | 2 | 3 | 4 | |
| 280Asn | 5 | 94 | — | 4 |
| 302Asn | 58 | 10 | 21 | — |
| 399Asn | 45 | 32 | 12 | 12 |
| 406Asn | 71 | — | 29 | — |
The structures are reported in Fig. 1.
Discussion
A detailed chemical, spectroscopic, and spectrometric analysis allowed us to determine the structure of the four Asn-linked glycoforms attached to the major capsid protein Vp54 from virus PBCV-1 (Fig. 1). As expected, the structure of the Vp54 glycoforms are unique and do not resemble any type of structure reported so far for any organism in the three domains of life.
MALDI MS experiments extended our knowledge on the site-specific glycosylation of Vp54, demonstrating that there is a preference for the type of oligosaccharide at the four glycosylation sites. 280Asn primarily has glycoform 2, whereas glycoform 1 is the main oligosaccharide at 406Asn. 302Asn and 399Asn are the positions where most of the major glycosylation heterogeneity occurs, even though glycoform 1 is still the main oligosaccharide.
Therefore, PBCV-1 glycosylation opens many questions regarding its biosynthetic pathway. PBCV-1 encodes five putative glycosyltransferases, A064R, A111/114R, A219/222/226R, A473L, and A546L, which are scattered throughout the PBCV-1 genome (4, 6). The A111/114R protein has two glycosyltransferase domains. However, clearly, there are more glycan linkages than virus-encoded glycosyltransferases, which may result from some of the viral-encoded glycosyltransferases serving more than one function or some host-encoded enzymes are involved in the process. Like Vp54, none of these PBCV-1–encoded glycosyltransferases has an identifiable signal peptide that would target them to the ER of the host. Furthermore, the cellular protein localization program PSORT predicts that all these proteins, with the exception of A473L, are located in the cytoplasm. None of these five PBCV-1–encoded putative glycosyltransferases have been enzymatically characterized until now because potential substrates were unknown. Solving the structure of the final biosynthetic products will allow functional characterization of these enzymes.
The most abundant oligosaccharide contains nine sugar residues (glycoform 1; Fig. 1): d- and l-rhamnose, 2,3-di-OMe-l-rhamnose, l-fucose, d-glucose, d-mannose, d-galactose, and d-xylose (two units), organized in a highly branched fashion. This glycoform can be dissected into a main chain, consisting of the five residues C-I-M-A-H (Fig. 1) further substituted with two monosaccharides, E and N, and one disaccharide, G-F. Glycoform 2 possesses an additional branch carrying a terminal l-arabinofuranose residue, unit D. The other two minor glycoforms, 3 and 4, differ from the first two by the lack of the mannose residue G.
The structures of these oligosaccharides have several remarkable features. The glycoprotein β-Glc-Asn linkage is rare in nature and has only been reported in glycoproteins from a few organisms. It was first described in surface glycopeptides and flagellins of the extreme halophilic archaea Halobacterium halobium (19, 20) and Halobacterium volcanii (21). Later, this glycoside linkage was reported in bacterial adhesion glycoproteins from Haemophilus influenzae (22, 23) and Actinobacillus pleuropneumoniae (13). In these cases, the Glc-Asn linkage was found in the typical sequon for N-glycosylation, and Glc was elongated with short linear chains of Glc or glucuronic acid. In eukaryotic organisms, a β-Glc-Asn linkage was reported in the B2 chain of rat kidney laminin, a major basement membrane glycoprotein, by using antibody that specifically recognizes this linkage (24). However, the glycoprotein has not been characterized further, leaving this N-glucosylation example as the only exception to the general N-glycosylation pattern in Eukarya.
Thus, the β-Glc-Asn linkage, although rare, has been reported in all three domains of life. However, the virus PBCV-1 glycosylation of Vp54 differs in many aspects from the systems previously mentioned. (i) The four N-linked glycans are not located in a typical N-X-(T/S) consensus site. Three of the four glycosylation sites share the sequence NTXT (X represents A, E, or G), whereas the fourth is NIPG; all of these asparagine flanking residues denote a new pattern of N-glycosylation. (ii) The PBCV-1 N-glycans are more complicated than those reported previously. In fact, the main Vp54 glycoform consists of nine different neutral sugars arranged in a highly branched fashion with several unusual features. (iii) The fucose unit is completely substituted and not terminal like in most N-linked glycans (25). Extending our view to all known carbohydrate structures, regardless of the type of glycosylation and source, Vp54 glycoforms differ from those reported in the literature. As far as we know, a hyperbranched fucose has been reported only in the complex phosphoglycan from Trypanosoma cruzi (26), but its substitution pattern does not resemble that of Vp54 glycoforms. (iv) The terminal residue at the nonreducing end of the main chain is a rhamnose capped with two methyl groups, which is a rare type of termination and so far only reported for glycolipids produced from Mycobacterium haemophilum (27) and Mycobacterium leprae (28). (v) The Vp54 oligosaccharide contains both d- and l-rhamnose residues, a feature that is rare, occurring only in bacteria and that returned 15 hits for natural oligosaccharides in the Bacterial Carbohydrates Structure Database (http://csdb.glycoscience.ru/bacterial). Interestingly, PBCV-1 encodes a functional “de novo” GDP-l-fucose biosynthetic pathway, which also produces GDP-d-rhamnose as a major product (29). Thus, PBCV-1 encodes a more complex glycosylation machinery, which comprises the glycosyltransferases and some of the enzymes required to produce the nucleotide-sugar substrates, which presumably are present in limited supply or absent in the host cell.
In summary, the structures of four glycans associated with virus PBCV-1 major capsid protein Vp54 are very unusual and provide additional support for the hypothesis that PBCV-1 and probably all of the chloroviruses glycosylate their major capsid proteins differently than all of the typical known viruses, and in a different fashion with respect to the other known glycosylation pathways. Chloroviruses and other members of the family Phycodnaviridae are believed to have a long evolutionary history, possibly dating back to the time when eukaryotes arose from prokaryotes (30–33), and we have suggested that the chlorovirus glycosylation pathway may be ancient, possibly existing before the development of the ER and Golgi (9).
In the past few years, increasing evidence has indicated that other giant viruses, in particular those belonging to the Mimiviridae family, encode enzymes involved in glycan production (34). Moreover, genome sequencing of several other members of the Phycodnaviridae family, which are closely related to the chloroviruses, indicate that they encode putative glycosyltransferases and other genes involved in glycan formation and remodeling (35). Some of these viruses are widely distributed in the oceans, where they are active participants in ecological and evolutionary events (36). Altogether, these data indicate that the presence of glycosylation machinery can no longer be considered as a hallmark solely of cellular organisms, but that viruses also encode unique and complex glycan systems, which are still mostly unknown. Identification of the Vp54 glycan structure represents a starting point to understanding these systems.
Materials and Methods
Glycan Isolation.
Procedures for growing and purifying virus PBCV-1 have been described (37–39). The Vp54 capsid protein was purified to near-homogeneity as reported previously (10). Seven milligrams of Vp54 were suspended in 1 mL of 100 mM Tris, 50 mM NaCl, 10 mM MgCl2, pH 7.5, and digested with proteinase K (P6556; Sigma) at 55 °C. To ensure complete digestion, proteinase K was added three times (0.7 mg each time) at 8-h intervals. The mixture was freeze-dried, suspended in water, and purified on a Bio-Gel P-10 chromatographic column (1.5 × 118 cm; 150-4144; Bio-Rad) eluted with distilled water. The chromatographic profile was monitored with an online refractive index detector (K-2310; Knauer), and fractions were pooled accordingly and monitored via 1H NMR. Carbohydrate containing fractions 1 (0.1 mg), 2 (0.26 mg), 3 (0.4 mg), and 4 (0.5 mg) were identified and eluted at 37.0%, 52.9%, 56.7%, and 61.5% of the column volume.
Glycan Analysis.
Monosaccharide analysis was performed on fractions 3 and 4 and required the preparation of the corresponding: acetylated alditols, partially methylated and acetylated alditols, acetylated methylglycosides, partially methylated and acetylated octylglycosides, or only acetylated octylglycosides, by using standard protocols (15). All GC-MS analyses were performed with an Agilent instrument (GC instrument Agilent 6850 coupled to an Agilent 5973 MS instrument), equipped with a SPB-5 capillary column (30 m × 0.25 i.d.; flow rate, 0.8 mL⋅min−1; Supelco) and He as carrier gas. Electron impact mass spectra were recorded at an ionization energy of 70 eV and an ionizing current of 0.2 mA. The temperature program used for the analyses was: 150 °C for 5 min, from 150 to 280 °C at 3 °C/min, 300 °C for 5 min.
NMR Spectroscopy.
All NMR experiments were carried out on a Bruker DRX-600 spectrometer equipped with a cryoprobe. Chemical shifts of spectra recorded in D2O are expressed in δ relative to internal acetone (2.225 and 31.45 ppm). Two-dimensional spectra [double-quantum filtered (DQ)-COSY, TOCSY, TROESY, HSQC, gHMBC, and HSQC-TOCSY] were measured by using standard Bruker software. For all experiments, 512 free induction decays (FIDs) of 2,048 complex data points were collected, 40 scans per FID were acquired for homonuclear spectra, and mixing times of 120 and 250 ms were used for TOCSY and for TROESY spectra acquisition, respectively. The spectral width was set to 10 ppm and the frequency carrier placed at the residual HOD peak. HSQC spectrum was acquired with 50 scans per FID, and the globally-optimized, alternating phase, rectangular pulses (GARP) sequence was used for 13C decoupling during acquisition. HSQC-TOCSY and gHMBC scans tripled and doubled, respectively, those of HSQC spectrum, and a mixing time of 100 ms was used for HSQC-TOCSY spectrum. Data processing was performed with the standard Bruker Topspin 3 program.
MS.
Purified Vp54 was digested using thermolysin (Promega) in 50 mM Tris⋅HCl, pH 7.8, 0.5 mM CaCl2, at 80 °C for 4 h. The peptides were then separated by reversed-phase HPLC on a Agilent 1200 system by using an Alltech C18 4.6 × 250-mm, 5-µM particle size column, with flow at 1 mL/min. Isocratic mobile phase A [acetonitrile/H2O (5:95) containing 0.1% TFA] was used for 5 min; then, a linear gradient from 0% to 100% mobile phase B [acetonitrile/H2O (90:10) containing 0.05% TFA] over 50 min was applied. Detection was performed by UV absorbance at 220 nm. Fractions, collected every 0.5 min, were dried by using a Speed-Vac and then directly used for MS analyses.
MALDI-TOF mass spectra were recorded in a linear mode on a PerSeptive Voyager STR equipped with a pulsed UV laser beam (nitrogen laser, λ = 337 nm) and in reflectron mode on a 4800 Proteomic Analyzer (Applied Biosystems) supplied with a Nd:yttrium–aluminum garnet laser at a wavelength of 355 nm. Mass spectra were acquired twice using 2,5-dihydroxybenzoic acid (DHB) oralphacyano-4-hydroxycinnamic acid, respectively, as the matrix. DHB has proved to be a better matrix for glycopeptides. MALDI TOF/TOF MS/MS spectra were also recorded on the 4800 system without any collision gas.
Supplementary Material
Acknowledgments
Work in the J.L.V.E Etten laboratory was partially supported by National Science Foundation-Experimental Program to Stimulate Competitive Research Grant EPS-1004094, Stanley Medical Research Institute Grant 11R-0001; National Center for Research and Resources Centers of Biomedical Research Excellence (COBRE) Program Grant P20-RR15635; Italian Ministry for University and for Scientific and Technology Research Grants PRIN 2009 and FIRB-MERIT RBNE08HWLZ; Regione Liguria; the National Council of Research; and Programma Operativo Regionale Campania Project Campania Research in Experimental Medicine Fondo Europeo Sviluppo 2007-2013 (to C.D.C. and R.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313005110/-/DCSupplemental.
References
- 1.Doms RW, Lamb RA, Rose JK, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193(2):545–562. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 2.Olofsson S, Hansen JES. Host cell glycosylation of viral glycoproteins—a battlefield for host defence and viral resistance. Scand J Infect Dis. 1998;30(5):435–440. doi: 10.1080/00365549850161386. [DOI] [PubMed] [Google Scholar]
- 3.Vigerust DJ, Shepherd VL. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007;15(5):211–218. doi: 10.1016/j.tim.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van Etten JL, Gurnon JR, Yanai-Balser GM, Dunigan DD, Graves MV. Chlorella viruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim Biophys Acta. 2010;1800(2):152–159. doi: 10.1016/j.bbagen.2009.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Van Etten JL. Unusual life style of giant chlorella viruses. Annu Rev Genet. 2003;37:153–195. doi: 10.1146/annurev.genet.37.110801.143915. [DOI] [PubMed] [Google Scholar]
- 6.Dunigan DD, Fitzgerald LA, Van Etten JL. Phycodnaviruses: A peek at genetic diversity. Virus Res. 2006;117(1):119–132. doi: 10.1016/j.virusres.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 7.Van Etten JL, Dunigan DD. Chloroviruses: Not your everyday plant virus. Trends Plant Sci. 2012;17(1):1–8. doi: 10.1016/j.tplants.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunigan DD, et al. Paramecium bursaria chlorella virus 1 proteome reveals novel architectural and regulatory features of a giant virus. J Virol. 2012;86(16):8821–8834. doi: 10.1128/JVI.00907-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graves MV, Bernadt CT, Cerny R, Van Etten JL. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology. 2001;285(2):332–345. doi: 10.1006/viro.2001.0937. [DOI] [PubMed] [Google Scholar]
- 10.Wang I-N, et al. Evidence for virus-encoded glycosylation specificity. Proc Natl Acad Sci USA. 1993;90(9):3840–3844. doi: 10.1073/pnas.90.9.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nandhagopal N, et al. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc Natl Acad Sci USA. 2002;99(23):14758–14763. doi: 10.1073/pnas.232580699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Que Q, et al. Protein glycosylation and myristylation in Chlorella virus PBCV-1 and its antigenic variants. Virology. 1994;203(2):320–327. doi: 10.1006/viro.1994.1490. [DOI] [PubMed] [Google Scholar]
- 13.Schwarz F, Fan Y-Y, Schubert M, Aebi M. Cytoplasmic N-glycosyltransferase of Actinobacillus pleuropneumoniae is an inverting enzyme and recognizes the NX(S/T) consensus sequence. J Biol Chem. 2011;286(40):35267–35274. doi: 10.1074/jbc.M111.277160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bock K, Pedersen S. Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv Carbohydr Chem Biochem. 1983;41:27–65. [Google Scholar]
- 15.De Castro C, Parrilli M, Holst O, Molinaro A. Microbe-associated molecular patterns in innate immunity: Extraction and chemical analysis of gram-negative bacterial lipopolysaccharides. Methods Enzymol. 2010;480:89–115. doi: 10.1016/S0076-6879(10)80005-9. [DOI] [PubMed] [Google Scholar]
- 16.De Castro C, Lanzetta R, Leone S, Parrilli M, Molinaro A. The structural elucidation of the Salmonella enterica subsp. enterica, reveals that it contains both O-factors 4 and 5 on the LPS antigen. Carbohydr Res. 2013;370:9–12. doi: 10.1016/j.carres.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 17.Gargiulo V, et al. Structural elucidation of the capsular polysaccharide isolated from Kaistella flava. Carbohydr Res. 2008;343(14):2401–2405. doi: 10.1016/j.carres.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Domon B, Costello CE. A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj J. 1988;5:397–409. [Google Scholar]
- 19.Wieland F, Heitzer R, Schaefer W. Asparaginylglucose: Novel type of carbohydrate linkage. Proc Natl Acad Sci USA. 1983;80(18):5470–5474. doi: 10.1073/pnas.80.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wieland F, Paul G, Sumper M. Halobacterial flagellins are sulfated glycoproteins. J Biol Chem. 1985;260(28):15180–15185. [PubMed] [Google Scholar]
- 21.Mengele R, Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J Biol Chem. 1992;267(12):8182–8185. [PubMed] [Google Scholar]
- 22.Gross J, et al. The Haemophilus influenzae HMW1 adhesin is a glycoprotein with an unusual N-linked carbohydrate modification. J Biol Chem. 2008;283(38):26010–26015. doi: 10.1074/jbc.M801819200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grass S, Lichti CF, Townsend RR, Gross J, St Geme JW., 3rd The Haemophilus influenzae HMW1C protein is a glycosyltransferase that transfers hexose residues to asparagine sites in the HMW1 adhesin. PLoS Pathog. 2010;6(5):e1000919. doi: 10.1371/journal.ppat.1000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreiner R, Schnabel E, Wieland F. Novel N-glycosylation in eukaryotes: Laminin contains the linkage unit beta-glucosylasparagine. J Cell Biol. 1994;124(6):1071–1081. doi: 10.1083/jcb.124.6.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley P, Schachter H, Taniguchi N. In: Essentials of Glycobiology. Varki A, et al., editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2009. pp. 101–114. [Google Scholar]
- 26.Allen S, Richardson JM, Mehlert A, Ferguson MAJ. Structure of a complex phosphoglycan epitope from gp72 of Trypanosoma cruzi. J Biol Chem. 2013;288(16):11093–11105. doi: 10.1074/jbc.M113.452763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Besra GS, et al. Structural elucidation and antigenicity of a novel phenolic glycolipid antigen from Mycobacterium haemophilum. Biochemistry. 1991;30(31):7772–7777. doi: 10.1021/bi00245a015. [DOI] [PubMed] [Google Scholar]
- 28.Daffé M, Lanéelle MA. Diglycosyl phenol phthiocerol diester of Mycobacterium leprae. Biochim Biophys Acta. 1989;1002(3):333–337. doi: 10.1016/0005-2760(89)90347-0. [DOI] [PubMed] [Google Scholar]
- 29.Tonetti M, et al. Paramecium bursaria Chlorella virus 1 encodes two enzymes involved in the biosynthesis of GDP-L-fucose and GDP-D-rhamnose. J Biol Chem. 2003;278(24):21559–21565. doi: 10.1074/jbc.M301543200. [DOI] [PubMed] [Google Scholar]
- 30.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol. 2001;75(23):11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117(1):156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Raoult D, et al. The 1.2-megabase genome sequence of Mimivirus. Science. 2004;306(5700):1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 33.Villarreal LP, DeFilippis VR. A hypothesis for DNA viruses as the origin of eukaryotic replication proteins. J Virol. 2000;74(15):7079–7084. doi: 10.1128/jvi.74.15.7079-7084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piacente F, et al. Giant DNA virus mimivirus encodes pathway for biosynthesis of unusual sugar 4-amino-4,6-dideoxy-D-glucose (Viosamine) J Biol Chem. 2012;287(5):3009–3018. doi: 10.1074/jbc.M111.314559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weynberg KD, Allen MJ, Gilg IC, Scanlan DJ, Wilson WH. Genome sequence of Ostreococcus tauri virus OtV-2 throws light on the role of picoeukaryote niche separation in the ocean. J Virol. 2011;85(9):4520–4529. doi: 10.1128/JVI.02131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5(10):801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 37.VAN Etten JL, Burbank DE, Kuczmarski D, Meints RH. Virus infection of culturable chlorella-like algae and dlevelopment of a plaque assay. Science. 1983;219(4587):994–996. doi: 10.1126/science.219.4587.994. [DOI] [PubMed] [Google Scholar]
- 38.Van Etten JL, Burbank DE, Xia Y, Meints RH. Growth cycle of a virus, PBCV-1, that infects Chlorella-like algae. Virology. 1983;126(1):117–125. doi: 10.1016/0042-6822(83)90466-x. [DOI] [PubMed] [Google Scholar]
- 39.Agarkova IV, Dunigan DD, Van Etten JL. Virion-associated restriction endonucleases of chloroviruses. J Virol. 2006;80(16):8114–8123. doi: 10.1128/JVI.00486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.