Abstract
Helicobacter pylori infects the stomachs of one in two humans and can cause sequelae that include ulcers and cancer. Here we sequenced the genomes of 97 H. pylori isolates from 52 members of two families living in rural conditions in South Africa. From each of 45 individuals, two H. pylori strains were isolated from the antrum and corpus parts of the stomach, and comparisons of their genomes enabled us to study within-host evolution. In 5 of these 45 hosts, the two genomes were too distantly related to be derived from each other and therefore represented evidence of multiple infections. From the remaining 40 genome pairs, we estimated that the synonymous mutation rate was 1.38 × 10−5 per site per year, with a low effective population size within host probably reflecting population bottlenecks and immune selection. Some individuals showed very little evidence for recombination, whereas in others, recombination introduced up to 100-times more substitutions than mutation. These differences may reflect unequal opportunities for recombination depending on the presence or absence of multiple infections. Comparing the genomes carried by distinct individuals enabled us to establish probable transmission links. Transmission events were found significantly more frequently between close relatives, and between individuals living in the same house. We found, however, that a majority of individuals (27/52) were not linked by transmission to other individuals. Our results suggest that transmission does not always occur within families, and that coinfection with multiple strains is frequent and evolutionarily important despite a fast turnover of the infecting strains within-host.
The bacterial pathogen Helicobacter pylori infects the stomach of about half of the human worldwide population. The pathogen is often carried asymptomatically for decades, but can also cause severe complications (1). It was first discovered by Marshall and Warren (2) to be a causative agent of stomach inflammation and both gastric and duodenal ulcers, and has since also been recognized as the cause of approximately 1 in 20 of all human cancers (3). A very fruitful approach to study H. pylori has been the creation of a Multilocus Sequence Analysis (MLSA) (4) scheme specific to this species (5). MLSA revealed much about the evolutionary history and global population structure of H. pylori, including that it infected anatomically modern humans at least 100,000 y ago, that it accompanied its host out of Africa 60,000 y ago, and that its current worldwide genetic variation reflects the human migrations that have happened since (6–8). This intimate relationship with humans combined with a fast rate of evolution make H. pylori a useful marker to trace the movements of human populations (9, 10).
A key evolutionary property of H. pylori is its very high recombination rate. This mechanism has been well studied through laboratory experiments (11–13). In vivo, extensive recombination was first detected by comparison of the phylogenetic signals of three gene fragments (14). MLSA studies also revealed high rates of admixture (6–10), although the relationships between strains sampled from different individuals are typically too complex to allow a complete reconstruction of the evolutionary events separating them. A simpler approach has been to compare isolates taken sequentially from the same patient to study within-host evolution. Such comparisons were first based on a handful of genes (15), later on extended panels of genes (16), and culminated with the use of whole genomes (17). These studies confirmed the prominent role played by recombination in the genomic evolution of H. pylori, with up to 40% of genes affected over 3 y of within-host evolution (17).
Despite its high medical importance, many questions remain unanswered about both the evolution and epidemiology of H. pylori. In terms of genomic evolution, there is a need to precisely quantify mutation and recombination rates, to investigate the effect of immune selection, and to describe the frequency and evolutionary role played by coinfections with multiple strains. In terms of epidemiology, H. pylori is thought to be transmitted either by oral-oral or fecal-oral route between close relatives within families (18), but this hypothesis needs to be formally tested and it is unclear whether infection can occasionally come from other sources. In the population genetic studies described above (6–10), precise inference about fine-scale evolution and epidemiology is typically impossible because the individual strains are too complexly related, partly as a result of the high recombination rate. On the other hand, in the within-host evolution studies described above (15–17), genomic evolution becomes much clearer but the ability to study other processes, such as transmission, is completely lost.
Here we take an intermediate strategy between these two types of previous studies. We present a whole-genome analysis of 97 H. pylori isolates from 52 members of two well-characterized extended families living in the same rural community in Ogies, Mpumalanga, South Africa. These individuals lived under comparable social circumstances with 100% of the households having a reticulated water supply, 87% having flushing toilets, and 98% of individuals having their own toothbrush. This community has been previously studied with regards to the role of dental plaque as a reservoir of H. pylori and for coinfections (19, 20). H. pylori isolates from these individuals have been analyzed based on the sequences of three gene fragments (21) and using MLSA (22). These studies suggested a more complex mode of transmission than previously thought, although definite inference about who infected whom could not be reached on the basis of such small fractions of the genome. Here we sequenced the whole genomes of two isolates from most participants (one from the antrum and one from the corpus part of the stomach), so that within-host evolution can be studied through a comparison of the genomes carried by the same individual. Furthermore, transmission routes can also be investigated through comparisons of the genomes carried by different individuals.
Results
Genomic Sequences.
We used 454 technology (23) to fully sequence 97 genomes of H. pylori carried by 52 members of two South African families living in the same rural community. Family 12 included 42 participants from four generations and family 13 included 10 participants from three generations (Fig. 1 and Table S1). A total of 786 genes from reference strain 26695 (24) were found to be present in all 97 genomes, with a concatenated length of 709 kbp, which represents ∼200 times more data per isolate than MLSA (Fig. S1 and Table S2). All of the analyses presented below are based on these core genes. The distribution across core genes of the ratio of substitution rates at nonsynonymous and synonymous sites, dN/dS (Table S2), had a mean of 0.14 and a 95% central range of 0.02–0.34, indicating that genes were subject to varying levels of purifying selection. There were three outliers to this distribution of dN/dS across genes: HP0411 (dN/dS = 2.50), HP1211 (dN/dS = 1.01), and HP1145 (dN/dS = 0.90). These three genes encoded unknown hypothetical proteins (24). These results are consistent with previously reported variations in the selective pressures acting between as well as within the genes of H. pylori (17, 25, 26). To guard against these variations, as well as against the relative strength of selection across time frames (16, 27) and the interaction between recombination and selection (28), we only used the synonymous substitutions in the remainder of the analysis.
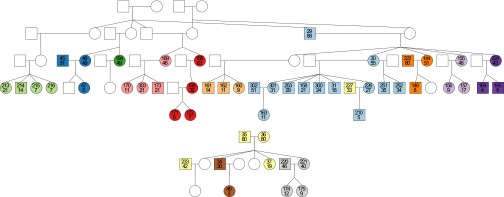
Fig. 1.
Pedigrees of families 12 (Upper) and 13 (Lower). Males and females are represented by squares and circles, respectively. Each participant is labeled with a unique identifier number (upper number) and his or her age (lower number). Individuals not included in the study (because they were dead, unwilling to participate, or did not carry H. pylori) are shown in white. Two participants are shown in the same color if and only if they share the same house. There are a total of 13 unique colors representing the 13 different houses in which the participants live.
Within-Host Evolution.
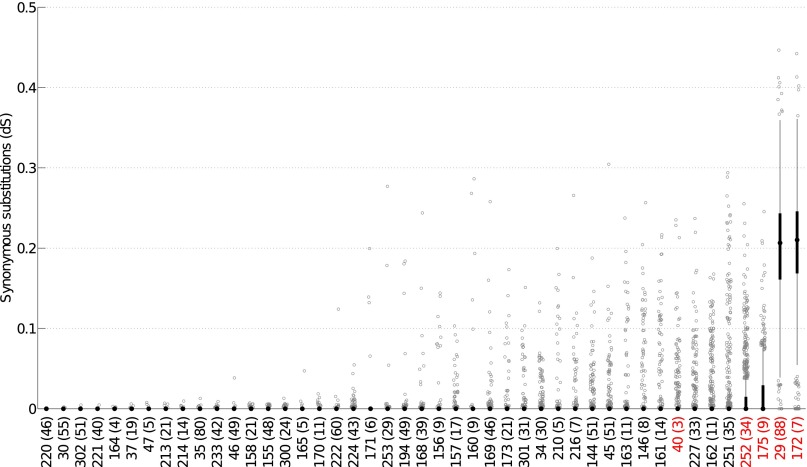
There were 45 individuals for whom two H. pylori genomes (one from the antrum and one from the corpus part of the stomach) were available, and we measured the synonymous distance, dS, for each core gene of each pair of genomes (Fig. 2). Based on these distances, we tested whether the two genomes were similar enough to have originated from the same infection. Pairs from 40 individuals were found to be compatible with a model of within-host evolution, where the differences between the genomes are explained by mutation and recombination events taking place during diversification from a common ancestor, which postdates a single colonization event. Among the five individuals for which within-host evolution was impossible (highlighted in red in Fig. 2), four had pairs of genomes with large numbers of nonidentical genes, therefore indicating multiple infection with at least two separate strains (individuals 29, 172, 175, and 252). In the last individual who was found to be incompatible with the within-host evolutionary model (individual 40), the two genomes had many identical genes, indicating that they were related (Fig. 2), but this individual was only 3-y-old, which did not leave enough evolutionary time to explain the high number of mutation and recombination events observed in other genes. This individual was therefore most probably infected twice with variants of the same strain, possibly from the same transmission donor, or had received multiple variants in the course of a single transmission.
Fig. 2.
Boxplots of the within-host synonymous distance (dS) for the 786 core genes of each of the 45 individuals in which two genomes were available. The individuals are sorted from left to right in increasing order of average dS across genes. Numbers in parentheses represent the age of the individual. The median dS is shown by a black dot, and the interquartile range (IQR) by a black rectangle. Boxplot whiskers have the standard maximum length of 1.5-times the IQR. In all but the last four individuals, the IQR spans from 0 to 0 and the black rectangle and whiskers are therefore not visible. Any gene with dS above or below the whiskers is shown as a gray open circle. The five individuals in red are the ones which were incompatible with evolution from a single infection.
Based on the pairs of genomes of the 40 individuals who were found to be compatible with within-host evolution, we estimated the synonymous mutation rate to be 1.38 × 10−5 per site per year, with a 95% credibility interval ranging from 9.14 × 10−6 to 1.85 × 10−5. This finding is in good agreement with previous genomic estimates based on serial isolation of H. pylori from the same host (17), and one-to-two orders-of-magnitude higher than estimates in other bacterial species (29). The time to the most recent common ancestor (TMRCA) for each patient was typically of the order of only a few years, with an average of 3.61 y (Table S3). This result was true even for older individuals, which was unexpected because our prior model stated that the TMRCA was equally likely to take any value between 0 and the age of the host (Materials and Methods).
Recombination Is a Major Driver of Diversification in Some Individuals Only.
The ratio r/m (30, 31) of the rates at which recombination and mutation introduce substitutions was found to vary widely from one individual to another. Some individuals (e.g., individuals 233, 158, 155, and 300 in Table S3) had very low r/m values, potentially even equal to zero, which would represent purely clonal evolution. In 14 individuals, no single gene was found to have a posterior probability of being recombined higher than 95% (Table S3). On the other hand, some individuals (e.g., individuals 146, 171, 160, and 161 in Table S3) had very high r/m values, similar or even higher than previously reported based on longitudinal isolates comparisons (16, 17). This result indicates that the effect of recombination varied significantly from one host to another.
Where recombination had taken place, the synonymous distance between donor and recipient was equal to 6.3% on average, with a central 95% range from 0.8 to 16.7%, which covers the full span of pairwise distances between unrelated strains (6). This result reflects the high promiscuity of H. pylori, and contrasts, for example, with Salmonella enterica, Escherichia coli, or Bacillus cereus, where recombination was found to happen preferentially between members of the same lineage (32–34). Across all 40 pairwise comparisons of genomes corresponding to within-host evolution, the number of recombinant genes (i.e., with posterior probability of recombination above 95%) was found to vary from 0 to 95, for a total of 665 recombination events (Table S3). For 432 (65%) of these, we were able to establish which of the two alleles was the recombinant one. We searched for putative origins of these recombinant alleles among all genomes, but only found possible donors for 54 (12.5%) of the imports, as summarized in Table S4. Only three instances were found with more than two genes putatively transferred from the same donor to the same recipient: these were from host 161 to 162 (10 genes), from host 166 to 144 (7 genes), and from host 161 to 216 (6 genes).
Transmission Analysis.
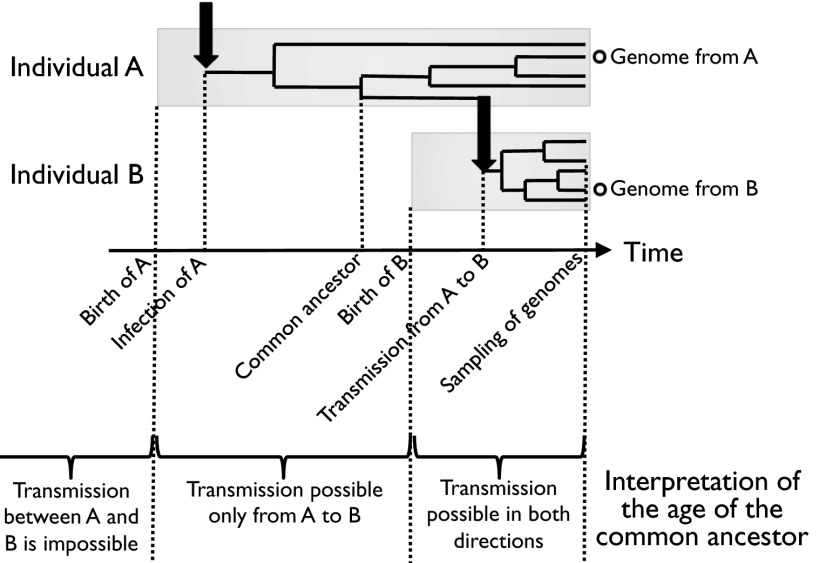
Having described the evolutionary dynamics of mutation and recombination from the relatively simple setting provided by within-host comparisons of genomes, we can now turn to the question of transmission between hosts. By measuring the TMRCA of two genomes from two distinct individuals, it is possible to determine who may have infected whom, assuming that no genomic variation from the infector is transmitted to the infected (35). Transmission from host A to host B is only possible if this TMRCA is smaller than the age of host A (Fig. 3). The results of this transmission analysis are shown in Fig. 4. The majority of the individuals (27 of 52) were completely unlinked, meaning that they were neither donor nor recipient of any transmission event. In family 13, the only link concerned individual 36, who could have infected or been infected by individual 166 of family 12. This transmission link between the two families may have been established via individual 227, who is a member of family 12 but lives in the same house as 36 and other members of family 13 (Fig. 1). Individual 227 may have acted as intermediate in the transmission between individuals 166 and 36, even if he was not found to carry the same strain (he could either have lost it since or it could have been unsampled).
Fig. 3.
Illustration of how the age of the common ancestor of a pair of genomes isolated from two distinct hosts is informative about the possibility of transmission between these two hosts. The Upper part shows an example of transmission from individual A to individual B. Within-host diversification is shown by the branching process, and transmission events are represented by the black arrows. Only one genome from each individual is considered at a time, and it is possible to estimate the date of their common ancestor. Because in this example the common ancestor is after the birth of A and before the birth of B, transmission from A to B is possible but not transmission from B to A. The Lower part of the figure gives the full scale of interpretation of possible dates for the common ancestor in term of transmission between individuals A and B.
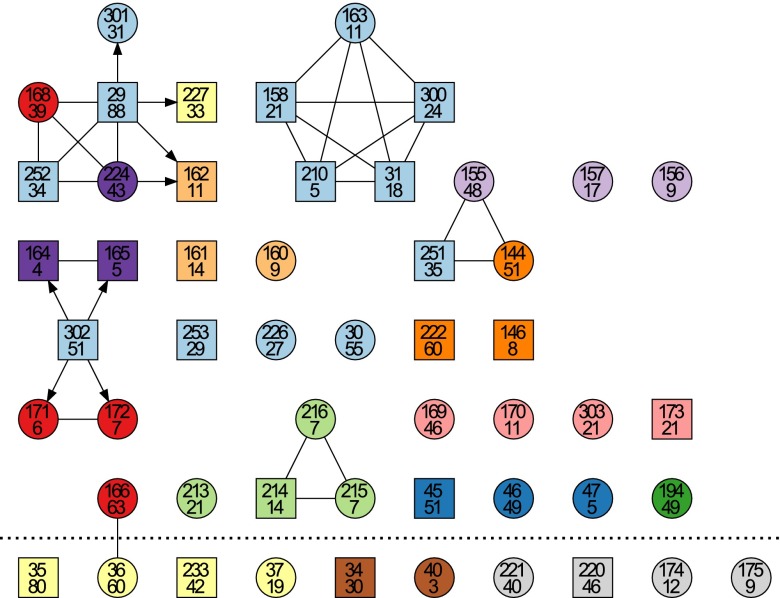
Fig. 4.
Results of the transmission analysis. Individuals are represented using the same symbols, labels, and colors as in Fig. 1. The members of family 12 are shown above the dotted line whereas the members of family 13 are shown on the last row, below the dotted line. An arrow between two individuals indicates unidirectional possibility of transmission from a donor (tail of the arrow) to a recipient (head of the arrow). An undirected link between two individuals indicates that transmission is possible in both directions.
Several transmission clusters were detected in family 12, including one cluster made of three siblings (individuals 214, 215, and 216) living in the same house, one cluster made of two siblings and their nephew (individuals 144, 155, and 251) living in three separate houses, a cluster where one individual (302) infected two young sisters (171 and 172) and two young brothers (164 and 165), spanning three different houses, and one cluster made of three brothers (158, 300, and 31) and their nephew (210) and niece (163), all of which live in the same house. Finally, the largest cluster was made of seven individuals (29, 301, 227, 162, 224, 252, and 168) spanning four houses, with at its center the oldest participant (29, who was 88-y-old). Among all these transmission clusters, there was only one possible instance of transmission from parent to offspring (from 29 to 224).
The inferred patterns of transmission between individuals (Fig. 4) were reconstructed solely on the basis of the homology of the H. pylori genomes they carry and the age of the individuals. We can therefore compare these results with other epidemiological variables that were not used in the analysis, such as the degree of relatedness between individuals or the memberships of households (Fig. 1). We found a strong positive association between transmission and kinship coefficient (r = 0.22, P = 5 × 10−6, simple Mantel test), indicating that transmission happened more often between close relatives. We also found a similarly strong positive association between transmission and house sharing (r = 0.24, P = 8 × 10−6, simple Mantel test). Because relatedness and house sharing are strongly correlated (Fig. 1), we tested the association of each variable while controlling for the other to disentangle their effects. We found a positive association of transmission with kinship when controlling for house sharing (r = 0.10, P = 7.9 × 10−3, partial Mantel test) and a positive association of transmission with house sharing when controlling for kinship (r = 0.12, P = 3.2 × 10−3, partial Mantel test). These results indicate that both kinship and house sharing have independent effects of roughly the same strength on transmission patterns.
Discussion
Variation in Recombination Rates Between Hosts.
H. pylori is often described as a highly recombinogenic species of bacteria (6, 14–17). Our study confirms that recombination can indeed be a potent force of genomic diversification, with several examples of within-host evolution where the ratio r/m of the effects of recombination and mutation may be over 100 (Table S3). However, we also found that in some infected individuals, recombination has played a much more modest role, if any. A first explanation for these differences could be that some lineages of H. pylori have higher rates of recombination than others. Such variations in recombination rates have been previously described between lineages of S. enterica (32), E. coli (36), or Chlamydia trachomatis (37). For H. pylori, laboratory experiments have shown that recombination rates depended significantly on the combination of recipient and donor strains (11, 13). Here, however, there seemed to be no relationship between infecting strain and effect of recombination. For example, the three individuals 155, 144, and 251 have infected each other (Fig. 4), but the effect of recombination was found to vary greatly between them (with r/m mean estimates of 0.5, 13.8, and 36.6, respectively) (Table S3). Another likely important source of variation in the effect of recombination is the presence or absence of multiple infections. Individuals with low r/m could correspond to individuals that were not multiply infected (so that recombination only happened between members of the same strain and therefore does not have much effect), whereas individuals with high r/m would be the ones where multiple infections were present. Our observation of strong variation of the rate of in vivo recombination, even within one community, reconciles previous controversial reports about the frequency of recombination in H. pylori, which included both very high (15, 17) as well as very low (38) values.
Multiple H. pylori Infections.
Infections with multiple strains of H. pylori have been reported in several studies, but their true incidence is unknown because analysis of multiple isolates from one individual is rarely performed (39, 40). A recent study found that infections with multiple strains of H. pylori were very common in India (41). We found definite evidence for multiple infections in 5 of 45 individuals, where the genome sequences of the H. pylori strains isolated from antrum and corpus shared only few identical genes. Table S3 shows the results for the remaining 40 individuals whose paired H. pylori genomes were highly related, such that for these individuals there was no direct evidence that multiple infections were present. It is, however, likely that some of them also hosted multiple infections that were not detected because only two isolates were studied in each case. Even if two infections were present in a host in equal proportions and complete mixture (giving the best chance to sample both), then the probability to take one isolate from each is only 50%. This finding would suggest that at least half of the multiple infections were undetected, giving an estimate for the total number of multiply infected individuals of at least 10 out of 45 (22%). It seems also likely that for some individuals, multiple infections were present in the past and acted as a source of recombined material, but had been removed by the time of sampling.
Diversity Bottleneck and Immune Selection.
In many individuals, the antrum and corpus genomes only differed at few genes (Fig. 2). Consequently, the TMRCA of pairs of genomes from the same infection were relatively low, with an average of 3.61 y (Table S3). This result is in good agreement with the previous observation that the genetic distance between serial isolates from a same host is correlated with the time separating the two isolations but uncorrelated with the age of the individuals (16). One explanation for these low TMRCA would be that colonization with H. pylori has been recent for most individuals, but this would go against the generally accepted idea that acquisition happens in early childhood (1, 18), as well as the high proportion of individuals who were found to be carriers of H. pylori in this setting (21, 22). More probably, this observation could be the result of strong genetic drift (42); for example, population bottlenecks caused by the selective pressure of the human immune system (43). Assuming an average of one cell division per day (44), the average TMRCA of 3.61 y would imply an effective population size of H. pylori equal to Ne = 1,318. This quantity indicates how quickly polymorphism is lost in the population. For example, in a multiple infection by two strains present in equal proportions (giving them the best chance to both persist for a long time) the expected time until one or other strain was lost would be 5 y (45). The two oldest individuals where multiple infections were detected (individuals 252 and 29, who were 34- and 88-y-old, respectively) are therefore unlikely to represent acquisition of multiple strains in early childhood. Instead, these two probably reflect that in highly endemic regions, even individuals who are already colonized are exposed to infection with new strains. Treatment with antibiotics could be another reason for the observed diversity bottlenecks. However, antibiotics can only be obtained through prescription in South Africa, and use of antibiotics in the Ogies community is low. Only five of the individuals for whom antrum-corpus strain pairs were available had reported recent antibiotic use, and there was no correlation with TMRCA. We thus conclude that antibiotics are an unlikely major reason for the observed low average TMRCA.
Missing Transmission Links.
To reconstruct putative transmission links between individuals, we compared the genomes of the H. pylori strains they carried while accounting for the within-host evolution that could have happened before and after the transmission event (Fig. 3). This approach revealed several instances of transmission, significantly overrepresented between closely related members of the same family or between individuals living in the same house (Fig. 4). We note that our genome-based approach identified far more inferred transmission links than a previous MLSA approach, which used the carriage of strains with an identical sequence type (ST) as evidence of likely recent transmission, and connected 14 individuals from the two families (22). Interestingly, all transmission links suggested by the “identical ST” criterion were supported by the whole-genome approach, indicating that because of the rapid diversification of H. pylori, ST identity is in fact a highly specific criterion for a transmission link. However, our current approach linked 25 individuals, and provided support for a direction of transmission for seven links, which is not possible using MLSA alone.
However, there remained a majority of individuals (27 of 52), who could have acted neither as donor nor recipient of any transmission event. Similarly, for a large proportion (378 of 432) of the genes that were found to have been imported through recombination during within-host evolution, we were unable to identify a possible origin among the 97 genomes we sequenced. These two observations, that donors of both transmission and recombination events could not be identified, are in good agreement with each other. The findings both suggest that some of the strains circulating presently or in the past have not been sampled in this study.
A first explanation may be that transmission of H. pylori sometimes happens outside of the familial context, and therefore members of other families living in the same South African rural community would have to be studied to find the missing links. The fact that we found a putative transmission link between members of the two families (between hosts 166 and 36) (Fig. 4) confirms the validity of this hypothesis. However, because transmission was found to predominantly happen between relatives and inhabitants of the same houses, the influence of other families seems insufficient to explain on its own the large numbers of missing transmission and recombination links. A second explanation would be that multiple infections are common, and we have discussed above the evidence supporting this claim. This finding would imply that not all infections have been sampled in the participating individuals, partly because only two genomes were sampled from each of them but also because they may have hosted other strains in the past that were not present anymore by the time of the endoscopy.
These two hypotheses, transmission outside of the familial setting and frequent turnover of infecting strains, are not mutually exclusive and could together explain the large fraction of missing donors of transmission and recombination. These hypotheses represent the two facets of what needs to be investigated to fully understand any infectious disease: within-host evolution on the one hand, and transmission from host to host on the other. Whole-genome sequencing holds great promise to elucidate these processes, not just for H. pylori as we have demonstrated here, but also for many other microbial pathogens (29). To fully exploit this potential requires researchers to consider within- and between-host dynamics jointly, because they can only be understood in light of each other.
Materials and Methods
Genome Sequencing and Assembly.
Draft genome sequences of H. pylori isolates from families 12 and 13 from Ogies, Mpumalanga, South Africa (19–22) were obtained using Roche 454 FLX technology (23). Library preparation was either done according to Genome Sequencer (GS) FLX General Library Preparation Method Manuals for FLX Chemistry or FLX Titanium chemistry (overall 26 isolates of family 12). Fifty-four isolates of family 12 and 19 isolates of family 13 were prepared according to the GS Rapid Library Preparation Method Manual (Roche). Emulsion PCR and 454 pyrosequencing were performed following the manufacturer’s instructions. The genomes of two isolates (antrum isolate of individual 303 and corpus isolate of individual 174) did not pass quality control, leaving a total of 97 genomes: 79 genomes from 42 members of family 12 and 18 genomes from 10 members of family 13 (Fig. 1 and Table S1). The genomes were assembled de novo using the Roche GS De Novo Assembler (v2.6), resulting in an average of 60 contigs per genomes (with a minimum of 26 and a maximum of 188) (Table S1).
Identification of Core Genes and Synonymous Polymorphisms.
The annotation of the previously sequenced reference genome H. pylori strain 26695 contains 1,590 predicted coding sequences (24). For each of these genes and each of our 97 genomes, we used BLAST (46) to look for homologs, following a similar approach to the one implemented by BIGSdb (47). If the best BLAST hit of a gene against a genome covered at least 90% of the positions of the query sequence, the gene was considered to be found. This is a conservative approach to finding gene homologs because it does not allow long indels or gene splits at contig ends. We found that 786 genes (49%) were present among all 97 genomes (Table S2). These core genes ranged in length from 201 bp up to 3,636 bp, with a mean of 902 bp and a total concatenated length of 709,155 bp, which represents 42.5% of the length of the 26695 genome (24). For each core gene, a query-anchored alignment was produced, so that each column of the alignment corresponded exactly to a position of the gene sequence in the reference genome. Consequently, our data were robust to insertions or deletions relative to the reference, which may be because of the known tendency of 454 to produce homopolymer frameshift errors. The core genes are well distributed around the reference genome 26695, except for a few regions of very low GC content (Fig. S1). All of the analyses presented are based on these core genes so that comparisons between genomes are always based on exactly the same data. Synonymous and nonsynonymous polymorphisms were distinguished using the method of Nei and Gojobori (48). The ratios dN/dS of rates of nonsynonymous and synonymous mutations were computed for each gene (Table S2). Only synonymous substitutions were used in the analyses of within-host genomic evolution and host-to-host transmission.
Evolutionary Model.
Let us consider a pair of genomes. Let t denote the time to their most recent common ancestor, so that in evolutionary terms 2t separate the two genomes. During this time, each gene indexed j may have recombined [with probability 1 − exp(−ρt), where ρ/2 is the recombination rate] or evolved clonally [with probability exp(−ρt)]. If the gene j evolved clonally, it has accumulated a number of mutations distributed as Binomial(Lj, tθs), where Lj is the number of synonymous sites in gene j, and θs/2 is the synonymous mutation rate. If the gene j recombined, it would have acquired a number of substitutions distributed as Binomial(Lj, ν), where ν is the distance between donor and recipient of the recombination event, which is distributed as Beta(α, β). This evolutionary model is related to the ClonalFrame model (49), but is more general in that ν takes a distribution instead of being a constant, as previously proposed (16). The parameters ρ and t are assumed to be specific to each pairwise comparison, which represents another generalization of the model, allowing us to capture variations in the recombination rate. The parameters θs, α and β are identical across comparisons. Unless otherwise stated, the prior distribution on any parameter is improper uniform over (0,∞).
Analysis of Within-Host Evolution.
There were 45 individuals for whom two genomes were sequenced (one isolated from the antrum and one from the corpus), and the synonymous distance dS was measured for each gene of these paired genomes (Fig. 2). For a given individual indexed i who is of age ai, if we assume that the two genomes are descended from the same ancestor within the host (i.e., are part of the same infection), then let ti denote the time to this common ancestor, which takes value between 0 (in the extreme case where the two genomes separated from each other just before isolation) and ai (in the opposite extreme case, where the individual was infected just after birth and the common ancestor dates from then). To reflect our initial ignorance about ti, we assume a uniform prior between 0 and ai for this parameter. The two genomes are assumed to be related according the model described in the previous paragraph, and inference was performed for all parameters using a Monte-Carlo Markov Chain (MCMC), initially considering that all pairs of genomes from the same individual had evolved within-host. Posterior-predictive distributions (50) on the number of substitutions between pairs of genomes were used to assess the fit of this model for each individual. In five individuals (29, 40, 172, 175, and 252), the number of differences was found to be incompatible with within-host evolution, which means that the two genomes result from two separate infection events. The MCMC was rerun with these individuals excluded to estimate all parameters (Tables S3 and S5).
Analysis of Recombination.
In the evolutionary model described above, the relative effect of recombination and mutation r/m (30, 31) is equal to ρα/[θ(α + β)] and this value is reported for each host in Table S3. For all recombination events with posterior probability above 95%, we attempted to establish which of the two alleles was the recombinant one by comparing them with the allele carried by the closest overall relative of the two genomes. If the distance of one allele to that of the closest relative was at least twice as high as that of the other allele, then we considered that the former was the recombinant allele. For each imported gene we searched for an exact match in another genome, which would therefore represent a possible source of recombination (32, 51).
Analysis of Transmission.
For each pair of genomes from different patients, we estimated the time t of their MRCA using the model above and setting the value of the global parameters θs, α, and β to their posterior mean from the within-host analysis above. Transmission from individual A to individual B was assessed to be possible only if the lower bound of the 95% credibility interval of t was smaller than the age aA of individual A. In other words, transmission from A to B is possible only if the age of the common ancestor of the genomes carried by A and B is smaller than the age of A, so that this common ancestor could have existed in individual A (Fig. 3). This process is assuming that only a single genomic variant is transmitted from A to B, or in other words that there is a strong bottleneck of diversity at the point of transmission. The correlation of transmission links (Fig. 4) with kinship coefficients and household memberships were measured using Mantel tests. Partial Mantel tests with permutation of residuals were used to test the association of transmission with kinship while controlling for house-sharing and vice-versa. These Mantel tests were performed only in family 12 because no transmission events were found within family 13, and they were conducted using a million random permutations in the statistical software zt (52). The analyses of within-host evolution, recombination and transmission described above were performed using Matlab code that was developed specifically for the purpose of this study, and which is available from the authors upon request.
Supplementary Material
Acknowledgments
We thank Mark Achtman, Daniel Falush, and Christine Josenhans for helpful comments on an earlier version of this manuscript, and Friederike Kops and Birgit Brenneke for excellent technical assistance. This study was supported by Grants SFB 900/A1 and SFB 900/Z1 from the Deutsche Forschungsgemeinschaft (to S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.J.F. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive (ENA) within the European Bioinformatics Institute database, www.ebi.ac.uk (study nos. PRJEB4021 and PRJEB4040–PRJEB4135).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304681110/-/DCSupplemental.
References
- 1.Suerbaum S, Josenhans C. Helicobacter pylori evolution and phenotypic diversification in a changing host. Nat Rev Microbiol. 2007;5(6):441–452. doi: 10.1038/nrmicro1658. [DOI] [PubMed] [Google Scholar]
- 2.Marshall BJ, Warren JR. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1(8336):1273–1275. [PubMed] [Google Scholar]
- 3.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 4.Maiden MC, et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95(6):3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Achtman M, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32(3):459–470. doi: 10.1046/j.1365-2958.1999.01382.x. [DOI] [PubMed] [Google Scholar]
- 6.Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582–1585. doi: 10.1126/science.1080857. [DOI] [PubMed] [Google Scholar]
- 7.Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moodley Y, et al. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8(5):e1002693. doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wirth T, et al. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: Lessons from Ladakh. Proc Natl Acad Sci USA. 2004;101(14):4746–4751. doi: 10.1073/pnas.0306629101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moodley Y, et al. The peopling of the Pacific from a bacterial perspective. Science. 2009;323(5913):527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulick S, et al. Mosaic DNA imports with interspersions of recipient sequence after natural transformation of Helicobacter pylori. PLoS ONE. 2008;3(11):e3797. doi: 10.1371/journal.pone.0003797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin EA, et al. Natural transformation of helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 2009;5(3):e1000337. doi: 10.1371/journal.ppat.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moccia C, et al. The nucleotide excision repair (NER) system of Helicobacter pylori: Role in mutation prevention and chromosomal import patterns after natural transformation. BMC Microbiol. 2012;12:67. doi: 10.1186/1471-2180-12-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suerbaum S, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci USA. 1998;95(21):12619–12624. doi: 10.1073/pnas.95.21.12619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falush D, et al. Recombination and mutation during long-term gastric colonization by Helicobacter pylori: Estimates of clock rates, recombination size, and minimal age. Proc Natl Acad Sci USA. 2001;98(26):15056–15061. doi: 10.1073/pnas.251396098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morelli G, et al. Microevolution of Helicobacter pylori during prolonged infection of single hosts and within families. PLoS Genet. 2010;6(7):e1001036. doi: 10.1371/journal.pgen.1001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennemann L, et al. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA. 2011;108(12):5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh K-L, Chan W-K, Shiota S, Yamaoka Y. Epidemiology of Helicobacter pylori infection and public health implications. Helicobacter. 2011;16(Suppl 1):1–9. doi: 10.1111/j.1523-5378.2011.00874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olivier BJ, et al. Absence of Helicobacter pylori within the oral cavities of members of a healthy South African community. J Clin Microbiol. 2006;44(2):635–636. doi: 10.1128/JCM.44.2.635-636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritz EL, Slavik T, Delport W, Olivier B, van der Merwe SW. Incidence of Helicobacter felis and the effect of coinfection with Helicobacter pylori on the gastric mucosa in the African population. J Clin Microbiol. 2006;44(5):1692–1696. doi: 10.1128/JCM.44.5.1692-1696.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delport W, Cunningham M, Olivier B, Preisig O, van der Merwe SW. A population genetics pedigree perspective on the transmission of Helicobacter pylori. Genetics. 2006;174(4):2107–2118. doi: 10.1534/genetics.106.057703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarz S, et al. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog. 2008;4(10):e1000180. doi: 10.1371/journal.ppat.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rothberg JM, Leamon JH. The development and impact of 454 sequencing. Nat Biotechnol. 2008;26(10):1117–1124. doi: 10.1038/nbt1485. [DOI] [PubMed] [Google Scholar]
- 24.Tomb JF, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388(6642):539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 25.Duncan SS, et al. Comparative genomic analysis of East Asian and non-Asian Helicobacter pylori strains identifies rapidly evolving genes. PLoS ONE. 2013;8(1):e55120. doi: 10.1371/journal.pone.0055120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olbermann P, et al. A global overview of the genetic and functional diversity in the Helicobacter pylori cag pathogenicity island. PLoS Genet. 2010;6(8):e1001069. doi: 10.1371/journal.pgen.1001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocha EPC, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239(2):226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- 28.Castillo-Ramírez S, et al. The impact of recombination on dN/dS within recently emerged bacterial clones. PLoS Pathog. 2011;7(7):e1002129. doi: 10.1371/journal.ppat.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Didelot X, Bowden R, Wilson DJ, Peto TEA, Crook DW. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet. 2012;13(9):601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feil EJ, et al. Recombination within natural populations of pathogenic bacteria: Short-term empirical estimates and long-term phylogenetic consequences. Proc Natl Acad Sci USA. 2001;98(1):182–187. doi: 10.1073/pnas.98.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3(2):199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 32.Didelot X, et al. Recombination and population structure in Salmonella enterica. PLoS Genet. 2011;7(7):e1002191. doi: 10.1371/journal.pgen.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Didelot X, Lawson DJ, Darling AE, Falush D. Inference of homologous recombination in bacteria using whole-genome sequences. Genetics. 2010;186(4):1435–1449. doi: 10.1534/genetics.110.120121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Didelot X, Méric G, Falush D, Darling AE. Impact of homologous and non-homologous recombination in the genomic evolution of Escherichia coli. BMC Genomics. 2012;13:256. doi: 10.1186/1471-2164-13-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Didelot X, et al. Microevolutionary analysis of Clostridium difficile genomes to investigate transmission. Genome Biol. 2012;13(12):R118. doi: 10.1186/gb-2012-13-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wirth T, et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol Microbiol. 2006;60(5):1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joseph SJ, et al. Population genomics of Chlamydia trachomatis: Insights on drift, selection, recombination, and population structure. Mol Biol Evol. 2012;29(12):3933–3946. doi: 10.1093/molbev/mss198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lundin A, et al. Slow genetic divergence of Helicobacter pylori strains during long-term colonization. Infect Immun. 2005;73(8):4818–4822. doi: 10.1128/IAI.73.8.4818-4822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taylor NS, et al. Long-term colonization with single and multiple strains of Helicobacter pylori assessed by DNA fingerprinting. J Clin Microbiol. 1995;33(4):918–923. doi: 10.1128/jcm.33.4.918-923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kersulyte D, Chalkauskas H, Berg DE. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31(1):31–43. doi: 10.1046/j.1365-2958.1999.01140.x. [DOI] [PubMed] [Google Scholar]
- 41.Patra R, et al. Multiple infection and microdiversity among Helicobacter pylori isolates in a single host in India. PLoS ONE. 2012;7(8):e43370. doi: 10.1371/journal.pone.0043370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charlesworth B. Fundamental concepts in genetics: Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10(3):195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 43.Young BC, et al. Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci USA. 2012;109(12):4550–4555. doi: 10.1073/pnas.1113219109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dumrese C, et al. The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype. FEBS Lett. 2009;583(10):1637–1643. doi: 10.1016/j.febslet.2009.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kimura M, Ota T. The average number of generations until extinction of an individual mutant gene in a finite population. Genetics. 1969;63(3):701–709. doi: 10.1093/genetics/63.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jolley KAA, Maiden MCJC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3(5):418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 49.Didelot X, Falush D. Inference of bacterial microevolution using multilocus sequence data. Genetics. 2007;175(3):1251–1266. doi: 10.1534/genetics.106.063305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gelman A, Meng X, Stern H. Posterior predictive assessment of model fitness via realized discrepancies. Statist Sinica. 1996;6:733–807. [Google Scholar]
- 51.Didelot X, Barker M, Falush D, Priest FG. Evolution of pathogenicity in the Bacillus cereus group. Syst Appl Microbiol. 2009;32(2):81–90. doi: 10.1016/j.syapm.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Bonnet E, Van de Peer Y. zt: A software tool for simple and partial Mantel tests. J Stat Softw. 2002;7:1–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.