Abstract
Many Proteobacteria use N-acyl-homoserine lactone (acyl-HSL) quorum sensing to control specific genes. Acyl-HSL synthesis requires unique enzymes that use S-adenosyl methionine as an acyl acceptor and amino acid donor. We developed and executed an enzyme-coupled high-throughput cell-free screen to discover acyl-HSL synthase inhibitors. The three strongest inhibitors were equally active against two different acyl-HSL synthases: Burkholderia mallei BmaI1 and Yersinia pestis YspI. Two of these inhibitors showed activity in whole cells. The most potent compound behaves as a noncompetitive inhibitor with a Ki of 0.7 µM and showed activity in a cell-based assay. Quorum-sensing signal synthesis inhibitors will be useful in attempts to understand acyl-HSL synthase catalysis and as a tool in studies of quorum-sensing control of gene expression. Because acyl-HSL quorum-sensing controls virulence of some bacterial pathogens, anti–quorum-sensing chemicals have been sought as potential therapeutic agents. Our screen and identification of acyl-HSL synthase inhibitors serve as a basis for efforts to target quorum-sensing signal synthesis as an antivirulence approach.
Keywords: bacterial communication, cell–cell signaling, enzyme inhibitors
Acyl-homoserine lactones (acyl-HSLs) are common intercellular quorum-sensing signals in Proteobacteria. Different bacterial species produce different acyl-HSLs. Most of the acyl-HSLs described are fatty acyl-HSLs. Synthesis of acyl-HSLs is typically catalyzed by members of the LuxI family of synthases. Acyl-HSL receptors are typically members of the LuxR family of transcription factors (1–3).
LuxI-type synthases transfer an acyl group from the fatty acid biosynthesis acyl carrier protein (ACP) to the methionyl amine of S-adenosyl-l-methionine (SAM); after which, cyclization of the methionyl moiety to homoserine lactone occurs. Most of our knowledge about the enzymology of acyl-HSL synthesis is from studies of RhlI from Pseudomonas aeruginosa. Product-inhibition kinetics show that RhlI catalysis follows a sequential ordered bi ter mechanism (4, 5). The transfer reaction occurs before lactonization with an acyl-SAM intermediate (5) and yields threeproducts: acyl-HSL, 5′-methylthioadenosine (MTA), and holo-ACP. LuxI-type synthases are considered as members of the Gcn5-related N-acetyltransferase superfamily (6), and yet they catalyze a unique reaction unlike other characterized enzymes in this superfamily (7).
Acyl-HSL synthases are unique enzymes, not present in Eukarya, and they are essential for quorum sensing. Measuring their activity has been cumbersome and not amenable to high-throughput inhibitor screening. Bioassays require considerable sample manipulation. A radiotracer has been developed, but this also requires sample manipulation and with respect to screening there are safety and regulatory issues. An assay in which the product holo-ACP is monitored with a thiol reagent has been developed, but it is not well suited to screening efforts because of limited sensitivity and interference with the absorbance readout by test compounds (4, 5, 8, 9).
Acyl-HSL quorum sensing controls different genes in different bacterial species, and, in some bacterial pathogens, virulence requires quorum sensing (1–3). For this reason, acyl-HSL quorum sensing has been considered as a potential therapeutic target and a variety of approaches have been used to identify quorum-sensing inhibitors (10, 11). By performing cell-based screens, or by synthesizing acyl-HSL analogs, investigators have identified a variety of inhibitors, which target the signal receptor. Enzymes that degrade acyl-HSLs (12) and end-product inhibition of acyl-HSL synthesis have been described (4), but there is very little information regarding acyl-HSL synthase inhibitors (12, 13). Acyl-HSL synthases are at least as inviting as therapeutic targets as are acyl-HSL receptors, and theoretical work suggests that effective therapeutic strategies may require inhibition of both signal synthases and reception simultaneously (14).
In an effort to better understand the enzymology of acyl-HSL synthases and perhaps exploit them as targets for quorum sensing inhibition, we developed a coupled enzyme assay with a fluorescent readout for use in a high-throughput inhibitor screen. By using this screen, we identified acyl-HSL synthase inhibitors, and we characterized the most potent of these compounds.
Results
A High-Throughput Screen for Acyl-HSL Synthase Inhibitors.
We chose to use BmaI1, an acyl-HSL synthase, from the pathogenic bacterium Burkholderia mallei as the primary target for our screen because the fatty acyl substrate for this enzyme, octanoyl-acyl carrier protein (C8-ACP) is relatively easy to synthesize in comparison with substituted acyl-ACPs. We developed a small volume (13.3-μL) assay with a fluorescent readout by modifying a commercially available S-adenosyl homocysteine assay (Fig. 1A). The first enzyme of the coupling assay, nucleoside hydrolase can use MTA, a product of acyl-HSL synthases, as a substrate (15). The resulting adenine is deamidated to give hypoxanthine, which is oxidized to give hydrogen peroxide. Hydrogen peroxide is oxidized by horseradish peroxidase, and the electrons are donated to the colorless and nonfluorescent 10-acetyl-3,7-dihydroxyphenoxazine (ADHP). This results in deacetylation of ADHP to finally give the pink and fluorescent resorufin (16). In the coupled assay, resorufin production is dependent on acyl-HSL synthesis (Fig. 1B). Octanoyl-CoA (C8-CoA) served as a poor acyl donor for BmaI1 (Fig. 1C). The reactions were stopped by addition of acetovanillone as an alternate electron acceptor for horseradish peroxidase (17).
Fig. 1.
The high-throughput screen from acyl-HSL synthase inhibitors. (A) The reaction of BmaI1 and coupling enzymes. The enzymes coupling the production of MTA to resorufin are shown in blue: nucleoside hydrolase (Nuh), adenine deaminase (Ade), xanthine oxidase (XO), and horseradish peroxidase (HRP). Products central to the coupling assay are highlighted in pink. The reactions results in the oxidation of ADHP to the fluorescent compound resorufin. A counter screen was used to eliminate inhibitors of the coupling reactions. (B) Two wells from a 384-well plate. The upper well shows the color, which develops as the reactions proceed. Acetovanillone was added to the lower well before the reaction was started. (C) The progress of 70-μL reactions with 50 μM C8-ACP (red), 500 μM octanoyl-CoA (blue), or the control with no acyl substrate (black).
We carried out a high-throughput screen for inhibitors of the coupled reaction in 384-well plates and screened 12,579 compounds at about 100 μg/mL. Because inhibition of BmaI1 or any of the other four enzymes in the reaction mixtures would affect resorufin production, we also performed a counter screen in which we initiated the coupling assay with MTA directly. Both the screen and the counter screen were reliable and robust (Z factor > 0.7) (18).
We performed the screen and the counter screen in duplicate and calculated Z scores for each compound tested. For both screens, the data fit a Gaussian distribution (SI Appendix, Fig. S1). We selected 171 compounds within two SDs of the average negative control for each plate in the primary screen. Of these, 47 had a counter screen Z score within 3 of the mean (Fig. 2A). We retested these compounds at 5 and 50 μg/mL by measuring the initial reaction velocity (Fig. 2B). The retesting confirmed 40 of the 47 as BmaI1 inhibitors.
Fig. 2.
High-throughput screen of inhibitors and verification. (A) The final raw fluorescence values for each of the 47 BmaI1 inhibitors with primary screen (boxes) and counter screen (triangles). The mean of two replicate assays is shown with the range as a vertical line. Acetovanillone-inhibited controls are gray and DMSO solvent controls are pink. (B) The percentage of inhibition of the initial velocity of reactions from retesting compounds. The pink box highlights the 15 compounds, which were selected for further analysis.
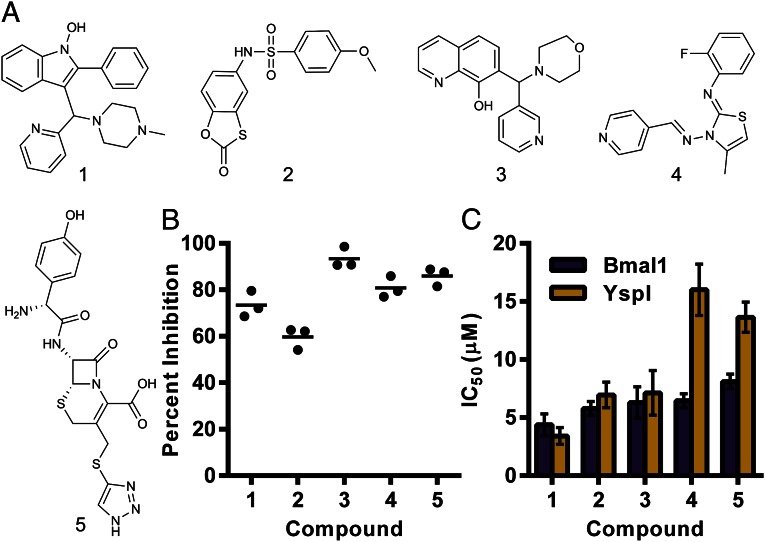
Of the 40 confirmed hits, 15 showed >65% inhibition of BmaI1 at 50 μg/mL. Twelve of the 15 compounds were commercially available and studied further. We tested the activity of the 12 compounds by using a previously established acyl-HSL synthase assay in which reduction of 2,6-dichlorophenolindophenol (DCPIP) by the thiol of holo-ACP is measured spectrophotometrically (19, 20). The reaction mixture for this secondary assay included detergent to eliminate aggregation-dependent false positives common in high-throughput screening (21). Five compounds showed >50% inhibition of the initial velocity of the reaction (Fig. 3). By using the coupled enzyme assay, we found the five compounds have IC50 values in the range of 4–10 μM (Fig. 3C).
Fig. 3.
Potency and specificity of the strongest inhibitors. (A) Structures of the five compounds showing greater than 50% inhibition in the DCPIP assay. Compound 5 is the antibiotic cefatrizine. (B) Efficacy of each compound at 100 µM in the DCPIP assay. The means of all treatments are significantly higher than the DMSO control (mean 0% inhibition) by Tukey’s multiple comparison test (multiplicity adjusted P < 0.0001). (C) Potency of each compound as measured by the enzyme coupled assay with B. mallei BmaI1 (purple) and Y. pestis YspI (gold). The error bars show the SD of the nonlinear regression analysis.
Do Inhibitors Affect Diverse Acyl-HSL Synthases?
There are hundreds of acyl-HSL synthase sequences available in public databases. Are the inhibitors identified in our screen specific for BmaI1, or do they show activity against distantly related acyl-HSL synthases? To address this question, we tested the ability of the five most potent compounds in the DCPIP reduction assay to inhibit a Yersinia pestis acyl-HSL synthase, YspI. YspI catalyzes synthesis of four acyl-HSLs, including C8-HSL (22), but is phylogenetically distant from BmaI1 (SI Appendix, Fig. S2), sharing only 19% amino acid sequence identity with BmaI1. All five of the compounds tested inhibited the activity of purified YspI, with compounds 1–3 showing equal potency against either BmaI1 or YspI (Fig. 3C), and compounds 4 and 5 showing reduced potencies with YspI.
Inhibition of BmaI1 in Whole Bacterial Cells.
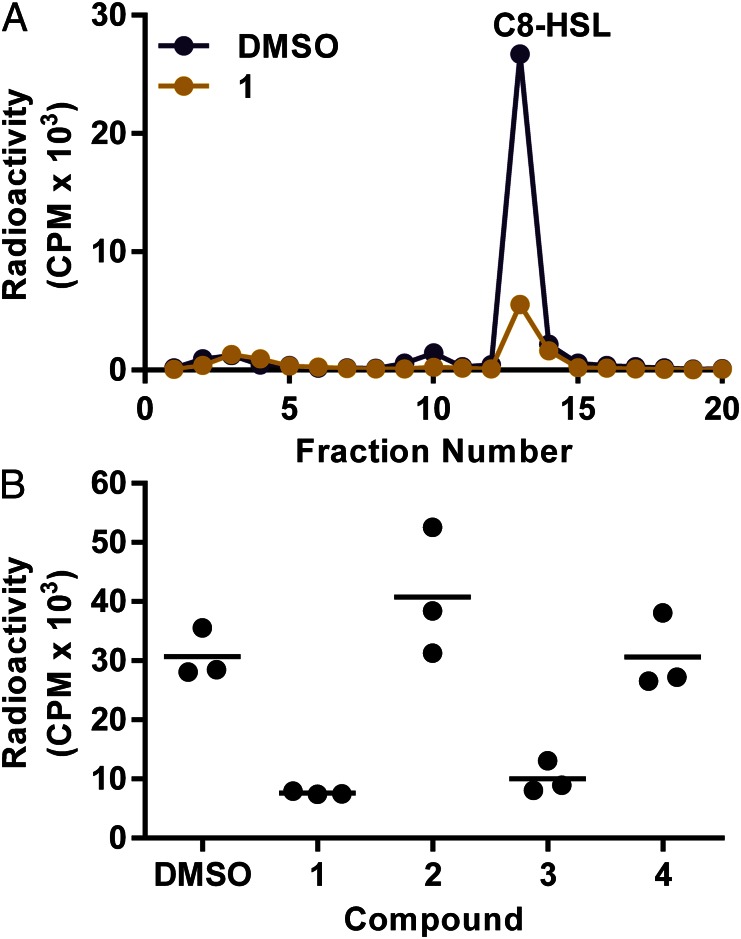
To begin to address the question of whether any of the potent inhibitors of BmaI1 targeted acyl-HSL synthase activity in whole cells, or showed any obvious off-target activities, we measured the effects of compounds 1–4 on C8-HSL production and growth in recombinant Escherichia coli containing the arabinose-inducible promoter bmaI1-expression vector pBD2 (23). We excluded compound 5, the β-lactam antibiotic, not for lack of interest but because it would be difficult to sort out antibiotic activity from direct inhibition of BmaI1. By using the recombinant E. coli, we were able to constitutively express bmaI1 and, thereby, avoid positive autoregulation (1), which can complicate inhibitor studies. We used a previously described acyl-HSL radiotracer assay (24, 25) to monitor the effects of inhibitors on BmaI1 activity (Fig. 4). We exposed the cells to 100 μM compound (about 30 μg/mL) for 10 min before incubating with [14C]methionine for 20 min. Compounds 1 and 3, but not compounds 2 and 4, caused the bacteria to produce substantially less C8-HSL than bacteria grown without inhibitors. None of the compounds affected the density of E. coli in the experiment. We also found that compounds 1 and 3 had little or no effect on growth E. coli (pBD2) over a range of concentrations (SI Appendix, Fig. S3).
Fig. 4.
Inhibition of C8-HSL synthesis in whole bacterial cells. BmaI1 activity in E. coli was followed by measuring [14C]methionine incorporation into acyl-HSL. Extracts from cultures incubated with 100 µM inhibitor for 10 min, followed incubation with inhibitor and [14C]methionine for 20 min were analyzed by HPLC and scintillation counting. Acyl-HSLs were solvent extracted and methionine remained in the aqueous phase. (A) HPLC profiles of extracts from a culture grown in the presence of compound 1 (gold) and the DMSO control culture (purple). Methionine standard was eluted in the void volume and C8-HSL in fraction 13. (B) Counts of extracted and separated acyl-HSL (pooled fraction 8–16) for three replicates with compounds 1–4. The means are shown as horizontal lines. The means of 1 and 3 are significantly different from the control by Tukey’s multiple comparison test (multiplicity adjusted P = 0.0036 and P = 0.0086, respectively).
Kinetics of Compound 1 Inhibition.
Because compound 1 was the most potent BmaI1 inhibitor tested (Fig. 3) and also showed strong activity in the cell-based assay (Fig. 4), we chose to study it further by performing kinetic analyses with BmaI1. We used the DCPIP assay for our kinetic analyses because it does not involve any coupling enzymes, rather it measures one of the reaction products, holo-ACP, directly (20). By using a pseudo–first-order kinetic analysis, we calculated a Michaelis constant (Ka) for SAM of 471 ± 39 μM. For C8-ACP, the Michaelis constant (Kb) was 14 ± 4 μM. The maximum catalytic constant (Kcat) was 0.081 ± 0.002 s−1 (SI Appendix, Fig. S4). To ensure accuracy in determining the apparent affinity of compound 1 (Kiapp), we used the Morrison equation for tight-binding inhibition (26). We determined the apparent affinity of compound 1 (Kiapp) for BmaI1 by fitting the velocity at different concentrations of inhibitor (Fig. 5). The data were fit to equations describing ideal inhibitor mechanisms as described in Methods. These equations assume the enzyme follows a sequential ordered mechanism, as was determined for RhlI (4, 20) and that the inhibitor binds to all relevant enzyme forms with equal affinity. We were unable to fit the data to the competitive model. Although it was possible to fit the data to both an uncompetitive and noncompetitive model, the noncompetitive model was found to be 99.99% correct by comparison using Akaike’s information criterion. The average fitted Ki from 30 to 90 μM C8-ACP and 100–500 μM SAM was 0.69 ± 0.05 μM.
Fig. 5.
Apparent affinity of compound 1 for BmaI1. The inhibited velocity (Vi) is plotted relative to the uninhibited velocity (V0) for different concentrations of compound 1. The data were fit to the Morrison equation for tight binding inhibition to determine Kiapp (see Methods).
Inhibitory Activity of Compound 1 Analogs.
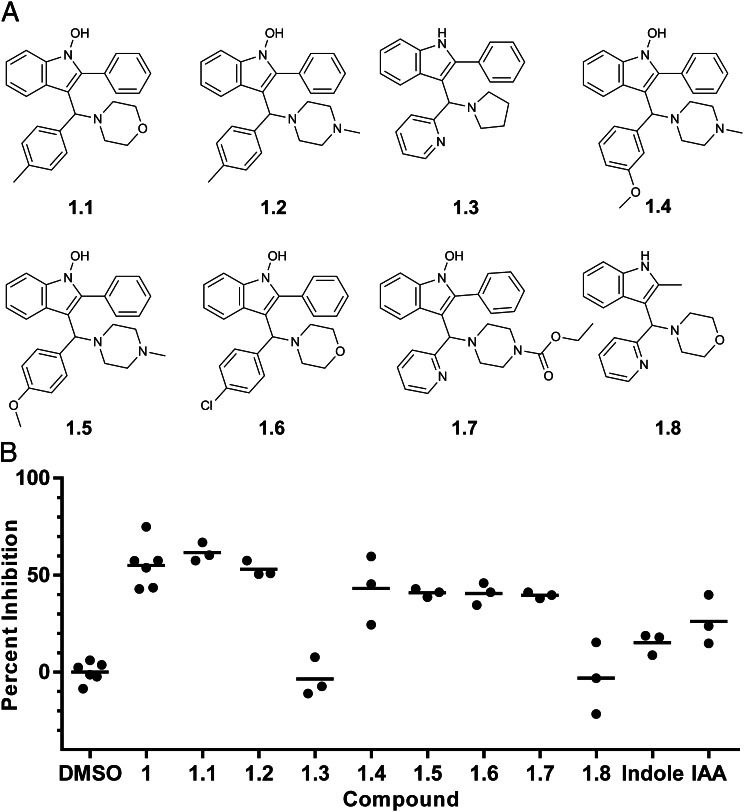
We measured BmaI1 inhibition by a series of compound 1 analogs in the DCPIP assay at concentrations close to the Kiapp for compound 1. Six of the eight compounds tested showed inhibitory activity comparable to compound 1, and two showed little or no activity (Fig. 6). We did not find a structure with significantly increased inhibitory activity. All compounds contained an invariant indole moiety, so we also tested indole and 3-indoleacrylic acid (IAA) for inhibition. Indole did not significantly inhibit BmaI1 at the concentration tested, but IAA did. Results with compounds 1.3 and 1.8 suggest the 1-hydroxy substitution of the indole core contributes to inhibition, although other changes are also made in these compounds. The results with the other compound analogs, especially 1.7, suggest there is flexibility in substitution and modification of the piperazine moiety.
Fig. 6.
Inhibitory activity of select compound 1 analogs. (A) The structure of each compound 1 analog tested. (B) The percentage of inhibition for each analog, indole, and IAA at 0.5 µM measured by the DCPIP method. DMSO and compound 1 are included as controls. Compounds 1–1.2 and 1.4–1.7 are significantly inhibitory compared with DMSO (multiple comparison P < 0.0002). Compounds 1.3 and 1.8 are significantly less inhibitory than compound 1 (multiple comparison P < 0.0001). Indole and IAA are significantly less inhibitory than compound 1 (multiple comparison P = 0.0001, P = 0.01). IAA shows significant inhibition compared with DMSO (multiple comparison P = 0.03).
Discussion
Acyl-HSL synthases are one of two potential targets for quorum-sensing inhibition in Proteobacteria. These enzymes carry out unique reactions (4, 5, 8, 9). We have been interested in identifying acyl-HSL synthase inhibitors to use as chemical probes for understanding the mechanism of enzyme activity, as tools to manipulate quorum sensing in the laboratory setting, and as potential scaffolds for therapeutic development. There has been little published on inhibitors of acyl-HSL synthases (4, 10, 12, 13), at least in part, because of the fact that inhibition is difficult to measure, particularly in cell-based assays. The unique product of acyl-HSL synthase activity is the acyl-HSL itself, which can be measured by using a bioassay (27, 28), by mass-spectrometric techniques (27, 29, 30), or by measuring incorporation of radiolabeled SAM into the product (24, 25). The previously described DCPIP assay, which measures the reactive thiol of the ACP product of the reaction, is not amenable to high-throughput screening because many compounds will affect absorbance and the assay lacks sensitivity (20). We overcame the obstacles to high-throughput screening by adapting a commercially available enzyme-coupled assay that can be used to measure one of the acyl-HSL synthase products, MTA. The reaction requires purified acyl-HSL synthase, acyl-ACP, and pure SAM, all of which are not available commercially. By screening over 12,000 compounds, we identified several inhibitors. The method serves as a basis for more extensive screening by those interested in developing quorum-sensing inhibitors as therapeutics.
We further studied several particularly strong inhibitors and found two potent compounds (1, 3), which showed activity not only in two different cell-free assays but also in a cell-based assay. We view these inhibitors as useful chemical biology probes but not necessarily good candidates as scaffolds for therapeutic development. Predicted absorption, distribution, metabolism, and excretion characteristics suggest they are more hydrophobic than desirable and will bind to serum protein (SI Appendix, Table S2).
The most potent compound, compound 1, was examined in more detail and behaved as a noncompetitive, tight-binding inhibitor with a submicromolar Ki. A noncompetitive inhibitor might be useful because the binding of the inhibitor is independent of substrate concentration (14). This is in contrast to competitive inhibitors or receptor agonists, which can be displaced by the natural ligands. It is of interest that compound 1 possesses a core indole moiety. Indole is produced by a number of bacterial species under a variety of conditions (31). We found that the tryptophan analog IAA inhibited BmaI1. Perhaps microbial production of indole-containing metabolites can influence quorum sensing.
It is of interest that one of the inhibitors identified in the high-throughput screen is a cephalosporin antibiotic, cefatrizine (Fig. 3; compound 5). A related antibiotic, ceftazidime, inhibits quorum sensing in the opportunistic pathogen P. aeruginosa by an unknown mechanism (32). Our finding suggests that cephalosporins might affect acyl-HSL synthases directly. Because cefatrizine has known antibiotic activity, we did not examine it in the cell-based assay, where we assume it would have off-target effects. With respect to therapeutic development, it is of interest to study cephalosporins further. An off-target activity against bacterial growth might be considered beneficial rather than a detriment for a therapeutic.
We believe acyl-HSL synthase inhibitors have several potential uses. They can be used as chemical biology probes for studies of acyl-HSL synthases. They can also serve as tools to study bacterial quorum sensing in whole cells. Finally, there has been considerable interest in developing quorum-sensing inhibitors as anti-bacterial virulence therapeutics. Most efforts to identify quorum-sensing inhibitors have focused on acyl-HSL signal receptors or have been unbiased screens, which, in the end, led to receptor inhibitors. Conceivably, noncompetitive acyl-HSL synthase inhibitors may be more efficacious than competitive receptor antagonists. Alternatively, theoretical considerations suggest inhibition of both signal production and reception may be required of a therapeutic modality (14).
Methods
Compound Library and Inhibitors.
The compound library for the high-throughput screen was derived from Enamine, Life Chemicals, and the National Institutes of Health Clinical Collections at the National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infectious Diseases at Harvard Medical School. For other studies, materials identified in the screen were purchased from commercial vendors who verified batch compound identity by NMR and liquid chromatography–mass spectrometry.
Enzyme Purification.
Bacterial strains used as sources of enzymes are described in the SI Appendix, Table S1. Protein purifications were carried out at 4 °C unless otherwise stated. Bacillus subtilis Sfp was purified by nickel affinity chromatography and precipitation as described previously (33). The gene bmaI1 (UniProt I1SB97_BURMA) was PCR-amplified from Burkholderia mallei American Type Culture Collection 23344 DNA, and the PCR product was cloned into pMCSG21 as described (34) to give pQC201. Hexahistidine-tagged BmaI1 was purified from E. coli strain Tuner DE3 containing the T7 promoter-driven expression plasmid, pQC201. Bacteria were grown overnight at 16 °C and harvested by centrifugation. BmaI1 was purified from lysed cells by using nickel affinity chromatography. The concentrated pure protein preparations were dialyzed against 100 mM sodium phosphate and 20% (vol/vol) glycerol (pH 7) to remove reducing agent and then flash-frozen in liquid nitrogen and stored at −80 °C. The identity and purity of BmaI1 was confirmed by electrophoresis and electrospray mass spectrometry (SI Appendix, Fig. S5). BmaI1 concentration was determined by using the calculated extinction coefficient of 29,450 M−1⋅cm−1 at 280 nm (35). YspI (UniProt Q7CGP3_YERPE) was PCR amplified from Y. pestis KIM6 DNA and cloned in pMCSG23 as described (34) to give pQC218. Maltose binding protein-tagged YspI was expressed in E. coli Tuner DE3 containing pQC218 and purified by using an amylose resin. Fractions containing pure YspI were pooled, dialyzed, and stored as described for BmaI1. We determined YspI concentration by using the calculated extinction coefficient of 103,710 M−1⋅cm−1 at 280 nm (35).
C8-ACP and SAM Preparation.
Fatty acyl carrier protein was purified from E. coli DK574 (pJT94) by adapting procedures described elsewhere (36–38) (SI Appendix, SI Methods). Over 500 mg of apo-ACP was obtained from 18 L of bacterial cell culture. C8-ACP was produced by using Bacillus subtilis Sfp to transfer the octanoyl group from C8-CoA (Sigma Chemical) to apo-ACP as described elsewhere (33). The conversion was confirmed by conformation-sensitive native PAGE (37) and mass spectrometry (SI Appendix, Fig. S5). For storage and to remove reducing agent, C8-ACP was dialyzed against 10 mM sodium 2-(N-morpholino)ethanesulfonate (pH 6.0) and 20% glycerol. The C8-ACP preparation was flash-frozen in liquid nitrogen and stored at −80 °C. C8-ACP concentration was measured by using the calculated extinction coefficient of 1,490 M−1⋅cm−1 at 280 nm (35, 38). SAM was synthesized enzymatically from methionine and ATP and was purified by flash chromatography as described (39). Purity was assessed by HPLC and mass spectrometry (SI Appendix, Fig. S5).
Microplate BmaI1 Activity Assays.
The primary and counter high-throughput screens were performed in low-volume, black, nonbinding, 384-well microplates. For the primary screen, each reaction mixture contained 1.5 μM BmaI1, 40 μM SAM, 25 μM C8-ACP, ∼1.5 μg of test compound, 0.47 μL of coupling-enzyme mixture (Cayman Chemical), 200 μM ADHP, 0.1 μM manganese sulfate, and 100 mM potassium phosphate (pH 7.0). The final volume was 13.3 μL. The enzymes and test compounds were preincubated together for at least 10 min, and the reactions were started by addition of substrates. Reactions were stopped at 40 min by addition of acetovanillone (3.3 μL) to a final concentration of 1.6 mM to give a 16.6 μL total volume. The counter screen was similar to the primary screen except we replaced BmaI1, C8-ACP, and SAM with MTA (12.5 μM) and adjusted the coupling-enzyme mixture per well to 0.4 μL. A control with addition of the solvent in which test compounds were dissolved (DMSO, 0.3 μL) and a control with acetovanillone in DMSO added before the reaction was started were used. The final fluorescence intensity was determined by using a Perkin-Elmer Envision plate reader with a 535-nm emission filter, a 590-nm excitation filter, and a 555-nm mirror. Fluorescence intensity was converted to a robust Z score with the equation Z = x − µ1/2/(1.4826 × σMAD), where x is the relative fluorescence value for a single well, µ1/2 is the median of all fluorescence values on a 384-well plate, and σMAD is the median average distribution. We scaled σMAD by 1.4826 such that one Z is equivalent to one SD.
To determine the IC50 of each inhibitor, we performed microtiter plate assays as described above, except that we used a range of inhibitor concentrations and we (vol/vol) measured reaction velocity. The change in velocity with respect to the log of inhibitor concentration was fit to a sigmoidal four-parameter dose–response equation (SI Appendix, SI Methods) to calculate each IC50.
Statistics and Regression Analysis.
All statistics and regression analyses were performed by using Prism 6 (GraphPad Software). Nonlinear regression analysis was carried out using a standard, least-squares, fit with 1,000 replicates. Variables determined by nonlinear regression are reported with the SD from 1,000 replicates. Replicate means were compared by using ANOVA with Tukey’s multiple comparison test, with P values being the lowest threshold for multiple comparisons where a given comparison would be considered significant.
The DCPIP Assay.
For compounds identified in the screen, we measured inhibition of BmaI1 and YspI activity by measuring holo-ACP with DCPIP as described elsewhere (19, 20). This assay has an advantage over the high-throughput screening assay in that it does not involve coupling enzymes. Reaction volumes were 50 µL in 384-well clear microplates at 25 °C. Unless otherwise specified, reaction mixtures contained 50 mM 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid (pH 7.5), 0.005% Nonidet P-40, 0.06 mM DCPIP, 50 µM C8-ACP, 1 mM SAM, 0.75 µM BmaI1, and inhibitors as specified. Reactions were started by addition of SAM after preincubation of the other reagents for 10 min. Absorbance (600 nm at 25 °C) was followed in a microplate reader (Tecan Genious Pro). The amount of holo-ACP produced was calculated from an extinction coefficient of 21,000 M−1⋅cm−1 and a path length of 0.35 cm. Percentage of inhibition of enzyme velocity was calculated by standard methods (26) and activated velocities are reported as zero percent inhibition.
Kinetic Analysis of Inhibitor Activity.
The apparent affinity (Kiapp) of compound 1 was determined by measuring the velocity of the secondary reaction at several inhibitor concentrations and fitting the data to a form of the Morrison equation (26): Vi/V0 = 1 − (((Et + It + Kiapp) − ((Et + It + Kiapp)2 − 4 × Et × It)0.5)/(2 × Et)). Velocities (Vi) were adjusted for the small change in absorbance attributable to compound 1. They were then normalized by the velocity of the DMSO control (V0) to give the dependent variable. The data were fit to the Morrison equation with total enzyme (Et) set to 0.75 μM, the total inhibitor concentration (It) as the independent variable, and Kiapp as an unknown constrained to positive numbers. The mode of inhibition of compound 1 was determined by using the Kiapp and fitting the data to competitive, uncompetitive, and noncompetitive models for a sequential ordered bisubstrate enzyme. The IC50 equations from Copeland (26) were used. They were solved for Kiapp after combining with the following equation (40): IC50 = Kiapp + Et/2. The equation used for noncompetitive binding is Kiapp = Ki × (1 + (Ka × B/(Kd × Kb + Kb × A + A × B))); the equation for uncompetitive is Kiapp = Ki × (1 + (Kd × Kb + Ka × B)/(Kb × A + A × B)); and the equation for competitive binding is Kiapp = Ki × (1 + (Ka × B + Kb × A + A × B)/(Kd × Kb)). The equations were simultaneously fit to two datasets, where the concentrations of C8-ACP (B) or SAM (A) were varied (SI Appendix, Fig. S4 C and D). The nonvaried substrate was set at 50 μM for C8-ACP (B) or 500 μM for SAM (A). The total enzyme concentration (Et) was set at 0.75 μM. The apparent affinity for SAM (Ka) and C8-ACP (Kb) were determined experimentally (SI Appendix, Fig. S3). The dissociation constant for SAM (Kd) and the enzyme affinity for the inhibitor (Ki) were constrained to positive numbers and Kd was shared between datasets.
Whole-Cell BmaI1-Activity Assays.
To monitor BmaI1 activity in whole cells, we measured C8-HSL production by using a radiolabel assay (24, 25). E. coli BW25113 containing pBD2, which contains an arabinose promoter-driven bmaI1, was grown at 37 °C with shaking in 3-(N-Morpholino)propanesulfonic acid minimal medium (41) with 0.4% glucose, 0.4% l-arabinose, and 100 µg/mL ampicillin for plasmid maintenance. When the culture reached a density of 0.8 at 600 nm, it was split into 600-µL volumes in 18-mm test tubes with 100 µM inhibitor in DMSO or DMSO alone. After 10 min at 37 °C with shaking, 60 µCi of [14C]methionine was added (55 mCi/mmol). After 20 min in the presence of the radiotracer, we extracted acyl-HSLs three times with 600-µL volumes of ethyl acetate. The pooled ethyl acetate extracts were dried and the dried material was dissolved in 0.1 mL of 50% methanol:water. The dissolved material was separated by HPLC (a 250 mm × 4.6 mm Nucleosil 5-µm C18 reverse-phase column with a 15-min gradient of 35–100% methanol:water at 1 mL/min). One-milliliter fractions were collected, and radioactivity in each fraction was measured by liquid scintillation counting.
Supplementary Material
Acknowledgments
We thank the staff at the National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infectious Diseases (NSRB), including Dr. Su Chiang, Doug Flood, Jen Nale, and David Wrobel, for advice and data handling and Dr. Charlotte Majerczyk for Burkholderia mallei American Type Culture Collection 23344 genomic DNA. This research was supported by National Institutes of Health Grants T32 AI055396 (to Q.H.C.), R01 GM101957 (to S.J.B.), U54 AI057141 (to E.P.G.), and U54 AI057159 (to the NSRB).
Footnotes
The authors declare no conflict of interest.
Data deposition: The compounds and the results of the screen have been deposited in the Pubchem database, http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=720554, (accession no. AID 720554).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313098110/-/DCSupplemental.
References
- 1.Fuqua C, Greenberg EP. Listening in on bacteria: Acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3(9):685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 2.Williams P. Quorum sensing: An emerging target for antibacterial chemotherapy? Expert Opin Ther Targets. 2002;6(3):257–274. doi: 10.1517/14728222.6.3.257. [DOI] [PubMed] [Google Scholar]
- 3.Waters CM, Bassler BL. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 4.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Natl Acad Sci USA. 1999;96(8):4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raychaudhuri A, Tullock A, Tipton PA. Reactivity and reaction order in acylhomoserine lactone formation by Pseudomonas aeruginosa RhlI. Biochemistry. 2008;47(9):2893–2898. doi: 10.1021/bi702009n. [DOI] [PubMed] [Google Scholar]
- 6.Watson WT, Minogue TD, Val DL, von Bodman SB, Churchill MEA. Structural basis and specificity of acyl-homoserine lactone signal production in bacterial quorum sensing. Mol Cell. 2002;9(3):685–694. doi: 10.1016/s1097-2765(02)00480-x. [DOI] [PubMed] [Google Scholar]
- 7.Vetting MW, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433(1):212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Moré MI, et al. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272(5268):1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 9.Schaefer AL, Val DL, Hanzelka BL, Cronan JE, Jr, Greenberg EP. Generation of cell-to-cell signals in quorum sensing: Acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93(18):9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaSarre B, Federle MJ. Exploiting quorum sensing to confuse bacterial pathogens. Microbiol Mol Biol Rev. 2013;77(1):73–111. doi: 10.1128/MMBR.00046-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellbye B, Schuster M. The sociomicrobiology of antivirulence drug resistance: A proof of concept. MBio. 2011;2(5):e00131–e11. doi: 10.1128/mBio.00131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fast W, Tipton PA. The enzymes of bacterial census and censorship. Trends Biochem Sci. 2012;37(1):7–14. doi: 10.1016/j.tibs.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung J, et al. Small-molecule inhibitor binding to an N-acyl-homoserine lactone synthase. Proc Natl Acad Sci USA. 2011;108(29):12089–12094. doi: 10.1073/pnas.1103165108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anand R, Rai N, Thattai M. Interactions among quorum sensing inhibitors. PLoS ONE. 2013;8(4):e62254. doi: 10.1371/journal.pone.0062254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorgan KM, et al. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal Biochem. 2006;350(2):249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: Applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal Biochem. 1997;253(2):162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 17.Heumüller S, et al. Apocynin is not an inhibitor of vascular NADPH oxidases but an antioxidant. Hypertension. 2008;51(2):211–217. doi: 10.1161/HYPERTENSIONAHA.107.100214. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 19.Hadler HI, Erwin MJ. The conjugation of thiols by 2,6-dichloroindophenol. Biochemistry. 1963;2:954–957. doi: 10.1021/bi00905a010. [DOI] [PubMed] [Google Scholar]
- 20.Raychaudhuri A, Jerga A, Tipton PA. Chemical mechanism and substrate specificity of RhlI, an acylhomoserine lactone synthase from Pseudomonas aeruginosa. Biochemistry. 2005;44(8):2974–2981. doi: 10.1021/bi048005m. [DOI] [PubMed] [Google Scholar]
- 21.Feng BY, et al. A high-throughput screen for aggregation-based inhibition in a large compound library. J Med Chem. 2007;50(10):2385–2390. doi: 10.1021/jm061317y. [DOI] [PubMed] [Google Scholar]
- 22.Kirwan JP, et al. Quorum-sensing signal synthesis by the Yersinia pestis acyl-homoserine lactone synthase YspI. J Bacteriol. 2006;188(2):784–788. doi: 10.1128/JB.188.2.784-788.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duerkop BA, Ulrich RL, Greenberg EP. Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1-BmaI1 quorum sensing. J Bacteriol. 2007;189(14):5034–5040. doi: 10.1128/JB.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407(6805):762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 25.Schaefer AL, Greenberg EP, Parsek MR. Acylated homoserine lactone detection in Pseudomonas aeruginosa biofilms by radiolabel assay. Methods Enzymol. 2001;336:41–47. doi: 10.1016/s0076-6879(01)36576-x. [DOI] [PubMed] [Google Scholar]
- 26.Copeland RA. Evaluation of enzyme inhibitors in drug discovery. A guide for medicinal chemists and pharmacologists. Methods Biochem Anal. 2005;46:1–265. [PubMed] [Google Scholar]
- 27.Pearson JP, et al. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91(1):197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaw PD, et al. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94(12):6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Charlton TS, et al. A novel and sensitive method for the quantification of N-3-oxoacyl homoserine lactones using gas chromatography-mass spectrometry: Application to a model bacterial biofilm. Environ Microbiol. 2000;2(5):530–541. doi: 10.1046/j.1462-2920.2000.00136.x. [DOI] [PubMed] [Google Scholar]
- 30.Cataldi TRI, Bianco G, Abate S. Profiling of N-acyl-homoserine lactones by liquid chromatography coupled with electrospray ionization and a hybrid quadrupole linear ion-trap and Fourier-transform ion-cyclotron-resonance mass spectrometry (LC-ESI-LTQ-FTICR-MS) J Mass Spectrom. 2008;43(1):82–96. doi: 10.1002/jms.1275. [DOI] [PubMed] [Google Scholar]
- 31.Madigan MT, Martinko JM, Parker J. Brock Biology of Microorganisms. 9th Ed. Upper Saddle River, NJ: Prentice Hall College Div; 2000. [Google Scholar]
- 32.Skindersoe ME, et al. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52(10):3648–3663. doi: 10.1128/AAC.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Christensen QH, Cronan JE. Lipoic acid synthesis: A new family of octanoyltransferases generally annotated as lipoate protein ligases. Biochemistry. 2010;49(46):10024–10036. doi: 10.1021/bi101215f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A, Mark ID. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol. 2009;498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 36.Repaske R. Lysis of gram-negative bacteria by lysozyme. Biochim Biophys Acta. 1956;22(1):189–191. doi: 10.1016/0006-3002(56)90240-2. [DOI] [PubMed] [Google Scholar]
- 37.Rock CO, Cronan JE., Jr Improved purification of acyl carrier protein. Anal Biochem. 1980;102(2):362–364. doi: 10.1016/0003-2697(80)90168-2. [DOI] [PubMed] [Google Scholar]
- 38.Cronan JE, Thomas J. Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 2009;459:395–433. doi: 10.1016/S0076-6879(09)04617-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iwig DF, Booker SJ. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-L-methionine. Biochemistry. 2004;43(42):13496–13509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
- 40.Yang J, Copeland RA, Lai Z. Defining balanced conditions for inhibitor screening assays that target bisubstrate enzymes. J Biomol Screen. 2009;14(2):111–120. doi: 10.1177/1087057108328763. [DOI] [PubMed] [Google Scholar]
- 41.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119(3):736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.