Abstract
Eukaryotic gene regulation usually involves sequence-specific transcription factors and sequence-nonspecific cofactors. A large effort has been made to understand how these factors affect the average gene expression level among a population. However, little is known about how they regulate gene expression in individual cells. In this work, we address this question by mutating multiple factors in the regulatory pathway of the yeast HO promoter (HOpr) and probing the corresponding promoter activity in single cells using time-lapse fluorescence microscopy. We show that the HOpr fires in an “on/off” fashion in WT cells as well as in different genetic backgrounds. Many chromatin-related cofactors that affect the average level of HO expression do not actually affect the firing amplitude of the HOpr; instead, they affect the firing frequency among individual cell cycles. With certain mutations, the bimodal expression exhibits short-term epigenetic memory across the mitotic boundary. This memory is propagated in “cis” and reflects enhanced activator binding after a previous “on” cycle. We present evidence that the memory results from slow turnover of the histone acetylation marks.
Keywords: gene expression noise, single-cell measurement, stochastic gene expression, HO regulation
Chromatin plays an essential role in gene regulation. Transcription regulation pathways usually involve recruitment or dissociation of multiple nucleosome-related factors, including histone modification enzymes and remodeling machineries. Mutations in these transcriptional “cofactors” lead to changes in the nucleosome occupancy, positioning, or structure (e.g., histone modifications and variants). These nucleosome configurations significantly affect gene expression (1). Nucleosomes may also contribute to the inheritance of gene expression across cell generations. For instance, it was proposed that some histone variants/modifications could be maintained in a mitotically heritable manner, although the nature and the strength of such inheritance are a matter of debate (2, 3).
Most gene expression studies rely on bulk assays to measure the ensemble average of a large quantity of cells. These assays are efficient and insightful, but they tend to mask cell-to-cell variability and asynchronized dynamics among a population. As a result, when a factor is found to affect gene expression, it is generally unknown whether it uniformly affects all cells, changes the fraction of cells that express this gene, or modulates the gene expression dynamics without affecting the actual expression level. It is also hard to use bulk assays to study cellular inheritance, which often requires tracking of cell pedigree.
In a previous study (4), we established a system to study the relationship between nucleosomes, gene expression, and its memory at the single-cell level. Using a cell cycle-regulated CLN2 promoter (CLN2pr) as a model, we found that a nucleosome-depleted region (NDR) over the activator SBF (Swi4/6) binding sites ensures reliable gene expression (4). In particular, when the SBF binding sites on CLN2pr are nucleosome-embedded, they induce “on/off” activation in individual cell cycles. Such bimodal activation displays short-term memory across cell cycles (i.e., when a CLN2pr fires in one cell cycle, the next cell cycle has a higher than average probability to fire). This work raised some important mechanistic questions. Why does the nucleosome vs. activator competition lead to bimodal gene expression? What determines the firing frequency of the promoter? How is the memory generated? Our strategy to address these questions is to perturb the gene activation pathway by mutations of related factors and to examine the resultant change in the bimodal gene expression and the memory. Because the HO promoter (HOpr) is one of the best-characterized promoters in yeast and a large group of factors participates in its regulation (5), we decided to carry out these experiments on the HOpr.
The HOpr is cell cycle-regulated by the activator SBF, a feature common with the CLN2pr. However, unlike the CLN2pr, where SBF can simply bind and activate, SBF binding on the HOpr is dependent on another transcription factor, Swi5 (6–8). The HOpr can be roughly divided into two regulatory regions: URS1 containing two Swi5 binding sites exposed in NDRs and URS2 containing ∼10 SBF binding sites embedded under a nucleosome array. As mother cells pass anaphase, Swi5 enters the nucleus; binds to the URS1; and recruits Swi/Snf, SAGA (Spt-Ada-Gcn5-acetyltransferase), and Mediator, leading to nucleosome loss over URS1. This event triggers a “wave” of nucleosome loss, first in the upstream part of URS2 and then moving downstream. The nucleosome eviction in URS2 depends on SBF and two histone chaperones, FACT (facilitates chromatin transcription) and Asf1, which, in turn, likely facilitate SBF binding. Finally, SBF recruits multiple factors, including Swi/Snf, SAGA, and Mediator, to promote the assembly of the transcription complex at TATA (9). This activation pathway does not occur in daughter cells due to the Ash1 inhibitor, which primarily accumulates in daughters (10). An Rpd3 histone deacetylase (HDAC) complex functions as a repressor of HO activation. Bulk assays have shown that mutations in any of the factors mentioned above, including Swi5, SBF, Ash1, Swi/Snf, SAGA, FACT, Asf1, Mediator, and Rpd3, significantly affect the average HO expression.
Despite extensive research on the HO regulation pathway, HO activity has never been probed in single cells. In this study, we applied time-lapse fluorescence microscopy to characterize and investigate the molecular mechanism of the stochastic HO expression.
Results
HOpr Induces On/Off Expression.
We constructed strains with the HOpr driving an unstable GFP reporter (11) containing a nuclear localization sequence (NLS) and monitored the GFP concentration using time-lapse fluorescence microscopy across multiple cell generations (12, 13) (Fig. 1 A and D). The half-life of the GFP is ∼40 min (13), allowing us to track the promoter activity dynamically. All the strains used in the single-cell analysis also contain Myo1-mCherry, which forms a ring at the bud neck between bud emergence and cytokinesis, and therefore serves as a marker for cell cycle progression (14). From the movie, we extracted two values: HOpr firing frequency [the fraction of cell cycles in which HOpr is turned on (Pon)] and amplitude (expression level calculated by the peak-to-trough difference in GFP intensity per cell cycle; Materials and Methods).
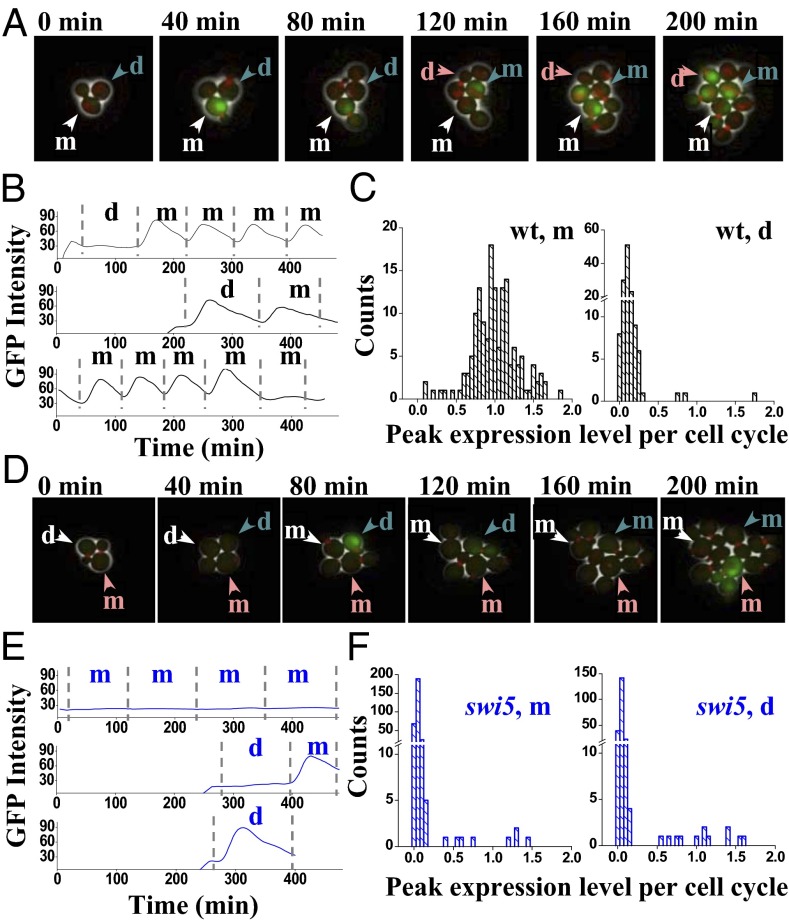
Fig. 1.
Stochastic expression of HOpr in WT (wt) and swi5 cells. (A) Movie clips of the growth of the wt cells (HOpr-GFP MYO1-mCherry). Daughter cycle (the first cell cycle after birth) and mother cycle (the subsequent cell cycles after separating with the first bud) can be clearly differentiated in the movie. The intensity of the GFP reflects the activity of the HOpr, and the red dot is the Myo1-mCherry. Three cells were tracked with different colored arrows. The white-labeled cell went through multiple mother cycles with GFP expressed once every cell cycle; the blue-labeled cell has one daughter cycle (no expression), followed by two mother cycles (strong expression). Both cells follow the “typical” GFP expression pattern. In contrast, the pink-labeled cell represents a rare case in which GFP is turned on during the daughter cycle. d, daughter cycle; m, mother cycle. (B) GFP intensity vs. time traces driven by HOpr in a single wt cell. The dashed vertical lines represent the cell division times measured by the disappearance of the Myo1 ring. (C) Histogram of the amplitude of GFP expression in wt mother and daughter cycles. The expression amplitudes are calculated as the peak-to-trough difference in the GFP signal during a given cell cycle, and they are normalized so that the average expression level in mother cycles is 1. Note the discontinuity in the y axis. (D–F) Same as in A–C, except in the swi5 strain (DY14800). In this case, HOpr is repressed in most of the cell cycles (e.g., the white-labeled cell in D). However, there are rare cells with sporadic HO expression in both mother and daughter cycles (e.g., the blue- and pink-labeled cells in D). Example traces of GFP intensity (E) and a histogram of GFP expression amplitude (F) are shown.
Fig. 1B (Top) shows a typical trace of GFP intensity as a function of time driven by the HOpr in WT yeast. The vertical bars represent the timing of cell division based on the disappearance of Myo1-mCherry. Consistent with the literature, this trace shows no GFP expression during the first cycle after birth (the daughter cycle) and shows only transient GFP expression in subsequent mother cycles. However, we also observed occasional HOpr activations in daughter cycles or “skipping” of activation in mother cycles (Fig. 1B, Middle and Bottom). These are rare events: Only 2% of mother cycles are “off,” and 2–3% of daughter cycles are “on” (Fig. 1C). In cells lacking the Swi5 inhibitor Ash1, HOpr was almost fully activated in daughters with a Pon of 94% (Table 1).
Table 1.
HO expression profile in WT and mutant cells
| Mother |
Daughter |
|||||||
| Strain | Pon, % | N | Amplitude (on)* | CV† | Pon, % | N | Amplitude (on)* | CV† |
| WT | 98.0 ± 1.1 | 154 | 1 ± 0.02 | 0.31 | 2.3 ± 1.3 | 131 | NA | 1.36 |
| swi5 | 2.7 ± 0.9 | 295 | NA | 2.40 | 6.3 ± 1.6 | 222 | 1.18 ± 0.11 | 2.49 |
| ash1 | 98.4 ± 1.1 | 127 | 1.0 ± 0.02 | 0.27 | 93.5 ± 3.1 | 62 | 1.11 ± 0.06 | 0.41 |
| swi5ash1 | 2.7 ± 0.8 | 408 | 0.86 ± 0.09 | 1.68 | 33.7 ± 2.5 | 356 | 1.13 ± 0.04 | 1.33 |
| swi5ace2 | ∼1 | ∼490 | NA | NA | ∼1 | ∼490 | NA | NA |
| swi5ash1ace2 | ∼1 | ∼440 | NA | NA | ∼1 | ∼440 | NA | NA |
| gcn5 | 32.3 ± 1.9 | 610 | 0.95 ± 0.03 | 1.42 | 3.4 ± 1.2 | 232 | NA | 1.85 |
| swi2-314 | 18.5 ± 1.8 | 453 | 0.84 ± 0.04 | 1.80 | 0 | 168 | NA | 0.66 |
| pob3, 30C | 78.6 ± 3.3 | 159 | 0.70 ± 0.02 | 0.59 | 1.9 ± 1.3 | 106 | NA | 0.76 |
| spt16-11, 30C | 71.6 ± 5.0 | 81 | 0.92 ± 0.05 | 0.74 | 7.4 ± 3.0 | 81 | NA | 1.17 |
| spt16-11, 32C | 58.1 ± 5.1 | 93 | 0.87 ± 0.05 | 0.87 | 6.5 ± 2.8 | 77 | NA | 1.49 |
| rpd3 | 99.1 ± 0.9 | 114 | 1.0 ± 0.03 | 0.32 | 53.8 ± 5.2 | 91 | 1.20 ± 0.06 | 0.90 |
| swi5rpd3 | 39.5 ± 3.2 | 228 | 0.99 ± 0.05 | 1.16 | 68.2 ± 3.8 | 151 | 1.29 ± 0.05 | 0.82 |
| swi5rpd3ace2 | 34.6 ± 4.2 | 130 | 1.09 ± 0.07 | 1.30 | 20.0 ± 3.7 | 115 | 1.09 ± 0.15 | 1.83 |
| gal11 | 92.5 ± 2.7 | 93 | 0.41 ± 0.01 | 0.28 | 16.9 ± 4.4 | 71 | 0.44 ± 0.03 | 0.94 |
| swi6 | 43.3 ± 9.0 | 30 | 0.56 ± 0.04 | 0.86 | 0 | 27 | NA | 0.67 |
Average expression amplitude during the “on” cell cycle normalized by that in the WT mother cells. The amplitude is listed as NA when the sample size is too small (on-cycle count <10). Error bar represents SE (same as in the Pon). NA, not available.
Coefficient of variance (CV) of the expression amplitude among all cell cycles, including on and off, defined as the ratio of the SD to the mean.
To test whether Swi5 is required for the stochastic HO expression in daughter cells, we performed the same measurements in swi5 cells (Fig. 1D and Movie S1). As shown in Fig. 1 D–F, the HOpr becomes largely inactive in the absence of Swi5, but there is still sporadic expression in both mother and daughter cycles with a Pon of 2.7% and 6.3%, respectively. In the swi5 ash1 double mutants, the Pon becomes ∼34% in daughters vs. ∼3% in mothers (Table 1). Therefore, the stochastic activation we observed is Swi5-independent. The higher Pon in daughters also indicates that there is a daughter-specific activator repressed by Ash1.
Ace2 is a factor specifically localized in daughter cells, and it contains a DNA-binding domain nearly identical to that of Swi5 (15). Swi5 and Ace2 bind to the same DNA motif in vitro, but they target different genes in vivo (16, 17). Only Swi5 effectively binds to and activates HO. We speculate that Ace2 can weakly activate the HOpr in the absence of Swi5. Indeed, in the swi5 ace2 double-mutant strain, the HOpr still shows “on/off” expression, but the Pon becomes ∼1% in both mother and daughter cycles. The same Pon was observed in the swi5 ace2 ash1 triple mutant (Table 1). This result confirmed that Ace2 can lead to stochastic firing of the HOpr and is largely responsible for the residual HO activation in the swi5 daughter cells.
Importantly, despite the large change in the Pon through the mutations of Swi5, Ace2, and/or Ash1, the actual expression level during the on-cycle remains comparable to that of the WT mother (Fig. 1 C and F and Table 1). In other words, these factors affect the firing frequency of the HOpr but not the firing amplitude.
Cofactors Affect the Firing Frequency and/or the Firing Amplitude of the HOpr.
The factors we studied above (Swi5, Ace2, and Ash1) are sequence-specific transcription factors. Next, we investigated the effect of sequence-nonspecific cofactors on HO expression.
Swi/Snf nucleosome remodeling enzyme and SAGA HDAC play critical roles in HO activation. When we deleted the catalytic subunit of SAGA (gcn5) or introduced a mutation into the catalytic subunit of SWI/SNF [swi2(E834K), a partially defective swi2 allele], the Pon in mother cycles drops to ∼32% and ∼19%, respectively (Fig. 2A and Table 1). The mutation in swi2 also slightly reduces the expression level during the on-cycle to ∼84% of the WT (Table 1). Averaging the GFP amplitudes in all cells of all cell cycles, the HOpr expression in gcn5 and swi2 strains is at ∼33% and ∼14% of the WT level, respectively, consistent with bulk measurements of HO mRNA (22% and 15%, respectively), confirming that the change in GFP intensity is mostly at the transcriptional level.
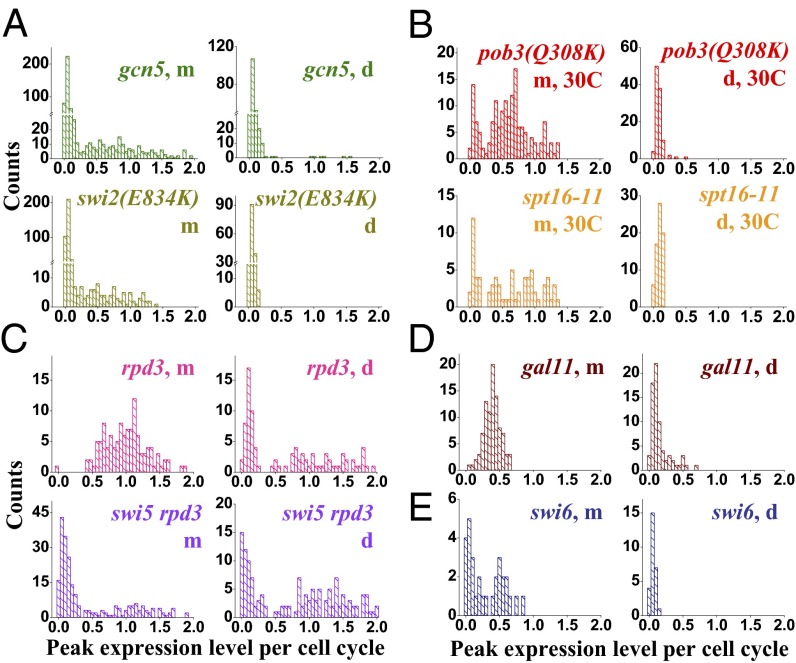
Fig. 2.
Effect of transcription cofactors on HOpr stochastic expression. Histograms of the HO expression amplitude in mother and daughter cycles in the strains of gcn5 and swi2(E834K) (A), pob3(Q308K) and spt16-11 at 30 °C (B), rpd3 and swi5 rpd3 (C), gal11 (D), and swi6 (E). Note the discontinuity in the y axis in A. Many of these factors significantly affect the frequency of HO firing. Statistics are provided in Table 1.
The FACT histone chaperone is recruited to the HOpr by SBF and facilitates nucleosome eviction over URS2 (7). FACT contains two subunits, Spt16 and Pob3, which are both essential for viability. We measured the HO expression in two temperature-sensitive strains, pob3 (Q308K) and spt16-11, at a semipermissive temperature of 30 °C, where FACT activity is partially impaired (7). The Pon in mother cycles is reduced to 79% and 72%, respectively, and the expression level during the on-cycles also mildly decreases (Fig. 2B). This effect becomes stronger when we further disabled FACT by slightly raising the temperature (Table 1).
Rpd3 is the catalytic subunit of the Sin3 complex, a highly conserved class I HDAC. Deletion of the SIN3 subunit leads to elevated histone acetylation across the HOpr and higher HO expression (18). Two Rpd3 complexes, large (Rpd3L) and small (Rpd3S), were proposed to function in transcription initiation and elongation, respectively (19). Rpd3L is recruited to the HOpr, and HO expression is affected by Rpd3L-specific but not Rpd3S-specific mutation (20, 21). These data indicate that Rpd3L plays a major role in HO regulation, likely targeting transcription initiation. Consistently, our single-cell measurement showed that the expression of mother cycles in the rpd3 strain remains the same as in WT but that the Pon in daughters dramatically increases from ∼2% to over 50% (Fig. 2C). In the rpd3 swi5 strain, HOpr expresses in ∼68% of the daughters and ∼40% in mothers (Fig. 2C). The higher expression in daughters is again due to the daughter-specific activator Ace2: If we further delete Ace2 on top of rpd3 and swi5, the Pon in the mother cycles remains at ∼35% but that in the daughter cycles is reduced to ∼20% (Table 1). The activation in the absence of Swi5 and Ace2 likely reflects the enhanced accessibility of SBF to its binding sites embedded under the acetylated URS2 histones.
All the above factors affect the firing frequency of the HOpr. In striking contrast, with the deletion of a mediator component, Gal11, the HO expression level decreases uniformly among the mother cells and its distribution remains monomodal (Fig. 2D). Among the mutant strains, gal11 is one of the very few that shows a smaller coefficient of variance than the WT in mothers (Table 1).
In summary, factors regulate HO expression by changing the HOpr firing frequency and/or the firing amplitude. These different effects are generated because factors participate in different parts of the activation pathway (Discussion). The two orthogonal effects provide high flexibility for fine-tuning of the promoter activity. Compared with mutants, the balance between these factors in WT cells seems to be carefully adjusted to achieve the maximum difference between mothers and daughters.
“Cis” Instead of “Trans” Memory of the Bimodal HO Expression in swi5 Cells.
The time-lapse movie provides information on cell pedigree, which we used to investigate the propagation (or “memory”) of the expression pattern. Note that we can only measure memory in strains exhibiting bimodal expression. For WT cells, where the HO expression occurs at a rate of almost 100% in mothers, memory is ill-defined. Instead, we evaluated memory in the swi5 strain by separating the on- and off-cycles and calculating the Pon in the subsequent cycles. Fig. 3A shows that HOpr is more likely to fire after an on-cycle than after an off-cycle, but this memory quickly disappears after one to two cell cycles.
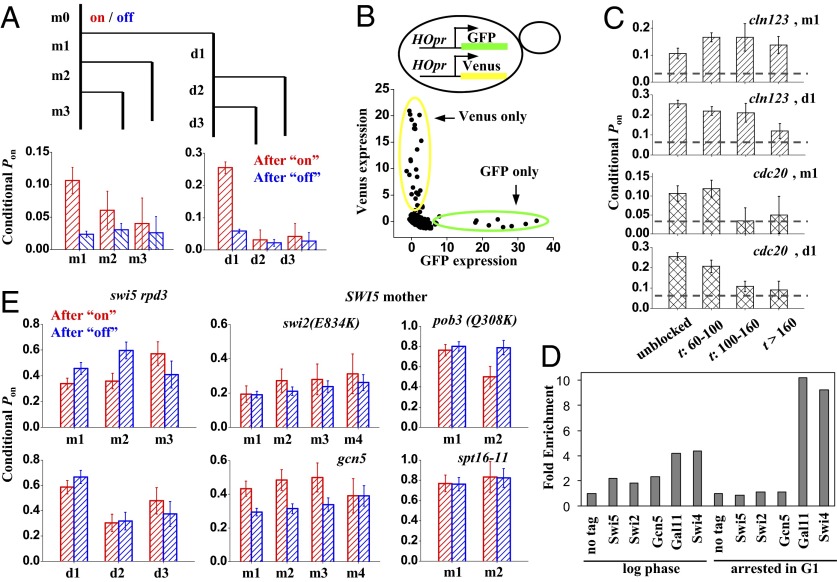
Fig. 3.
Memory of HO expression in the swi5 strain. (A) HO expression in swi5 cells exhibits short-term memory. We divided all cell cycles [mother cycle 0 (m0)] as either “on” (red) or “off” (blue). At the end of m0, the cell divided into mother (m; left branch) and daughter (d; right branch). We followed both cells up to three cycles until the end of the movie (m1–m3 and d1–d3) and recorded the “on/off” status of the HOpr in each of them. Finally, we calculated the Pon in m1–m3 and d1–d3 with either an on or off m0. After an on-cycle, the probability to be on is significantly higher than that after an off-cycle. (B) Coexpression pattern of HOpr-GFP and HOpr-Venus in swi5−/− diploid cells (DY15910). The x axis and y axis for each dot in the plot correspond to the GFP vs. Venus expression level in the same cell cycle (including both mother and daughter). The dots are completely off-diagonal, indicating that the two alleles are activated independently. (C) Memory of HO expression in cell cycles delayed in early G1 (yLB79; cln123, MET-CLN2) or early M (yLB80; cdc20::MET-CDC20). The plots show the Pon in m1 and d1 following a previous on-cycle in either unblocked cells (first bin) or blocked cells with prolonged cell cycle time t (budding-to-budding time). The horizontal lines represent the average Pon, and the existence of memory is indicated by bars significantly higher than this baseline. Memory is quickly lost with a delay in G2/M but not in G1. (D) ChIP experiments show stable Mediator (Gal11) and SBF binding to HOpr following prolonged G1 arrest. Binding of the indicated factors to HOpr was measured by ChIP in growing (log-phase) or arrested cdc28 cells. (E) Memory of HO expression in different genetic backgrounds. In swi5 rpd3 and all SWI5 strains (with the exception of gcn5), the Pon following an on-cycle (red) or an off-cycle (blue) is not significantly different, showing no detectable memory.
There are two general mechanisms, “trans” vs. “cis,” for such memory. Trans means that memory is generated by the variation in a global factor, such as a transcription activator. For instance, some cells may have high concentrations of free SBF or Ace2 in the nucleus, which promote HOpr activation. As these cells divide, the subsequent cycles may inherit the high concentrations, leading to a consecutive firing. In the case of the cis mechanism, memory is stored in the local chromatin environment. One possible scenario is that after the first activation event, some factors or histone marks are left on the HOpr that facilitate its activation in the next cycle. These two possibilities can be differentiated by using two copies of the same promoter driving different fluorescence reporters in the same cell. If the memory is generated in trans, the activities of the two promoters in the same cell should be affected simultaneously and correlate with each other; if the memory is generated in cis, they should be independent.
We generated a swi5−/− diploid strain with two copies of HOpr, one driving GFP and the other driving Venus, both integrated at the native HO loci (Fig. 3B; SI Materials and Methods). This strain also contains a MATa allele and a MAT-deletion allele so that HOpr will not be repressed by the a1-α2 repressor. Both GFP and Venus are turned on sporadically with a Pon of ∼1% in the mother and 3–5% in the daughter, similar to the swi5 haploid cells. Importantly, with our limited statistics of 33 activation events, we did not find a single cycle where GFP and Venus are both activated (Fig. 3B). We carried out statistical tests to assess if the expression of GFP and Venus is independent (SI Materials and Methods;Tables S1–S3). Both the Fisher’s exact test and a Bayesian model comparison showed that it is highly probable for GFP and Venus to be activated independently. This result strongly suggests a cis mechanism of the memory generation.
Memory Is Caused by Enhanced SBF Binding After an On-Cycle.
To probe the memory mechanism further, we investigated whether the memory loss occurs at a certain cell cycle stage. Our strategy is to lengthen the cell cycle at different phases and examine whether the memory is affected.
We generated two swi5 strains that go through conditional cell cycle arrest in G1 or G2/M due to methionine-regulated shutoff of either a G1 cyclin or the APC regulator CDC20. In a flow cell that can rapidly switch between positive and negative methionine media, we grew individual cells into a microcolony, stalled them at a specific cell cycle stage, and released them into the next cell cycle while monitoring GFP expression in this process (SI Materials and Methods and Movies S2 and S3). During the uninterrupted cycles, both strains displayed the same on/off expression pattern and short-term memory as observed previously (Fig. 3C; “unblocked”), showing that the constitutive expression of G1 cyclin or CDC20 did not perturb the HOpr activation. For cells with prolonged cell cycles, we grouped them according to the budding-to-budding cycle time and quantified the memory for each group. For the cells arrested in G1, the memory does not change significantly with time (Fig. 3C). In contrast, for the cells arrested at G2/M, memory sharply decays to the basal level as the block duration increases (Fig. 3C). This result shows that the memory is quickly lost during early mitosis, before SBF association (22), but not during G1, after SBF association.
The persistent memory in G1 is consistent with an earlier observation that cells can be arrested for days in G1 by nutrient starvation and can still express HO when they reenter the mitotic cell cycle (23). We performed ChIP experiments to examine factors potentially defining memory (Materials and Methods). Fig. 3D clearly shows that there is no Swi5 bound to HO after a prolonged G1 arrest; neither are the chromatin factors, including Swi/Snf, Gcn5/SAGA, and FACT. In contrast, both SBF and Gal11 are present at the HOpr during the arrest, and their binding is Swi5-dependent. These observations suggest that after initial SBF binding, SBF and a mediator can stay on the promoter for a long time.
The data in Fig. 3 C and D support the following scenario: After the first round of activation, there is some local “mark” that facilitates the second round of SBF binding, but this mark has a finite lifetime. As a consequence, the memory of the previous activation can be erased by delaying the next round of SBF binding (the case of G2/M-arrested cells). Once SBF binds, it remains bound to the HOpr so that the cell is committed to HO expression despite a long G1 arrest.
Memory May Be Related to Histone Acetylation.
To understand the molecular nature of the mark that facilitates a second round of SBF binding, we evaluated the HOpr memory in different genetic backgrounds. Surprisingly, memory we observed in swi5 cells completely disappears with the further disruption of Rpd3: In swi5 rpd3 background, the Pon rates following a previous on-cycle are comparable, if not lower, than those after an off-cycle (Fig. 3E). We also quantified memory for strains with intact SWI5 and some other mutations that lead to variegated HO expression in mothers, including gcn5, swi2(E834K), pob3(Q308K), and spt16-11. There is no detectable memory in these strains except in the SWI5+ gcn5 strain (Fig. 3E). The change of memory is unlikely due to the variation of G2/M length: All the mutants here have a comparable doubling time, and some strains showing memory [e.g., gcn5 (doubling time of 106 min)] divide more slowly than those with no memory, [e.g., swi5rpd3, swi2(E834K) (doubling time of 92 and 84 min, respectively)]. Because Rpd3 is a histone deacetylase and Gcn5 is a histone acetylase, their effect on the memory suggests that the memory may be related to histone acetylation (Discussion).
Discussion
Two Orthogonal Effects on HO Expression.
From our single-cell analysis, we found that there are two orthogonal regulation modes of the HOpr: Some factors only affect the firing frequency, whereas others only affect the firing amplitude. These different effects can be explained by the model in Fig. 4A. The HO activation pathway can be divided into two parts: SBF binding and activation. The former involves a double-negative feedback loop (Fig. 4A, green arrow): Nucleosomes prevent efficient SBF binding, and SBF can recruit factors to evict nucleosomes. This network structure can lead to bimodality in gene expression (24, 25). When we mutate factors exclusively involved in this part of the pathway (Fig. 4A, red), such as Swi5 and Ace2, we are effectively shifting the balance between the two competitive sides, resulting in a change in on/off probability. In contrast, mutations in the factors working downstream of the SBF binding (Fig. 4A, blue) will not change firing frequency; instead, they may affect the recruitment of transcription machinery, and thus the expression level. Some of the factors, including Swi/Snf and FACT, contribute to both parts of the pathway (Fig. 4A, orange). Mutations in these factors generate a mixed effect on both the firing frequency and amplitude.
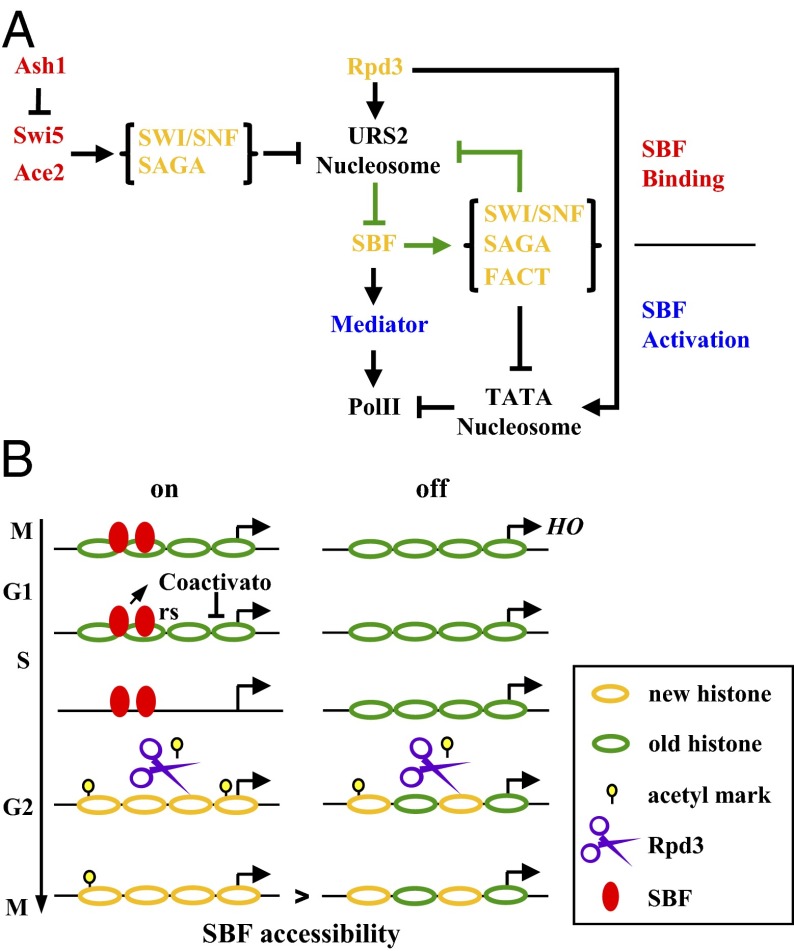
Fig. 4.
Model for on/off HO expression and its memory. (A) Model for bimodal HO expression and the effect from different factors. We propose that the on/off HO expression is caused by the mutual exclusion of nucleosome and SBF (green arrow). Factors solely working upstream the SBF binding (red) would regulate the HO firing frequency, factors solely working downstream (blue) would regulate the firing amplitude, and factors working both up and downstream (orange) would have a mixed effect on HO expression. (B) Model for the memory of HOpr expression in swi5 cells. Based on our experimental results, we hypothesized that differential concentrations of histone acetylation following an on- or off-cycle would lead to the apparent memory. A detailed explanation of the model is provided in the main text. The model in SWI5 cells is shown in Fig. S2.
This model is further supported by the two following experiments. First, we examined the HO expression in the swi6 strain. SBF contains two subunits, Swi4 and Swi6. In the absence of Swi6, Swi4 can act as a weaker activator for the target genes (26). The model predicts that a partially active SBF would reduce both the firing frequency and amplitude. Indeed, in the swi6 cells, the Pon is reduced to 43% and the expression level during the on-cycle is also reduced to 56% (Fig. 2D). Second, we examined the expression from another promoter, the CLN2pr, with SBF binding sites situated in a constitutive NDR. Based on the model, we expected that the CLN2pr would be on in all the cell cycles in WT or mutant background. Also, factors that affect both the firing frequency and amplitude of HOpr, such as Pob3, Spt16, and Swi6, will only affect the firing amplitude of the CLN2pr. Consistently, in pob3(Q308K), spt16-11, and swi6 strains, the CLN2pr is activated in 100% of the cell cycles but the expression amplitude is uniformly reduced to 71%, 79%, and 90%, respectively, of the WT level (Fig. S1).
“Intrinsic” vs. “Extrinsic” Component of Gene Expression Noise.
In Fig. 3B, our two-color experiment showed that the two copies of the HOpr in the same cell operate independently. This is a case where “intrinsic noise” is significantly higher than “extrinsic noise” (27). Similar observations were made with genes inserted at silent loci (28). However, study of another stochastic promoter in yeast, GAL1-10, have shown that intrinsic noise only contributes 2–20% of the total noise (29, 30). This discrepancy is unlikely due to methodology, because we reach the same conclusion with time-lapse measurements of GAL1 promoter-GFP/Venus induction. Therefore, the main source of gene expression noise varies from gene to gene. For the PHO5 and GAL1-10 promoters, we speculate that their activator concentration in the nucleus has large cell-to-cell variation. In contrast, in the case of HOpr and silent loci, the expression noise most likely comes from variable local interactions between the activator and chromatin.
Model for the Memory in HO Expression.
HOpr in swi5 cells exhibits short-lived memory. We found that (i) the memory is caused by a cis mechanism; (ii) the memory reflects enhanced SBF binding after a previous on-cycle; and (iii) deletion of Rpd3 eliminates the memory in swi5 cells, and gcn5 restores the memory in SWI5 cells.
The memory can be explained by the model shown in Fig. 4B. After an on-cycle, the nucleosomes over HO URS2 are completely turned over due to their dissociation during the transcriptional activation (7, 31). For an off-cycle, this dissociation does not occur; however, H3H4 tetramers become one-half new and one-half old during DNA replication (32). Newly synthesized H3 and H4 are acetylated at a number of lysine residues (e.g., K5/K12 of H4), and this acetylation is gradually removed after nucleosome assembly (33–35). Compared with an off-cycle, the HOpr that just experienced an on-cycle likely carries more newly deposited histones, and thus more acetylation. This higher histone acetylation may persist by the time SBF is available in the nucleus, leading to a higher SBF binding probability. If the cell cycle is artificially delayed before the presence of SBF, the acetylation level on the HOpr will eventually reach its equilibrium level regardless of the initial condition and the memory will be lost. In the absence of Rpd3, all the HOprs, whether or not they have been recently transcribed, will be highly acetylated and, again, the memory is eliminated.
This model could also explain the observations in SWI5 strains. In the presence of both Swi5 and Gcn5, Gcn5 will be recruited to the HOpr and acetylates the nearby histones in almost every cell cycle. Therefore, the original histone acetylation marks are “overwritten,” and any influence from the previous cycle is erased (Fig. S2). That is why there is no memory in the SWI5 GCN5 swi2(E834K), pob3(Q308K), and spt16-11 strains. In contrast, in gcn5 cells, the histone acetylation pattern cannot be reset. The higher acetylation level on HOpr following an on-cycle likely facilitates SWI/SNF remodeling and SBF binding, leading to an apparent transcriptional memory.
In our model, the memory is a “passive” consequence of slow turnover of acetylated histones. Together with previous results on CLN2pr (4), it suggests that expression-permissive chromatin may have an intrinsic potential for memory. Unlike previously proposed histone-based epigenetic inheritance mechanisms (2), the model in Fig. 4B does not involve self-replication or spreading of certain histone modifications or variants.
Physiological Relevance of the Memory in HO Expression.
The memory is not present in SWI5 GCN5 backgrounds. Physiologically, the elimination of the memory in HO expression is important for WT yeast. Because of the differential regulation in mother and daughter cycles, regulation of the HO gene has evolved to switch its expression mode in one of the progenies on cell division in a largely deterministic fashion. To achieve such a pattern, previous cell cycles should have a minimal effect on subsequent ones, such that each time there is a new and independent decision. We propose that the loss of memory is due to Swi5 binding and the recruitment of Gcn5, which “overwrites” the acetylation mark. This scenario is probably quite general: transcription factors bound in NDR can recruit downstream factors to reshape the chromatin landscape and start a new regulatory program.
For HO, memory appears to be counteradaptive; however, for some genes, memory can be beneficial. For instance, for a gene under selection for continuous expression, if the transcription factors temporarily dissociate due to stochastic fluctuation or during mitosis, certain histone marks may encourage them to reassociate with the same locus, thus contributing to the stability of the expression pattern. The memory mechanism we have elucidated here has a short time scale. There are other mechanisms that will generate memory, some of which may have much longer time scales (2, 3, 36).
Materials and Methods
Strains.
The strain list is provided in Table S4. All strains were constructed using standard methods, and they are all W303-congenic. The HO-GFP-NLS-PEST reporter consists of an in-frame fusion of GFP, the SV40 T-antigen NLS, and the Cln2 destabilization sequence (PEST), all replacing the HO ORF. A HIS3 or NatMX4 marker was inserted downstream of the HO 3′ UTR.
Time-Lapse Fluorescence Microscopy and Microfluidic Device.
The time-lapse assay was performed using samples growing between a coverslip and an agar pad (11) or in an Onix microfluidic device, which provides the flexibility of changing up to six media. With the agar pad, we typically made movies for ∼8 h, beyond which cells become too clustered and start to grow out of focus. The microfluidic device uses a silicone ceiling with a height similar to that of yeast cells to hold them in place and restrict their growth in a single focal plane so that movies can go on for much longer time (15–20 h). Images were typically acquired every 4 min (5 min for some slow-growing strains). The movies were analyzed using data analysis software written in MATLAB (MathWorks) (12). To get the curve in Fig. 1 B and E, we tracked each single cell along the movie frames, calculated the average GFP intensity within the cell boundary, plotted it as a function of time, and smoothed the curve. For a given cell cycle, the expression amplitude is calculated as the peak-to-trough difference in the GFP signal, and it is normalized by the average expression amplitude in WT mother cells.
ChIP Experiment.
We used strains with a temperature-sensitive mutation in the cyclin-dependent kinase to arrest cells in G1. Compared with nutrient starvation, this method gives much better cell cycle synchrony during the release from arrest (Fig. S3). Cells were grown at a permissive temperature to midlog phase and shifted to the nonpermissive temperature of 37 °C to arrest cells in G1. After 6–10 h at 37 °C, we treated the cells with formaldehyde to cross-link proteins to DNA and performed ChIP experiments to investigate what proteins are bound to HOpr at this arrest. ChIPs with Myc-tagged proteins and RNA measurements from bulk cultures were performed as described (37). Strains DY13565 (no tag, cdc28), DY12843 (Swi5-Myc cdc28), DY8844 (Swi2-Myc cdc28), DY13561 (Gcn5-Myc cdc28), DY13563 (Gal11-Myc cdc28), and DY13576 (Swi4-Myc cdc28) were used.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM039067 (to D.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13701.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306113110/-/DCSupplemental.
References
- 1.Rando OJ, Winston F. Chromatin and transcription in yeast. Genetics. 2012;190(2):351–387. doi: 10.1534/genetics.111.132266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yuan G, Zhu B. Histone variants and epigenetic inheritance. Biochim Biophys Acta. 2012;1819(3-4):222–229. doi: 10.1016/j.bbagrm.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17(7):R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 4.Bai L, Charvin G, Siggia ED, Cross FR. Nucleosome-depleted regions in cell-cycle-regulated promoters ensure reliable gene expression in every cell cycle. Dev Cell. 2010;18(4):544–555. doi: 10.1016/j.devcel.2010.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasmyth K. Regulating the HO endonuclease in yeast. Curr Opin Genet Dev. 1993;3(2):286–294. doi: 10.1016/0959-437x(93)90036-o. [DOI] [PubMed] [Google Scholar]
- 6.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97(3):299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 7.Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34(4):405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mitra D, Parnell EJ, Landon JW, Yu Y, Stillman DJ. SWI/SNF binding to the HO promoter requires histone acetylation and stimulates TATA-binding protein recruitment. Mol Cell Biol. 2006;26(11):4095–4110. doi: 10.1128/MCB.01849-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosma MP, Panizza S, Nasmyth K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol Cell. 2001;7(6):1213–1220. doi: 10.1016/s1097-2765(01)00266-0. [DOI] [PubMed] [Google Scholar]
- 10.Sil A, Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84(5):711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 11.Mateus C, Avery SV. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16(14):1313–1323. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 12.Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle start examined at single-cell resolution. Mol Cell. 2006;21(1):3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 13.Charvin G, Cross FR, Siggia ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE. 2008;3(1):e1468. doi: 10.1371/journal.pone.0001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448(7156):947–951. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 15.Colman-Lerner A, Chin TE, Brent R. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell. 2001;107(6):739–750. doi: 10.1016/s0092-8674(01)00596-7. [DOI] [PubMed] [Google Scholar]
- 16.Dohrmann PR, Voth WP, Stillman DJ. Role of negative regulation in promoter specificity of the homologous transcriptional activators Ace2p and Swi5p. Mol Cell Biol. 1996;16(4):1746–1758. doi: 10.1128/mcb.16.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dohrmann PR, et al. Parallel pathways of gene regulation: homologous regulators SWI5 and ACE2 differentially control transcription of HO and chitinase. Genes Dev. 1992;6(1):93–104. doi: 10.1101/gad.6.1.93. [DOI] [PubMed] [Google Scholar]
- 18.Krebs JE, Kuo MH, Allis CD, Peterson CL. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13(11):1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carrozza MJ, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 20.Takahata S, Yu Y, Stillman DJ. Repressive chromatin affects factor binding at yeast HO (homothallic switching) promoter. J Biol Chem. 2011;286(40):34809–34819. doi: 10.1074/jbc.M111.281626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biswas D, Takahata S, Stillman DJ. Different genetic functions for the Rpd3(L) and Rpd3(S) complexes suggest competition between NuA4 and Rpd3(S) Mol Cell Biol. 2008;28(14):4445–4458. doi: 10.1128/MCB.00164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10(2):129–141. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 23.Nasmyth K. A repetitive DNA sequence that confers cell-cycle START (CDC28)-dependent transcription of the HO gene in yeast. Cell. 1985;42(1):225–235. doi: 10.1016/s0092-8674(85)80118-5. [DOI] [PubMed] [Google Scholar]
- 24.Ferrell JE, Jr, et al. Simple, realistic models of complex biological processes: Positive feedback and bistability in a cell fate switch and a cell cycle oscillator. FEBS Lett. 2009;583(24):3999–4005. doi: 10.1016/j.febslet.2009.10.068. [DOI] [PubMed] [Google Scholar]
- 25.Ingolia NT, Murray AW. Positive-feedback loops as a flexible biological module. Curr Biol. 2007;17(8):668–677. doi: 10.1016/j.cub.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breeden L, Mikesell GE. Cell cycle-specific expression of the SWI4 transcription factor is required for the cell cycle regulation of HO transcription. Genes Dev. 1991;5(7):1183–1190. doi: 10.1101/gad.5.7.1183. [DOI] [PubMed] [Google Scholar]
- 27.Swain PS, Elowitz MB, Siggia ED. Intrinsic and extrinsic contributions to stochasticity in gene expression. Proc Natl Acad Sci USA. 2002;99(20):12795–12800. doi: 10.1073/pnas.162041399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu EY, Zawadzki KA, Broach JR. Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell. 2006;23(2):219–229. doi: 10.1016/j.molcel.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 29.Biggar SR, Crabtree GR. Cell signaling can direct either binary or graded transcriptional responses. EMBO J. 2001;20(12):3167–3176. doi: 10.1093/emboj/20.12.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304(5678):1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Removal of promoter nucleosomes by disassembly rather than sliding in vivo. Mol Cell. 2004;14(5):667–673. doi: 10.1016/j.molcel.2004.05.013. [DOI] [PubMed] [Google Scholar]
- 32.Xu M, et al. Partitioning of histone H3-H4 tetramers during DNA replication-dependent chromatin assembly. Science. 2010;328(5974):94–98. doi: 10.1126/science.1178994. [DOI] [PubMed] [Google Scholar]
- 33.Jackson V, Shires A, Tanphaichitr N, Chalkley R. Modifications to histones immediately after synthesis. J Mol Biol. 1976;104(2):471–483. doi: 10.1016/0022-2836(76)90282-5. [DOI] [PubMed] [Google Scholar]
- 34.Allis CD, Chicoine LG, Richman R, Schulman IG. Deposition-related histone acetylation in micronuclei of conjugating Tetrahymena. Proc Natl Acad Sci USA. 1985;82(23):8048–8052. doi: 10.1073/pnas.82.23.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc Natl Acad Sci USA. 1998;95(12):6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voth WP, et al. Forkhead proteins control the outcome of transcription factor binding by antiactivation. EMBO J. 2007;26(20):4324–4334. doi: 10.1038/sj.emboj.7601859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.