Abstract
A strategy was developed to extend the lifetime of an peptide-based substrate for Abl kinase in the cytosolic environment. Small β-turn structures were added to the peptide’s N-terminus to block entry into peptidase catalytic sites. The influence of the size of the β-turn and two covalent cross-linking strategies on the rate of hydrolysis was assessed. The most peptidase-resistant substrate was degraded at a rate of 0.6 pmol mg−1 s−1 and possessed a half-life of 20.3 ± 1.7 min in a Baf/BCR-ABL cytosolic lysate, representing 16- and 40-fold improvements, respectively, over that of a control peptide lacking the β-turn structure. Furthermore, the kcat/KM value of this peptide was 432 μM−1 min−1, a 1.25X increase over the unmodified control, verifying that the added β-turn did not hinder the substrate properties of the peptide. This improved peptide was microinjected into single Baf/BCR-ABL cells and substrate phosphorylation measured. Zero to forty percent of the peptide was phosphorylated in the single cells. In contrast, when the control peptide without a β-turn was loaded into cells, the peptide was too rapidly degraded to detect phosphorylation. This work demonstrates that small β-turn structures can render peptides more resistant to hydrolysis while retaining substrate efficacy and shows that these stabilized peptides have the potential to be of high utility in single-cell enzyme assays.
Introduction
Peptides are increasingly used as intracellular probes, sensors, and modifiers of cellular protein function. However, their utility can be limited by a short lifetime within the cytosol due to metabolism by aminopeptidases and endopeptidases which remove N-terminal amino acids and hydrolyze peptide bonds, resulting in peptide fragments which are recycled into individual amino acids or utilized for antigen presentation.1–2 Evidence suggests that carboxypeptidases do not participate in cytosolic peptide metabolism since peptides are not observed to be cleaved from the carboxy-terminal end.3 A limited set of aminopeptidases and endopeptidases appear to exist free in the cytosol of cells, including tripeptidylpeptidase II (TPPII),4–6 thimet oligopeptidase (TOP),2 prolyl oligopeptidase (POP),7 and leucine aminopeptidase (LAP).8–9 A common structural feature of these cytosolic peptidases is a very narrow and deep cleft or tunnel wherein lies the catalytic site, and available crystallographic and experimental evidence suggest that the N-terminus of the peptide initially enters into the cleft or tunnel in order to access the active site.4– 9 These structural features suggest that addition of a well-folded N-terminal domain on a peptide could serve to restrict its access to the catalytic site in cytosolic peptidases.
Folded proteins and unstructured loops are poorly metabolized by peptidases within cells most likely due to their bulky size which restricts the access of these molecules to the catalytic sites of peptidases.7 Similarly, large N-terminal groups on a peptide can dramatically extend the lifetime of the peptide when exposed to model proteases. For example, a cyclized, 10-residue peptide (RES-701-1) conjugated to the N-terminus of a biologically active peptide possessed a lifetime 2- to 4-fold greater than that of the linear control peptide upon exposure to trypsin, chymotrypsin, and prolylendopeptidase.10 Small, well-folded peptides, such as those based on a β-hairpin, also exhibited significant resistance to model proteases with their resistance correlated to their folding stability.11–12 A β-hairpin peptide stabilized by an aromatic pocket and cation-π interactions possessed a half-life 7- to 43-fold greater than a control peptide when exposed to chymostrypsin, trypsin, or pronase E.11 In addition, the stability of peptides in the presence of isolated proteases was increased when the peptides contained cyclized or folded domains.13–16
In this work, we describe the use of a β-turn structure appended to the N-terminus of an Abl substrate to extend peptide lifetime in cytosolic lysates and single cells while preserving its ability to act as an intracellular reporter of Abl kinase activity. Abl kinase was selected based on its importance in chronic myelogenous leukemia (CML). In most CML patients, the proximate origin of the tumor is a constitutively active Abl enzyme formed as a result of fusion of the Bcr protein to the Abl protein.17 The average rates of hydrolysis in a cytosolic lysate and phosphorylation via purified kinase were measured with capillary electrophoresis coupled to laser-induced fluorescence detection (CE-LIF). Substrate efficacy was assessed by measuring KM and kcat values for each construct and comparing to the unmodified control. Finally, the substrate possessing the best-performing β-turn structure was microinjected into single, living cells, and substrate fate was assessed.
Materials and Methods
Chemicals
Complete peptides 1 and 6, un-crosslinked peptides 4 and 5 still on the resin, 6-carboxyfluorescein (6-FAM), and Fmoc-Lys(6-FAM) were purchased from AnaSpec (Torrence, CA). 9-Fluorenylmethoxycarbonyl (Fmoc) amino acids, Fmoc-PAL-PEG-PS resin, and Fmoc-(PEG)2-OH were obtained from EMD Biosciences (Darmstadt, Germany). Hydroxybenzotriazole (HOBt), 2-(1H-Benzotriazole-1-yl)-1,1,3,3-Tetramethyluronium Hexafluorophosphate (HBTU), N,N-Diisopropylethylamine (DIPEA), dimethylformamide (DMF), dichloromethane (DCM), dimethyl sulphoxide (DMSO), Trizma hydrochloride (Tris-HCl), bromoacetic acid, diisopropylcarbodiimide (DIC), and protease and phosphatase inhibitor cocktails were received from Sigma (Carlsbad, CA). Trifluoroactic acid (TFA), triisopropylsilane (TIS), diethyl ether, triethylamine, ethane dithiol (EDT), Tris(2-carboxyethyl)-phosphine (TCEP), β-mercaptoethanol (BME), acetonitrile and methanol were purchase from Fisher Scientific (Fair Lawn, NJ). Fluorescein Arsenical Hairpin binding reagent (FlAsH) was obtained from Life Technologies (Grand Island, NY). Bovine serum albumin (BSA) was procured from Calbiochem. Roswell Park Memorial Institute Media (RPMI-1640) was purchased from Cellgro. Penicillin/streptomycin was obtained from Gibco. Fetal bovine serum (FBS) was procured from Atlanta Biologicals. Active Abl-1 enzyme was purchased from Invitrogen.
Peptide Synthesis
Peptides 2 and 3 were synthesized via standard Fmoc solid phase peptide synthesis using Fmoc-PAL-PEG-PS resin (Tetras Peptide Synthesizer, CreoSalus, Louisville, KY). Coupling of Fmoc amino acids was performed in DMF with 4 equivalents (eq) amino acid, 4 eq HOBt, 4 eq HBTU, and 8 eq DIPEA. 2 eq Fmoc-(PEG)2-OH was coupled in DMF with 4 eq HOBt, 4 eq HBTU, and 8 eq DIPEA. Peptides labeled on the lysine side chain were prepared by coupling 2 eq of Fmoc-Lys(6-FAM)-OH in DMF with 4 eq HOBt, 4 eq HBTU, and 8 eq DIPEA. Fmoc deprotection was accomplished in 20% piperidine in DMF. Cleavage and deprotection was accomplished with TFA:TIS:H2O in a ratio of 95: 2.5: 2.5 for 3 h.
Peptide Cyclization
Cyclization of peptides 4 and 5 (Figure S1) was completed utilizing a modification of the procedure described by Ivanov et al.18 To bromoacetylate the N-terminus, 2 eq of bromoacetic acid and 1 eq DIC were mixed for 30 min at 0 °C in a glass round bottom flask with constant stirring. Resin containing the peptide was swelled for 2–3 h in DMF and washed thoroughly with DMF. 4 eq DIPEA and the bromoacetic acid/DIC mixture were added to the resin and reacted until a Kaiser test yielded no color change, indicating there were no free amines present. Resin was successively washed 3X each with DMF, acetic acid, and DCM prior to a final methanol rinse. Peptides were deprotected and cleaved using a 95: 1.5: 2.5: 1 ratio of TFA:TIS:H2O:EDT. The cleaving solution was evaporated by nitrogen gas and the peptides precipitated in cold diethyl ether. Peptides were dissolved in deionized (DI) H2O and lyophilized to produce a yellow powder. After lyophilization, peptides were re-dissolved in 10 mL DI H2O with minimal DMSO to facilitate solubilization. 6 eq triethylamine was added to the mixture and reacted overnight to complete cyclization.
Peptide 6 was crosslinked utilizing a procedure described previously by Kottegoda et al.19 1 μM FlAsH binding reagent and 10 μM EDT were mixed in DMSO. Un-crosslinked peptide 6 and the reducing agent TCEP were dissolved in a 1: 25 mole ratio in phosphate buffered saline (PBS; 137 mM NaCl, 10 mM Na2HPO4, 27 mM KCl, 1.74 mM KH2PO4, pH 7.4). The peptide 6-TCEP mixture was reacted with 1 mM BME and the FlAsH-EDT mixture for 15 – 90 min at room temperature to form the crosslinked peptide. Reaction progress was monitored with CE-LIF as the fluorescence emission of crosslinked peptide 6 is dramatically increased over that of the un-crosslinked form.
Peptide Purification and Characterization
All peptides were purified via reversed-phase HPLC with a 250 mm × 10 mm semiprep column (Jupiter 5 μM C18 particles, Phenomenex, Torrance, CA). Poorly soluble peptides were dissolved in H2O with up to 50% acetonitrile prior to HPLC purification. Peptides were eluted using a linear gradient of mobile phase A (95% H2O and 5% acetonitrile with 0.1% TFA) and mobile phase B (95% acetonitrile and 5% H2O with 0.1% TFA) in a reversed-phase separation. Un-crosslinked peptides were purified using a linear gradient of 100% A to 100% B over 30 min. Each crosslinked peptide was purified with a unique gradient optimized for each separation, ranging from 45–120 min linear gradients. To increase purity, peptides 4 and 5 were HPLC purified prior to the covalent cross-linking step as well as after. Crude sample was subjected to two rounds of HPLC purification before crosslinking and an additional two rounds of purification after crosslinking. Absorbance detection at 220 nm and fluorescence detection (480 nm excitation and 530 nm emission) was used to detect the fluorescently labeled peptides.
Mass spectrometric analysis was performed via electrospray ionization-time of flight mass spectrometry (Bruker, Billerica, MA) or via matrix-assisted laser desorption ionization-time of flight (Applied Biosystems, Carlsbad, CA) to verify peptide molecular weight (Figure S-2 and Table S-1). Peptide concentration was determined with amino acid analysis in the presence of a standard by the Molecular Structure Facility at the University of California, Davis (Davis, CA).
Circular dichroism spectroscopy (Figure S3) was performed using a ChirascanTM plus Circular Dichroism/Fluorescence spectrometer. Spectra were obtained at 25 °C in deionized water (pH 7.38) with 20 μM peptide. A wavelength scan (180 – 260 nm) was done with a 0.5 nm step size with 1 × 105 scans/nm and a bandwidth of 1 nm.
Cell Culture
Baf/BCR-ABL cells, a mouse B-cell lymphoma line that was stably transfected with and overexpresses Bcr-Abl,20 was cultured in RPMI 1640 medium supplemented with 10% FBS, penicillin (100 units mL−1), and streptomycin (100 μg mL−1). Cells were maintained at 37 °C with 5% CO2 and placed into fresh media when the culture reached a density of 1×106 cells mL−1. For cells utilized in single cell studies, poly(dimethyl siloxane) (PDMS, Sylgard 184) was used to glue a silicon O-ring (McMaster-Carr) to a CELLocate coverslip (Eppendorf). The coverslip was coated with Cell-Tak (BD Biosciences) and 500 μL of a dilute Baf/BCR-ABL suspension in RPMI 1640 media was added to the chamber and incubated for 20 min at 37 °C. Immediately prior to experiment start, the media in the chamber was exchanged with 37 °C extracellular buffer (ECB; 135 mM NaCl, 15 mM KCl, 10 mM HEPES, 1 mM MgCl2, 1 mM CaCl2, pH 7.4).
Lysate Degradation Assay
A Baf/BCR-ABL cell pellet was washed with and resuspended in cold ECB. Cells were lysed by incubating in liquid nitrogen for 1 min followed by thawing at 37 °C, repeated for a total of 3 freeze-thaw cycles. The mixture was centrifuged at 14,000 × g at 4 °C for 5 min. The supernatant was reserved and kept at 4 °C until use. Total protein in the lysate was determined as described by Proctor et al.21
Assessment of peptide degradation was performed in triplicate for each peptide 1–6 by incubating peptide (2 μM) with the Baf/BCR-ABL cell lysate (3 mg mL−1 total cell protein) and incubating at 37 °C. Aliquots were removed over time and stopped by adding HCl to a final concentration of 100 mM and heating at 95 °C for 4 min. Samples were diluted 20–50X in electrophoretic buffer prior to separation and detection with CE-LIF. Intact peptide was identified by adding standard peptide to the HCl-terminated aliquots and comparing electropherograms with and without the added standard. Peaks appearing at migration times differing from the intact peptide were identified as degradation products (Figures S4–S6). The percentage of undegraded peptide at each time point was determined from the ratio of the peak area of intact peptide to that under all peaks. The initial degradation rates were calculated utilizing the 0 min and 1 min time points and are expressed in terms of pmol of peptide per mg of protein per sec (pmol mg−1 s−1).
Lysate Phosphorylation Assay
A Baf/BCR-ABL pellet of 2–3 million cells was washed 3–4X with PBS buffer. The pellet was resuspended in water with 10X protease and phosphatase inhibitor cocktails and incubated until cells were ruptured upon visual observation under a microscope. The mixture was centrifuged at 14,000 × g for 10 min at 4 °C. The supernatant was reserved and kept at 4 °C until use. Total protein in the lysate was determined as described above in the Lysate Degradation Assay section.
Assessment of peptide phosphorylation was performed in triplicate for select peptides 1, 5, and 6 by mixing peptide (10 μM) with the Baf/BCR-ABL cell lysate (3 mg mL−1 total cell protein) and assay buffer [50 mM Tris (pH 7.4), 5 mM MgCl2, 2 mM DTT, and 1 mM ATP] and incubating at 30 °C for varying times. Aliquots were removed and reactions stopped by heating the mixture to 95 °C for 4 min. Samples were diluted 20X in electrophoretic buffer prior to separation and detection with CE-LIF (Figure S7).
Measurement of Kinetic Parameters
Kinase assays were performed at 30 °C in assay buffer [50 mM Tris (pH 7.4), 5 mM MgCl2, 2 mM DTT, and 1 mM ATP] with Abl-1 kinase (400 nM) and peptide (concentrations ranging from 5 – 60 μM). Aliquots were removed from the reaction mixture at various timepoints and terminated by incubating at 95 °C for 4 min. The immobilized metal ion affinity-based fluorescent polarization (IMAP) assay (Molecular Devices Corp., Sunnyvale, CA) was used to measure the amount of phosphorylated peptide in reaction mixtures. A calibration curve was constructed by measuring the anisotropy of solutions of known ratios of phosphorylated to non-phosphorylated peptide. The standards of 100% phosphorylated peptides were prepared using Abl-1 kinase and phosphorylation was verified with CE-LIF. Samples were diluted to the final concentration of 100 nM for the IMAP assay with a buffer containing 10 mM Tris-HCl (pH 7.2), 10 mM MgCl2, and 0.01% Tween-20. Anisotropy was measured using a fluorescence plate reader (SpectraMax M5, Molecular Devices, Sunnyvale, CA) with an excitation of 485 nm (bandwidth of 9 nm) and emission of 525 nm (bandwidth of 15 nm).
Capillary Electrophoresis
For samples not involving single cells, CE was performed on a custom-built CE system and laser induced fluorescence (473 nm, Lasermate Group, Inc, Pomona, CA) was used for detection of fluorescent compounds. Fused silica capillaries [50 μM inner diameter, 360 μM outer diameter (Polymicro Technologies, Phoenix, AZ)] had a total length of 40 cm and an effective length of 21 cm. Capillaries were conditioned prior to use with 0.1 M NaOH for 12 h, H2O for 1 h, 0.1 M HCl for 6 h, and H2O again for 12 h. After each sample, the capillary was rinsed with approximately 10 column volumes each of 0.1 M NaOH, H2O, and electrophoretic buffer by applying pressure to the capillary outlet. A sample plug was hydrodynamically loaded into the capillary by raising the capillary inlet relative to the outlet for a set period of time. Volume injected was determined with Poiseuille’s equation.22 Electrophoresis was initiated by applying a negative voltage to the capillary outlet while the inlet was held at ground. Electrophoretic buffer for separations involving peptides 1 or 6 was 100 mM tris and 100 mM tricine, pH 8.2. Electrophoretic buffer for separations involving peptides 2 – 5 was 100 mM tris and 100 mM tricine, pH 8.2, with 5% EOTrol HR (Target Discovery, Palo Alto, CA) as an additive.
For single cell experiments (Figure S8), CE was performed on a previously described custom built CE system mounted on the stage of an inverted microscope and LIF was utilized for detection.23 Fused silica capillaries [50 μM inner diameter, 360 μM outer diameter (Polymicro Technologies, Phoenix, AZ)] had a total length of 43 cm and an effective length of 19.5 cm. Capillaries were conditioned and rinsed between samples as described above. Cells on CELLocate coverslips were microinjected (Transjector 5246, Eppendorf, Hamburg, Germany) with peptide 1 or 5, rinsed 10X with ECB, and incubated for 0 – 30 min at 37 °C. The microinjector tip (Femtotips® from Eppendorf, Hamburg, Germany) contained stock solutions of 1 mM peptide 1 or 5. A compensation pressure of 80 hPa, an injection pressure of 200 hPa, and an injection time of 0.5 s were utilized for microinjection. Laser-based cell lysis was achieved with a focused Nd:YAG laser as previously described.23 To calibrate the amount of peptide on the electropherograms, the area under the peak of a hydrodynamically loaded, known concentration of peptide was measured and compared to the area of the peptide peak on the experimental electropherogram. The data presented represent all cells successfully microinjected with peptide.
Results and Discussion
Design of peptide constructs
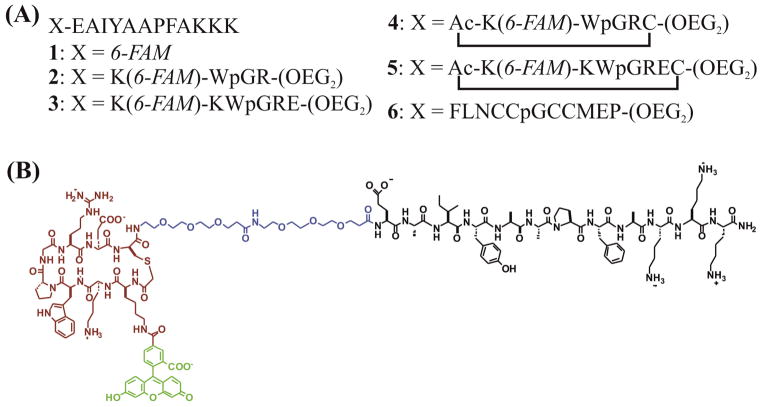
A series of peptide-based substrates were designed for Abl kinase (Figure 1). The substrates were comprised of four domains: an N-terminal β-turn peptide, an oligoethylene glycol (OEG) linker, a fluorophore, and a C-terminal Abl substrate peptide based on a previously published sequence.24 A control peptide possessed only the fluorophore and substrate peptide. The C-terminus of all peptides was amidated and the carboxyfluorescein was either attached to the N-terminus (as in the control substrate peptide 1) or to the lysine side chain at the N-terminus of the peptide (peptides 2 – 5). The β-turn sequences in peptides 2 – 5 were designed to form a type IIprime; D-Pro-Gly turn25 flanked by a Trp-Arg cross-strand pair and an additional cross-strand salt bridge in peptides 3 and 5. Cyclization of peptides 4 and 5 (Supporting Information Figure S1) was accomplished by bromo-acetylation of the N-terminus followed by crosslinking to the sulfhydryl side chain of the cysteine residue after deprotection and cleavage.18 Peptide 6 possessed an N-terminal tetracysteine motif that binds the biarsenical dye FlAsH [4′,5prime;-bis(1,3,2,-dithioarsolan-2-yl)fluorescein] with high affinity to form a fluorescent cyclic β-turn.26 All peptides were synthesized utilizing standard Fmoc solid-phase peptide synthesis and characterized by MALDI mass spectrometry. Circular dichroism spectroscopy was used to verify that the N-terminal domains of peptides 2–3 form the predicted type IIprime; β-turn (Supporting Information Figure S3).
Figure 1.
(A) The linear sequences of the β-turn peptide constructs, amidated on the carboxy terminus. The amino acids are represented by their standard single letter codes with p representing D-proline, 6-FAM is 6-carboxyfluorescein, Ac is an acetylated terminus, and OEG is 3-(2-(2-(2-aminoethoxy)ethoxy)ethoxy)propanoic acid. Peptides 2 – 5 are fluorescently labeled on the lysine side chain. Peptides 4 and 5 are cross-linked from the N-terminus to the cysteine sulfhydryl group. (B) The line bond structure of peptide 5. Substrate is shown in black, the OEG2 linker in blue, the cross-linked β-bend in red, and 6-FAM in green.
Analysis of peptide hydrolysis in a cytosolic lysate
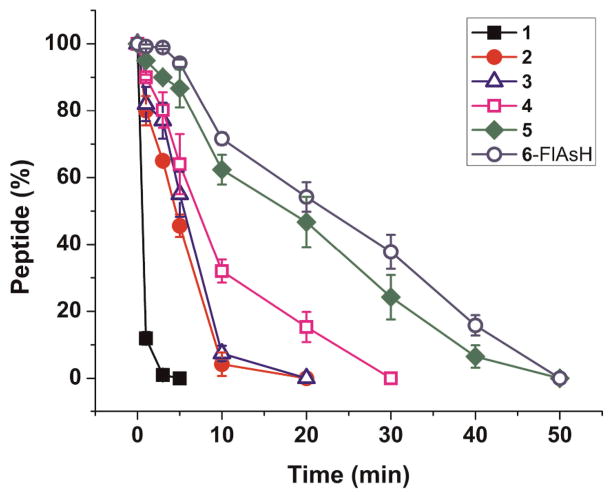
The stability of the kinase substrates in the presence of cellular peptidases was assessed by incubating the peptides in a cell lysate formed from Baf/BCR-ABL cells which over express BCR-ABL. Aliquots of the reaction mixture were removed at various times and the amount of intact peptide at each time point was determined using CE-LIF (Figure 2 and Supporting Information Figures S4–S6). The control peptide 1 possessed a half-life of 0.5 ± 0.1 min and 90% of the peptide was degraded within 3 min at an initial rate of 9.8 pmol mg−1 s−1. Peptides 2 and 3, with an appended β-turn sequence, displayed an approximate 10-fold increase in half-life of 4.8 ± 0.4 and 5.3 ± 0.7 min, respectively, yet both of these peptides were largely metabolized (> 90%) within 10 min. Peptide 2 was degraded at an initial rate of 2.2 pmol mg−1 s−1 and peptide 3 at a rate of 2 pmol mg−1 s−1. The longer half-life of peptides 2 and 3 relative to 1 were likely due to the β-turn structure which would need to be unfolded in order to be accessed by the active site of a peptidase. β-hairpins and β-turns typically fold and unfold on the ms times scale. Thus, during the brief time that they are unfolded, the peptidases may gain access to the peptides. For this reason, peptides with a stably cross-linked β-hairpin (4 and 5) structure were synthesized. When the β-hairpin structure was stabilized by covalent crosslinking, the half-life of the peptide increased to 9.2 ± 1.2 and 20.3 ± 1.7 min for peptides 4 and 5, respectively. These peptides were 90% degraded by 25 and 40 min, demonstrating an increased lifetime for cross-linked peptides which are unable to linearize. The initial rates of degradation were also slowed, to 1.1 pmol mg−1 s−1 and 0.6 pmol mg−1 s−1 for peptides 4 and 5, respectively. Peptide 6 bound with FlAsH displayed the longest half-life at 24.6 ± 1.0 min and required 45 min for 90% degradation in the cytosolic lysate, and was degraded at an initial rate of 0.1 pmol mg−1 s−1. Thus, it appears that increasing the diameter of the hairpin structure also correlates with increased half-life, suggesting these peptides create the greatest steric hindrance to entrance into the restricted cavity surrounding the peptidase catalytic sites.27
Figure 2.
Metabolism of the peptide constructs in a Baf/BCR-ABL cytosolic lysate. Data points are the average of triplicate measurements and error bars represent one standard deviation.
Determination of substrate efficacy in vitro
To assess the suitability of peptides 1–5 and peptide 6-FlAsH to act as substrates for Abl, the peptides were incubated with purified kinase at 37 °C and phosphorylated peptide was quantified over time (Table 1). The KM of the peptides appended to the β-hairpin structures was 1 – 1.5X greater than that of the linear control peptide, but within the range of previously published Abl peptide substrates.28–29 The OEG linker appeared to be effective in displacing the folded structure away from the phosphorylation site so that kinase-substrate binding was not substantially hampered. The Vmax was also increased by a factor of 1 – 1.5 relative to that of the linear control peptide, though the kcat remained similar for all 6 peptides (Table 1).
Table 1.
Peptide properties.
| Peptide | MW (Da) | β-hairpin size (Å) | t1/2 (min) | KM (μM) | Vmax (μM min−1) | kcat (min−1) | kcat/KM (μM−1 min−1) |
|---|---|---|---|---|---|---|---|
| 1 | 1692.3 | N/A | 0.5 ± 0.1 | 61 ± 9 | 2.1 ± 0.2 | 21,000 | 344 |
| 2 | 2723.4 | 10 × 16 | 4.8 ± 0.4 | 62 ± 6 | 2 ± 0.1 | 20,000 | 323 |
| 3 | 2981.4 | 14 × 16 | 5.3 ± 0.7 | 90 ± 8 | 3 ± 0.2 | 30,000 | 333 |
| 4 | 2867.3 | 14 × 16 | 9.2 ± 1.2 | 62 ± 15 | 2.4 ± 0.4 | 24,000 | 387 |
| 5 | 3124.2 | 17 × 22 | 20.3 ± 1.7 | 74 ± 23 | 3.2 ± 0.6 | 32,000 | 432 |
| 6 | 3401.3 | 18 × 25 | 24.6 ± 1.0 | 95 ± 21 | 2.7 ± 0.4 | 27,000 | 284 |
t1/2 is the time required for 50% of the peptide to be degraded in a cytosolic lysate. KM and Vmax are with respect to Abl kinase.
As peptide 5 and peptide 6-FlAsH possessed the longest lifetimes in a cell lysate yet displayed efficiencies similar to that of the control substrate, the phosphorylation of peptide 5 and peptide 6-FlAsH in a cytosolic lysate was measured. Baf/BCR-ABL cells are a murine B cell leukemia line stably transfected with Bcr-Abl, a fusion protein that overexpresses catalytically active Abl kinase as a fusion with the Bcr protein. As the substrate preference for Abl and Bcr-Abl are similar,28–29 phosphorylation of peptides 5 and 6-FlAsH by Bcr-Abl was expected. Peptides 1, 5, and 6-FlAsH were incubated at 30 °C in a Baf/BCR-ABL cytosolic lysate containing ATP and phosphatase and protease inhibitors. Aliquots were removed over time and phosphorylation was monitored with CE-LIF (Supporting Information Figure S7). After 20 min incubation in the Baf/BCR-ABL lysate, peptides 1, 5, and 6-FlAsH were 3.1 ± 0.5%, 9.8 ± 0.1%, and 5.8 ± 0.4% phosphorylated, respectively. No phosphorylation of any of the peptides was observed when the Bcr-Abl inhibitor Gleevec was added to the lysate.
Behavior of peptides in single Baf/BCR-ABL cells
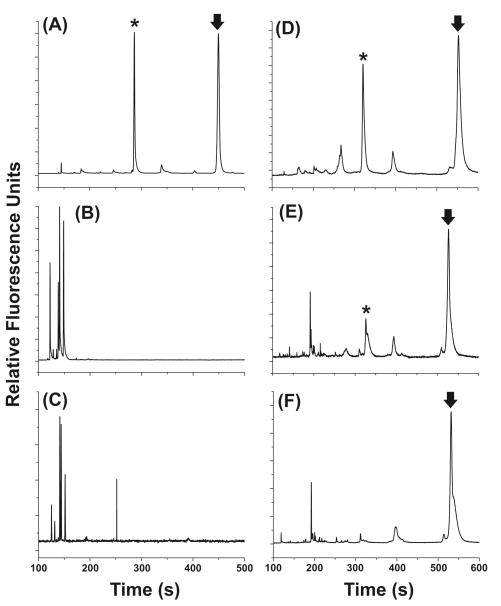
Based on its longer lifetime in a cytosolic lysate coupled with the highest kcat/KM value (Table 1), peptide 5 was selected to assess the performance of a peptide construct within intact, living cells. To accomplish this, peptide 1 or 5 was microinjected into single Baf/BCR-ABL cells and the cells incubated at 37 °C. The cells were then lysed with a brief pulse from an Nd:YAG laser and the cellular contents loaded into a capillary followed by electrophoretical separation of the substrate and product forms of the peptide. Intact non-phosphorylated and phosphorylated peptides were identified by their migration times compared to standard mixtures and the amount of phosphorylation was quantified based on the relative area under each peak (Figure 3). After a 20-min incubation of the control peptide 1 in single cells (n = 3), neither intact substrate nor phosphorylated product was detected. However, after a 20-min incubation of peptide 5 in single cells (n = 12), intact, non-phosphorylated peptide 5 accounted for 41 ± 17% of all species present and phosphorylated peptide 5 accounted for 8 ± 7% of all species present. To determine if Bcr-Abl was responsible for phosphorylation of the substrate peptides, cells were incubated with the Bcr-Abl inhibitor Gleevec prior to peptide incubation and subsequent CE-LIF. In cells incubated with Gleevec prior to microinjection with peptide 1 (n = 5), neither intact substrate nor phosphorylated product was detected after 20 min. In cells incubated with Gleevec prior to microinjection with peptide 5 (n = 5), 60 ± 12% of peptide present was intact, non-phosphorylated peptide, and no phosphorylated product was detected after 20 min (Figure S8). These results indicated that Bcr-Abl was likely responsible for phosphorylation of peptide 5 and that peptide 5 was substantially more resistant to degradation than peptide 1 in the cytosol of single cells. To further confirm that the phosphorylation peak was not a fragment peptide, the intact parent peptide was incubated with Pronase E, an aggressive protease mixture derived from S. griseus, to generate fluorescent peptide fragments. The resulting mixture of fragments was hydrodynamically loaded into the capillary following loading of a single cell and the contents were simultaneously electrophoresed. None of the fragment peaks co-migrated with the intact, non-phosphorylated or phosphorylated substrate, further indicating that the observed peak was phosphorylated substrate.
Figure 3.
Measurement of Bcr-Abl activity in single Baf/BCR-ABL cells using peptide 1 (A–C) or peptide 5 (D–F). The arrow represents intact non-phosphorylated peptide and the asterisk represents the phosphorylated peptide. Electropherograms of non-phosphorylated and phosphorylated standards of peptide 1 (A) or peptide 5 (D). Electropherograms from a single, untreated Baf/BCR-ABL cell 20 min after microinjection with peptide 1 (B) or peptide 5 (E). Electropherograms from a single, Gleevec-treated Baf/BCR-ABL cell 20 min after microinjection with peptide 1 (C) or peptide 5 (F).
Conclusions
In summary, we report the design and validation of intracellular peptidase-resistant reporters for Abl kinase and demonstrate their application in single, living cells. Significantly, the attachment of a cyclic β-turn to the N-terminus of substrate peptides yielded a peptide with a prolonged lifetime relative to that of a control linear peptide in cytosolic lysates and single cells. The peptides with β-turns maintained their ability to act as kinase substrates, making them viable intracellular kinase reporters. This strategy for peptide substrate protection should be generally applicable to measurement of the activity of other kinases in single cells. This strategy is therefore expected to serve as a valuable tool in understanding kinase activities in single CML cells as well as single cells from other cancer cell types.
Supplementary Material
Acknowledgments
This work was supported by the NIH (EB011763). We thank Dr. Robert M. Hughes for providing us with FlAsH.
References
- 1.Yewdell JW, Reits E, Neefjes J. Nat Rev Immunol. 2003;3:952–961. doi: 10.1038/nri1250. [DOI] [PubMed] [Google Scholar]
- 2.Saric T, Beninga J, Graef CI, Akopian TN, Rock KL, Goldberg AL. J Biol Chem. 2001;276:36474–36481. doi: 10.1074/jbc.M105517200. [DOI] [PubMed] [Google Scholar]
- 3.Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- 4.Geier E, Pfeifer G, Wilm M, Lucchiari-Hartz M, Baumeister W, Eichmann K, Niedermann G. Science. 1999;283:978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 5.Macpherson E, Tomkinson B, Bålöw R, Höglund S, Zetterqvist Ö. Biochem J. 1987;248:259–263. doi: 10.1042/bj2480259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomkinson B. Trends Biochem Sci. 1999;24:355–359. doi: 10.1016/s0968-0004(99)01435-8. [DOI] [PubMed] [Google Scholar]
- 7.Fülöp V, Böcskei Z, Polgár L. Cell. 1998;94:161–170. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 8.Turzynski A, Mentlein R. Eur J Biochem. 1990;190:509–515. doi: 10.1111/j.1432-1033.1990.tb15603.x. [DOI] [PubMed] [Google Scholar]
- 9.Beninga J, Rock KL, Goldberg AL. J Biol Chem. 1998;273:18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 10.Shibata K, Suzawa T, Soga S, Mizukami T, Yamada K, Hanai N, Yamasaki M. Bioorg Med Chem Lett. 2003;13:2583–2586. doi: 10.1016/s0960-894x(03)00476-1. [DOI] [PubMed] [Google Scholar]
- 11.Cline LL, Waters ML. Biopolymers. 2009;92:502–507. doi: 10.1002/bip.21266. [DOI] [PubMed] [Google Scholar]
- 12.Cheng Z, Miskolzie M, Campbell RE. ChemBioChem. 2007;8:880–883. doi: 10.1002/cbic.200600565. [DOI] [PubMed] [Google Scholar]
- 13.Matsoukas J, Apostolopoulos V, Kalbacher H, Papini A, Tselios T, Chatzantoni K, Biagioli T, Lolli F, Deraos S, Papathanassopoulos P, Troganis A, Mantzourani E, Mavromoustakos T, Mouzaki A. J Med Chem. 2005;48:1470–1480. doi: 10.1021/jm040849g. [DOI] [PubMed] [Google Scholar]
- 14.Sako Y, Goto Y, Murakami H, Suga H. ACS Chem Biol. 2008;3:241–249. doi: 10.1021/cb800010p. [DOI] [PubMed] [Google Scholar]
- 15.Fung S, Hruby VJ. Curr Opin Chem Biol. 2005;9:352–358. doi: 10.1016/j.cbpa.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh CT. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 17.Bartram CR, de Klein A, Hagemeijer A, van Agthoven T, van Kessel AG, Bootsma D, Grosveld G, Ferguson-Smith MA, Davies T, Stone M, Heisterkamp N, Stephenson JR, Groffen J. Nature. 1983;306:277–280. doi: 10.1038/306277a0. [DOI] [PubMed] [Google Scholar]
- 18.Ivanov B, Grzesik W, Robey FA. Bioconjugate Chem. 1995;6:269–277. doi: 10.1021/bc00033a006. [DOI] [PubMed] [Google Scholar]
- 19.Kottegoda S, Aoto PC, Sims CE, Allbritton NL. Anal Chem. 2008;80:5358–5366. doi: 10.1021/ac8003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahon FX, Deininger MWN, Schultheis B, Chabrol J, Reiffers J, Goldman JM, Melo JV. Blood. 2000;96:1070–1079. [PubMed] [Google Scholar]
- 21.Proctor A, Wang Q, Lawrence DS, Allbritton NL. Analyst. 2012;137:3028–3038. doi: 10.1039/c2an16162a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weinberger R. Practical Capillary Electrophoresis. 1. Academic Press; London: 1993. [Google Scholar]
- 23.Sims CE, Meredith GD, Krasieva TB, Berns MW, Tromberg BJ, Allbritton NL. Anal Chem. 1998;70:4570–4577. doi: 10.1021/ac9802269. [DOI] [PubMed] [Google Scholar]
- 24.Songyang Z, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantley LC. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 25.Haque TS, Gellman SH. JACS. 1997;119:2303–2304. [Google Scholar]
- 26.Adams SR, Tsien RY. Nat Protoc. 2008;3:1527–1534. doi: 10.1038/nprot.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varshavsky A. PNAS. 1996;93:12142–12149. doi: 10.1073/pnas.93.22.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griswold IJ, MacPartlin M, Bumm T, Goss VL, O’Hare T, Lee KA, Corbin AS, Stoffregen EP, Smith C, Johnson K, Moseson EM, Wood LJ, Polakiewicz RD, Druker BJ, Deininger MW. Mol Cell Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu J, Phan H, Lam KS. Bioorg Med Chem. 1998;8:2279–2284. doi: 10.1016/s0960-894x(98)00413-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.