Summary
Host inflammation alters the availability of nutrients such as iron to limit microbial growth. However, Salmonella enterica serovar Typhimurium thrives in the inflamed gut by scavenging for iron with siderophores. By administering Escherichia coli strain Nissle 1917, which assimilates iron by similar mechanisms, we show that this non-pathogenic bacterium can outcompete and reduce S. Typhimurium colonization in mouse models of acute colitis and chronic persistent infection. This probiotic activity depends on E. coli Nissle iron acquisition as mutants deficient in iron uptake colonize the intestine but do not reduce S. Typhimurium colonization. Additionally, the ability of E. coli Nissle to overcome iron restriction by the host protein lipocalin-2, which counteracts some siderophores, is essential as S. Typhimurium is unaffected by E. coli Nissle in lipocalin-2-deficient mice. Thus, iron availability impacts S. Typhimurium growth and E. coli Nissle reduces S. Typhimurium intestinal colonization by competing for this limiting nutrient.
Introduction
Salmonella enterica serovar Typhimurium (S. Typhimurium) is one of the leading causes of acute gastroenteritis characterized by inflammatory diarrhea. While the normal intestine is largely inhabited by commensal microbes, which largely include Bacteroides and Firmicutes, inflammation enhances the colonization of S. Typhimurium and other Enterobacteriaceae (Barman et al., 2008; Lawley et al., 2008; Lupp et al., 2007; Stecher et al., 2007). Recent studies have shown that S. Typhimurium thrives in the inflamed gut because it can utilize unique carbon and energy sources (Thiennimitr et al., 2011; Winter et al., 2010) and is resistant to antimicrobial proteins that are secreted by the host as part of the nutritional immune response (Liu et al., 2012; Raffatellu et al., 2009). S. Typhimurium employs specialized transporters to acquire essential micronutrient metals (Liu et al., 2012; Raffatellu et al., 2009), one of the most important being iron (Crouch et al., 2008; Raffatellu et al., 2009). Levels of free iron are extremely low in the host environment due to sequestration by host proteins including heme, ferritin, transferrin and lactoferrin (Andrews and Schmidt, 2007). Additional mechanisms are employed by the host to further limit iron availability during inflammation (Weinberg, 1984), including secretion of the hormone hepcidin, which prevents the gut from absorbing iron from the bloodstream by inhibiting the iron transporter ferroportin-1 (Ganz, 2003).
When starved for iron, bacteria synthesize and export small-molecule high-affinity iron chelators termed siderophores. Enterochelin is a catecholate-type siderophore secreted by all Enterobacteriaceae, including Salmonella and commensal E. coli (Raymond et al., 2003), which is sufficient to overcome the host’s iron limitation in a normal (non-inflamed) environment. However, during inflammatory responses, the host secretes lipocalin-2, an antimicrobial peptide that sequesters ferric enterochelin, thereby limiting the growth of strains like commensal E. coli that rely solely upon enterochelin for siderophore-based iron acquisition (Berger et al., 2006; Flo et al., 2004). Some pathogens evade this response by synthesizing additional siderophores that are not sequestered by lipocalin-2 (Fischbach et al., 2006a). For instance, Salmonella can synthesize and secrete salmochelin (Muller et al., 2009), a C-glucosylated derivative of enterochelin that is too large to fit into the enterochelin-binding pocket of lipocalin-2 (Fischbach et al., 2006b; Hantke et al., 2003).
Probiotics are commensal microorganisms that are believed to exert beneficial effects on the host. Escherichia coli Nissle 1917 (E. coli Nissle, serotype O6:K5:H1) is a probiotic strain that was originally isolated from a soldier who appeared resistant to an outbreak of diarrhea (Nissle, 1959). E. coli Nissle has been shown to establish persistent colonization of the intestine and has been used to treat or prevent a variety of intestinal disorders (Cukrowska et al., 2002; Kruis et al., 2004; Lodinova-Zadnikova and Sonnenborn, 1997; Mollenbrink and Bruckschen, 1994; Nissle, 1959), including acute enteritis (Henker et al., 2007), but the mechanistic basis for its protective actions is unknown. As salmochelin-mediated iron acquisition during inflammation enhances S. Typhimurium colonization (Raffatellu et al., 2009), we hypothesized that E. coli Nissle might protect the host by utilizing similar mechanisms to compete with S. Typhimurium for essential micronutrients.
A snapshot analysis of the E. coli Nissle genome revealed that it shares many fitness properties found in uropathogenic E. coli (UPEC) strains of the same serotype (Grozdanov et al., 2004). Intriguingly, the E. coli Nissle genome appears to encode for as many iron uptake systems as UPEC (Garcia et al., 2011), an armament that notably includes salmochelin, the hydroxamate-type siderophore aerobactin, the mixed-type siderophore yersiniabactin and the hemin uptake transporter ChuA. As redundancy in iron uptake promotes the growth of UPEC in the bladder and the kidney (Garcia et al., 2011), we reasoned that it may also contribute to E. coli Nissle colonization of the inflamed gut. To test the hypothesis that iron uptake mechanisms are important for E. coli Nissle probiotic activity, we set out to examine the effect of administering wild-type E. coli Nissle or mutant derivatives deficient in iron uptake on the course of S. Typhimurium infection.
Results
Probiotic E. coli Nissle 1917 reduces S. Typhimurium fecal shedding
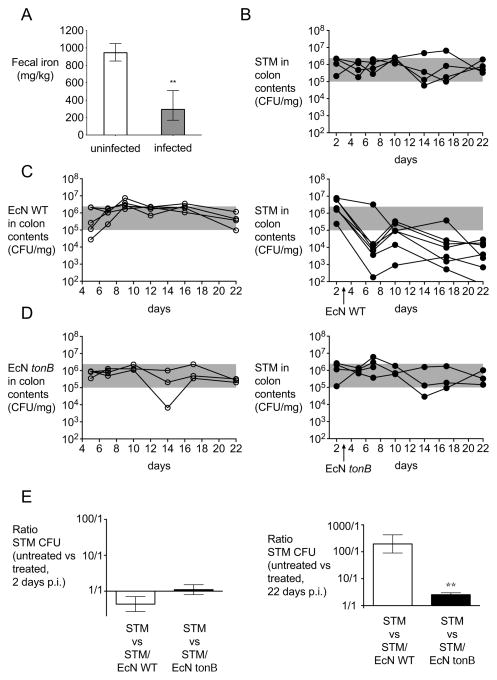
Our prior studies indicated that S. Typhimurium must overcome iron limitation to successfully infect the host (Crouch et al., 2008; Raffatellu et al., 2009). To confirm that iron limitation occurs during S. Typhimurium infection, we first measured the concentration of iron in the feces of mice four days after infection in an S. Typhimurium colitis model by inductively coupled plasma mass spectrometry (ICP-MS). We found that the concentration of fecal iron in the absence of infection was approximately 950 mg/kg (Fig. 1A). In contrast, the concentration of fecal iron was significantly reduced on average to 300 mg/kg in mice infected with S. Typhimurium (Fig. 1A), confirming that infection results in limitation of this metal in the colonic environment. To gain insight on the mechanism behind the lower levels of intestinal iron, we determined the expression of hepcidin and ferroportin-1 (Fig S1). While hepcidin transcripts were highly abundant in the liver of both mock-infected and S. Typhimurium-infected mice, the expression of hepcidin was not upregulated during S. Typhimurium infection (Fig S1). Nonetheless, we observed a modest but significant downregulation of ferroportin-1 in the liver and in the intestine of mice infected with S. Typhimurium, which should have resulted in higher levels of intestinal iron due to its reduced intestinal absorption. However, mice infected with S. Typhimurium also showed a significant weight loss and reduced food uptake (Fig S1 and data not shown), which may contribute to the lower concentration of iron in the intestine of infected mice (Fig 1A).
Figure 1. Probiotic E. coli Nissle 1917 reduces S. Typhimurium fecal shedding.
(A) The concentration of iron in fecal samples collected from mock-infected (n = 4) or S. Typhimurium-infected (n = 4) C57BL/6 mice four days post-infection. Bars represent geometric means ± standard deviation. (B–E) 129X1/SvJ mice were infected with S. Typhimurium and either untreated (B) or treated with one dose of E. coli Nissle wild-type (C) or tonB mutant (D) three days after infection. S. Typhimurium (black circles), E. coli Nissle wild-type or tonB mutant (white circles) were enumerated in the colonic contents. (E) Ratio of colony forming units (CFU) recovered from fecal samples of mice infected with S. Typhimurium that were untreated compared with mice treated with one dose of either E. coli Nissle wild-type or tonB mutant three days after infection. Ratios 2 days after infection (i.e., 1 day before treatment) and 22 days after infection are shown. Bars represent geometric means ± standard deviation. Data are representative of n=2 experiments. STM=S. Typhimurium; EcN=E. coli Nissle. Significant difference is indicated by ** (P value ≤ 0.01). (See also Figure S1 and Table S1).
Once we established that intestinal iron is indeed limited during S. Typhimurium infection, we next assessed the effects of E. coli Nissle administration on S. Typhimurium infection. First, we utilized 129X1/SvJ mice, which develop chronic Salmonella colitis with persistent infection (Lawley et al., 2008). To ensure that all mice became highly colonized, streptomycin was administered prior to infection as previously described (Barthel et al., 2003; Lawley et al., 2008). S. Typhimurium-infected mice were then followed for three weeks post-infection and exhibited consistent high levels of fecal shedding of Salmonella (Fig. 1B) as well as high levels of inflammation (Fig. 2). Remarkably, a single therapeutic dose of wild-type E. coli Nissle administered three days after inoculation with S. Typhimurium was able to establish persistent colonization (Fig. 1C) and significantly reduce S. Typhimurium colonization by more than 2 logs for the duration of the study (Fig. 1C, 1E, and S1). To determine whether the beneficial effect of E. coli Nissle is dependent upon its ability to acquire iron, a mutation was constructed in the tonB gene of E. coli Nissle.
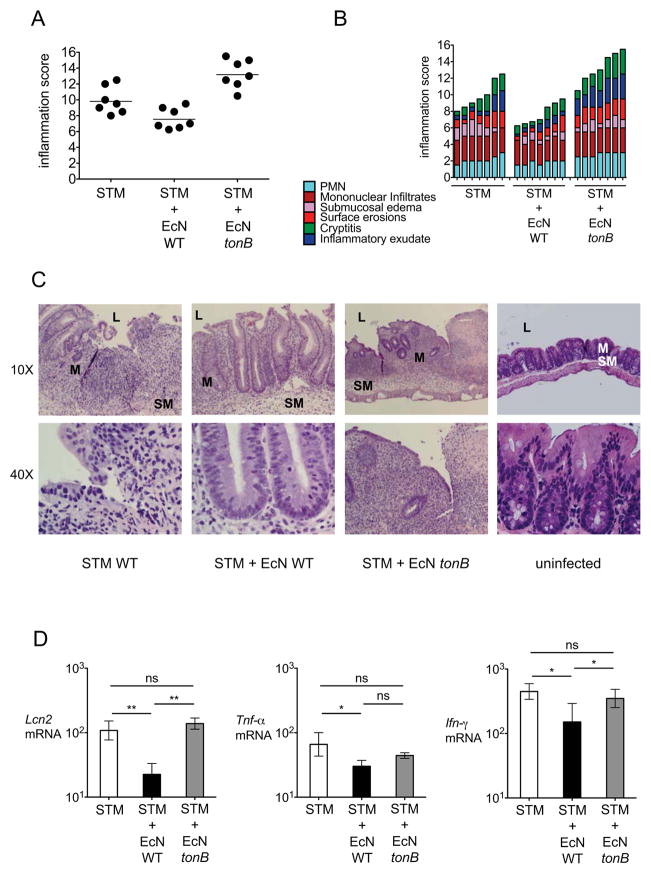
Figure 2. Intestinal host response in mice with persistent S. Typhimurium infection.
129X1/SvJ mice were infected with S. Typhimurium and were either left untreated or were treated with one dose of E. coli Nissle wild-type or tonB mutant three days after infection. Cecal samples were collected at 6 or 22 days after infection. (A) Blinded histopathology scores of cecal samples at 22 days after infection. The score of individual mice (circles) and the geometric mean for each group (bars) are indicated. (B) A detailed scoring for the animals shown in (A) is provided. Each stacked column represents an individual mouse. (C) H&E stained sections from representative animals for each group. Although E. coli Nissle wild-type had no effect on inflammatory cell infiltration, mucosal integrity (assessed by cryptitis and surface erosions) was modestly improved. (D) Transcript levels of Lcn2, Tnf-α and Inf-γ were determined in the ceca of 129X1/SvJ mice infected with S. Typhimurium (white bars), infected with S. Typhimurium and treated with either one dose of E. coli Nissle wild-type (black bars) or tonB mutant (gray bars) 3 days after infection. Samples were collected 6 days after infection. Data are expressed as fold-increase over mock-infected mice. Bars represent the geometric means ± standard deviation. STM=S. Typhimurium; EcN=E. coli Nissle; L; lumen, M, mucosa, SM; submucosa. Significant difference is indicated by * (P value ≤ 0.05) or ** (P value ≤ 0.01).
TonB provides the energy necessary for active transport of iron-laden siderophores and heme (Braun and Hantke, 2011). As expected, in vitro growth of E. coli Nissle tonB was equivalent to wild-type in an iron rich media (Fig. 3A and S2) or when tonB was complemented in trans (Fig. S2), but not in media when iron was limited by the addition of serum (Fig. 3C) or of by iron chelation with 2,2′-dipyridyl (Fig. 3E). Strikingly, E. coli Nissle tonB was still able to establish persistent colonization when administered to 129X1/SvJ mice three days after S. Typhimurium infection but was unable to reduce the S. Typhimurium burden in the feces (Fig. 1D, 1E, S1). Moreover, only wild-type E. coli Nissle was able to ameliorate the chronic inflammation observed in these S. Typhimurium-challenged mice at 22 days post-infection (Fig. 2). Consistent with this observation, transcript levels of Lcn2, which encodes for lipocalin-2, and levels of the pro-inflammatory cytokines Tnf-α and Ifn-γ were reduced in the cecum by wild-type E. coli Nissle but not by the tonB mutant (Fig. 2D). Therefore, our results suggested that iron acquisition is important for the probiotic activity of E. coli Nissle and for its ability to compete with S. Typhimurium in the inflamed gut during chronic infection.
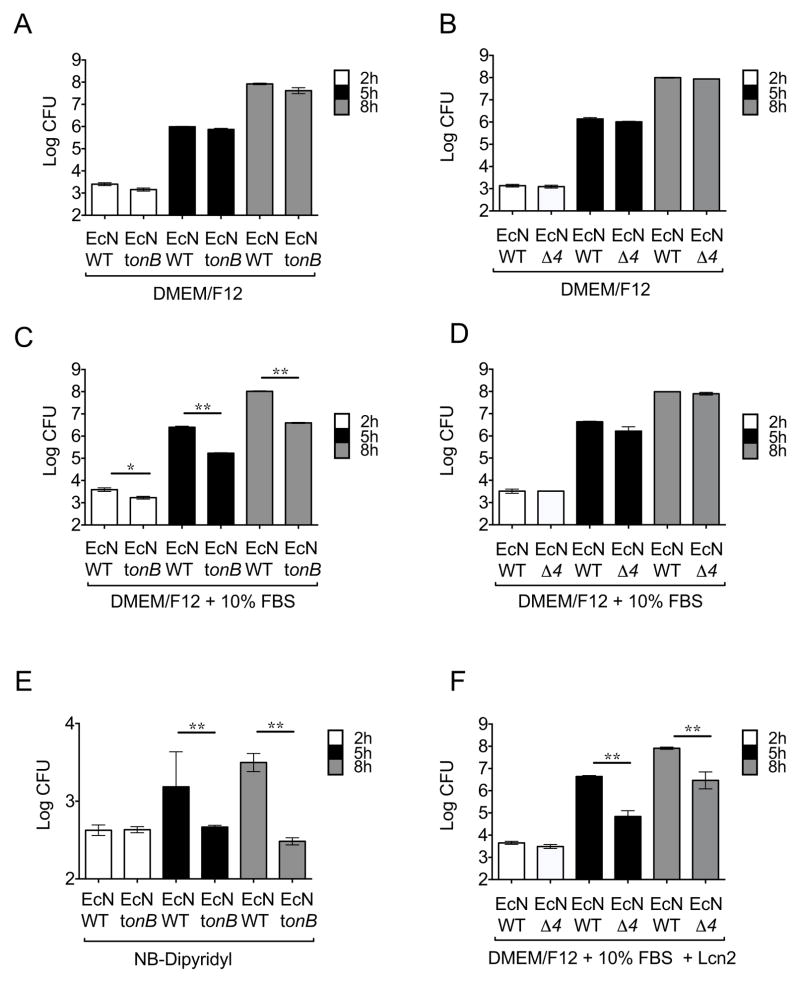
Figure 3. Growth of E. coli Nissle 1917 strains in iron-rich and iron-limited media.
Growth of E. coli Nissle wild-type and the mutants in iron uptake tonB or iroN fuyA iutA chuA (Δ4) was determined. (A,C,E) Growth of E. coli Nissle wild-type and the tonB mutant in DMEM/F12 (A) or DMEM/F12 supplemented with 10% fetal bovine serum (C) or nutrient broth (NB) supplemented with Dipyridyl (E). (B,D,F) Growth of E. coli Nissle wild-type and the Δ4 mutant in DMEM/F12 (B) or DMEM/F12 supplemented with 10% fetal bovine serum with the absence (D) or presence (F) of 1μg/ml lipocalin-2 (Lcn2). Bacteria were enumerated at 2h, 5h, and 8h after inoculation. Bars represent the geometric means ± standard deviation of at least three experiments. STM=S. Typhimurium; EcN=E. coli Nissle. Significant differences are indicated by * (P value ≤ 0.05) or ** (P value ≤ 0.01). (See also Figure S2 and Table S2)
Growth of E. coli Nissle strains in iron-rich and iron-limited media
As tonB mutations impede multiple iron acquisition mechanisms, a mutant strain was constructed which lacks four separate tonB-dependent iron transport systems that have been shown to contribute to iron acquisition during UPEC urinary tract infection (Garcia et al., 2011). The resulting strain lacks the salmochelin receptor IroN, the aerobactin receptor IutA, the yersiniabactin receptor FuyA and the heme receptor ChuA (E. coli Nissle iroN fyuA iutA chuA) and it was termed E. coli Nissle Δ4. Notably, in contrast to the tonB mutant, E. coli Nissle Δ4 may acquire iron via ferric enterochelin through the FepA receptor. However, blockade of ferric enterochelin via lipocalin-2 should result in growth inhibition. Therefore, we determined the growth of the E. coli Nissle Δ4 in rich media supplemented with serum and/or the enterochelin scavenger lipocalin-2, as we previously described (Raffatellu et al., 2009). As expected, the E. coli Nissle Δ4 grew as well as wild-type in iron rich media (Fig. 3B and S2) and – in contrast to the E. coli Nissle tonB (Fig. 3C) - in rich media supplemented with serum (Fig. 3D), consistent with effective iron acquisition via ferric enterochelin. However, this strain exhibited a significant growth impairment in medium supplemented with the iron scavenger lipocalin-2 (Fig. 3F). Furthermore, the growth inhibition by lipocalin-2 on the E. coli Nissle Δ4 mutant was no longer observed if ferric iron was added to the media in the form of iron citrate (Fig. S2) (Pressler et al., 1988). Promisingly, the growth of wild-type E. coli Nissle was not impaired by supplementation with lipocalin-2 (Fig. 3, S2). Taken together, these results suggested that lipocalin-2 resistance mediated by non-enterochelin high-affinity iron uptake mechanisms might allow E. coli Nissle to grow in the inflamed gut and reduce S. Typhimurium colonization.
E. coli Nissle 1917 requires iron uptake systems to reduce S. Typhimurium intestinal colonization
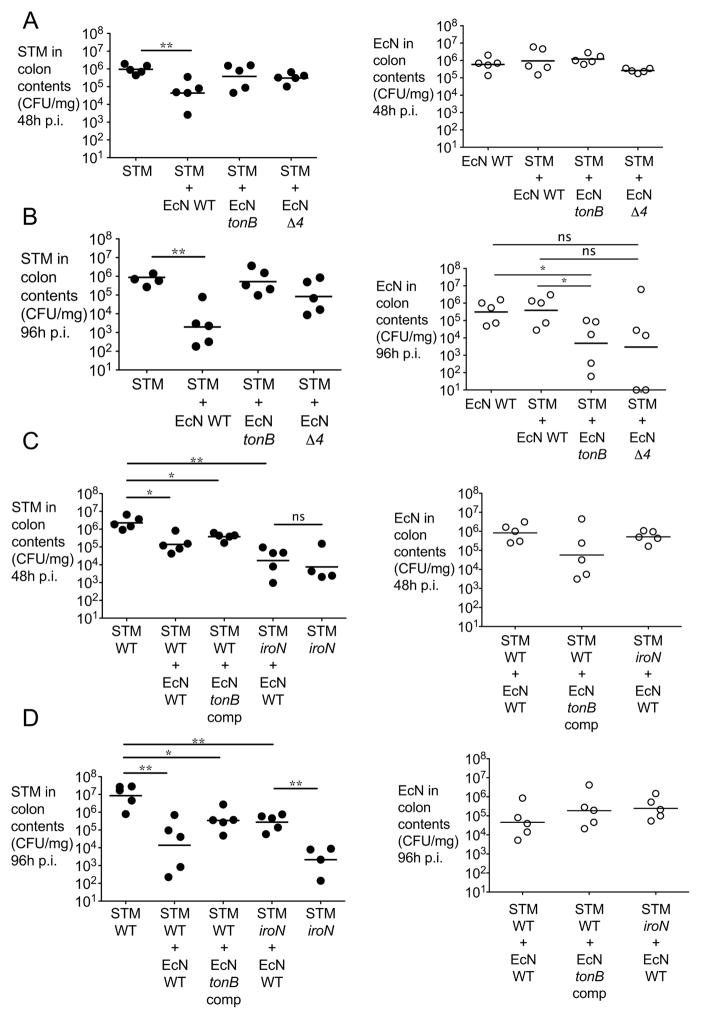
To further assess whether high affinity iron transporters may contribute to the probiotic effect of E. coli Nissle, we administered wild-type E. coli Nissle or isogenic mutant strains deficient in iron uptake to mice infected with S. Typhimurium in a model of acute colitis. Specifically, C57BL/6 mice were administered streptomycin one day prior to infection as previously described(Barthel et al., 2003; Raffatellu et al., 2009). Mice were then infected with S. Typhimurium alone or co-administered with an equal dose of E. coli Nissle wild-type or mutants (Figure 4 and S3). Colonization in the colon content was then determined at 48, 72 and 96 hours after infection. In this model of acute colitis, the administration of E. coli Nissle significantly reduced intestinal colonization with S. Typhimurium at every time point (Fig. 4A, 4B, and S3). In contrast, neither the E. coli Nissle tonB nor the Δ4 mutants were able to reduce intestinal S. Typhimurium colonization (Fig. 4A, 4B, and S3) despite successfully establishing intestinal colonization (Fig. 4A, 4B, and S3). Of note, while E. coli Nissle wild-type outgrew S. Typhimurium, both E. coli Nissle tonB and the Δ4 mutant were outcompeted by S. Typhimurium at 96 hours after infection (see also Fig. 7). Moreover, complementation of E. coli Nissle tonB in trans partly rescued this strain’s capability to reduce S. Typhimurium colonization (Fig. 4C, 4D, and S3).
Figure 4. E. coli Nissle 1917 requires iron uptake systems to reduce S. Typhimurium intestinal colonization.
(A,B) C57BL/6 mice were infected with S. Typhimurium alone or co-administered with either wild-type E. coli Nissle, the tonB mutant or the iroN fuyA iutA chuA mutant (Δ4). (C,D) C57BL/6 mice were infected with S. Typhimurium alone (wild-type or iroN mutant) or co-administered with either wild-type E. coli Nissle, the tonB mutant or the tonB mutant complemented in trans when indicated. CFU in colonic contents were enumerated at 48h (A,C) and 96h (B,D) after infection. S. Typhimurium (black circles) and E. coli Nissle (white circles) counts are shown. Representative experiments of n=2 are shown. STM=S. Typhimurium; EcN=E. coli Nissle. Significant difference is indicated by * (P value ≤ 0.05) or ** (P value ≤ 0.01). (See also Figure S3)
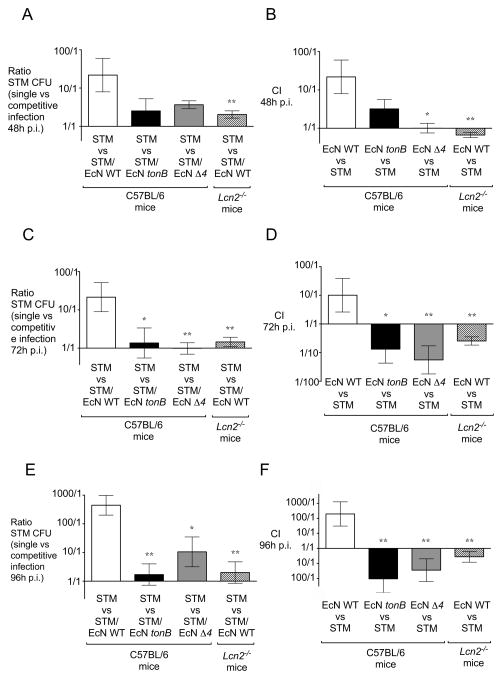
Figure 7. Ratios of S. Typhimurium and E. coli Nissle in the acute colitis model.
Ratio of the CFU recovered from the fecal samples of mice infected with S. Typhimurium that were untreated in comparison with mice that were administered one dose of either E. coli Nissle wild-type, the tonB mutant, or the iroN fuyA iutA chuA (Δ4) mutant at the time of infection. (A, C, E) Ratio of S. Typhimurium CFU at 48 hours (A), 72 hours (C), 96 hours (E) post infection. (B, D, F) Competitive index (CI) in the indicated mixed infection was calculated by dividing the output ratio (E. coli Nissle CFU / S. Typhimurium CFU in the colonic contents of mice at the indicated time points by the input ratio (E. coli Nissle CFU / S. Typhimurium CFU). CI of the indicated E. coli Nissle strain versus S. Typhimurium at 48 hours (B), 72 hours (D), 96 hours (F) post infection. Bars represent geometric means ± standard deviation. Significant difference is indicated by * (P value ≤ 0.05) or ** (P value ≤ 0.01).
To gain further insight into the competition for iron between S. Typhimurium and E. coli Nissle in the inflamed gut, we tested whether E. coli Nissle would affect the colonization of an S. Typhimurium strain lacking the IroN receptor (iroN mutant), which cannot acquire iron via salmochelin ((Raffatellu et al., 2009) and Fig. S2). We have previously shown that an S. Typhimurium iroN mutant has a colonization defect in the inflamed gut that is dependent on the expression of lipocalin-2 (Raffatellu et al., 2009). Consistent with our earlier study, we found that an S. Typhimurium iroN mutant showed a defect in colonization when compared to S. Typhimurium wild-type (Fig. 4C, 4D, and S3). Consistent with our present study, administration of E. coli Nissle did not further reduce colonization of the S. Typhimurium iroN mutant, supporting the notion that competition for iron is an essential trait of its probiotic activity (Fig. 4C, 4D, and S3). Of note, at 96 hours post-infection, the colonization of the S. Typhimurium iroN mutant further decreased because of the higher levels of lipocalin-2 (Fig. 4D and S3C). In contrast, the downregulation of lipocalin-2 caused by the administration of E. coli Nissle partly rescued the iroN mutant, which however is still significantly lower than wild-type S. Typhimurium at this time point; this is consistent with our previously published results showing that the iroN mutant is rescued in Lcn2−/− mice (Raffatellu et al., 2009). Taken together, these results demonstrate that iron uptake is crucial for the beneficial effect of E. coli Nissle in reducing the colonization of S. Typhimurium.
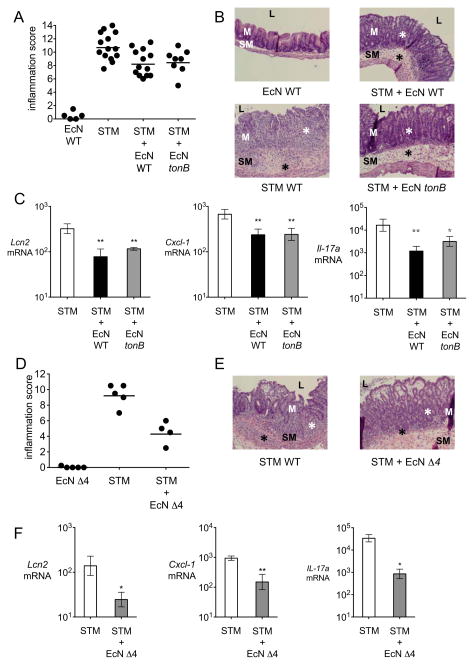
As E. coli Nissle was able to diminish the intestinal burden of S. Typhimurium, we next analyzed whether the probiotic strain also ameliorated the intestinal inflammation caused by S. Typhimurium in the acute colitis model (Fig. 5, S4). Administration of E. coli Nissle alone did not cause intestinal inflammation (Fig. 5, S4). Intriguingly, the addition of wild-type E. coli Nissle, E. coli Nissle tonB or E. coli Nissle Δ4 each resulted in similar reductions of intestinal inflammation in S. Typhimurium-infected C57BL/6 mice (Fig. 5A, B, D, E, S4). In accordance with the reduction in inflammation, decreased transcript levels of Lcn2, of the neutrophil chemoattractant Cxcl-1, and of the pro-inflammatory cytokine Il-17a were also observed (Fig. 5C, F). To determine whether E. coli Nissle has direct anti-inflammatory effects, mice treated with dextran sodium sulfate (DSS) in the drinking water as a means to induce colitis in the absence of a bacterial infection were administered a single dose of E. coli Nissle (Wirtz et al., 2007). Similar to E. coli Nissle administration during S. Typhimurium infection, treatment with E. coli Nissle reduced the expression of pro-inflammatory cytokines in DSS-treated mice (Fig S4). Based on these results, it appears that E. coli Nissle exerts beneficial effects on cecal inflammation during acute Salmonella colitis that are independent of its antagonism of S. Typhimurium colonization.
Figure 5. E. coli Nissle 1917 ameliorates intestinal inflammation during acute S. Typhimurium infection.
(A) Blinded histopathology scores of cecal samples 4 days after infection of C57BL/6 mice administered wild-type E. coli Nissle, S. Typhimurium, or a mixture of S. Typhimurium and E. coli Nissle wild-type or tonB mutant. Scores of individual mice (circles) and geometric means for each group (bars) are indicated. (B) H&E stained sections from representative animals for each group. E. coli Nissle wild-type or tonB co-administration with S. Typhimurium reduced the density of inflammatory infiltrates (black asterisks) and the degree of crypt injury (white asterisks), compared to S. Typhimurium infection alone. (C) Transcript levels of Lcn2, Cxcl-1 and Il-17a were determined in the ceca of C57BL/6 mice infected with S. Typhimurium (white bars), a mixture of S. Typhimurium and wild-type E. coli Nissle (black bars), or a mixture of S. Typhimurium and E. coli Nissle tonB (gray bars). Data are expressed as fold-increase over mock-infected mice. Bars represent the geometric means ± standard deviation. (D) Blinded histopathology scores of cecal samples 4 days after infection of mice administered E. coli Nissle iroN fuyA iutA chuA (Δ4), S. Typhimurium, or a mixture of S. Typhimurium and E. coli Nissle Δ4. Scores of individual mice (circles) and geometric means for each group (bars) are indicated. (E) H&E stained sections from representative animals from each group. E. coli Nissle Δ4 co-administration with S. Typhimurium greatly reduced chronic inflammatory infiltrates (black asterisks) and the degree of crypt injury (white asterisks), compared to S. Typhimurium infection alone. (F) Transcript levels of Lcn2, Cxcl-1 and Il-17a were determined in the ceca of mice infected with S. Typhimurium (white bars) or a mixture of S. Typhimurium and E. coli Nissle Δ4 (gray bars). Data are expressed as fold-increase over mock-infected mice. Bars represent geometric means ± standard deviation. STM=S. Typhimurium; EcN=E. coli Nissle. L; lumen, M, mucosa, SM; submucosa. Significant difference is indicated by * (P value ≤ 0.05) or ** (P value ≤ 0.01). (See also Figure S4 and Table S3).
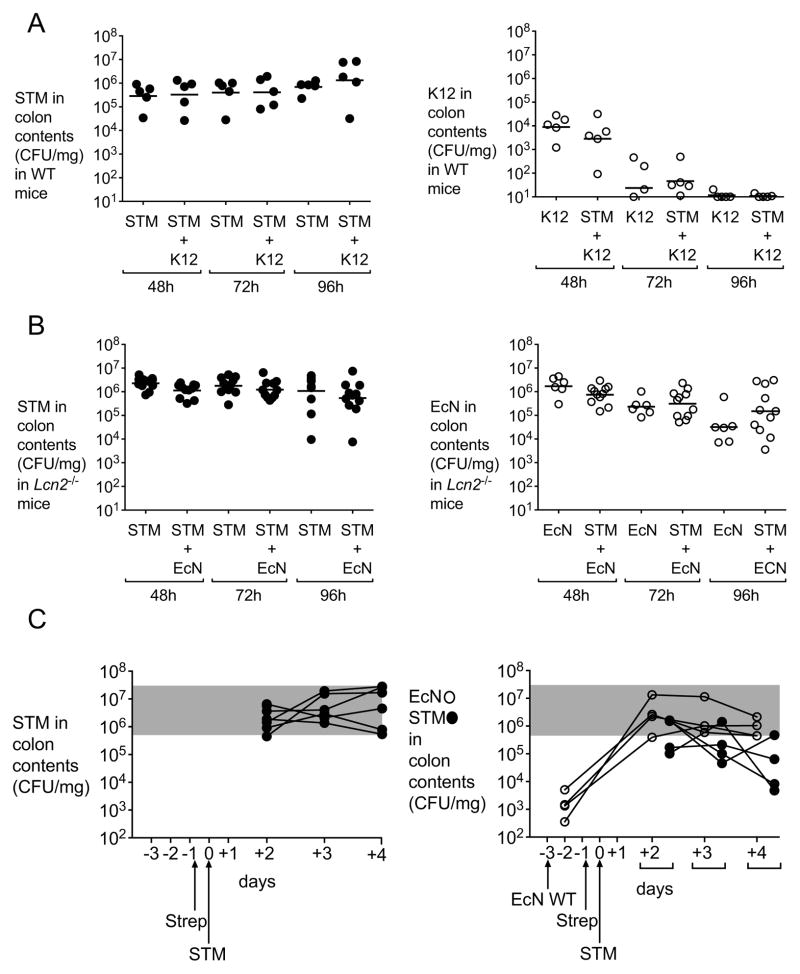
The host inflammatory response limits iron availability to invading microbes in what is known as nutritional immunity (Cassat and Skaar, 2013). One arm of the nutritional immune response during infection is the upregulation of lipocalin-2, which, as described earlier, sequesters iron-bound enterochelin. We previously demonstrated that the ability to acquire iron through salmochelin provides a colonization advantage to S. Typhimurium wild-type in competition with an iroN mutant in mice that express lipocalin-2, but not in Lcn2−/− mice, likely because enterochelin alone is sufficient to overcome other mechanisms of ferric iron restriction in the host (Raffatellu et al., 2009). Consistent with this notion, intestinal S. Typhimurium colonization in wild-type mice was not affected by administration of a non-probiotic commensal E. coli strain (Fig. 6A) that relies on enterochelin for ferric iron acquisition and is thus susceptible to lipocalin-2 (Berger et al., 2006; Flo et al., 2004). We therefore hypothesized that E. coli Nissle would lose its competitive advantage over S. Typhimurium in the absence of lipocalin-2. As predicted, intestinal S. Typhimurium colonization in Lcn2−/− mice was not decreased by the administration of E. coli Nissle, despite successful colonization of the mouse intestine by this probiotic strain (Fig. 6B). As we previously showed, wild-type and Lcn2−/− mice infected with S. Typhimurium did not show significant differences in S. Typhimurium colonization (Fig. 4, S3, and 6B) or inflammation (Fig. S4 and S5). Furthermore, the expression of pro-inflammatory cytokines was similar between wild-type mice and Lcn2−/− mice, with the exception of Cxcl-1, which was reduced in Lcn2−/− mice, as previously shown for Cxcl-8 (Bachman et al., 2009). Nonetheless, reduction in the expression of Cxcl-1 did not result in differences in the neutrophil influx observed by the pathology, likely because of redundant mechanisms of neutrophil migration to the gut (light blue bars, Fig. S4 and S5). Of note, administration of E. coli Nissle to Lcn2−/− mice infected with S. Typhimurium also ameliorated intestinal inflammation, despite the minimal differences observed in the expression of pro-inflammatory cytokines. These results may indicate that lipocalin-2 has additional immunomodulatory effects that are independent of its iron acquisition mechanisms(Bachman et al., 2009).
Figure 6. E. coli Nissle 1917 reduction of S. Typhimurium intestinal colonization requires functional lipocalin-2 and is independent of the time of administration.
(A) C57BL/6 mice were infected with S. Typhimurium alone or co-administered with commensal E. coli K-12. CFU in colonic contents were enumerated at 48h, 72h and 96h post-infection. STM (black circles) and E. coli K-12 (white circles) counts are shown. (B) C57BL/6 Lcn2−/− mice were infected with S. Typhimurium alone or co-administered with wild-type E. coli Nissle. CFU in colonic contents were enumerated at 48h, 72h and 96h post-infection. S. Typhimurium (black circles) and E. coli Nissle (white circles) counts are shown. (C) C57BL/6 mice were administered a single dose of E. coli Nissle wild-type or mock, three days before being infected with S. Typhimurium. Streptomycin treatment was performed one day prior to S. Typhimurium infection. CFU in colonic contents were enumerated at 2, 3 and 4 days post-infection. S. Typhimurium (black circles) and E. coli Nissle (white circles) counts are shown. K12=E. coli K12; STM=S. Typhimurium; EcN=E. coli Nissle. (See also Figure S5).
While we had administered E. coli Nissle as a therapeutic either during or after S. Typhimurium administration, this strain was originally isolated as a normal commensal of the gut flora from a healthy soldier who did not acquire Shigella during an outbreak. To test whether colonization of mice with E. coli Nissle would confer protection to infection, C57BL/6 mice were administered E. coli Nissle three days prior to S. Typhimurium infection. As shown in Fig. 6C, mice that were pre-colonized with E. coli Nissle showed a significant reduction in colonization from S. Typhimurium, indicating that colonization with this probiotic offers at least partial protection to infection with a gastrointestinal pathogen.
Next, we determined the ratio of the S. Typhimurium colony forming units detected in the colonic content of C57BL/6 mice when S. Typhimurium was administered alone versus in competition with E. coli Nissle. Overall, there was a marked reduction (up to 445-fold) in S. Typhimurium fecal colonization in wild-type mice that were co-administered wild-type E. coli Nissle at the time of infection relative to mice that were infected with S. Typhimurium only, a difference which was not observed in either Lcn2−/− mice or wild-type mice that were administered either the E. coli Nissle tonB or the E. coli Nissle Δ4 mutants (Fig. 7A, C, E). Furthermore, only wild-type E. coli Nissle co-administered with S. Typhimurium to wild-type mice was able to outcompete S. Typhimurium by up to 195-fold (Fig. 7B, D, F). Taken together, our results show that iron acquisition in the inflamed gut is a critical mechanism for the ability of the probiotic E. coli Nissle to limit Salmonella intestinal colonization.
Discussion
E. coli Nissle was isolated from a healthy soldier during a Shigella outbreak in 1917 under the hypothesis that a protective commensal strain must have colonized the gut of that soldier (Nissle, 1959). In the years since, E. coli Nissle has had a long history of use in clinical settings to treat gastrointestinal disorders, though the molecular mechanism of its beneficial activity is not well understood (Schultz, 2008), much like all natural probiotics (reviewed in (Balakrishnan and Floch, 2012)).
Probiotics may exert their beneficial effects by either direct interaction with the host, or by competition with pathogenic species (mechanisms frequently grouped under the term “colonization resistance”; reviewed in (Lawley and Walker, 2013)). A large focus of research on probiotics has been on their interactions with the immune system rather than their competition with other microbes. Regarding E. coli Nissle, several studies have proposed that selective immune modulation may contribute to its activity(reviewed in (Behnsen et al., 2013; Jacobi and Malfertheiner, 2011), including the activation of γδ T cells (Guzy et al., 2008), the reduction of the secretion of pro-inflammatory cytokines in the mucosa (Grabig et al., 2006), the enhanced secretion of IgA and IgM (Cukrowska et al., 2002), and the production of tight junction proteins (Ukena et al., 2007) and human beta-defensin-2 (Wehkamp et al., 2004). While E. coli Nissle has been proposed to have both pro- and anti-inflammatory effects, the net result of these alterations is an overall enhancement of the mucosal barrier. Consistent with these findings, as well as with other studies that have shown E. coli Nissle to ameliorate the disease caused by an infection (Hockertz, 1997; Splichalova et al., 2011), we also observed a reduction in both intestinal pathology and expression of pro-inflammatory cytokines when S. Typhimurium-infected mice were administered E. coli Nissle in both a chronic and an acute model of infection, as well as when we induced inflammation independent of infection.
The two mouse models that we employed (129X1/SvJ for the chronic model; C57BL/6 for the acute model) develop intestinal inflammation when infected with S. Typhimurium after streptomycin treatment. However, the host response to S. Typhimurium is not identical in these mice because of genetic differences as well as differences in the composition of the microbiota. At present, the best studied genetic distinction is that 129X1/SvJ have a functional Nramp1 (Slc11a1) allele, which renders the mice more resistant to infection with S. Typhimurium. However, functional Nramp1 alone does not explain differences in the host response to S. Typhimurium (Brown et al., 2013), and altered expression or function of other genes (for instance, caspase-11) may also play a role (Kayagaki et al., 2011). These variances may explain why in the chronic model of infection only wild-type E. coli Nissle mediated a reduction of inflammation, while in the acute model of infection the reduction was independent of iron acquisition. Nevertheless, our study indicates that the mild-to-moderate reduction in intestinal inflammation only partly explains the beneficial effects of administering E. coli Nissle during infection.
Intestinal inflammation of infectious and non-infectious origins results in an alteration of the normal flora and a significant microbial dysbiosis, including the loss of Bacteroidetes and Firmicutes and the proliferation of Enterobacteriaceae (reviewed in (Fava and Danese, 2011; Mukhopadhya et al., 2012)). A mechanism for the proliferation of Enterobacteriaceae in the inflamed gut was recently provided by Winter and colleagues, who showed that this family can utilize host-derived nitrate for respiration (Winter et al., 2013). The fact that E. coli Nissle also benefits from inflammation could also explain why its anti-inflammatory effects are mild to moderate: a complete reduction of inflammation would be detrimental to its own colonization, as indicated in our mice that were pre-colonized with E. coli Nissle prior to S. Typhimurium infection. A change in the composition of the normal flora, with loss of Bacteroidetes and Firmicutes, is also observed during infection caused by S. Typhimurium as this pathogen has been shown to thrive during inflammation and successfully compete with the microbiota (reviewed in (Thiennimitr et al., 2012)). While the host inflammatory response limits the availability of essential nutrients, including metal ions, to invading microbes in what is known as nutritional immunity, pathogens including Salmonella have evolved many mechanisms to evade this response and acquire essential metals necessary to mount a successful infection(Thiennimitr et al., 2012).
Our previous studies indicated that iron uptake via salmochelin, which confers resistance to iron sequestration mediated by lipocalin-2, is essential for efficient colonization of S. Typhimurium (Crouch et al., 2008; Raffatellu et al., 2009). As evasion of lipocalin-2-mediated iron withholding has also been found to be essential to the virulence of many other Gram-negative enteric pathogens (Bachman et al., 2009; Caza et al., 2008; Garcia et al., 2011; Himpsl et al., 2010; Payne et al., 2006), this mechanism has come to be viewed as a virulence trait. In contrast to this trend, our research demonstrates that lipocalin-2-resistant iron acquisition is not a property unique to virulence as it is essential for the probiotic activity of E. coli Nissle.
Deletion of up to three iron receptors did not result in a growth defect in media supplemented with lipocalin-2 (data not shown), indicating that E. coli Nissle possesses redundant lipocalin-2-resistant iron uptake systems. Integrating this finding with our observation that E. coli Nissle outcompetes S. Typhimurium in vivo along with our previous finding that S. Typhimurium benefits from acquiring iron in a lipocalin-2 resistant fashion during inflammation, we hypothesized that Nissle’s multiple iron uptake systems provide a competitive advantage against S. Typhimurium when the intestine is inflamed and, as we observed, fecal iron is significantly reduced. Consistent with our hypothesis, our results show that E. coli Nissle’s ability to displace the highly evolved pathogen from its intestinal niche is dependent on iron acquisition. Furthermore, E. coli Nissle was only able to reduce the colonization of S. Typhimurium when lipocalin-2 was expressed. While lipocalin-2 is one of the host defenses exploited by S. Typhimurium to colonize the inflamed gut and compete with the microbiota, E. coli Nissle also subverts this host defense mechanism to thrive in the same inflamed and iron-starved environment. By scavenging for iron more effectively than S. Typhimurium, both with its own armament of siderophores and with its ability to compete with S. Typhimurium for uptake of salmochelin, E. coli Nissle tips the scales back in favor of the host, effectively augmenting the host’s innate immune response by acting as a surrogate of sorts for lipocalin-2. It is along these lines that we propose that E. coli Nissle – and possibly other beneficial components of the microbiota – may provide colonization resistance in part by boosting the host’s nutritional immunity, sequestering nutrients from pathogens when the host fails to do so.
Taken together, our results show that iron acquisition in the inflamed gut is a critical mechanism for the ability of the probiotic E. coli Nissle to limit Salmonella intestinal colonization. Furthermore, we have demonstrated that this action of E. coli Nissle results from its resistance to lipocalin-2, previously considered a mechanism of virulence but now also seen as an essential property of a protective commensal organism. As antibiotic treatment is contraindicated for uncomplicated Salmonella infections due to prolongation of fecal shedding, the administration of E. coli Nissle may be a feasible alternative to diminish Salmonella colonization and ameliorate symptoms. As microbial dysbiosis is apparent in a variety of intestinal disorders (DuPont and DuPont, 2011), iron acquisition may also contribute to other probiotic actions attributed to E. coli Nissle. The ability of E. coli Nissle to withstand inflammation and outcompete a highly evolved pathogen for an essential micronutrient may be seen as a paradigm for understanding the protective actions of commensal microorganisms and a foundation upon which to build future probiotics tailored to the treatment of different diseases.
Experimental Procedures
Bacterial Strains and Culture Conditions
All strains used in this study are listed in Table S1. S. Typhimurium strain IR715 is a fully virulent, nalidixic acid resistant derivative of wild-type isolate ATCC 14028. Escherichia coli Nissle 1917 wild-type is a non-pathogenic human E. coli isolate that we obtained from Ulrich Sonnenborn, Ardeypharm. An IR715 derivative carrying a mutation in iron, and E. coli Nissle derivatives carrying mutations in tonB or iroN, iutA, fyuA, and chuA were used for this study. Mutant construction is described in the supporting information and the plasmids and primers used are detailed in Tables S1 and S2. For animal infections, all strains were grown in Miller Luria-Bertani (LB) media at 37°C with aeration overnight.
In vitro growth assays
S. Typhimurium and E. coli Nissle strains were tested for the ability to grow in iron limiting conditions (Nutrient Broth supplemented with 0.2 mM 2,2-Dipyridyl, Sigma) at 37°C with aeration overnight. To test lipocalin-2 sensitivity, approximately 103 CFU from an overnight culture were inoculated into tissue culture medium comprising DMEM/F12 (Invitrogen) plus 10% fetal bovine serum (FBS, Invitrogen) or in the same medium containing human lipocalin-2 (1μg/ml, R&D Systems) as previously published (Raffatellu et al., 2009). To compare general growth rates, a 1:1 mixture of E. coli Nissle wild-type and either the tonB or the Δ4 mutants containing 1×107 CFU was inoculated in M9 minimal media at 37°C with aeration. When indicated, iron (III) citrate and iron (III) sulfate were added at a final concentration of 1mM and 200 μM, respectively. CFU were enumerated by plating serial dilutions at 2h, 5h and 8h after inoculation.
Animal infections
For acute infections, female C57BL/6 (Taconic) and lipocalin-2-deficient (Lcn2−/−) mice were orally gavaged with a dose of 109 CFU in 100μl of phosphate-buffered saline (PBS) 24 hours after pre-treatment with streptomycin (100μl of a 200 mg/ml solution in sterile water) (Barthel et al., 2003; Raffatellu et al., 2009). Mice were infected with either S. Typhimurium alone or with a 1:1 mixture of strains as indicated. Fecal pellets were collected at 48 and 72 hours post-infection and weighed for CFU determination, homogenized in 1 ml of sterile PBS, and serial dilutions were plated on LB agar containing appropriate antibiotics. At 96 hours post-infection, mice were euthanized and the cecum was collected for isolation of mRNA and for histopathology; the liver was collected as indicated to measure hepatic gene expression. Bacteria were enumerated in the colon content on agar plates containing the appropriate antibiotics. To render all strains equally resistant to streptomycin, either pACYomega or pHP45omega (Table S1) were introduced by electroporation. When noted, the competitive indices were calculated by dividing the output ratio (E. coli Nissle CFU / S. Typhimurium CFU) by the input ratio (E. coli Nissle CFU / S. Typhimurium CFU). In some groups of mice, a single dose of 109 CFU of E. coli Nissle was administered three days prior to S. Typhimurium infection, as indicated. In other groups, colitis was induced by administration of dextran sodium sulfate (Wirtz et al., 2007); some of these mice were administered a single dose of 109 CFU E. coli Nissle the same day DSS treatment was started (day 1). DSS-treated mice were sacrificed at day 6 and the cecum was harvested for RNA purification and analysis.
For chronic infections, female 129X1/SvJ mice (Jackson Laboratories, Bar Harbor, ME) were orally gavaged as described above with S. Typhimurium 24 hours after pre-treatment with streptomycin (Lawley et al., 2008). 72 hours post-infection, groups of mice were administrated a single dose of 109 CFU E. coli Nissle wild-type or E. coli tonB mutant in 100 μl of LB by oral gavage. Individual mice were followed for the duration of the experiment (up to 22 days). Fecal pellets and colon contents were collected and processed as described above.
Quantitative Real-Time PCR
For analysis of gene expression by quantitative real-time PCR, total RNA was extracted from cecal and hepatic tissues with TRI Reagent (Molecular Research Center; Cincinnati, OH). For DSS-treated mice, oligo(dT) purification of mRNA was performed using the Dynabeads mRNA Purification kit (Invitrogen). Real-time PCR was performed using SYBR Green (Roche, Indianapolis, IN) and the Roche Lightcycler 480 system (Roche, Indianapolis, IN). Data were analyzed using the comparative ΔΔ-Ct method. Target gene transcription of each sample was normalized to the respective levels of mRNA β-actin. A list of the real-time primers used in this study is provided in Table S3.
Histopathology
Tissue samples were fixed in formalin, processed according to standard procedures for paraffin embedding, sectioned at 5 μm, and stained with hematoxylin and eosin. Blinded examination by a board-certified pathologist was used to score the pathology of cecal samples using previously published methods (Barthel et al., 2003; Raffatellu et al., 2009). Each section was evaluated for the presence of neutrophils, mononuclear infiltrate, submucosal edema, surface erosions, inflammatory exudates, and cryptitis. Inflammatory changes were scored from 0 to 4 according to the following scale: 0 = none; 1 = low; 2 = moderate; 3 = high; 4 = extreme. The inflammation score for each mouse was calculated by adding the score for each parameter and was interpreted as follows: 0–2 = within normal limit; 3–5 = mild; 6–8 = moderate; 8+ = severe.
Measurement of Iron in Fecal Samples by ICP-MS
The amount of iron in mouse fecal samples was measured by ICP-MS as described previously (Corbin et al., 2008; Liu et al., 2012) and is detailed in the Supplemental Experimental Procedures.
Statistical analysis
Differences between treatment groups were analyzed by ANOVA followed by Student’s t test. A P value equal to or below 0.05 was considered statistically significant.
Supplementary Material
Highlights.
Probiotic E. coli Nissle reduces Salmonella intestinal colonization and persistence
E. coli Nissle outcompetes Salmonella for iron in the inflamed gut
Specialized iron transporters are essential for E. coli Nissle probiotic activity
E. coli Nissle overcomes lipocalin-2-mediated iron sequestration
Acknowledgments
We would like to acknowledge Ulrich Sonnenborn and Ardeypharm for the gift of the E. coli Nissle wild-type strain, Sean-Paul Nuccio for help with editing the manuscript, Takeshi Haneda and Andreas Bäumler for the gift of plasmids, Eric Skaar and Russell Gerards for their help with ICP-MS, Alfredo Torres and Harry Mobley for their help with mutant construction, Elizabeth Nolan for helpful discussions, and Kelly Smith for the original gift of Lcn2−/− mice. This work was supported by Public Health Service Grants AI083663 (to M.R.) and AI77629 (to F.C.F.), and an IDSA ERF/NIFID Astellas Young Investigator Award to M.R.. J.Z.L. was supported by the NIH Immunology Research Training Grant T32 AI60573 and then by a predoctoral fellowship from the American Heart Association.
Footnotes
Supplemental information includes five figures, three tables, Supplemental Experimental Procedures, and Supplemental References.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews NC, Schmidt PJ. Iron homeostasis. Annu Rev Physiol. 2007;69:69–85. doi: 10.1146/annurev.physiol.69.031905.164337. [DOI] [PubMed] [Google Scholar]
- Bachman MA, Miller VL, Weiser JN. Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog. 2009;5:e1000622. doi: 10.1371/journal.ppat.1000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balakrishnan M, Floch MH. Prebiotics, probiotics and digestive health. Curr Opin Clin Nutr Metab Care. 2012;15:580–585. doi: 10.1097/MCO.0b013e328359684f. [DOI] [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infect Immun. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnsen J, Deriu E, Sassone-Corsi M, Raffatellu M. Probiotics: properties, examples, and specific applications. Cold Spring Harb Perspect Med. 2013;3:a010074. doi: 10.1101/cshperspect.a010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, Fong HE, Cheung CC, Mak TW. Lipocalin 2-deficient mice exhibit increased sensitivity to Escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci U S A. 2006;103:1834–1839. doi: 10.1073/pnas.0510847103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Hantke K. Recent insights into iron import by bacteria. Curr Opin Chem Biol. 2011;15:328–334. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- Brown DE, Libby SJ, Moreland SM, McCoy MW, Brabb T, Stepanek A, Fang FC, Detweiler CS. Salmonella enterica Causes More Severe Inflammatory Disease in C57/BL6 Nramp1G169 Mice Than Sv129S6 Mice. Vet Pathol. 2013 doi: 10.1177/0300985813478213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caza M, Lepine F, Milot S, Dozois CM. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect Immun. 2008;76:3539–3549. doi: 10.1128/IAI.00455-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Seeley EH, Raab A, Feldmann J, Miller MR, Torres VJ, Anderson KL, Dattilo BM, Dunman PM, Gerads R, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. [DOI] [PubMed] [Google Scholar]
- Crouch ML, Castor M, Karlinsey JE, Kalhorn T, Fang FC. Biosynthesis and IroC-dependent export of the siderophore salmochelin are essential for virulence of Salmonella enterica serovar Typhimurium. Molecular microbiology. 2008;67:971–983. doi: 10.1111/j.1365-2958.2007.06089.x. [DOI] [PubMed] [Google Scholar]
- Cukrowska B, LodInova-ZadnIkova R, Enders C, Sonnenborn U, Schulze J, Tlaskalova-Hogenova H. Specific proliferative and antibody responses of premature infants to intestinal colonization with nonpathogenic probiotic E. coli strain Nissle 1917. Scand J Immunol. 2002;55:204–209. doi: 10.1046/j.1365-3083.2002.01005.x. [DOI] [PubMed] [Google Scholar]
- DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8:523–531. doi: 10.1038/nrgastro.2011.133. [DOI] [PubMed] [Google Scholar]
- Fava F, Danese S. Intestinal microbiota in inflammatory bowel disease: friend of foe? World J Gastroenterol. 2011;17:557–566. doi: 10.3748/wjg.v17.i5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nature chemical biology. 2006a;2:132–138. doi: 10.1038/nchembio771. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Lin H, Zhou L, Yu Y, Abergel RJ, Liu DR, Raymond KN, Wanner BL, Strong RK, Walsh CT, et al. The pathogen-associated iroA gene cluster mediates bacterial evasion of lipocalin 2. Proceedings of the National Academy of Sciences of the United States of America. 2006b;103:16502–16507. doi: 10.1073/pnas.0604636103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, Akira S, Aderem A. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. [DOI] [PubMed] [Google Scholar]
- Ganz T. Hepcidin, a key regulator of iron metabolism and mediator of anemia of inflammation. Blood. 2003;102:783–788. doi: 10.1182/blood-2003-03-0672. [DOI] [PubMed] [Google Scholar]
- Garcia EC, Brumbaugh AR, Mobley HL. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect Immun. 2011;79:1225–1235. doi: 10.1128/IAI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabig A, Paclik D, Guzy C, Dankof A, Baumgart DC, Erckenbrecht J, Raupach B, Sonnenborn U, Eckert J, Schumann RR, et al. Escherichia coli strain Nissle 1917 ameliorates experimental colitis via toll-like receptor 2- and toll-like receptor 4-dependent pathways. Infection and immunity. 2006;74:4075–4082. doi: 10.1128/IAI.01449-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. Journal of bacteriology. 2004;186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzy C, Paclik D, Schirbel A, Sonnenborn U, Wiedenmann B, Sturm A. The probiotic Escherichia coli strain Nissle 1917 induces gammadelta T cell apoptosis via caspase- and FasL-dependent pathways. Int Immunol. 2008;20:829–840. doi: 10.1093/intimm/dxn041. [DOI] [PubMed] [Google Scholar]
- Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proc Natl Acad Sci U S A. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henker J, Laass M, Blokhin BM, Bolbot YK, Maydannik VG, Elze M, Wolff C, Schulze J. The probiotic Escherichia coli strain Nissle 1917 (EcN) stops acute diarrhoea in infants and toddlers. European journal of pediatrics. 2007;166:311–318. doi: 10.1007/s00431-007-0419-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himpsl SD, Pearson MM, Arewang CJ, Nusca TD, Sherman DH, Mobley HL. Proteobactin and a yersiniabactin-related siderophore mediate iron acquisition in Proteus mirabilis. Mol Microbiol. 2010;78:138–157. doi: 10.1111/j.1365-2958.2010.07317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockertz S. Augmentation of host defence against bacterial and fungal infections of mice pretreated with the non-pathogenic Escherichia coli strain Nissle 1917. Arzneimittelforschung. 1997;47:793–796. [PubMed] [Google Scholar]
- Jacobi CA, Malfertheiner P. Escherichia coli Nissle 1917 (Mutaflor): new insights into an old probiotic bacterium. Dig Dis. 2011;29:600–607. doi: 10.1159/000333307. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Kruis W, Fric P, Pokrotnieks J, Lukas M, Fixa B, Kascak M, Kamm MA, Weismueller J, Beglinger C, Stolte M, et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut. 2004;53:1617–1623. doi: 10.1136/gut.2003.037747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infect Immun. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodinova-Zadnikova R, Sonnenborn U. Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biology of the neonate. 1997;71:224–232. doi: 10.1159/000244421. [DOI] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Mollenbrink M, Bruckschen E. Treatment of chronic constipation with physiologic Escherichia coli bacteria. Results of a clinical study of the effectiveness and tolerance of microbiological therapy with the E. coli Nissle 1917 strain (Mutaflor) Med Klin (Munich) 1994;89:587–593. [PubMed] [Google Scholar]
- Mukhopadhya I, Hansen R, El-Omar EM, Hold GL. IBD-what role do Proteobacteria play? Nat Rev Gastroenterol Hepatol. 2012;9:219–230. doi: 10.1038/nrgastro.2012.14. [DOI] [PubMed] [Google Scholar]
- Muller SI, Valdebenito M, Hantke K. Salmochelin, the long-overlooked catecholate siderophore of Salmonella. Biometals : an international journal on the role of metal ions in biology, biochemistry, and medicine. 2009;22:691–695. doi: 10.1007/s10534-009-9217-4. [DOI] [PubMed] [Google Scholar]
- Nissle A. Explanations of the significance of colonic dysbacteria & the mechanism of action of E. coli therapy (mutaflor) Medizinische. 1959;4:1017–1022. [PubMed] [Google Scholar]
- Payne SM, Wyckoff EE, Murphy ER, Oglesby AG, Boulette ML, Davies NM. Iron and pathogenesis of Shigella: iron acquisition in the intracellular environment. Biometals. 2006;19:173–180. doi: 10.1007/s10534-005-4577-x. [DOI] [PubMed] [Google Scholar]
- Pressler U, Staudenmaier H, Zimmermann L, Braun V. Genetics of the iron dicitrate transport system of Escherichia coli. J Bacteriol. 1988;170:2716–2724. doi: 10.1128/jb.170.6.2716-2724.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell host & microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond KN, Dertz EA, Kim SS. Enterobactin: an archetype for microbial iron transport. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1012–1018. doi: 10.1002/ibd.20377. [DOI] [PubMed] [Google Scholar]
- Splichalova A, Trebichavsky I, Rada V, Vlkova E, Sonnenborn U, Splichal I. Interference of Bifidobacterium choerinum or Escherichia coli Nissle 1917 with Salmonella Typhimurium in gnotobiotic piglets correlates with cytokine patterns in blood and intestine. Clin Exp Immunol. 2011;163:242–249. doi: 10.1111/j.1365-2249.2010.04283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS biology. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Bäumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012;15:108–114. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Bäumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, Bleich A, Bruder D, Franzke A, Rogler G, et al. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One. 2007;2:e1308. doi: 10.1371/journal.pone.0001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J, Harder J, Wehkamp K, Wehkamp-von Meissner B, Schlee M, Enders C, Sonnenborn U, Nuding S, Bengmark S, Fellermann K, et al. NF-kappaB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infection and immunity. 2004;72:5750–5758. doi: 10.1128/IAI.72.10.5750-5758.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ED. Iron withholding: a defense against infection and neoplasia. Physiol Rev. 1984;64:65–102. doi: 10.1152/physrev.1984.64.1.65. [DOI] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2:541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.