Abstract
The tight junction is a multi-protein complex and is the apical most junctional complex in certain epithelial and endothelial cells. A great deal of attention has been devoted to the understanding of these proteins in contributing to the barrier function - that is, regulating the paracellular flux or permeability between adjacent cells. However, tight junction proteins are now recognized as having functions beyond the barrier. The focus of this review is to discuss the barrier function of the tight junction and to summarize the literature with a focus on the role of tight junction proteins in proliferation, transformation, and metastasis.
Keywords: occludin, claudin, tight junction, cancer, metastasis
1. Introduction
The tight junction (TJ) complex is the apical most junctional complex in many types of epithelial and endothelial cells. The TJ can be sub-divided into the integral membrane and cytoplasmic proteins. Occludin, tricellulin, marvelD3, and the claudins (of which there are 27 members [1]) are tetra-spanning membrane proteins whose N- and C-termini reside in the cytosol and each possesses two extracellular loop regions. Occludin, tricellulin, and marvelD3 each contain a MARVEL (MAL-related proteins for vesicle trafficking and membrane link) domain, whereas the claudins do not. The junctional adhesion molecules (JAMs) are single pass membrane proteins with two IgG-like motifs. The cytoplasmic adaptor proteins are the zonula occludens or ZO proteins, and are designated ZO-1, -2, and -3. These proteins link the membrane proteins to the actin cytoskeleton. Collectively, the TJ imparts two functions in the cell: a barrier function, namely regulating the permeability of solutes between adjacent cells, and a fence function, controlling the lateral diffusion of proteins within the lipid bilayer [2, 3]. Traditionally, research efforts focused on the barrier and fence functions; however, there is a new movement in the field, which is to understand how TJ proteins participate in cell proliferation, transformation, and metastasis suppression. The focus of this review shall be to briefly orient the reader with TJ proteins by describing their traditional roles followed by a summary of the aforementioned novel functions of this class of proteins. In addition to the aspects of TJ proteins to be discussed in this review, these proteins are also key components to cell signaling events; for a review of the role of TJ proteins with regards to signaling and gene expression, please see Balda and Matter [4].
2. Traditional functions of tight junction proteins
2.1. ZO proteins
ZO-1 was the first TJ protein described and ZO-2 and -3 were subsequently identified by co-immunoprecipitation studies [5–9]. ZO proteins are classified as members of the membrane associated guanylate kinase (MAGuK) family and are composed of three postsynaptic density 95/disc-large/zona occludens (PDZ) domains and one Src homology (SH3) and GuK domain [10]. Via fusion of biotin ligate to either the N- or C-terminus of ZO-1, it was found that the ends of the ZO-1 protein are embedded in different functional sub-compartments of the TJ [11]. The manner in which the ZO proteins interact with the membrane proteins appears to be specific to each type of membrane protein. The unique-5 (U5) region of ZO-1, located between the SH3 and GuK domains, is responsible for ZO-1 localization to the TJ and for the interaction with the distal C-terminus of occludin [12–14]. With the exception of claudin-12, the conserved C-terminal YV motif of claudins interact with the PDZ domains of ZO proteins through a conserved C-terminal YV motif [15]. Moreover, the C-terminus of JAMs contains a PDZ domain-binding motif, which interacts with ZO-1 [16]. ZO-1 and ZO-2 are critical to junction assembly [17, 18] and permeability [19], respectively, and in the absence of ZO-1 and -2, cells fail to form TJs [20]. Both ZO-1 and -2 knockouts are embryonic lethal in mice due to apoptosis and reduced yolk sac angiogenesis and proliferation [21, 22].
2.2. Occludin
Occludin was the first transmembrane TJ protein discovered [23] and its function in the TJ remains to be completely understood. Occludin overexpression increases electrical resistance, implying a pro-barrier phenotype; yet, occludin overexpression increases permeability to small molecule tracers [24]. While the occludin-null mouse forms intact TJs, the mice exhibited a variety of abnormal phenotypes including postnatal growth retardation, thinning of compact bone, calcification in the brain, testicular atrophy, male infertility, loss of cytoplasmic granuoles in salivary epithelial cells, females not suckling their young, and gastric inflammation and hyperplasia [25]. Silencing of occludin in vitro increases permeability to divalent organic cations and also to small molecules under hydrostatic pressure [26, 27]. Furthermore, in the microvascular endothelial cells of the retina, occludin regulates vascular endothelial growth factor (VEGF)-induced permeability through its phosphorylation and subsequent ubiquitination in vitro and in vivo [28, 29]. Shen and Turner demonstrated a clear network between the actin cytoskeleton and the TJ as actin depolymerization results in a loss of barrier function mediated through caveolae-dependent occludin internalization [30]. Furthermore, caveolin-1-dependent occludin endocytosis is necessary for the tumor necrosis factor-1 mediated loss of barrier function [31]. Clearly, occludin is a dynamic protein at the TJ.
2.3. Claudins
The claudin family is regarded as the backbone of the TJ [32]. Interestingly, multiple claudin family members are able to co-exist in the same tight junction strand while other combinations of claudins fail to do so [33]. Claudins interact with other claudins in the same cell through their N-terminal extracellular loops (cis-interactions) while claudins interact with claudins in adjacent cells through their C-terminal extracellular loops (trans-interactions) [34]. This cis- and trans-interaction leads to the formation of a “zipper”-like structure, thus describing the claudin-driven barrier.
Support for claudins as the major drivers of TJ formation is derived largely in part from the fact that the occludin null mouse is viable and genetic ablation of claudin proteins results in deleterious barrier-specific phenotypes. Deletion of claudin-1 compromises the epidermal barrier and is lethal within one day due to excessive water loss [35]. The claudin-5 knockout mouse has severe brain hemorrhaging and dies within 10 hours after birth [36]. The claudin-11 knockout is viable; however, the mouse has hind limb weakness, slowed conductive velocities of the central nervous system, and male sterility [37]. In humans, mutations in claudins-16 and 19 are associated with hypomagnesemia [38] and renal magnesium wasting [39], respectively. Analysis of claudin-11 [40] and -14 [41] knockout mice revealed that these proteins are indispensable for maintenance of endocochlear potential and cochlear hair cells, respectively, and loss of either leads to deafness. The claudin-15 knockout will be discussed in the following section. Undeniably, claudins are essential to the formation of the TJ and proper TJ function is of the upmost importance.
2.4. Junctional adhesion molecules (JAMs)
The JAMs (~40kDa) are a group of proteins that are subdivided as JAM-1 (or A), -2 (or B), and -3 (or C). JAM-A is a mediator of barrier formation [42, 43] and function [44, 45]. JAM-A is also crucial to polarity [46], potentially through interactions with the polarity protein PAR-3 [47, 48]. Further, Laukoetter et al. found that JAM-A knockout mice exhibit increased polymorphonuclear leukocyte infiltration and, consistent with the in vitro study, increased mucosal permeability [49].
2.5. Tricellulin
Tricellulin is concentrated at regions where three cells form a contact or at the tricellular TJ (tTJ), thus the name tricellulin. Silencing tricellulin disrupts the tTJ and reduces barrier integrity to small molecule tracers [50]. In humans with nonsyndromic deafness, there is an association between hearing loss and four recessive mutations at splice sites of the tricellulin gene [51, 52]. Furthermore, loss of occludin shifts tricellulin from the tTJ to the bicellular TJ (bTJ) to compensate for the loss of occludin at bTJ. Thus, these proteins collectively support the epithelial barrier at bi- and tricellular points [53]. Importantly, tricellulin integrates into claudin-based TJs independent of binding with ZO-1 [53], although tricellulin can interact with ZO-1 [51]. Finally, when localized at TJs, tricellulin expression increases electrical resistance values and decreases permeability; while when expressed exclusively at tTJs, tricellulin decreases solute permeability to macromolecules but not ions [54].
2.6. MarvelD3
Recently, the third member of the MARVEL containing proteins, MarvelD3, was described. To date, very little is known about marvelD3. Two independent studies using RNAi demonstrate phenotypes for marvelD3. Steed and colleagues show that silencing of marvelD3 does not affect TJs as assessed by immunofluorescence of occludin and ZO-1 nor does it affect the kinetics of TJ assembly as measured by Ca2+ switch assay. However, in the Ca2+ switch assay and under normal Ca2+ conditions, marvelD3 silencing eventually resulted in higher resistance readings [55]. Conversely, Raleigh and colleagues found that silencing of marvelD3 delays the assembly of TJs [56]. In a recent genome-wide association study, an intergenic single nucleotide polymorphism near the MARVELD3 gene was linked to resistance to severe malaria [57]. The interactions between marvelD3 with occludin and tricellulin is influenced by claudins and these interactions further modulate the function of claudins [58].
3. Tight junctions and proliferation
3.1. ZO proteins
Matter and Balda initially conceived the notion that TJ proteins could participate in cell cycle regulation upon their discovery of the ZO-1 interacting protein ZONAB (ZO-1-associated nucleic acid-binding protein) [59]. ZONAB interacts with the promoters of cell cycle regulatory proteins [59] and regulates the ErbB-2 promoter activity and endogenous ErbB-2 expression [59]. Silencing ZONAB or expressing ZO-1 peptides that bind ZONAB reduces proliferation rates while ZONAB overexpression increases cell density [5]. ZONAB also regulates the cell cycle through a direct interaction with PCNA and cyclin D1 [60]. Finally, ZONAB impedes differentiation by direct binding and repression of the megalin and cubilin promoters [61], whose gene products are large (~600 and 460kDa, respectively) glycoproteins involved in the absorption of glomerular-filtered substrates in differentiated tubules (reviewed in [62]). Clearly, the ZO-1-ZONAB protein complex is critical to regulation of cell proliferation and differentiation.
Intriguingly, ZO-2 appears to participate in proliferation control as a result of its nuclear accumulation in sub-confluent cultures [63, 64]. ZO-2 interacts with the DNA-binding protein scaffold attachment factor-B (SAF-B) [64] along with the AP-1 transcription factors Jun and Fos, and the CCAAT/enhancer binding protein (C/EBP). ZO-2, but not ZO-1, negatively regulates the promoters of AP-1 target genes [65]. Huerta and colleagues identified a complex consisting of ZO-2 and c-Myc in which c-Myc binds directly to an E-box within the cyclin D1 promoter and this complex recruits histone deacetylase 1, thus repressing cyclin D1 [66]. A follow-up study supports ZO-2 suppression of cyclin D1 through the finding that ZO-2 inhibits the cell cycle at G1/S and shuttles into the nucleus during G1 and leaves during mitosis, thus providing a model whereby ZO-2 is present in the nucleus in sub-confluent (i.e. proliferating) cells, but absent from the nucleus and at the TJ in confluent (i.e. quiescent) cells [67, 68]. Conversely, ZO-2 nuclear accumulation causes an increase in the M2 type of pyruvate kinase, which is associated with increased proliferation [69].
3.2. Occludin
While the occludin null mouse exhibited no gross barrier abnormalities, the finding that these mice exhibit mucus cell hyperplasia [70] suggests that occludin may be involved in cell proliferation. This supposition is supported by the observations of Phillips et al, who noted that loss of occludin increases proliferation rates [27]. Surprisingly, occludin was identified in centrosomes and mutational analysis revealed that occludin may regulate mitotic entry via centrosome separation in a phosphorylation-dependent manner [71]. Finally, in a cell culture model of uveal melanoma, blood vessel epicardial substance overexpression lead to an increase in ZO-1 and occludin, which correlates with decreased cell proliferation [72].
3.3. Claudins
Gene deletion studies in claudin-15 null mice revealed that these mice, which were viable and developed normally, exhibited a phenotype described as megaintestine [73]. Tamura and colleagues reported that, when compared with normal littermates, the claudin-15 (−/−) mouse was found to have an approximate two-fold increase in the size of the upper small intestine. The authors further characterize this phenomenon and demonstrate an increase in proliferation of the crypts of the upper small intestine with no changes in apoptosis. Importantly, there were no changes in the expression of other claudins and the mice did not exhibit any disease phenotype such as cancer. In ovarian cancer, miR-155 inhibits the proliferation of ovarian tumor initiating cells by targeting CLAUDIN-1 3′ untranslated region (UTR) [74]. However in hepatoma cells, miR-198 upregulates the expression of claudin-1 and E-cadherin and this regulation contributes to cell growth retardation conferred by miR-198 overexpression [75].
4. Tight junctions and tumorigenesis
4.1. Epithelial to mesenchymal transition
Epithelial to mesenchymal transition, or EMT, like many physiological processes, is an essential feature to both pathological and physiological events [76]. The EMT is an important feature of development, cancer, fibrosis, and pathology [77] and there are a number of features that distinguish epithelial and mesenchymal cells. Epithelial cells are characterized by well-developed junctions, an apical-basolateral polarization which is seen at the cell-cell junction, and the ability to become motile. However, motility is a feature that is rarely seen under normal physiological conditions [77, 78]. On the contrary, mesenchymal cells lack polarization due to the loss of an organized junctional layer. Reorganization of the cytoskeleton and organelles is generally not associated with a lamina [77–80]. Furthermore, transforming growth factor-beta (TGF-β) treated cells undergo EMT and are subsequently resistant to apoptosis [81].
In MDCK cells, TGF-β treatment induces EMT concurrent with the loss of claudin-1, -2, occludin, and the adherens junction protein E-cadherin [82]. Moreover, expression of the homeodomain protein HOXB7 in MCF10A and MDCK cells represses claudins-1 and -7 and mis-localizes claudin-4 while HOXB7 expressing MDCK cells form tumors in mice [83]. The pro-EMT repressor Snail directly interacts with E-boxes within the promoters of occludin and claudins-3, -4, and -7, but not ZO-1, and suppresses their promoter activities thus reducing mRNA and protein content [84]. The mechanistic manner in which TJ proteins are repressed during EMT was clarified through the finding that the TGF-β effector proteins SMAD-3 and -4 complex with the EMT repressor protein SNAIL1 and this SNAIL1-SMAD-3/4 complex represses occludin and claudin-3 in breast epithelial cells. The repression of TJ molecules is relieved upon SNAIL-1 or SMAD-4 siRNA-mediated silencing [85]. While it does not appear that ZO-1 is modified in the same manner that the claudins and occludin are during EMT, there is evidence that ZO-1 is involved in dedifferentiation and tumor formation. Reichert and colleagues expressed the PDZ domains of ZO-1 leading to a lack of TJ localization [86]. Functionally, MDCK cells stably expressing the ZO-1 PDZ domains fail to differentiate in collagen type I gel cultures, form tumors in nude mice, and decrease epithelial markers (with a corresponding increase in mesenchymal markers) [86]. Finally, recent studies have unveiled the findings that tricellulin [87] and marvelD3 [88] are silenced in EMT settings of gastric carcinoma and pancreatic cancer cells, respectively.
4.2 Transformation and metastasis
4.2.1. Occludin
Occludin emerged as a critical mediator of transformation from the discovery that it is transcriptionally repressed following constitutive Raf-1 expression and subsequent re-expression sufficiently rescued the transformed phenotype [89]. The Raf-1 induced occludin repression is mediated through a direct interaction between activated Slug and the E-box in the occludin promoter [90]. Congruently, in addition to occludin, claudin-1, -2 and ZO-1 are repressed by the active Ras signaling cascade, and chemical inhibition of MEK restores TJs [91–93]. Domain analyses of occludin revealed that the N- and C-terminal halves along with the second extracellular loop are essential for occludin reversal of Raf-1 transformation in vitro and tumor formation in vivo, whereas the first extracellular loop is dispensable to repression of transformation [94]. Loss of the tumor suppressor von Hippel-Lindau (VHL) down-regulates occludin and claudin-1, independent of E-cadherin. In clear cell renal cell carcinoma (CCRCC) cell lines, occludin is reduced in early lesions in patients with germline VHL mutations [95]. Occludin is epigenetically silenced through promoter hypermethylation in murine melanoma cells and forced expression of occludin also reduces migration in melanoma cells. Stable occludin expression in melanoma and breast cancer cells followed by injection into the craniolateral thorax and mammary fat pad, respectively, reduces the size of lung metastases [96]. Further, occludin induces premature senescence in breast cancer cells, which was blocked by chemical inhibition of the MEK pathway [97].
4.2.2. Claudins
While the current literature suggests that occludin would be an anti-transformation protein, the verdict on the claudin family is less clear. A great deal of effort in regards to understanding changes in claudin content in a wide range of cancers is well-documented in clinically based association studies. In the interest of brevity, the focus of this section shall be only on those studies where claudin content has been experimentally manipulated. For a comprehensive review of claudins and cancer, including association studies, please see Singh et al. [98]. Increased claudin-1 content was reported in human primary colon carcinomas and metastases as well as in cell lines derived from primary and metastatic tumors while genetic manipulation of claudin-1 in vitro and in vivo followed by cellular and rodent based assays supported these observations [99]. On the other hand, claudin-1 positivity is associated with better patient outcome in lung adenocarcinomas and genetic manipulation in lung carcinoma cell lines supports claudin-1 as a negative regulator of metastatic phenotypes and metastasis in vitro and in vivo [100]. Claudin-4 over-expression in invasive pancreatic cancer cells reduces invasion and survival in soft agar growth assays and reduces the number of lung metastases following tail vein injection in mice [101]. Surprisingly, claudins-3 and -4 are overexpressed in ovarian cancer [102, 103] and these findings are substantiated in human ovarian surface epithelial cells where overexpression of claudins-3 and -4 increases cell invasion and motility and siRNA studies support the invasion findings [104]. In ductal carcinoma in situ and invasive ductal carcinoma of the breast, claudin-7 is down regulated, which correlates with promoter hypermethylation in breast cancer cell lines; however, this was not the case in invasive ductal carcinomas [105]. Claudin-2 is increased in colorectal cancer and inflammatory bowel disease-associated colorectal cancer and expression of claudin-2 in colon cells which lack claudin-2 expression increases cell proliferation, anchorage-independent growth, and tumor volume in vivo [106].
4.2.3. Connection of occludin and claudins to stem cell-like phenotype and lineage/cancer genes
Breast cancers with low gene expression of claudins 3/4/7, occludin, and E-cadherin were termed “claudinlow” breast cancer by Herschkowitz et al. [107]. This subtype of breast cancer is mostly triple negative (HER2−, ER−, and PR−) and shows poor prognosis; furthermore, the claudinlow breast cancer loosely resembles the mammary epithelial stem cell [108]. With regard to other cancer types, it remains to be determined if the claudinlow phenotype also correlates with stem cell-like potential.
Thyroid transcription factor 1 (TTF-1 or NKX2-1) is a lung lineage gene that controls pulmonary development and maturation [109, 110]; it is also the most recurrently amplified gene in lung adenocarcinomas [111–114]. The gene amplification of TTF-1 suggests a pro-oncogenic function for TTF-1. However, mouse models implicate tumor-suppressive and anti-metastatic activities of TTF-1 [115–117]. Taken together, TTF-1 is a cancer gene with context-dependent functional multiplicity. We recently discovered that both occludin and claudin-1 are under direct transcriptional control by TTF-1 [118]. TTF-1 knockdown conferred human lung cancer cells resistance to anoikis, and expression of occludin restored cellular sensitivity to anoikis. Furthermore, overexpression of occludin impeded migration and induced anoikis in lung carcinoma cells [118]. Interestingly, analysis of metastatic and non-metastatic lung cancer cells from mice revealed that occludin content is associated with TTF-1 content, whereas loss of TTF-1 has no effect on claudin-1 protein levels. Collectively, we suggest that the TJ proteins mediate the anti-metastatic activity of TTF-1 and predict that other tissue lineage master regulators may also be functionally linked to the TJ constituents.
4.3. Genetic evidence linking tight junction molecules to cancer
While the literature is replete with data demonstrating the expression changes of occludin and claudins in a wide range of cancer types, expression alterations may or may not be indicative of genetic causes. The experimental observations reviewed so far convincingly suggest that TJ proteins play positive and negative functional roles in the tumorigenic process. However, direct evidence of genetic alterations of TJ genes in human cancers has not been explicitly noted in the literature. In view of the robust cancer genomic studies in the recent years, we wonder if an inkling of TJ gene mutations could be detected in the cancer genomic data. To this end, we limit our analysis to the two human TJ genes (OCCLUDIN and CLAUDIN-1). Using the cBio Cancer Genomics Portal [119], we probed 28 datasets for mutations and DNA copy number alterations (CNAs) of these two TJ genes. The results, as shown in Fig. 1, are rather intriguing. First of all, CNAs are the predominant form of genetic alterations associated with both genes. In the case of occludin, the general trend is a loss of DNA copy number. This is in line with the common decrease of occludin expression seen in multiple tumor types [120]. However, for claudin-1, it appears that CLAUDIN-1 undergoes DNA copy number increases frequently in cervical squamous cell carcinoma and endocervical adenocarcinoma (30.6%) and lung squamous cell carcinoma (29.2%). These observations may be counter-intuitive initially. However, a significant overexpression of claudin-1 was indeed reported in cervical [121] and lung squamous cell carcinomas [122]. Therefore, gene dosage alterations are likely a factor shaping the expression patterns of TJ molecules.
Fig. 1. Cancer genetic alteration profiles of OCCLUDIN and CLAUDIN-1.
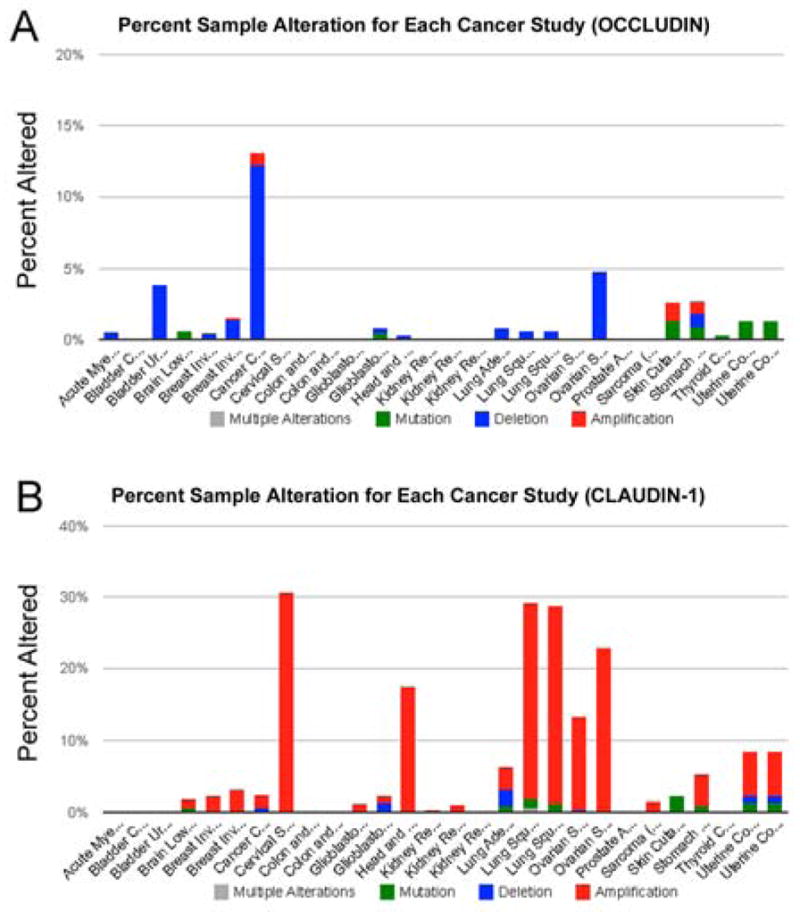
(A) The OCCLUDIN gene profile was interrogated in these studies: Acute Myeloid Leukemia (TCGA, Provisional) altered in 0.5% of 187 cases; Bladder Cancer (MSKCC, JCO 2013) altered in 0% of 97 cases; Bladder Urothelial Carcinoma (TCGA, Provisional) altered in 3.8% of 26 cases; Brain Lower Grade Glioma (TCGA, Provisional) altered in 0.6% of 169 cases; Breast Invasive Carcinoma (TCGA [125]) altered in 0.4% of 482 cases; Breast Invasive Carcinoma (TCGA, Provisional) altered in 1.6% of 760 cases; Cancer Cell Line Encyclopedia [126] altered in 13.2% of 882 cases; Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA, Provisional) altered in 0% of 36 cases; Colon and Rectum Adenocarcinoma [127] altered in 0% of 212 cases; Colon and Rectum Adenocarcinoma (TCGA, Provisional) altered in 0% of 221 cases; Glioblastoma [128] altered in 0% of 91 cases; Glioblastoma Multiforme (TCGA, Provisional) altered in 0.8% of 236 cases; Head and Neck Squamous Cell Carcinoma (TCGA, Provisional) altered in 0.3% of 302 cases; Kidney Renal Clear Cell Carcinoma (TCGA, Provisional) altered in 0% of 290 cases; Kidney Renal Clear Cell Carcinoma (TCGA, in revision) altered in 0% of 418 cases; Kidney Renal Papillary Cell Carcinoma (TCGA, Provisional) altered in 0% of 100 cases; Lung Adenocarcinoma (TCGA, Provisional) altered in 0.8% of 129 cases; Lung Squamous Cell Carcinoma [129] altered in 0.6% of 178 cases; Lung Squamous Cell Carcinoma (TCGA, Provisional) altered in 0.6% of 177 cases; Ovarian Serous Cystadenocarcinoma [130] altered in 0% of 316 cases; Ovarian Serous Cystadenocarcinoma (TCGA, Provisional) altered in 4.8% of 311 cases; Prostate Adenocarcinoma [131] altered in 0% of 103 cases; Sarcoma [132] altered in 0% of 207 cases; Skin Cutaneous Melanoma (TCGA, Provisional) altered in 2.7% of 225 cases; Stomach Adenocarcinoma (TCGA, Provisional) altered in 2.6% of 115 cases; Thyroid Carcinoma (TCGA, Provisional) altered in 0.3% of 318 cases; Uterine Corpus Endometrioid Carcinoma [133] altered in 1.2% of 240 cases; Uterine Corpus Endometrioid Carcinoma (TCGA, Provisional) altered in 1.2% of 240 cases. (B) The Claudin-1 gene profile was interrogated in these studies: Acute Myeloid Leukemia (TCGA, Provisional) altered in 0% of 187 cases; Bladder Cancer (MSKCC, JCO 2013) altered in 0% of 97 cases; Bladder Urothelial Carcinoma (TCGA, Provisional) altered in 0% of 26 cases; Brain Lower Grade Glioma (TCGA, Provisional) altered in 1.8% of 169 cases; Breast Invasive Carcinoma (TCGA [125]) altered in 2.1% of 482 cases; Breast Invasive Carcinoma (TCGA, Provisional) altered in 3% of 760 cases; Cancer Cell Line Encyclopedia [126] altered in 2.3% of 882 cases; Cervical Squamous Cell Carcinoma and Endocervical Adenocarcinoma (TCGA, Provisional) altered in 30.6% of 36 cases; Colon and Rectum Adenocarcinoma [127] altered in 0% of 212 cases; Colon and Rectum Adenocarcinoma (TCGA, Provisional) altered in 0% of 221 cases; Glioblastoma [128] altered in 1.1% of 91 cases; Glioblastoma Multiforme (TCGA, Provisional) altered in 2.1% of 236 cases; Head and Neck Squamous Cell Carcinoma (TCGA, Provisional) altered in 17.5% of 302 cases; Kidney Renal Clear Cell Carcinoma (TCGA, Provisional) altered in 0.3% of 290 cases; Kidney Renal Clear Cell Carcinoma (TCGA, in revision) altered in 1% of 418 cases; Kidney Renal Papillary Cell Carcinoma (TCGA, Provisional) altered in 0% of 100 cases; Lung Adenocarcinoma (TCGA, Provisional) altered in 6.2% of 129 cases; Lung Squamous Cell Carcinoma [129] altered in 29.2% of 178 cases; Lung Squamous Cell Carcinoma (TCGA, Provisional) altered in 28.8% of 177 cases; Ovarian Serous Cystadenocarcinoma [130] altered in 13.3% of 316 cases; Ovarian Serous Cystadenocarcinoma (TCGA, Provisional) altered in 22.8% of 311 cases; Prostate Adenocarcinoma [131] altered in 0% of 103 cases; Sarcoma [132] altered in 1.4% of 207 cases; Skin Cutaneous Melanoma (TCGA, Provisional) altered in 2.2% of 225 cases; Stomach Adenocarcinoma (TCGA, Provisional) altered in 5.2% of 115 cases; Thyroid Carcinoma (TCGA, Provisional) altered in 0% of 318 cases; Uterine Corpus Endometrioid Carcinoma [133] altered in 8.3% of 240 cases; Uterine Corpus Endometrioid Carcinoma (TCGA, Provisional) altered in 8.3% of 240 cases. The analyses were conducted using the cBio Cancer Genomics Portal [119]. TCGA, The Cancer Genome Atlas.
By functional cancer genomics, TJ-related molecules as a whole are considered a significant pathway. The evidence came from a transposon-directed mutagenesis study to search for cooperating mutations with an oncogenic K-Ras allele in promoting murine pancreatic adenocarcinomas [123]. The results identify the TJ signaling pathway as a cellular process that is enriched in candidate cancer genes scored positive in the screen. Since the majority of the genes identified in the transposon insertional mutagenesis screen (90%) are predicted to be disrupted based on the orientation of the transposon with respect to the gene, an implication of this study is that most of the candidate cancer genes found by the study to be associated with the TJ pathway are putative tumor suppressors. Combining this observation and the CNAs affecting OCCLUDIN and CLAUDIN-1, we suggest that there is putative genetic evidence linking TJ genes to tumorigenesis. Our simplistic analyses via the cBio Portal are only meant to provoke researchers to initiate further studies to examine all the TJ genes for genetic and epigenetic aberrations.
5. Conclusion
The last several years have detailed new insights into the role for TJ proteins in cell proliferation, transformation, and metastasis. In view of the considerable functional data and putative genetic evidence connecting TJ factors to the tumorigenic process, we have come a long way from the early demonstration of TJ attenuation in tumors more than 30 years ago [124]. While instinctively one would expect TJ dissolution and concomitant downregulation of TJ factors as a prerequisite for cellular transformation, the reality is that TJ molecule expression patterns, claudins in particular, are more complex than originally anticipated. In terms of the intricate balance of regulation of individual TJ factors, we are only beginning to build a fundamental framework for a holistic understanding. Nevertheless, experimental evidence increasingly suggests that TJ proteins are players in initiation and progression of cancers. Still, a multitude of questions remain. For example, while there have been advances as to how occludin and claudins come to be lost in highly proliferative settings or during transformation, little is known with regards to the precise mechanism that these proteins utilize to intervene on the transformation process. Similarly, it is not yet known if cellular localization is critical. The ZO proteins have been described at the TJ, but also in the nucleus where several regulatory events are occurring [67, 68]. A recent study did demonstrate that occludin is present at centrosomes [71], but this study lacked a mechanistic understanding of the function of occludin’s presence at these organelles. The use of cell culture models has been extremely valuable to solidify the claim that TJ proteins participate in cellular events beyond barrier regulation. Clearly, it is imperative to revisit the knockout models, perhaps in an inducible setting, to definitively attribute these protein functions to transformation and metastasis.
Acknowledgments
D.M. was supported in part by the National Institutes of Health (CA127547).
Footnotes
Conflict of Interest Statement
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mineta K, Yamamoto Y, Yamazaki Y, Tanaka H, Tada Y, Saito K, Tamura A, Igarashi M, Endo T, Takeuchi K, Tsukita S. Predicted expansion of the claudin multigene family. FEBS Lett. 2011;585:606–612. doi: 10.1016/j.febslet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Meer G, Gumbiner B, Simons K. The tight junction does not allow lipid molecules to diffuse from one epithelial cell to the next. Nature. 1986;322:639–641. doi: 10.1038/322639a0. [DOI] [PubMed] [Google Scholar]
- 4.Balda MS, Matter K. Tight junctions and the regulation of gene expression. Biochim Biophys Acta. 2009;1788:761–767. doi: 10.1016/j.bbamem.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Balda MS, Garrett MD, Matter K. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J Cell Biol. 2003;160:423–432. doi: 10.1083/jcb.200210020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gumbiner B, Lowenkopf T, Apatira D. Identification of a 160-kDa polypeptide that binds to the tight junction protein ZO-1. Proc Natl Acad Sci U S A. 1991;88:3460–3464. doi: 10.1073/pnas.88.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haskins J, Gu L, Wittchen ES, Hibbard J, Stevenson BR. ZO-3, a novel member of the MAGUK protein family found at the tight junction, interacts with ZO-1 and occludin. J Cell Biol. 1998;141:199–208. doi: 10.1083/jcb.141.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jesaitis LA, Goodenough DA. Molecular characterization and tissue distribution of ZO-2, a tight junction protein homologous to ZO-1 and the Drosophila discs-large tumor suppressor protein. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevenson BR, Siliciano JD, Mooseker MS, Goodenough DA. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J Cell Biol. 1986;103:755–766. doi: 10.1083/jcb.103.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woods DF, Bryant PJ. ZO-1, DlgA and PSD-95/SAP90: homologous proteins in tight, septate and synaptic cell junctions. Mech Dev. 1993;44:85–89. doi: 10.1016/0925-4773(93)90059-7. [DOI] [PubMed] [Google Scholar]
- 11.Van Itallie CM, Aponte A, Tietgens AJ, Gucek M, Fredriksson K, Anderson JM. The N- and C- termini of ZO-1 are surrounded by distinct proteins and functional protein networks. J Biol Chem. 2013 doi: 10.1074/jbc.M113.466193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fanning AS, Little BP, Rahner C, Utepbergenov D, Walther Z, Anderson JM. The unique-5 and -6 motifs of ZO-1 regulate tight junction strand localization and scaffolding properties. Mol Biol Cell. 2007;18:721–731. doi: 10.1091/mbc.E06-08-0764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Fanning AS, Anderson JM, Lavie A. Structure of the conserved cytoplasmic C-terminal domain of occludin: identification of the ZO-1 binding surface. J Mol Biol. 2005;352:151–164. doi: 10.1016/j.jmb.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Sundstrom JM, Tash BR, Murakami T, Flanagan JM, Bewley MC, Stanley BA, Gonsar KB, Antonetti DA. Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach. Journal of proteome research. 2009;8:808–817. doi: 10.1021/pr7007913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ebnet K, Schulz CU, Meyer Zu Brickwedde MK, Pendl GG, Vestweber D. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. J Biol Chem. 2000;275:27979–27988. doi: 10.1074/jbc.M002363200. [DOI] [PubMed] [Google Scholar]
- 17.McNeil E, Capaldo CT, Macara IG. Zonula occludens-1 function in the assembly of tight junctions in Madin-Darby canine kidney epithelial cells. Mol Biol Cell. 2006;17:1922–1932. doi: 10.1091/mbc.E05-07-0650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umeda K, Matsui T, Nakayama M, Furuse K, Sasaki H, Furuse M, Tsukita S. Establishment and characterization of cultured epithelial cells lacking expression of ZO-1. J Biol Chem. 2004;279:44785–44794. doi: 10.1074/jbc.M406563200. [DOI] [PubMed] [Google Scholar]
- 19.Hernandez S, Chavez Munguia B, Gonzalez-Mariscal L. ZO-2 silencing in epithelial cells perturbs the gate and fence function of tight junctions and leads to an atypical monolayer architecture. Exp Cell Res. 2007;313:1533–1547. doi: 10.1016/j.yexcr.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 20.Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 21.Katsuno T, Umeda K, Matsui T, Hata M, Tamura A, Itoh M, Takeuchi K, Fujimori T, Nabeshima Y, Noda T, Tsukita S. Deficiency of zonula occludens-1 causes embryonic lethal phenotype associated with defected yolk sac angiogenesis and apoptosis of embryonic cells. Mol Biol Cell. 2008;19:2465–2475. doi: 10.1091/mbc.E07-12-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu J, Kausalya PJ, Phua DC, Ali SM, Hossain Z, Hunziker W. Early embryonic lethality of mice lacking ZO-2, but Not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol Cell Biol. 2008;28:1669–1678. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita S. Occludin: a novel integral membrane protein localizing at tight junctions. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balda MS, Whitney JA, Flores C, Gonzalez S, Cereijido M, Matter K. Functional dissociation of paracellular permeability and transepithelial electrical resistance and disruption of the apical-basolateral intramembrane diffusion barrier by expression of a mutant tight junction membrane protein. J Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou M, Furuse M, Sasaki H, Schulzke JD, Fromm M, Takano H, Noda T, Tsukita S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol Biol Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu AS, McCarthy KM, Francis SA, McCormack JM, Lai J, Rogers RA, Lynch RD, Schneeberger EE. Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. American journal of physiology Cell physiology. 2005;288:C1231–1241. doi: 10.1152/ajpcell.00581.2004. [DOI] [PubMed] [Google Scholar]
- 27.Phillips BE, Cancel L, Tarbell JM, Antonetti DA. Occludin independently regulates permeability under hydrostatic pressure and cell division in retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2008;49:2568–2576. doi: 10.1167/iovs.07-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami T, Felinski EA, Antonetti DA. Occludin phosphorylation and ubiquitination regulate tight junction trafficking and vascular endothelial growth factor-induced permeability. J Biol Chem. 2009;284:21036–21046. doi: 10.1074/jbc.M109.016766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murakami T, Frey T, Lin C, Antonetti DA. Protein Kinase Cbeta Phosphorylates Occludin Regulating Tight Junction Trafficking in Vascular Endothelial Growth Factor-Induced Permeability In Vivo. Diabetes. 2012 doi: 10.2337/db11-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L, Turner JR. Actin depolymerization disrupts tight junctions via caveolae-mediated endocytosis. Mol Biol Cell. 2005;16:3919–3936. doi: 10.1091/mbc.E04-12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchiando AM, Shen L, Graham WV, Weber CR, Schwarz BT, Austin JR, 2nd, Raleigh DR, Guan Y, Watson AJ, Montrose MH, Turner JR. Caveolin-1-dependent occludin endocytosis is required for TNF-induced tight junction regulation in vivo. J Cell Biol. 2010;189:111–126. doi: 10.1083/jcb.200902153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukita S, Furuse M. Occludin and claudins in tight-junction strands: leading or supporting players? Trends Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- 33.Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piontek J, Winkler L, Wolburg H, Muller SL, Zuleger N, Piehl C, Wiesner B, Krause G, Blasig IE. Formation of tight junction: determinants of homophilic interaction between classic claudins. FASEB J. 2008;22:146–158. doi: 10.1096/fj.07-8319com. [DOI] [PubMed] [Google Scholar]
- 35.Furuse M, Hata M, Furuse K, Yoshida Y, Haratake A, Sugitani Y, Noda T, Kubo A, Tsukita S. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. J Cell Biol. 2002;156:1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. 2003;161:653–660. doi: 10.1083/jcb.200302070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gow A, Southwood CM, Li JS, Pariali M, Riordan GP, Brodie SE, Danias J, Bronstein JM, Kachar B, Lazzarini RA. CNS myelin and sertoli cell tight junction strands are absent in Osp/claudin-11 null mice. Cell. 1999;99:649–659. doi: 10.1016/s0092-8674(00)81553-6. [DOI] [PubMed] [Google Scholar]
- 38.Konrad M, Hou J, Weber S, Dotsch J, Kari JA, Seeman T, Kuwertz-Broking E, Peco-Antic A, Tasic V, Dittrich K, Alshaya HO, von Vigier RO, Gallati S, Goodenough DA, Schaller A. CLDN16 genotype predicts renal decline in familial hypomagnesemia with hypercalciuria and nephrocalcinosis. J Am Soc Nephrol. 2008;19:171–181. doi: 10.1681/ASN.2007060709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, Vitzthum H, Suzuki Y, Luk JM, Becker C, Schlingmann KP, Schmid M, Rodriguez-Soriano J, Ariceta G, Cano F, Enriquez R, Juppner H, Bakkaloglu SA, Hediger MA, Gallati S, Neuhauss SC, Nurnberg P, Weber S. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79:949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kitajiri S, Miyamoto T, Mineharu A, Sonoda N, Furuse K, Hata M, Sasaki H, Mori Y, Kubota T, Ito J, Furuse M, Tsukita S. Compartmentalization established by claudin-11-based tight junctions in stria vascularis is required for hearing through generation of endocochlear potential. J Cell Sci. 2004;117:5087–5096. doi: 10.1242/jcs.01393. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Yosef T, Belyantseva IA, Saunders TL, Hughes ED, Kawamoto K, Van Itallie CM, Beyer LA, Halsey K, Gardner DJ, Wilcox ER, Rasmussen J, Anderson JM, Dolan DF, Forge A, Raphael Y, Camper SA, Friedman TB. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12:2049–2061. doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Nusrat A, Schnell FJ, Reaves TA, Walsh S, Pochet M, Parkos CA. Human junction adhesion molecule regulates tight junction resealing in epithelia. J Cell Sci. 2000;113(Pt 13):2363–2374. doi: 10.1242/jcs.113.13.2363. [DOI] [PubMed] [Google Scholar]
- 43.Mandell KJ, McCall IC, Parkos CA. Involvement of the junctional adhesion molecule-1 (JAM1) homodimer interface in regulation of epithelial barrier function. J Biol Chem. 2004;279:16254–16262. doi: 10.1074/jbc.M309483200. [DOI] [PubMed] [Google Scholar]
- 44.Mandell KJ, Babbin BA, Nusrat A, Parkos CA. Junctional adhesion molecule 1 regulates epithelial cell morphology through effects on beta1 integrins and Rap1 activity. J Biol Chem. 2005;280:11665–11674. doi: 10.1074/jbc.M412650200. [DOI] [PubMed] [Google Scholar]
- 45.Vetrano S, Rescigno M, Cera MR, Correale C, Rumio C, Doni A, Fantini M, Sturm A, Borroni E, Repici A, Locati M, Malesci A, Dejana E, Danese S. Unique role of junctional adhesion molecule-a in maintaining mucosal homeostasis in inflammatory bowel disease. Gastroenterology. 2008;135:173–184. doi: 10.1053/j.gastro.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 46.Rehder D, Iden S, Nasdala I, Wegener J, Brickwedde MK, Vestweber D, Ebnet K. Junctional adhesion molecule-a participates in the formation of apico-basal polarity through different domains. Exp Cell Res. 2006;312:3389–3403. doi: 10.1016/j.yexcr.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 47.Ebnet K, Suzuki A, Horikoshi Y, Hirose T, Meyer Zu Brickwedde MK, Ohno S, Vestweber D. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) EMBO J. 2001;20:3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S. Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions. J Cell Biol. 2001;154:491–497. doi: 10.1083/jcb.200103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laukoetter MG, Nava P, Lee WY, Severson EA, Capaldo CT, Babbin BA, Williams IR, Koval M, Peatman E, Campbell JA, Dermody TS, Nusrat A, Parkos CA. JAM-A regulates permeability and inflammation in the intestine in vivo. J Exp Med. 2007;204:3067–3076. doi: 10.1084/jem.20071416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol. 2005;171:939–945. doi: 10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri S, Ramzan K, Khan SN, Chattaraj P, Friedman PL, Anderson JM, Belyantseva IA, Forge A, Friedman TB. Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet. 2006;79:1040–1051. doi: 10.1086/510022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chishti MS, Bhatti A, Tamim S, Lee K, McDonald ML, Leal SM, Ahmad W. Splice-site mutations in the TRIC gene underlie autosomal recessive nonsyndromic hearing impairment in Pakistani families. J Hum Genet. 2008;53:101–105. doi: 10.1007/s10038-007-0209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikenouchi J, Sasaki H, Tsukita S, Furuse M. Loss of occludin affects tricellular localization of tricellulin. Mol Biol Cell. 2008;19:4687–4693. doi: 10.1091/mbc.E08-05-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krug SM, Amasheh S, Richter JF, Milatz S, Gunzel D, Westphal JK, Huber O, Schulzke JD, Fromm M. Tricellulin forms a barrier to macromolecules in tricellular tight junctions without affecting ion permeability. Mol Biol Cell. 2009;20:3713–3724. doi: 10.1091/mbc.E09-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steed E, Rodrigues NT, Balda MS, Matter K. Identification of MarvelD3 as a tight junction-associated transmembrane protein of the occludin family. BMC Cell Biol. 2009;10:95. doi: 10.1186/1471-2121-10-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y, Long M, Turner JR. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell. 2010;21:1200–1213. doi: 10.1091/mbc.E09-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Timmann C, Thye T, Vens M, Evans J, May J, Ehmen C, Sievertsen J, Muntau B, Ruge G, Loag W, Ansong D, Antwi S, Asafo-Adjei E, Nguah SB, Kwakye KO, Akoto AO, Sylverken J, Brendel M, Schuldt K, Loley C, Franke A, Meyer CG, Agbenyega T, Ziegler A, Horstmann RD. Genome-wide association study indicates two novel resistance loci for severe malaria. Nature. 2012;489:443–446. doi: 10.1038/nature11334. [DOI] [PubMed] [Google Scholar]
- 58.Cording J, Berg J, Kading N, Bellmann C, Tscheik C, Westphal JK, Milatz S, Gunzel D, Wolburg H, Piontek J, Huber O, Blasig IE. In tight junctions, claudins regulate the interactions between occludin, tricellulin and marvelD3, which, inversely, modulate claudin oligomerization. J Cell Sci. 2013;126:554–564. doi: 10.1242/jcs.114306. [DOI] [PubMed] [Google Scholar]
- 59.Balda MS, Matter K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000;19:2024–2033. doi: 10.1093/emboj/19.9.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sourisseau T, Georgiadis A, Tsapara A, Ali RR, Pestell R, Matter K, Balda MS. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol Cell Biol. 2006;26:2387–2398. doi: 10.1128/MCB.26.6.2387-2398.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lima WR, Parreira KS, Devuyst O, Caplanusi A, N’Kuli F, Marien B, Van Der Smissen P, Alves PM, Verroust P, Christensen EI, Terzi F, Matter K, Balda MS, Pierreux CE, Courtoy PJ. ZONAB promotes proliferation and represses differentiation of proximal tubule epithelial cells. J Am Soc Nephrol. 2010;21:478–488. doi: 10.1681/ASN.2009070698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaseda R, Hosojima M, Sato H, Saito A. Role of megalin and cubilin in the metabolism of vitamin D(3), Therapeutic apheresis and dialysis : official peer-reviewed journal of the International Society for Apheresis. the Japanese Society for Apheresis, the Japanese Society for Dialysis Therapy. 2011;15(Suppl 1):14–17. doi: 10.1111/j.1744-9987.2011.00920.x. [DOI] [PubMed] [Google Scholar]
- 63.Islas S, Vega J, Ponce L, Gonzalez-Mariscal L. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp Cell Res. 2002;274:138–148. doi: 10.1006/excr.2001.5457. [DOI] [PubMed] [Google Scholar]
- 64.Traweger A, Fuchs R, Krizbai IA, Weiger TM, Bauer HC, Bauer H. The tight junction protein ZO-2 localizes to the nucleus and interacts with the heterogeneous nuclear ribonucleoprotein scaffold attachment factor-B. J Biol Chem. 2003;278:2692–2700. doi: 10.1074/jbc.M206821200. [DOI] [PubMed] [Google Scholar]
- 65.Betanzos A, Huerta M, Lopez-Bayghen E, Azuara E, Amerena J, Gonzalez-Mariscal L. The tight junction protein ZO-2 associates with Jun, Fos and C/EBP transcription factors in epithelial cells. Exp Cell Res. 2004;292:51–66. doi: 10.1016/j.yexcr.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 66.Huerta M, Munoz R, Tapia R, Soto-Reyes E, Ramirez L, Recillas-Targa F, Gonzalez-Mariscal L, Lopez-Bayghen E. Cyclin D1 is transcriptionally down-regulated by ZO-2 via an E box and the transcription factor c-Myc. Mol Biol Cell. 2007;18:4826–4836. doi: 10.1091/mbc.E07-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tapia R, Huerta M, Islas S, Avila-Flores A, Lopez-Bayghen E, Weiske J, Huber O, Gonzalez-Mariscal L. Zona occludens-2 inhibits cyclin D1 expression and cell proliferation and exhibits changes in localization along the cell cycle. Mol Biol Cell. 2009;20:1102–1117. doi: 10.1091/mbc.E08-03-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gonzalez-Mariscal L, Tapia R, Huerta M, Lopez-Bayghen E. The tight junction protein ZO-2 blocks cell cycle progression and inhibits cyclin D1 expression. Ann N Y Acad Sci. 2009;1165:121–125. doi: 10.1111/j.1749-6632.2009.04024.x. [DOI] [PubMed] [Google Scholar]
- 69.Traweger A, Lehner C, Farkas A, Krizbai IA, Tempfer H, Klement E, Guenther B, Bauer HC, Bauer H. Nuclear Zonula occludens-2 alters gene expression and junctional stability in epithelial and endothelial cells. Differentiation. 2008;76:99–106. doi: 10.1111/j.1432-0436.2007.00227.x. [DOI] [PubMed] [Google Scholar]
- 70.Schulzke JD, Gitter AH, Mankertz J, Spiegel S, Seidler U, Amasheh S, Saitou M, Tsukita S, Fromm M. Epithelial transport and barrier function in occludin-deficient mice. Biochim Biophys Acta. 2005;1669:34–42. doi: 10.1016/j.bbamem.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 71.Runkle EA, Sundstrom JM, Runkle KB, Liu X, Antonetti DA. Occludin localizes to centrosomes and modifies mitotic entry. J Biol Chem. 2011;286:30847–30858. doi: 10.1074/jbc.M111.262857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jayagopal A, Yang JL, Haselton FR, Chang MS. Tight junction-associated signaling pathways modulate cell proliferation in uveal melanoma. Invest Ophthalmol Vis Sci. 2011;52:588–593. doi: 10.1167/iovs.10-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tamura A, Kitano Y, Hata M, Katsuno T, Moriwaki K, Sasaki H, Hayashi H, Suzuki Y, Noda T, Furuse M, Tsukita S. Megaintestine in claudin-15-deficient mice. Gastroenterology. 2008;134:523–534. doi: 10.1053/j.gastro.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 74.Qin W, Ren Q, Liu T, Huang Y, Wang J. MicroRNA-155 is a novel suppressor of ovarian cancer-initiating cells that targets CLDN1. FEBS Lett. 2013 doi: 10.1016/j.febslet.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 75.Elfimova N, Sievers E, Eischeid H, Kwiecinski M, Noetel A, Hunt H, Becker D, Frommolt P, Quasdorff M, Steffen HM, Nurnberg P, Buttner R, Teufel A, Dienes HP, Drebber U, Odenthal M. Control of mitogenic and motogenic pathways by miR-198, diminishing hepatoma cell growth and migration. Biochim Biophys Acta. 2013;1833:1190–1198. doi: 10.1016/j.bbamcr.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 76.Rastaldi MP. Epithelial-mesenchymal transition and its implications for the development of renal tubulointerstitial fibrosis. J Nephrol. 2006;19:407–412. [PubMed] [Google Scholar]
- 77.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol. 2006;172:973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 79.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 80.Thiery JP. Epithelial-mesenchymal transitions in development and pathologies. Curr Opin Cell Biol. 2003;15:740–746. doi: 10.1016/j.ceb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 81.Valdes F, Alvarez AM, Locascio A, Vega S, Herrera B, Fernandez M, Benito M, Nieto MA, Fabregat I. The epithelial mesenchymal transition confers resistance to the apoptotic effects of transforming growth factor Beta in fetal rat hepatocytes. Mol Cancer Res. 2002;1:68–78. [PubMed] [Google Scholar]
- 82.Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol Biol Cell. 2006;17:1871–1879. doi: 10.1091/mbc.E05-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu X, Chen H, Parker B, Rubin E, Zhu T, Lee JS, Argani P, Sukumar S. HOXB7, a homeodomain protein, is overexpressed in breast cancer and confers epithelial-mesenchymal transition. Cancer Res. 2006;66:9527–9534. doi: 10.1158/0008-5472.CAN-05-4470. [DOI] [PubMed] [Google Scholar]
- 84.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. J Cell Sci. 2003;116:1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 85.Vincent T, Neve EP, Johnson JR, Kukalev A, Rojo F, Albanell J, Pietras K, Virtanen I, Philipson L, Leopold PL, Crystal RG, de Herreros AG, Moustakas A, Pettersson RF, Fuxe J. A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-beta mediated epithelial-mesenchymal transition. Nat Cell Biol. 2009;11:943–950. doi: 10.1038/ncb1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reichert M, Muller T, Hunziker W. The PDZ domains of zonula occludens-1 induce an epithelial to mesenchymal transition of Madin-Darby canine kidney I cells. Evidence for a role of beta-catenin/Tcf/Lef signaling. J Biol Chem. 2000;275:9492–9500. doi: 10.1074/jbc.275.13.9492. [DOI] [PubMed] [Google Scholar]
- 87.Masuda R, Semba S, Mizuuchi E, Yanagihara K, Yokozaki H. Negative regulation of the tight junction protein tricellulin by snail-induced epithelial-mesenchymal transition in gastric carcinoma cells. Pathobiology. 2010;77:106–113. doi: 10.1159/000278293. [DOI] [PubMed] [Google Scholar]
- 88.Kojima T, Takasawa A, Kyuno D, Ito T, Yamaguchi H, Hirata K, Tsujiwaki M, Murata M, Tanaka S, Sawada N. Downregulation of tight junction-associated MARVEL protein marvelD3 during epithelial-mesenchymal transition in human pancreatic cancer cells. Exp Cell Res. 2011;317:2288–2298. doi: 10.1016/j.yexcr.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 89.Li D, Mrsny RJ. Oncogenic Raf-1 disrupts epithelial tight junctions via downregulation of occludin. J Cell Biol. 2000;148:791–800. doi: 10.1083/jcb.148.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Z, Wade P, Mandell KJ, Akyildiz A, Parkos CA, Mrsny RJ, Nusrat A. Raf 1 represses expression of the tight junction protein occludin via activation of the zinc-finger transcription factor slug. Oncogene. 2007;26:1222–1230. doi: 10.1038/sj.onc.1209902. [DOI] [PubMed] [Google Scholar]
- 91.Chen Y, Lu Q, Schneeberger EE, Goodenough DA. Restoration of tight junction structure and barrier function by down-regulation of the mitogen-activated protein kinase pathway in ras-transformed Madin-Darby canine kidney cells. Mol Biol Cell. 2000;11:849–862. doi: 10.1091/mbc.11.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lan M, Kojima T, Osanai M, Chiba H, Sawada N. Oncogenic Raf-1 regulates epithelial to mesenchymal transition via distinct signal transduction pathways in an immortalized mouse hepatic cell line. Carcinogenesis. 2004;25:2385–2395. doi: 10.1093/carcin/bgh248. [DOI] [PubMed] [Google Scholar]
- 93.Xie L, Law BK, Chytil AM, Brown KA, Aakre ME, Moses HL. Activation of the Erk pathway is required for TGF-beta1-induced EMT in vitro. Neoplasia. 2004;6:603–610. doi: 10.1593/neo.04241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Z, Mandell KJ, Parkos CA, Mrsny RJ, Nusrat A. The second loop of occludin is required for suppression of Raf1-induced tumor growth. Oncogene. 2005;24:4412–4420. doi: 10.1038/sj.onc.1208634. [DOI] [PubMed] [Google Scholar]
- 95.Harten SK, Shukla D, Barod R, Hergovich A, Balda MS, Matter K, Esteban MA, Maxwell PH. Regulation of renal epithelial tight junctions by the von Hippel-Lindau tumor suppressor gene involves occludin and claudin 1 and is independent of E-cadherin. Mol Biol Cell. 2009;20:1089–1101. doi: 10.1091/mbc.E08-06-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N. Epigenetic silencing of occludin promotes tumorigenic and metastatic properties of cancer cells via modulations of unique sets of apoptosis-associated genes. Cancer Res. 2006;66:9125–9133. doi: 10.1158/0008-5472.CAN-06-1864. [DOI] [PubMed] [Google Scholar]
- 97.Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N. Occludin-mediated premature senescence is a fail-safe mechanism against tumorigenesis in breast carcinoma cells. Cancer Sci. 2007;98:1027–1034. doi: 10.1111/j.1349-7006.2007.00494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Singh AB, Sharma A, Dhawan P. Claudin family of proteins and cancer: an overview. Journal of oncology. 2010;2010:541957. doi: 10.1155/2010/541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dhawan P, Singh AB, Deane NG, No Y, Shiou SR, Schmidt C, Neff J, Washington MK, Beauchamp RD. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115:1765–1776. doi: 10.1172/JCI24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chao YC, Pan SH, Yang SC, Yu SL, Che TF, Lin CW, Tsai MS, Chang GC, Wu CH, Wu YY, Lee YC, Hong TM, Yang PC. Claudin-1 is a metastasis suppressor and correlates with clinical outcome in lung adenocarcinoma. Am J Respir Crit Care Med. 2009;179:123–133. doi: 10.1164/rccm.200803-456OC. [DOI] [PubMed] [Google Scholar]
- 101.Michl P, Barth C, Buchholz M, Lerch MM, Rolke M, Holzmann KH, Menke A, Fensterer H, Giehl K, Lohr M, Leder G, Iwamura T, Adler G, Gress TM. Claudin-4 expression decreases invasiveness and metastatic potential of pancreatic cancer. Cancer Res. 2003;63:6265–6271. [PubMed] [Google Scholar]
- 102.Rangel LB, Agarwal R, D’Souza T, Pizer ES, Alo PL, Lancaster WD, Gregoire L, Schwartz DR, Cho KR, Morin PJ. Tight junction proteins claudin-3 and claudin-4 are frequently overexpressed in ovarian cancer but not in ovarian cystadenomas. Clin Cancer Res. 2003;9:2567–2575. [PubMed] [Google Scholar]
- 103.Zhu Y, Brannstrom M, Janson PO, Sundfeldt K. Differences in expression patterns of the tight junction proteins,claudin 1, 3, 4 and 5, in human ovarian surface epithelium as compared to epithelia in inclusion cysts and epithelial ovarian tumours. Int J Cancer. 2006;118:1884–1891. doi: 10.1002/ijc.21506. [DOI] [PubMed] [Google Scholar]
- 104.Agarwal R, D’Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005;65:7378–7385. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 105.Kominsky SL, Argani P, Korz D, Evron E, Raman V, Garrett E, Rein A, Sauter G, Kallioniemi OP, Sukumar S. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033. doi: 10.1038/sj.onc.1206199. [DOI] [PubMed] [Google Scholar]
- 106.Dhawan P, Ahmad R, Chaturvedi R, Smith JJ, Midha R, Mittal MK, Krishnan M, Chen X, Eschrich S, Yeatman TJ, Harris RC, Washington MK, Wilson KT, Beauchamp RD, Singh AB. Claudin-2 expression increases tumorigenicity of colon cancer cells: role of epidermal growth factor receptor activation. Oncogene. 2011;30:3234–3247. doi: 10.1038/onc.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cardoso WV, Lu J. Regulation of early lung morphogenesis: questions, facts and controversies. Development. 2006;133:1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 110.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87:219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 111.Kendall J, Liu Q, Bakleh A, Krasnitz A, Nguyen KC, Lakshmi B, Gerald WL, Powers S, Mu D. Oncogenic cooperation and coamplification of developmental transcription factor genes in lung cancer. Proc Natl Acad Sci U S A. 2007;104:16663–16668. doi: 10.1073/pnas.0708286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kwei KA, Kim YH, Girard L, Kao J, Pacyna-Gengelbach M, Salari K, Lee J, Choi YL, Sato M, Wang P, Hernandez-Boussard T, Gazdar AF, Petersen I, Minna JD, Pollack JR. Genomic profiling identifies TITF1 as a lineage-specific oncogene amplified in lung cancer. Oncogene. 2008;27:3635–3640. doi: 10.1038/sj.onc.1211012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tanaka H, Yanagisawa K, Shinjo K, Taguchi A, Maeno K, Tomida S, Shimada Y, Osada H, Kosaka T, Matsubara H, Mitsudomi T, Sekido Y, Tanimoto M, Yatabe Y, Takahashi T. Lineage-specific dependency of lung adenocarcinomas on the lung development regulator TTF-1. Cancer Res. 2007;67:6007–6011. doi: 10.1158/0008-5472.CAN-06-4774. [DOI] [PubMed] [Google Scholar]
- 114.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maeda Y, Tsuchiya T, Hao H, Tompkins DH, Xu Y, Mucenski ML, Du L, Keiser AR, Fukazawa T, Naomoto Y, Nagayasu T, Whitsett JA. Kras(G12D) and Nkx2-1 haploinsufficiency induce mucinous adenocarcinoma of the lung. J Clin Invest. 2012;122:4388–4400. doi: 10.1172/JCI64048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Snyder EL, Watanabe H, Magendantz M, Hoersch S, Chen TA, Wang DG, Crowley D, Whittaker CA, Meyerson M, Kimura S, Jacks T. Nkx2-1 Represses a Latent Gastric Differentiation Program in Lung Adenocarcinoma. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, Yoon S, Crowley D, Bronson RT, Chiang DY, Meyerson M, Jacks T. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Runkle EA, Rice SJ, Qi J, Masser D, Antonetti DA, Winslow MM, Mu D. Occludin is a direct target of thyroid transcription factor-1 (TTF-1/NKX2-1) J Biol Chem. 2012;287:28790–28801. doi: 10.1074/jbc.M112.367987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tsukita S, Yamazaki Y, Katsuno T, Tamura A. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene. 2008;27:6930–6938. doi: 10.1038/onc.2008.344. [DOI] [PubMed] [Google Scholar]
- 121.Szabo I, Kiss A, Schaff Z, Sobel G. Claudins as diagnostic and prognostic markers in gynecological cancer. Histol Histopathol. 2009;24:1607–1615. doi: 10.14670/HH-24.1607. [DOI] [PubMed] [Google Scholar]
- 122.Jung JH, Jung CK, Choi HJ, Jun KH, Yoo J, Kang SJ, Lee KY. Diagnostic utility of expression of claudins in non-small cell lung cancer: different expression profiles in squamous cell carcinomas and adenocarcinomas. Pathol Res Pract. 2009;205:409–416. doi: 10.1016/j.prp.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 123.Mann KM, Ward JM, Yew CC, Kovochich A, Dawson DW, Black MA, Brett BT, Sheetz TE, Dupuy AJ, Chang DK, Biankin AV, Waddell N, Kassahn KS, Grimmond SM, Rust AG, Adams DJ, Jenkins NA, Copeland NG I. Australian Pancreatic Cancer Genome. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109:5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Alroy J. Ultrastructure of canine urinary bladder carcinoma. Vet Pathol. 1979;16:693–701. doi: 10.1177/030098587901600608. [DOI] [PubMed] [Google Scholar]
- 125.N. Cancer Genome Atlas. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.N. Cancer Genome Atlas. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.N. Cancer Genome Atlas Research. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.N. Cancer Genome Atlas Research. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.N. Cancer Genome Atlas Research. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, Antipin Y, Mitsiades N, Landers T, Dolgalev I, Major JE, Wilson M, Socci ND, Lash AE, Heguy A, Eastham JA, Scher HI, Reuter VE, Scardino PT, Sander C, Sawyers CL, Gerald WL. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, Chiang DY, Reva B, Mermel CH, Getz G, Antipin Y, Beroukhim R, Major JE, Hatton C, Nicoletti R, Hanna M, Sharpe T, Fennell TJ, Cibulskis K, Onofrio RC, Saito T, Shukla N, Lau C, Nelander S, Silver SJ, Sougnez C, Viale A, Winckler W, Maki RG, Garraway LA, Lash A, Greulich H, Root DE, Sellers WR, Schwartz GK, Antonescu CR, Lander ES, Varmus HE, Ladanyi M, Sander C, Meyerson M, Singer S. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA N. Cancer Genome Atlas Research. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]


