Abstract
Local protein synthesis and its activity-dependent modulation via dopamine receptor stimulation play an important role in synaptic plasticity - allowing synapses to respond dynamically to changes in their activity patterns. We describe here the metabolic labeling, enrichment and mass spectrometry-based identification of candidate proteins specifically translated in intact hippocampal neuropil sections upon treatment with the selective D1/D5 receptor agonist SKF81297. Using the non-canonical amino acid azidohomoalanine and click chemistry we identified over 300 newly synthesized proteins specific to dendrites and axons. Candidates specific for the SKF81297-treated samples were predominantly involved in protein synthesis and synapse-specific functions. Furthermore, we demonstrate a dendrite-specific increase in proteins synthesis upon application of SKF81297. This study provides the first snapshot in the dynamics of the dopaminergic hippocampal neuropil proteome.
Keywords: MUDPIT, click chemistry, metabolic labeling, BONCAT, local protein synthesis, dopaminergic signaling
Introduction
Information storage in the nervous system occurs, at least in part, by altering the strength of synaptic connections. Many studies have shown that both long-term behavioral memories and synaptic plasticity require new protein synthesis (see [1]). Unlike most cells, neurons possess the capacity to synthesize proteins in compartments remote from the cell body. Following the discovery of polyribosomes at or near synapses [2] and the demonstration of synaptic plasticity that requires local translation [3, 4] many additional studies have shown that local translation near synapses is coupled to ongoing synaptic transmission [5, 6] and plasticity [7]. While there are many examples of plasticity that require protein synthesis, the identification of specific proteins that are synthesized, particularly in dendrites, is thus far limited to a few proteins such as Arc [8] and CAMKIIα [9].
Neuromodulators play a key role in controlling information flow among brain areas [10]. The axons of dopaminergic neurons project to the hippocampus [11] and release dopamine (DA) after animals are exposed to a novel environment [12]. In addition a number of studies have indicated that DA plays an important role in hippocampus-dependent learning [13, 14]. In addition, stimulation of D1-like dopaminergic receptors has been reported to be necessary and sufficient for long-term plasticity at synapses [15-19], which is a protein synthesis-dependent process. Previous work by Smith and colleagues [20] demonstrated an increase in dendritic protein synthesis after the application of a D1/D5 dopamine receptor agonist, SKF81297. They also observed the upregulation in surface expression of GluA1, an AMPA receptor subunit. This, in turn, resulted in an increased frequency of miniature excitatory postsynaptic currents (mEPSCs, or minis), which are able to regulate local protein synthesis [5].
In order to discover the proteomic response proximal to an inducing event, a method for the pulse-labeling of newly synthesized proteins is required. Although radioactive amino acids can, in principle, be used for this purpose, practically speaking the recovery of labeled proteins from gels and their subsequent identification via mass spectrometry is not an option since it would contaminate not only the mass spectrometer but also the environment through the exhausts of the vacuum system. We have therefore developed a non-radioactive method for the labeling, visualization and identification of newly synthesized proteins using non-canonical amino acids [21-23]. Bioorthogonal non-canonical amino acid tagging (BONCAT) makes use of methionine derivatives that are functionalized with either an azide (azidohomoalanine, AHA) or alkyne group (homopropargylglycine, HPG). Bath application of the amino acids to cells or tissues results in their uptake and subsequent charging onto methionyl tRNAs via the cell's own methionyl tRNA synthetase. During protein synthesis AHA or HPG are incorporated into proteins and can then be tagged with an alkyne- or azide-bearing molecule using a copper-catalyzed reaction [23-26]. Tagged newly synthesized proteins can be purified and subsequently identified using mass spectrometry [21, 22] or visualized in situ[23, 27] when a fluorescent tag is used. The visualization of newly synthesized proteins using the above technique is referred to as Fluorescent Non-Canonical Amino Acid Tagging, or FUNCAT [21]. In this study we examined the proteins that are rapidly synthesized in the dendrites of the hippocampus under control and dopamine-stimulated conditions. Using BONCAT, we purified and enriched over 300 newly synthesized proteins.
Materials and Methods
Reagents
All reagents were ACS grade and purchased from Sigma unless noted otherwise. SKF81297 was purchased from Tocris Bioscience and used at a final concentration of 40 μM in BONCAT experiments and 25-75 μM in FUNCAT experiments. Triazole ligand (Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine) was purchased from Sigma and used at a stock concentration of 200 mM in DMSO. The following primary antibodies (Ab) were used: mouse anti-Bassoon mAb (clone SAP7F407; WB 1:1,000; StressGen Biotechnologies), rabbit anti-Piccolo 44a pAb (WB 1:2,000; [28]), mouse anti-Pan-Cadherin mAb (clone CH-19; WB 1:1,000, Sigma-Aldrich), mouse anti-(X000DF)-actin mAb (clone AC-15; WB 1:2,000; Sigma Aldrich), rabbit anti-biotin pAb (WB 1:10,000; Bethyl Laboratories Inc), mouse anti-CaM Kinase II mAB (clone 6G9; WB 1:1,000; Thermo Scientific Pierce Antibodies), mouse anti-FMRP mAb (WB 1:1,000; Chemicon International), rabbit anti-Homer 1 pAb (WB 1:1,000; Synaptic Systems), mouse anti-MAP2 mAb (WB 1:1,000; Sigma-Aldrich). The following secondary antibodies were used at a dilution of 1:10,000 for WB: HRP-conjugated anti-rabbit Ab, HRP-conjugated anti-mouse Ab, HRP-conjugated anti-guinea pig Ab (Jackson Immuno Research) and anti-rabbit Alexa 488 Ab (IF: 1:1,000; Invitrogen).
Organic synthesis
AHA was prepared as described previously [29, 30]. TexasRed-PEO2-Alkyne and Biotin-PEO-propargylamide were prepared as described before [21, 23]. The disulfide biotin alkyne tag (DST-alkyne) was prepared according to a recently published protocol [31].
Preparation and maintenance of cultured neurons
Dissociated hippocampal neurons were prepared and maintained as previously described [32]. Briefly, hippocampi from postnatal day 0 to 2 rat pups (strain Sprague-Dawley) were dissected out and dissociated by either trypsin or papain and plated at a density of 40,000 cells/cm2 onto poly-D-lysine-coated glass-bottom Petri dishes (Mattek). Cultures were maintained in Neurobasal A medium containing B-27 and Glutamax supplements (Invitrogen) at 37°C for 18–24 days before use.
Visualization of new protein synthesis via FUNCAT
Monitoring of SKF81297-dependent protein synthesis in dissociated hippocampal neurons was done as described previously [23]. Briefly, growth medium was removed from neuronal cultures and replaced with methionine-free Hibernate A (HibA) medium (BrainBits LLC.) for 30 min to deplete endogenous methionine. For AHA labeling, HibA was supplemented with 2 mM AHA in the presence or absence of 25-75 μM SKF 81297. After incubation at 37°C, 5% CO2 for 1 hour, cells were incubated in HibA supplemented with 2 mM methionine for 10 min and subsequently washed with chilled PBS-MC (1mM MgCl2, 0.1 mM CaCl2 in PBS pH = 7.4) on ice followed by immediate fixation with chilled 4% paraformaldehyde, 4% sucrose in PBS-MC for 20 min at room temperature (RT).
For CuAAC, in order to avoid copper bromide-derived precipitates, TCEP in combination with copper sulfate was used to generate the Cu(I) catalyst during the CuAAC reaction. Briefly, a CuAAC reaction mix composed of 200 μM triazole ligand (stock solution dissolved at 200 mM in DMSO), 2 μM fluorescent alkyne tag, 400 μM TCEP and 200 μM CuSO4 was mixed in PBS (pH 7.6) with vigorous vortexing after addition of each reagent. Hippocampal primary cultures were incubated overnight at 20°C with the CuAAC reaction mix in a humid box under gentle agitation. Following incubation, cells were washed three times for 10 min each at RT with 1% Tween-20, 0.5 mM EDTA in 1x PBS pH 7.4 followed by three rinses with 1x PBS pH 7.4 prior to immunostaining with rabbit polyclonal anti-MAP2 antibody (1:1000, Chemicon) and anti-rabbit Alexa 488 secondary antibody (1:1000, Invitrogen) using standard conditions.
Microdissected slices
Hippocampal slices (500 μm-thick) were prepared from adult male (Sprague Dawley) rats, as previously described [33]. Dendrites were physically separated from the cell bodies by a transection along the border between the stratum pyrimidale and stratum radiatum in the CA1 region of the hippocampus [3]. We made three additional cuts to generate mini slices that include the synaptic neuropil of stratum radiatum in area CA1, but do not include principal cell bodies. Dendritic slices were recovered on artificial cerebrospinal fluid (aCSF) for 2 hours prior to noncanonical amino acid incubation and SKF81297 stimulation. In some experiments the isotopic amino acid, 13C6-arginine (13C6-Arg) was used as an additional validation for new protein synthesis.
Sample preparation and processing for MS and Western Blot analyses
For samples to be processed via MS we used a DST-alkyne probe for the click-reaction. Briefly, AHA-labeled microdissected and transectioned slices were homogenized in PBS pH 7.6 with protease inhibitors without EDTA (“PI w/o EDTA”; Roche). Benzonase (Sigma) and SDS (1% final concentration) were added. Samples were boiled for five minutes, then cooled before they were centrifuged at 3000 rpm at 4°C for 5 minutes. The resulting supernatants were treated with β-mercaptoethanol (2% final concentration) for 1 hour in the dark at room temperature. The proteins were acetone precipitated and the resulting pellet resuspended in PBS pH 7.6 with PI w/o EDTA, 0.1% SDS and 0.05% Triton X-100. The CuAAC reaction, consisting of 200 μM Triazole ligand, 25 μM DST-alkyne probe, 10 μg/ml Cu(I) bromide suspension, was incubated at 4°C overnight under agitation. The CuAAC reaction was centrifuged at 3000 rpm at 4°C for 5 minutes and the supernatant was desalted using PD-10 columns (GE Healthcare) and eluted in 0.05% SDS in PBS pH 7.6. A dot blot analysis was performed to determine the newly synthesized protein concentration for the NeutrAvidin purification. The desalted samples were then boiled for 8 minutes and cooled to room temperature. In the meantime, the NeutrAvidin resin (Pierce/Thermo) was washed three times with PBS pH 7.6. To the cooled, desalted sample PBS pH 7.6 containing PI w/o EDTA were added to achieve a final concentration of 0.05% SDS and 1% Nonidet P-40. The sample was then applied to the washed NeutrAvidin slurry and incubated at room temperature with agitation for 2 hours. After binding reaction was complete, the NeutrAvidin beads were washed twice with 1% NP-40 in PBS pH 7.6, twice with PBS pH 7.6, and once with 50 mM ammonium bicarbonate. The bound proteins were released via reduction by twice incubating the beads with 1% β-mercaptoethanol in 50 mM ammonium bicarbonate for 30 minutes in the dark under agitation. The proteins were lyophilized to 100-150 μl final volume and methanol/chloroform precipitated. The protein pellet was resuspended in 8 M urea in 100 mM Tris-HCl pH 8.5. Samples were prepared for MS as described by Dieterich et al. [22]. After trypsin digestion, samples were desalted using an HPLC coupled to a reverse-phase resin peptide macrotrap column (Microm Bioresources, Auburn, CA). MS/MS was performed on an LTQ-FT instrument (Thermo Fisher Scientific) using standard conditions. The mass spectrometry analysis was performed as described by Bell et al. [34].
For Western blot analysis of particular proteins microdissected and transectioned slices were homogenized and lysed as described above. Lysates were diluted to 0.1% SDS, 0.2% Triton X-100 in PBS, pH 7.8 w/o EDTA before addition of 200 μM triazole ligand, 50 μM Biotin-PEO-propargylamide tag and 10 μg/ml Cu (I) bromide. The reaction was allowed to proceed overnight at 4°C under constant agitation, and excess reagents were removed by gel filtration through PD-10 columns (GE Healthcare). Identical sample concentrations were calculated based on amido black protein concentration assay and re-checked via silver stained gels [35]. Adjusted clicked protein samples were purified on NeutrAvidin affinity resin (Pierce/Thermo). After extensive washing in incubation buffer, followed by washes in 1% Nonidet P-40 as well as 0.05% SDS in PBS, proteins were eluted from the resins with SDS sample buffer supplemented with 10 mM Biotin.
Bioinformatics processing and criteria
The RAW files were converted to mgf files using ReAdW4Mascot2 (http://peptide.nist.gov/metric). Spectra were searched using SEQUEST on a Sorcerer computer (Sage-N Research). Spectra were searched against the IPI rat database (v. 3.71, 39,611 rat entries plus 262 contaminant entries appended to an equal number of decoy sequences constructed as described in Cox and Mann, Nat Biotech, 2008, 26, 1367-72). Precursor mass tolerance was 10ppm and fragment ion tolerance was 1.0 Da. Carboxyamidomethylation of Cys (+57.021) was set as a fixed modification. Oxidation of Met (+15.995), Met to AHA (-4.986), DST tagged Met (+195.076), heavy labeled Arg (+6.020), heavy labeled Pro (+5.017), and carbamylation of peptide N-terminus (+43.006) were set as variable modifications. Search results were analyzed with PeptideProphet and ProteinProphet and then loaded into Scaffold (v. 3.00.06, Proteome Software, Portland, Oregon, USA) to validate MS/MS identifications.
After this processing, we evaluated our data using two different sets of criteria: (1) 80% peptide and protein probabilities, and at least one peptide for each protein identification (0.3% protein FDR, <0.1% peptide FDR); and (2) 90% peptide and 95% protein probabilities and at least two peptides for each protein identification (0% protein and peptide FDR). This allowed us to assess both medium and high stringency data sets.
A total of three independent experimental sets (both SKF81297 treatment and vehicle control) were processed via MS/MS. Ultimately, we only included proteins on the candidate lists (‘SKF81297’, ‘control’ and ‘common’) that were present in at least two sample groups. To identify these proteins we examined the low-confidence protein lists and the high confidence protein lists within themselves, and also the low and high lists to one another. The two programs Scaffold and Cytoscape [36] were used for comparison and visualization of the data sets, GOEAST (Gene Ontology Enrichment Analysis Software Toolkit; http://omicslab.genetics.ac.cn/GOEAST/) for gene ontology (GO) assessment. For systematic pathway analysis we used Ingenuity IPA Software. In-house developed scripts were used to compare the three candidate lists with a database of the neuropil transcriptome [37], and with SynProt (www.synprot.de), a proteomics database derived from detergent-resistant synaptic junctions fractions. SynProt currently contains 2133 non-redundant entries of rat, mouse and some human proteins, which have been manually extracted from twelve proteomic studies and verified for synaptic subcellular localization. Note, that this database contains not only PSD scaffold proteins but also everything that is biochemically or morphologically associated with synaptic junctions (extracellular matrix, presynaptic cytomatrix of the active zone, glial endfeet, etc).
Results
Visualization of newly synthesized proteins in hippocampal neurons
To study the dopaminergic hippocampal subproteome, we first visualized the newly synthesized proteome using FUNCAT. We examined whether a change in newly synthesized proteins could be detected in dendrites after stimulation of D1-like dopaminergic receptors. Primary hippocampal cultures were first incubated with methionine-free media for 30 minutes and then incubated with AHA for 1 hour, with and without the D1/D5-agonist SKF81297 (25-75 μM). This incubation was immediately chased with a methionine incubation for 10 minutes to terminate the charging of AHA onto tRNAs. Then, the neurons were immediately fixed, tagged with a fluorescent probe (see methods), and immunostained for MAP2, a dendritic protein marker. In dissociated hippocampal cultures treated with the D1/D5-agonist, there was a concentration-dependent 25-60% increase in FUNCAT fluorescence in the distal dendrites (Figure 1).
Figure 1.
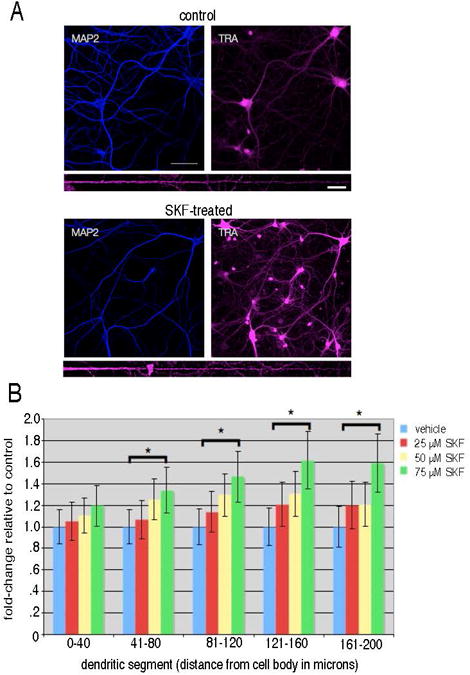
SKF81297 stimulation produced an increase in newly synthesized proteins in the distal dendrites. (A) Representative images from dissociated hippocampal cultures (DIV 21) incubated with 2 mM AHA in HibA for 1 hour (control) and stimulated with 40 μM SKF81297 (SKF-treated). Following the click reaction to visualize newly synthesized proteins (labeled TRA) immunostaining for MAP2 was conducted to visualize the dendrites (blue signal). Below each set of images a straightened representative dendrite is shown for each condition. Scalebar = 10 μm. (B) Summary graph showing the effects of dopamine agonist stimulation on new protein synthesis detected with FUNCAT. Dissociated hippocampal cultures were incubated with different concentrations (25 μM, 50 μM, 75μM) of the dopamine receptor agonist SKF81297 for 1 hour in 2 mM AHA in HibA. The data were analyzed by binning 40 micron segments of the dendrite as a function of distance from the cell body. Treatment of neurons with 75 μM SKF 81297 resulted in a significant enhancement of the newly synthesized protein signal in all dendritic segments greater than 40 microns from the cell body. * denotes p <0.05.
Identification of newly synthesized proteins in hippocampal neurons
To determine the proteins that are synthesized in the neuropil, we microdissected acute hippocampal slices to obtain sections of the neuropil from area CA1 (Figure 2). After recovery in aCSF for 2 hours, the dendritic slices were incubated with 4 mM AHA and 4 mM 13C6-Arg in aCSF with or without 40 μM SKF81297 for 2.5 hours before they were harvested and flash-frozen until further processing. Newly synthesized proteins were tagged and affinity-purified, as previously described [21, 22] with the following modification: newly synthesized proteins were tagged with a DST-alkyne probe [31] in order to facilitate their reduction and release from the NeutrAvidin column using β-mercaptoethanol. Released proteins were then digested with trypsin to generate the peptides for proteomic analysis via MS/MS (Figure 3). We conducted three biological replicates of the experimental (dopamine-agonist SKF81297-treated) and four biological replicates of the control (vehicle-treated) group.
Figure 2.

Microdissection of a dendritic segment from an acute hippocampal slice. A. 500μm-thick acute hippocampal slices isolated from Sprague-Dawley rats (P24-26) were microdissected in order to obtain CA1 tissue enriched in synaptic neuropil (outlined in red; stratum radiatum) and de-enriched for principal cell bodies (outlined in green; stratum pyramidale). Scalebar = 1 mm. B. Cartoon depicting the dimensions and dendritic orientation of a typical microdissected slice.
Figure 3.
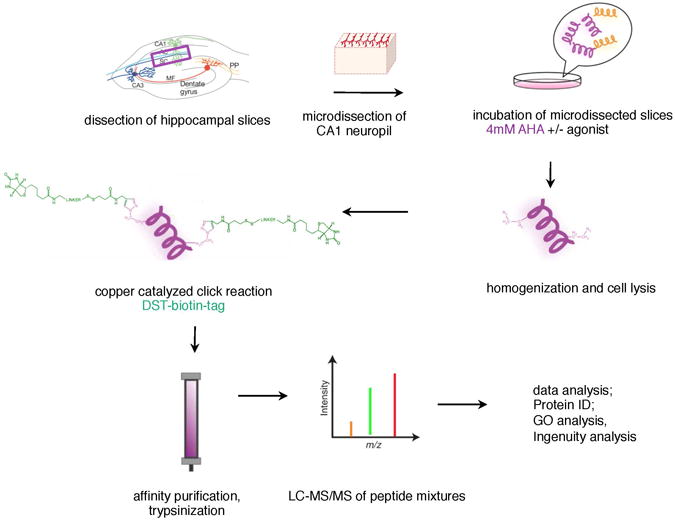
Scheme of work-flow for BONCAT experiments. Acute hippocampal slices were prepared and microdissected neuropil sections cut. Mini slices were incubated in 4mM AHA in the presence or absence of the dopamine receptor agonist for 2.5 hrs. Then slices were homogenized and cells were lysed. The click reaction was carried out (see methods) and biotin-conjugated proteins were affinity purified, eluted from the column with β-mercaptoethanol and then trypsinized. Trypsinized peptides were analyzed via LC-MS/MS and then proteins and protein networks were analyzed using a variety of software packages.
The hippocampal dopaminergic subproteome
Across the three complete datasets that we acquired and analyzed, we identified a total of 891 unique proteins (Spectrum Report, Table 1; Protein Table, Table 2) spanning a multitude of different gene ontology (GO) categories of biological processes, molecular function and cellular compartments (see Supplementary Table S1). Six hundred and sixteen proteins were common to both the vehicle and D1/D5-agonist-treated samples; there were 175 proteins that were unique to the vehicle group and 100 proteins that were unique to the agonist-treated sample (Figure 4A; Table 2). Four hundred forty-nine of these proteins are associated with a synaptic localization as derived from SynProt, a comprehensive synapse-associated protein database. In the SKF81297 group 38/100 proteins, and from the vehicle group 53/175 proteins, and from the common group 358/616 candidate proteins have an association with synapses (Table 3, Figure 4B). Since the neuropil sections from area CA1 contain also astrocytes and interneurons, we refined our candidate list for specifically dendritically and axonally synthesized proteins by using a neuropil transcriptome database [37]. Out of a total of 2550 transcripts potentially translated in hippocampal neurites, we found 224 candidate proteins in both groups (Table 4), 30 proteins in the D1-agonist-treated group (Table 5) and 57 in the vehicle-treated group (Table 6, Figure 4C). For our subsequent analyses on the hippocampal dopaminergic proteome we considered only these neuropil transcriptome filtered candidates. Taken together all candidate proteins span 276 GO categories for biological processes, 129 GO categories for cellular compartments, and 86 categories for molecular function (Supplementary Table S2). Interestingly, candidate proteins unique to the SKF81297-treated sample belonged to GO categories specific for synaptic localization (synapse, synaptosome, synapse part) and protein transport, signaling and synaptic transmission processes (Figure 5A+B, Supplementary Table S3), whereas vehicle-treated candidate proteins did not show synapse-specific association (Figure 5C, Supplementary Table S4).
Figure 4.

Venn diagram summary of three complete MudPIT experiments. (A) The two circles represent the 891 unique proteins that were found in the three experiments. The 175 proteins that were found solely in the control group are represented in green, the 100 proteins unique to the SKF81297-stimulated group in magenta, and the 616 proteins found in both groups in grey. (B) 449 candidate proteins of the three experiments as shown in the overlap area of all candidates (blue circle) with the database SynProt (see main text; orange circle) can be assigned to a synaptic localization or association. (C) The two circles represent the candidates after filtering using a transcriptome database of the hippocampal neuropil mRNAs. Thirty proteins were found solely in the D1-agonist treated group (magenta circle), 57 proteins were found in the control group only (green circle). The remainder 224 proteins were found in both groups (grey overlap area).
Figure 5.
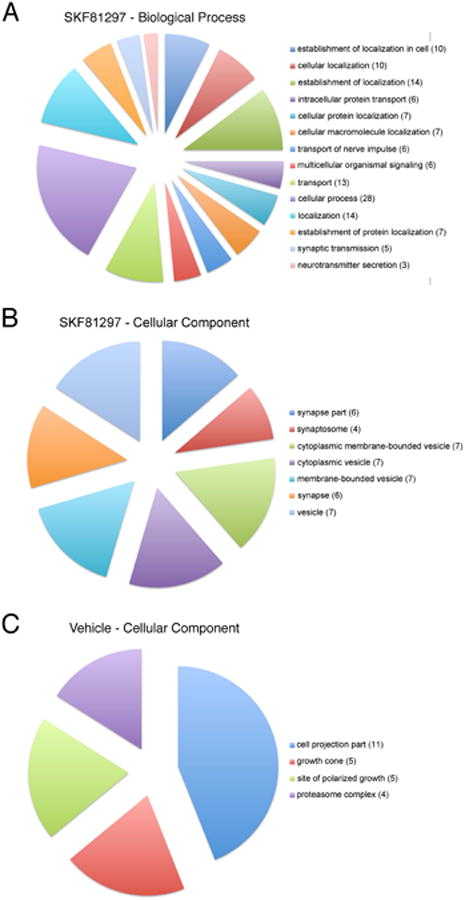
Gene ontology analysis of the dopaminergic hippocampal subproteome. (A+B) Pie charts depicting the representations of the two GO categories biological processes (A) and cellular compartments (B) for the candidates of the SKF81297-treated samples. (C) Vehicle-treated candidates, although larger in total number, can be attributed only to a single GO category (cellular compartment) with 4 GO subcategories. Numbers in brackets indicate the number of candidate proteins within the GO subcategory.
Using Ingenuity, we analyzed the protein networks that are represented by the hits we obtained for both D1/D5-agonist- and vehicle-treated samples in more detail. In addition to standard housekeeping proteins, such as those found in glycolysis, there were a number of proteins associated with synaptic plasticity such as gephyrin [38], CaMKIIa [39], Calnexin [40], PSD95 [41], Kalirin [42], and several glutamate receptors. A candidate protein that was identified in previous work [20], GluR1, was also present in this data as a protein that was expressed in both conditions, but with greater sequence coverage in the agonist-treated sample. Notably, protein networks involved in cellular assembly and organization, nervous system development and function, cell-to-cell signaling and interaction were well represented by the candidates identified in the SKF81297-treated samples but not among the candidates from the vehicle-treated samples (Figure 6A). Here, the presynaptic protein Munc13-1 (UNC13A [43]), Rabphilin3A (RPH3A, [44]), the voltage-gated potassium channel subunit beta-1 (KCNAB1, [45]), Ras and Rab interactor 1 (RIN1, [46]) and cell adhesion molecule1 (CADM1, [47]) were present in the D1-agonist-treated but not in the control samples. In addition, we identified many ribosomal subunits specifically in the D1/D5 agonist-treated sample, including the 40S ribosomal protein S25 (RPS25, [48]), rho guanine nucleotide exchange factor 2 (ARHGEF2, [49]), splicing factor, arginine/serine-rich 1 (SRSF1, [50]), chromosome segregation 1-like protein (CSE1L, [51]), RAN [52]), Importin5 (IPO5, [53]), Adenosylhomocysteinase (AHCY, [54]), 60S ribosomal protein L13a (RPL13A, [55]) as represented in protein network depicted in Figure 6B. This, along with the identification of some translation elongation and initiation factors, suggests that dopamine signaling enhances the translational capacity of dendrites, consistent with data we obtained using FUNCAT (Figure 1). Though cytoskeletal proteins are widely abundant, we noticed a marked increase in the number of identified structural proteins, perhaps to accommodate an increase in the number of synaptic contacts induced by the stimulation of the D1/D5 receptors [20]. Additional proteins that we observed were from the presynaptic terminal or for cell adhesion. Consistent with the GO analysis, proteins characteristic of the vehicle-treated sample include proteasome-associated proteins (Figure 7), such as PSMD12 [56], PSMA1 [57], PSMB5 [58], PSMC3 [59], the presenilin-1 associated protein MTCH1 [60] and α-Synuclein (SNCA, [61]). The complete list of identified protein networks for both the D1-agonist and vehicle-treated dendritically synthesized subproteomes can be found in Tables 7 and 8.
Figure 6.
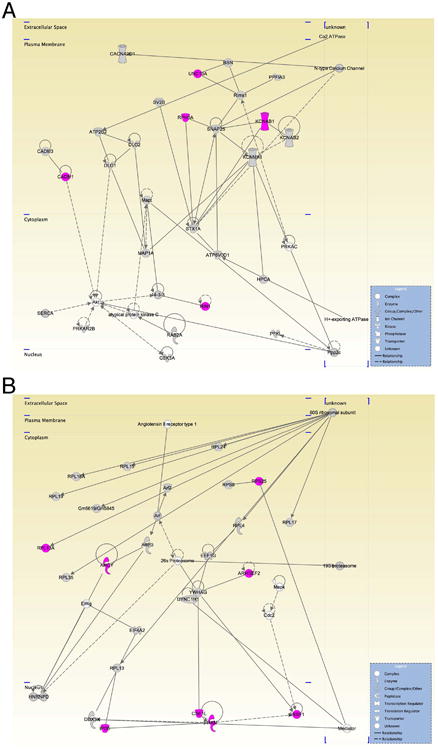
Protein networks represented by the candidate proteins identified in the SKF81297-treated samples. (A) Protein network involved in cellular assembly and organization, nervous system development and function, cell-to-cell signaling and interaction consisting of the following molecules: ATPase, Ca2+ transporting, plasma membrane 2 (ATP2B2); ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 (ATP6V0D1); atypical protein kinase C; bassoon (BSN); Ca2 ATPase; calcium channel, voltage-dependent 2/δ subunit 1 (CACNA2D1); cell adhesion molecule 1 (CADM1); cell adhesion molecule 3 (CADM3); discs, large homolog 1 (Drosophila) (DLG1); discs, large homolog 2 (Drosophila) (DLG2); glycogen synthase kinase 3 α(GSK3); H+-exporting ATPase; hippocalcin (HPCA); potassium voltage-gated channel, shaker-related subfamily, B member 1 (KCNAB1); potassium voltage-gated channel, shaker-related subfamily, B member 2 (KCNAB2); potassium large conductance calcium-activated channel, subfamily M, α member 1 (KCNMA1); microtubule-associated protein 1A (MAP1A); microtubule-associated protein τ (Mapt); N-type Calcium Channel; phosphofructokinase, liver, (PFKL); protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF) interacting protein (liprin), α 3, (PPFIA3); Ppp2c; PRKAC; protein kinase, cAMP-dependent, regulatory, type II, B (PRKAR2B); RAB2A, member RAS oncogene family (RAB2A); regulating synaptic membrane exocytosis 1 (Rims1); Ras and Rab interactor 1 (RIN1); rabphilin 3A homolog (mouse) (RPH3A); SERCA; synaptosomal-associated protein, 25kDa (SNAP25); syntaxin 1A (brain) (STX1A); synaptic vesicle glycoprotein 2B (SV2B); unc-13 homolog A (C. elegans) (UNC13A). (B) Protein network involved in protein synthesis, cellular assembly and organization, genetic disorder: 19S proteasome; 26S Proteasome; 60S ribosomal subunit; adenosylhomocysteinase (AHCY); Angiotensin II receptor type 1; ADP-ribosylation factor 2 (Arf2); ADP-ribosylation factor 3 (ARF3); Arf; Rho/Rac guanine nucleotide exchange factor (GEF) 2 (ARHGEF2); Cdc2; CSE1; chromosome segregation 1-like (yeast) (CSE1L); DEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (DDX3X); dynein, cytoplasmic 1, heavy chain 1 (DYNC1H1); eukaryotic translation elongation factor 1 γ (EEF1G); eukaryotic translation initiation factor 4A2 (EIF4A2); Eif4g; ribosomal protein L7A pseudogene (Gm5619/Gm5845); heterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (HNRNPD); importin 5 (IPO5); Mapk; Mediator; RAN, member RAS oncogene family (RAN); ribosomal proteins RPL4; RPL13; RPL15; RPL17; RPL19; RPL24; RPL35; RPL13A; RPL18A; RPS8; RPS25; serine/arginine-rich splicing factor 1 (SRSF1); tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, γ polypeptide (YWHAG). Icons in grey, molecules present in both SKF81297- and vehicle-treated samples; icons in magenta, proteins present in SKF81297-treated samples only; icons in white, molecules not identified in either sample.
Figure 7.
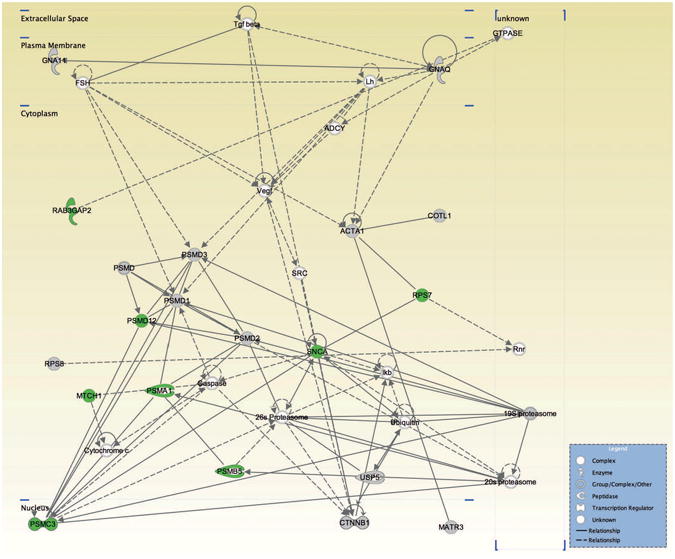
Protein network represented by the candidate proteins identified in the vehicle-treated samples: 19S proteasome; 20S proteasome; 26S Proteasome; actin, α 1, skeletal muscle (ACTA1); ADCY; Caspase; coactosin-like 1 (Dictyostelium) (COTL1); catenin (cadherin-associated protein), B 1, 88kDa (CTNNB1); Cytochrome c; FSH; guanine nucleotide binding protein (G protein), α 11 (Gq class) (GNA11); guanine nucleotide binding protein (G protein), q polypeptide (GNAQ); GTPASE; Ikb; Lh; matrin 3 (MATR3); mitochondrial carrier 1 (MTCH1); proteasome (prosome, macropain) subunit, alpha type 1 (PSMA1), B type 5 (PSMB5), ATPase, 3 (PSMC3), non-ATPase 1 (PSMD1); non-ATPase 2 (PSMD2), non-ATPase 3 (PSMD3), non-ATPase 12 (PSMD12); PSMD; RAB3 GTPase activating protein subunit 2 (non-catalytic) (RAB3GAP2); Rnr; ribosomal protein S7 (RPS7); ribosomal protein S8 (RPS8); synuclein, alpha (non A4 component of amyloid precursor) (SNCA); SRC; Tgf B; Ubiquitin; ubiquitin specific peptidase 5 (isopeptidase T) (USP5); vEGF. Icons in grey, molecules present in both vehicle- and SKF81297-treated samples; icons in green, proteins present in vehicle-treated samples only; icons in white, molecules not identified in either sample.
In the last set of experiments, we validated a subset of newly synthesized protein candidates using Western blots. Samples were prepared as above for the proteomic analysis with the following modifications: following purification but prior to trypsin treatment, the proteins were precipitated for Western blot analysis. Using this approach, we confirmed the presence of newly synthesized MAP2, CAMKIIα, β-actin and Pan-Cadherin. Furthermore, we identified Homer 1 as newly synthesized (Figure 8). A number of other proteins were specific to SKF81297-stimulation, such as PKA and CREB, both to be expected due to the D1-like receptors' role in the cAMP pathway. Interestingly, there were also proteins involved in synaptic plasticity, such as the presynaptic proteins bassoon and piccolo as well as those involved in protein translation, Eef1/2 and Eif4b/e (data not shown).
Figure 8.
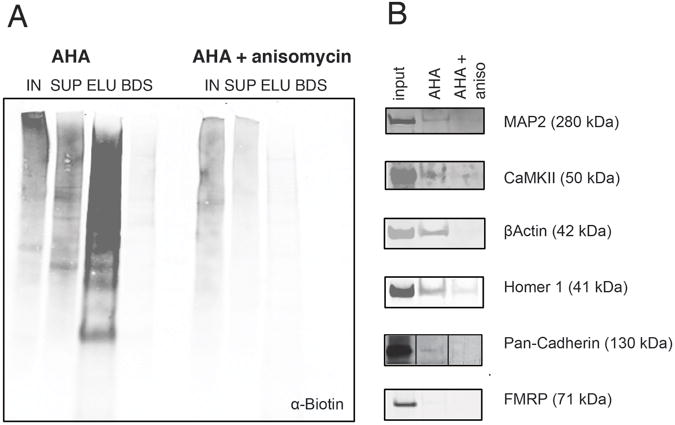
Validation of newly synthesized candidate proteins via Western blot analysis. Newly synthesized proteins were isolated from dendritic segments incubated with aCSF and 4 mM AHA +/- SKF81297 stimulation (40 μM) or +/- anisomycin (40 μM) for 2.5 hours and then Western blot analysis was conducted. (A) Western blot analysis using an anti-biotin antibody to detect newly synthesized proteins that successfully incorporated AHA. In the lanes shown IN =1% input; SUP =1% supernatant; ELU =10% of the eluate; BDS = 10% beads (after elution). In the AHA-treated samples, the majority of the newly synthesized proteins were detected in the eluate and very little signal was left on the beads. In experiments in which anisomycin was included, there was much less biotinylated signal present in the entire blot, and the amount of biotinylated protein present in the eluate was substantially reduced. (B) Following elution of biotinylated proteins, Western blot analysis for individual proteins of interest was conducted. Clear evidence for newly synthesized MAP2, CAMKIIα, β-actin, Pan-Cadherin and Homer 1 was obtained. MAP2, CAMKIIα, β-actin, and Protocadherin were candidates identified by MS. FMRP served as a negative control.
Discussion
In this study we paired the FUNCAT and BONCAT techniques with dopaminergic stimulation to examine whether dopamine can modulate the hippocampal proteome. Both techniques introduce the small bioorthogonal azide-handle into nascent proteins using the cell's own protein synthesis machinery. This metabolic labeling enabled us to visualize D1/D5 dopamine receptor agonist-dependent global proteome alterations in primary hippocampal neurons when using a fluorescent alkyne tag. In addition, we specifically identified dendritically synthesized proteins in hippocampal neuropil sections via the use of an affinity tag to enrich and affinity purify newly synthesized proteins prior to tandem MS analysis. Consistent with previous work (Smith et al., 2005) we observed an upregulation in protein translation upon the activation of the D1/D5 dopaminergic receptors in hippocampal primary neurons (Figure 1). Moreover, we isolated the hippocampal neuropil subproteome by using microdissected acute hippocampal slices. Using BONCAT we identified a total of 891 unique proteins in the neuropil layer of which 50% have a synaptic association. As these neuropil sections contain not only neuronal processes but also astrocytes and the somata of interneurons, we filtered our candidate lists using a recently published neuropil transcriptome database (Cajigas et al., 2012) to focus only on candidates originating from dendrites and axons. Interestingly, the remaining candidates of the D1/D5 dopamine receptor agonist–treated group show a remarked association with synaptic localization and function compared to the candidate list unique to the control (unstimulated) group. In agreement with previously published work (Smith et al., 2005), we found GluA1 among the identified candidates as well as components of the translation machinery demonstrating the dendritic protein synthesis-dependent response of dopaminergic signaling. This study is the first proteomic snapshot into the neurite-specific contribution to the overall neuronal proteome. Over 300 candidate proteins were synthesized over the course of 2.5 hours in dendrites and axons, emphasizing the importance of immediate translational responses in both dendrites and axons. Finally, we also provide a series of new candidates that were separate from the previously identified PKA/cAMP pathway that is implicated in D1-like receptor signaling. Notably, presynaptic proteins, cell adhesion molecules and constituents of the protein synthesis machinery were readily synthesized upon stimulation of D1/D5 receptors.
In summary, the candidates that we have identified provide greater insight into the wide range of proteins that are potentially involved not only in the maintenance of late long-term potentiation (L-LTP), but also possibly integral to dopaminergic signaling and the treatment of neurodegenerative disease. Future experiments will be aided by the use of neuron-specific tagging methods [62] in which a mutant methionyl-tRNA synthetase is expressed in pyramidal neurons. In addition, as other agonists, such as BDNF [3] and DHPG [4] have been shown to elicit protein-synthesis-dependent changes at synapses, it will be interesting to compare these proteomes with the SKF81297-proteome to identify common and distinct plasticity proteins.
Supplementary Material
Acknowledgments
This work was supported by the German Academy for Natural Scientists Leopoldina and the DFG (D.C.D.), the NIH (E.M.S. and D.A.T. 5R01GM62523-9), HHMI (E.M.S.), Gordon and Betty Moore Foundation (M.J.S, S.H.) and the Beckman Institute (M.J.S, S.H.).
Abbreviations
- AHA
Azidohomoalanine
- BONCAT
bioorthogonal non-canonical amino acid tagging
- FUNCAT
fluorescent non-canonical amino acid tagging
- HPG
homopropargylglycine
Footnotes
Conflict of interest statement. The authors declare no competing financial and commercial interests.
References
- 1.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci. 1982;2:284–291. doi: 10.1523/JNEUROSCI.02-03-00284.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang H, Schuman EM. A requirement for local protein synthesis in neurotrophin-induced hippocampal synaptic plasticity. Science. 1996;273:1402–1406. doi: 10.1126/science.273.5280.1402. [DOI] [PubMed] [Google Scholar]
- 4.Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1256. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- 5.Sutton MA, Wall NR, Aakalu GN, Schuman EM. Regulation of dendritic protein synthesis by miniature synaptic events. Science. 2004;304:1979–1983. doi: 10.1126/science.1096202. [DOI] [PubMed] [Google Scholar]
- 6.Sutton MA, Ito HT, Cressy P, Kempf C, et al. Miniature neurotransmission stabilizes synaptic function via tonic suppression of local dendritic protein synthesis. Cell. 2006;125:785–799. doi: 10.1016/j.cell.2006.03.040. [DOI] [PubMed] [Google Scholar]
- 7.Cajigas IJ, Will T, Schuman EM. Protein homeostasis and synaptic plasticity. Embo Journal. 2010;29:2746–2752. doi: 10.1038/emboj.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niere F, Wilkerson JR, Huber KM. Evidence for a fragile x mental retardation protein-mediated translational switch in metabotropic glutamate receptor-triggered arc translation and long-term depression. J Neurosci. 2012;32:5924–5936. doi: 10.1523/JNEUROSCI.4650-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang Y, Rosenstein A, Kreiman G, Schuman EM, Kennedy MB. Tetanic stimulation leads to increased accumulation of Ca(2+)/calmodulin-dependent protein kinase II via dendritic protein synthesis in hippocampal neurons. J Neurosci. 1999;19:7823–7833. doi: 10.1523/JNEUROSCI.19-18-07823.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito HT, Schuman EM. Frequency-dependent signal transmission and modulation by neuromodulators. Front Neurosci. 2008;2:138–144. doi: 10.3389/neuro.01.027.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gasbarri A, Sulli A, Packard MG. The dopaminergic mesencephalic projections to the hippocampal formation in the rat. Prog Neuropsychopharmacol Biol Psychiatry. 1997;21:1–22. doi: 10.1016/s0278-5846(96)00157-1. [DOI] [PubMed] [Google Scholar]
- 12.Ihalainen JA, Riekkinen P, Jr, Feenstra MG. Comparison of dopamine and noradrenaline release in mouse prefrontal cortex, striatum and hippocampus using microdialysis. Neurosci Lett. 1999;277:71–74. doi: 10.1016/s0304-3940(99)00840-x. [DOI] [PubMed] [Google Scholar]
- 13.Gasbarri A, Sulli A, Innocenzi R, Pacitti C, Brioni JD. Spatial memory impairment induced by lesion of the mesohippocampal dopaminergic system in the rat. Neuroscience. 1996;74:1037–1044. doi: 10.1016/0306-4522(96)00202-3. [DOI] [PubMed] [Google Scholar]
- 14.El-Ghundi M, Fletcher PJ, Drago J, Sibley DR, et al. Spatial learning deficit in dopamine D(1) receptor knockout mice. Eur J Pharmacol. 1999;383:95–106. doi: 10.1016/s0014-2999(99)00573-7. [DOI] [PubMed] [Google Scholar]
- 15.Frey U, Matthies H, Reymann KG. The effect of dopaminergic D1 receptor blockade during tetanization on the expression of long-term potentiation in the rat CA1 region in vitro. Neurosci Lett. 1991;129:111–114. doi: 10.1016/0304-3940(91)90732-9. [DOI] [PubMed] [Google Scholar]
- 16.Frey U, Huang YY, Kandel ER. Effects of cAMP simulate a late stage of LTP in hippocampal CA1 neurons. Science. 1993;260:1661–1664. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- 17.Huang YY, Kandel ER. D1/D5 receptor agonists induce a protein synthesis-dependent late potentiation in the CA1 region of the hippocampus. Proc Natl Acad Sci U S A. 1995;92:2446–2450. doi: 10.1073/pnas.92.7.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthies H, Becker A, Schroeder H, Kraus J, et al. Dopamine D1-deficient mutant mice do not express the late phase of hippocampal long-term potentiation. Neuroreport. 1997;8:3533–3535. doi: 10.1097/00001756-199711100-00023. [DOI] [PubMed] [Google Scholar]
- 19.Swanson-Park JL, Coussens CM, Mason-Parker SE, Raymond CR, et al. A double dissociation within the hippocampus of dopamine D1/D5 receptor and beta-adrenergic receptor contributions to the persistence of long-term potentiation. Neuroscience. 1999;92:485–497. doi: 10.1016/s0306-4522(99)00010-x. [DOI] [PubMed] [Google Scholar]
- 20.Smith WB, Starck SR, Roberts RW, Schuman EM. Dopaminergic stimulation of local protein synthesis enhances surface expression of GluR1 and synaptic transmission in hippocampal neurons. Neuron. 2005;45:765–779. doi: 10.1016/j.neuron.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Dieterich DC, Link AJ, Graumann J, Tirrell DA, Schuman EM. Selective identification of newly synthesized proteins in mammalian cells using bioorthogonal noncanonical amino acid tagging (BONCAT) Proc Natl Acad Sci. 2006;103:9482–9487. doi: 10.1073/pnas.0601637103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dieterich DC, Lee JJ, Link AJ, Graumann J, et al. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–540. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 23.Dieterich DC, Hodas JJ, Gouzer G, Shadrin IY, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nat Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Link AJ, Tirrell DA. Cell Surface Labeling of Escherichia coli via Copper(I)-Catalyzed [3+2] Cycloaddition. J Am Chem Soc. 2003;125:11164–11165. doi: 10.1021/ja036765z. [DOI] [PubMed] [Google Scholar]
- 25.Link AJ, Mock ML, Tirrell DA. Non-canonical amino acids in protein engineering. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 26.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. P Natl Acad Sci USA 2002. 99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beatty KE, Liu JC, Xie F, Dieterich DC, et al. Fluorescence visualization of newly synthesized proteins in mammalian cells. Angew Chem Int Ed Engl. 2006;45:7364–7367. doi: 10.1002/anie.200602114. [DOI] [PubMed] [Google Scholar]
- 28.Dick O, Hack I, Altrock WD, Garner CC, et al. Localization of the presynaptic cytomatrix protein Piccolo at ribbon and conventional synapses in the rat retina: comparison with Bassoon. Journal of Comparative Neurology. 2001;439:224–234. doi: 10.1002/cne.1344. [DOI] [PubMed] [Google Scholar]
- 29.Link AJ, Vink MK, Tirrell DA. Preparation of the functionalizable methionine surrogate azidohomoalanine via copper-catalyzed diazo transfer. Nat Protoc. 2007;2:1879–1883. doi: 10.1038/nprot.2007.268. [DOI] [PubMed] [Google Scholar]
- 30.Link AJ, Vink MK, Tirrell DA. Synthesis of the functionalizable methionine surrogate azidohomoalanine using Boc-homoserine as precursor. Nat Protoc. 2007;2:1884–1887. doi: 10.1038/nprot.2007.269. [DOI] [PubMed] [Google Scholar]
- 31.Szychowski J, Mahdavi A, Hodas JJ, Bagert JD, et al. Cleavable biotin probes for labeling of biomolecules via azide-alkyne cycloaddition. J Am Chem Soc. 2010;132:18351–18360. doi: 10.1021/ja1083909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 33.Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- 34.Bell C, Smith GT, Sweredoski MJ, Hess S. Characterization of the Mycobacterium tuberculosis Proteome by Liquid Chromatography Mass Spectrometry-based Proteomics Techniques: A Comprehensive Resource for Tuberculosis Research. J Proteome Res. 2012;11:119–130. doi: 10.1021/pr2007939. [DOI] [PubMed] [Google Scholar]
- 35.Heukeshoven J, Dernick R. Characterization of a solvent system for separation of water-insoluble poliovirus proteins by reversed-phase high-performance liquid chromatography. J Chromatogr. 1985;326:91–101. doi: 10.1016/s0021-9673(01)87434-3. [DOI] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cajigas IJ, Tushev G, Will TJ, tom Dieck S, et al. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high resolution imaging. Neuron. 2012 doi: 10.1016/j.neuron.2012.02.036. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prior P, Schmitt B, Grenningloh G, Pribilla I, et al. Primary structure and alternative splice variants of gephyrin, a putative glycine receptor-tubulin linker protein. Neuron. 1992;8:1161–1170. doi: 10.1016/0896-6273(92)90136-2. [DOI] [PubMed] [Google Scholar]
- 39.Lin CR, Kapiloff MS, Durgerian S, Tatemoto K, et al. Molecular cloning of a brain-specific calcium/calmodulin-dependent protein kinase. Proc Natl Acad Sci U S A. 1987;84:5962–5966. doi: 10.1073/pnas.84.16.5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galvin K, Krishna S, Ponchel F, Frohlich M, et al. The major histocompatibility complex class I antigen-binding protein p88 is the product of the calnexin gene. Proc Natl Acad Sci U S A. 1992;89:8452–8456. doi: 10.1073/pnas.89.18.8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho K, Brown MW, Bashir ZI. Mechanisms and physiological role of enhancement of mGlu5 receptor function by group II mGlu receptor activation in rat perirhinal cortex. J Physiol. 2002;540:895–906. doi: 10.1113/jphysiol.2001.013920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai T, Sanjo H, Akira S. Duet is a novel serine/threonine kinase with Dbl-Homology (DH) and Pleckstrin-Homology (PH) domains. Gene. 1999;227:249–255. doi: 10.1016/s0378-1119(98)00605-2. [DOI] [PubMed] [Google Scholar]
- 43.Rossner S, Fuchsbrunner K, Lange-Dohna C, Hartlage-Rubsamen M, et al. Munc13-1-mediated vesicle priming contributes to secretory amyloid precursor protein processing. J Biol Chem. 2004;279:27841–27844. doi: 10.1074/jbc.C400122200. [DOI] [PubMed] [Google Scholar]
- 44.Inagaki N, Mizuta M, Seino S. Cloning of a mouse Rabphilin-3A expressed in hormone-secreting cells. J Biochem. 1994;116:239–242. doi: 10.1093/oxfordjournals.jbchem.a124512. [DOI] [PubMed] [Google Scholar]
- 45.England SK, Uebele VN, Shear H, Kodali J, et al. Characterization of a voltage-gated K+ channel beta subunit expressed in human heart. Proc Natl Acad Sci U S A. 1995;92:6309–6313. doi: 10.1073/pnas.92.14.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han L, Wong D, Dhaka A, Afar D, et al. Protein binding and signaling properties of RIN1 suggest a unique effector function. Proc Natl Acad Sci U S A. 1997;94:4954–4959. doi: 10.1073/pnas.94.10.4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masuda M, Yageta M, Fukuhara H, Kuramochi M, et al. The tumor suppressor protein TSLC1 is involved in cell-cell adhesion. J Biol Chem. 2002;277:31014–31019. doi: 10.1074/jbc.M203620200. [DOI] [PubMed] [Google Scholar]
- 48.Li ML, Latoud C, Center MS. Cloning and sequencing a cDNA encoding human ribosomal protein S25. Gene. 1991;107:329–333. doi: 10.1016/0378-1119(91)90335-9. [DOI] [PubMed] [Google Scholar]
- 49.Ren Y, Li R, Zheng Y, Busch H. Cloning and characterization of GEF-H1, a microtubule-associated guanine nucleotide exchange factor for Rac and Rho GTPases. J Biol Chem. 1998;273:34954–34960. doi: 10.1074/jbc.273.52.34954. [DOI] [PubMed] [Google Scholar]
- 50.Ge H, Zuo P, Manley JL. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 51.Brinkmann U, Brinkmann E, Gallo M, Pastan I. Cloning and characterization of a cellular apoptosis susceptibility gene, the human homologue to the yeast chromosome segregation gene CSE1. Proc Natl Acad Sci U S A. 1995;92:10427–10431. doi: 10.1073/pnas.92.22.10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren M, Drivas G, D'Eustachio P, Rush MG. Ran/TC4: a small nuclear GTP-binding protein that regulates DNA synthesis. J Cell Biol. 1993;120:313–323. doi: 10.1083/jcb.120.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deane R, Schafer W, Zimmermann HP, Mueller L, et al. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differently with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arredondo-Vega FX, Charlton JA, Edwards YH, Hopkinson DA, Whitehouse DB. Isozyme and DNA analysis of human S-adenosyl-L-homocysteine hydrolase (AHCY) Ann Hum Genet. 1989;53:157–167. doi: 10.1111/j.1469-1809.1989.tb01780.x. [DOI] [PubMed] [Google Scholar]
- 55.Price SR, Nightingale MS, Bobak DA, Tsuchiya M, et al. Conservation of a 23-kDa human transplantation antigen in mammalian species. Genomics. 1992;14:959–964. doi: 10.1016/s0888-7543(05)80117-x. [DOI] [PubMed] [Google Scholar]
- 56.Saito A, Watanabe TK, Shimada Y, Fujiwara T, et al. cDNA cloning and functional analysis of p44.5 and p55, two regulatory subunits of the 26S proteasome. Gene. 1997;203:241–250. doi: 10.1016/s0378-1119(97)00524-6. [DOI] [PubMed] [Google Scholar]
- 57.Tamura T, Lee DH, Osaka F, Fujiwara T, et al. Molecular cloning and sequence analysis of cDNAs for five major subunits of human proteasomes (multicatalytic proteinase complexes) Biochim Biophys Acta. 1991;1089:95–102. doi: 10.1016/0167-4781(91)90090-9. [DOI] [PubMed] [Google Scholar]
- 58.Akiyama K, Yokota K, Kagawa S, Shimbara N, et al. cDNA cloning and interferon gamma down-regulation of proteasomal subunits X and Y. Science. 1994;265:1231–1234. doi: 10.1126/science.8066462. [DOI] [PubMed] [Google Scholar]
- 59.Nelbock P, Dillon PJ, Perkins A, Rosen CA. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science. 1990;248:1650–1653. doi: 10.1126/science.2194290. [DOI] [PubMed] [Google Scholar]
- 60.Xu X, Shi Y, Wu X, Gambetti P, et al. Identification of a novel PSD-95/Dlg/ZO-1 (PDZ)-like protein interacting with the C terminus of presenilin-1. J Biol Chem. 1999;274:32543–32546. doi: 10.1074/jbc.274.46.32543. [DOI] [PubMed] [Google Scholar]
- 61.Ueda K, Fukushima H, Masliah E, Xia Y, et al. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:11282–11286. doi: 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, et al. Cell-selective metabolic labeling of proteins. Nat Chem Biol. 2009;5:715–717. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


