Abstract
Objective
To determine whether variations in gene expression exist at multiple subsites along the sinonasal tract in patients with chronic sinusitis with polyps and in healthy controls.
Study Design
Prospective, controlled study.
Setting
Academic medical center.
Subjects and Methods
Tissue expression levels of 5 genes, previously found to be characteristic of ethmoid polyps, were measured using real-time quantitative polymerase chain reaction in 100 sinonasal tissue samples. Specimens harvested from 5 regions—the ethmoid sinus, septum, inferior turbinate, middle turbinate, and lateral nasal wall—in 10 patients with chronic sinusitis and ethmoid polyps were compared to tissue from similar regions in 10 control patients without sinusitis. Western blot analysis was performed to validate differential gene expression at the protein level.
Results
Gene expression levels of ethmoid polyps differed significantly from those of healthy ethmoid mucosa, as well as tissue from 4 surrounding anatomical sites in both patients with chronic sinusitis and controls. Alterations specific to the polyp tissue included downregulated genes, prolactin-induced protein (fold change 377.2 ± 169.0, P < .0001), and zinc α2-glycoprotein (fold change 72.1 ± 26.5, P < .0001), as well as upregulated genes, met proto-oncogene (fold change 2.5 ± 0.7, P = .029), and periostin (fold change 7.5 ± 3.4, P = .003). No significant differences in gene expression was found for neurabin 2 (fold change 1.0, P = .99).
Conclusion
The transcriptional pattern of ethmoid polyps appears to be unique compared with other subsites in the sinonasal cavity of patients with chronic sinusitis. Care must be taken when collecting specimens for molecular studies of the sinonasal tract to differentiate polyp from nonpolyp tissue in chronic sinusitis.
Keywords: sinonasal polyposis, nasal polyp, chronic rhinosinusitis, sinusitis, periostin, met proto-oncogene, prolactin-inducible protein, zinc α2-glycoprotein, protein phosphatase 1
Chronic sinusitis with polyps has long remained a challenging disease for both patients and their treating physicians. Although medical and surgical therapies can temporarily alleviate symptoms, there is no cure for most patients with this disorder. Regrowth of polyps and recurrent infections have been demonstrated to have an adverse impact on both healthcare expenditures1 and quality of life.2
Although nasal polyps are most commonly reported to arise in the regions of the middle meatus3 and anterior ethmoid sinus,4 they may originate in the other paranasal sinuses, the turbinates, or even the nasal septum. It remains unknown whether the regional variations observed in polyp development are secondary to site-specific genetic factors, the location of disease onset, or differential exposure to environmental stimuli.
To better understand the underlying pathophysiology, a number of research groups have examined gene expression profiles of sinus polyps. The expression profile of a diseased tissue, however, has little meaning unless it can be compared to a control tissue with presumed “normal” expression. Published studies have used a variety of different locations as controls for polyp tissue, including adjacent nonpolyp mucosa from the same patient,5 ethmoid tissue from patients without chronic rhinosinusitis (CRS),6 and most commonly inferior turbinate biopsies of patients without CRS.7 If regional variations in sinonasal gene expression do in fact exist, this finding would be critically important for future study design and interpretation. The purpose of the present study was to investigate whether variations in gene expression exist at multiple subsites along the sinonasal tract in patients with chronic sinusitis with polyps and healthy controls.
Materials and Methods
Study Population
The study group consisted of patients undergoing endoscopic sinus surgery for CRS with polyps (polyp group n = 10), as defined by accepted diagnostic criteria of the American Academy of Otolaryngology–Head and Neck Surgery. To ensure as homogeneous a study population as possible, enrollment required the presence of bilateral frank polyps extending from the middle meatus as seen on rigid endoscopy as well as pansinus opacification on computed tomography (CT) scan. Exclusion criteria included age younger than 18 years, aspirin-exacerbated respiratory disease, cystic fibrosis, antrochoanal polyps, and use of oral steroids within 1 month of surgery. The control group (n = 10) consisted of patients undergoing endoscopic dacryocystorhinostomy for epiphora or orbital decompression for Graves disease, with no evidence of sinusitis or nasal polyps on history, nasal endoscopy, or CT scan. Informed consent was obtained from all subjects according to the institutional review board (IRB) study protocol approved by the Human Studies Committee of the Massachusetts Eye and Ear Infirmary.
Tissue Collection
A total of 100 sinonasal tissue samples were collected from 5 distinct anatomical sites. Study specimens were obtained under general anesthesia at the start of the patient’s surgical procedure. In patients with chronic sinusitis, polyp tissue emanating from the ethmoid sinus was harvested. Separate mucosal biopsies from the same side were taken from non-polyp tissue of the septum, inferior turbinate, middle turbinate, and lateral nasal wall (overlying the lacrimal sac). From control patients, mucosa was obtained from the ethmoid sinus (ethmoid bulla), septum, inferior turbinate, middle turbinate, and lateral nasal wall. Tissue samples were collected, rinsed in normal saline, placed in RNAlater (Ambion, Austin, Texas), and stored at −80°C until time of RNA extraction.
RNA Extraction
Total RNA was purified using RNeasy spin columns (QIAGEN, Valencia, California) according to the manufacturer’s protocol and a modification for hypocellular, dense connective tissues as previously described.8,9 Quantification and quality assessment of the RNA were performed using Agilent 2100 Bioanalyzer and RNA Pico Kit (Agilent Technologies, Santa Clara, California). Only samples that yielded clean and undegraded RNA based on the appearance of electropherograms or RNA integrity numbers greater than 7 were used. The RNA was reverse transcribed with the Taqman Reverse Transcription Reagents kit (Applied Biosystems, Foster City, California).
Studied Genes
Expression levels were determined for 5 genes previously shown to be differentially expressed by sinus polyps: mesenchymal-epithelial transition factor (MET), periostin, protein phosphatase 1 regulatory subunit 9B (PPP1R9B), prolactin-inducible protein (PIP), and zinc α2-glycoprotein (AZGP1). These genes were chosen based on the results of prior genome-wide expression profiling where these 5 genes were found to be most characteristic of ethmoid polyps by bioinformatic analysis, including supervised and unsupervised clustering and gene set enrichment analysis.6 For each gene, expression in nasal polyps was either upregulated (MET, periostin, PPP1R9B) or downregulated (PIP, AZGP1) compared to control tissue by both polymerase chain reaction (PCR) and immunohistochemistry (IHC).
RT-PCR Quantification of Relative mRNA
Real-time quantitative PCR (RT-qPCR) was performed for each gene in each location using 6-FAM linked fluorescent probes and primers designed and optimized by Applied Biosystems. The measurements were carried out in duplicate or triplicate on an Applied Biosystems 7700 Sequence Detector using 96-well plates and conditions as previously described.10 Gene expression levels were quantified relative to the 18S rRNA gene and compared between groups using the comparative threshold cycle (CT) method (ie, the ΔΔCT method).11
Western Blot
Protein was extracted from ethmoid polyps (chronic sinusitis, n = 6) and ethmoid mucosa (control, n = 6) from independent samples in each group. Tissue was homogenized using RIPA buffer (Sigma-Aldrich, St Louis, Missouri). Protein concentration was determined by absorbance spectroscopy, and equal volumes were loaded onto 4% to 20% Tris glycine gel with SeaBlue Plus2 marker and Novex SDS buffer (Invitrogen, Carlsbad, California). Electrophoresis was performed at 120 mV until proteins separated in the gel. The gel was transferred to a PDVF membrane (Millipore, Billerica, Massachusetts) at 300 milliamps for 60 minutes. Membranes were blocked with 5% bovine serum albumin and incubated for 16 hours with primary antibodies to MET (Santa Cruz Biotechnology, Santa Cruz, California), periostin (Abcam, Cambridge, Massachusetts), PIP (Santa Cruz Biotechnology), AZGP1 (Biovendor, Candler, North Carolina), and β-actin (Cell Signaling Technologies, Danvers, Massachusetts). Membranes were washed and then incubated for 1 hour with horseradish peroxidase–conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, Pennsylvania) and developed using the ECL Western blot detection kit and photographic film (GE Healthcare, Piscataway, New Jersey).
Statistical Analysis
A repeated-measures analysis was used to compare the overall regional differences between gene expression levels within nasal polyp patients and controls. Two-sample t tests were used to compare gene expression between nasal polyp and control groups by both subsite and individual genes. To account for multiple comparisons, a P value of .005 (.05/10) was considered statistically significant, based on the Bonferroni adjustment, when comparing subsites within each study group (a total of 10 comparisons). Similarly, a P value of .002 (.05/25) was considered significant when directly comparing between the 2 study groups (a total of 25 comparisons).
Results
Demographic and medical data for study patients are listed in Table 1. In patients with polyps, a statistically significant alteration in gene expression was seen between polyp tissue and each of the other anatomical subsites for 4 of 5 genes (MET, P = .002; periostin, P < .0001; PIP, P < .0001; AZGP1, P < .0001). There were no differences in gene expression between nonpolyp mucosa of the septum, inferior turbinate, middle turbinate, or lateral nasal wall. Within this group, the effects of asthma (P = .37), patient age (P = .28), number of prior surgeries (P = .13), and gender (P = .24) showed no significant differences among study subjects for all genes. In control subjects, levels of gene expression at the 5 anatomical subsites did not show significant variation for the 5 genes studied (P > .005).
Table 1.
Patient Demographics
| Patient Group | No. | Men, % | Age, Mean ± SD, y (Range) | LM CT Stage, Mean ± SD (Range) | Asthma, No. (%) | Prior Sinus Surgery, Mean ± SD (Range) |
|---|---|---|---|---|---|---|
| CRS | 10 | 7 (70) | 53.4 ± 10.5 (39–73) | 19.8 ± 3.2 (14–24) | 5 (50) | 1.9 ± 1.9 (0–5) |
| Control | 10 | 4 (40) | 56.0 ± 12.6 (38–80) | 0.7 ± 1.1a (0–3) | 0 (0) | 0 |
Abbreviations: CRS, chronic sinusitis; CT, computed tomography; LM, Lund-Mackay CT scan scoring system.
Three patients had incidental findings of mucous retention cysts that were not associated with active sinusitis.
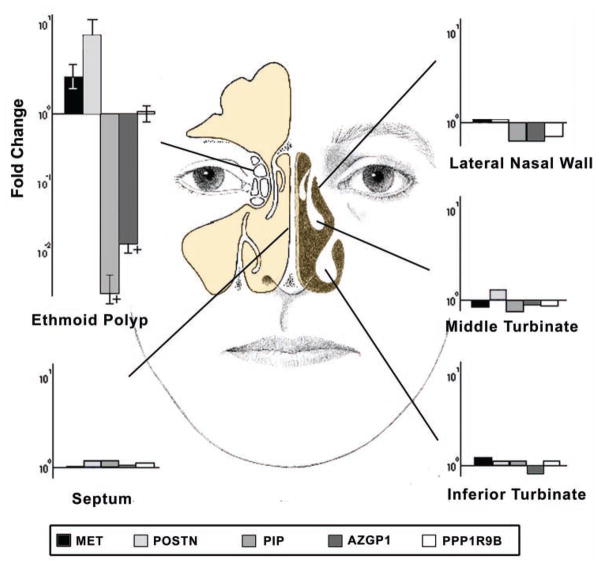
When comparing polyp patients to control patients, gene expression levels for ethmoid polyp tissue were different from each control subsite evaluated, including normal ethmoid mucosa, septum, inferior turbinate, middle turbinate, and lateral nasal wall (Figure 1). As expected, statistically significant downregulation of PIP (fold change 377.2 ± 169.0, P < .0001) and AZGP1 (fold change 72.1 ± 26.5, P < .0001) was found in polyp tissue compared to each control subsite, whereas MET (fold change 2.5 ± 0.7, P = .029) and periostin (fold change 7.5 ± 3.4, P = .003) were upregulated in polyp tissue. In polyp patients, nonpolyp mucosal subsites demonstrated equivalent gene expression compared to each similar subsite in control patients (P = .21–.95; Figure 1). PPP1R9B expression did not show any difference between subsites in polyp patients (fold change 1.0, P = .99) vs controls. Western blot analysis of MET, periostin, PIP, and AZGP1 confirmed differential protein expression between ethmoid polyps and control mucosa (Figure 2).
Figure 1.
Comparison of gene expression of 5 genes at 5 distinct anatomical subsites within the sinonasal cavity in patients with chronic sinusitis and nasal polyps (n = 10) relative to expression in mucosa from identical subsites in control patients without sinusitis. All specimens were collected from the same side in each patient; + indicates statistically significant differences at P ≤ .002). MET indicates met proto-oncogene; POSTN, periostin; PIP, prolactin-induced protein; AZGP1, zinc α2-glycoprotein; PPP1R9B, protein phosphatase 1 regulatory subunit 9B. Bars indicate +/− standard error of the mean.
Figure 2.
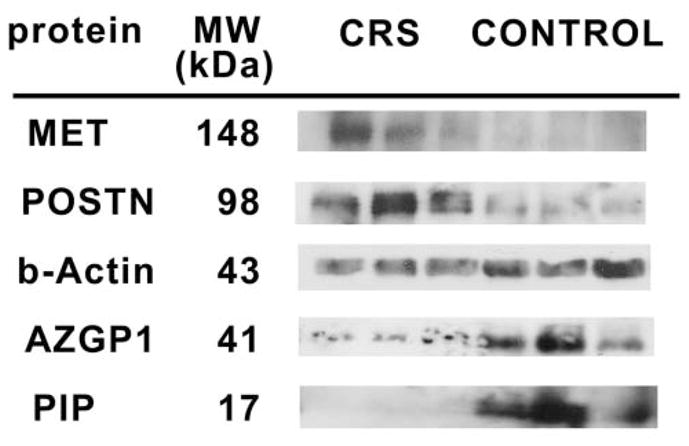
Western blot analysis of protein expression of genes found to be significantly dysregulated in ethmoid polyp tissue from patients with chronic sinusitis (CRS; n = 3) compared to control ethmoid mucosa in patients without sinusitis (n = 3). MET indicates met proto-oncogene; POSTN, periostin; PIP, prolactin-induced protein; AZGP1, zinc α2-glycoprotein. β-Actin serves as a loading control.
Discussion
This is the first report that demonstrates a unique gene expression pattern for polyps compared to multiple subsites in the sinonasal cavity. Previous studies have demonstrated similarities among different regions in the airway. The unified airway theory recognizes the similar linings of the upper and lower respiratory tracts and parallel disease processes, such as asthma and allergic rhinitis.12 Similar patterns of mucosal inflammation have been observed in nasal polyps and middle turbinates in patients with chronic sinusitis, suggesting a diffuse mucosal involvement in the disease process.13 Regional alterations in mucosal function have been examined by comparing ciliary beat frequency in sinonasal epithelial explants from the inferior turbinate, uncinate process, and sphenoethmoid recess.14 No differences were found in ciliary beat frequency, and it was concluded that the inferior turbinate is a valid location for study of ciliary motility within the sinonasal cavity.14 The regional alterations in sinonasal gene expression seen in the present study also have implications for the study of upper and lower airway diseases.
The inferior turbinate is a common location to obtain biopsy15 and brushings16 of epithelial cells for research purposes in a number of disorders, including nasal polyps, CRS without polyps, and lower airway diseases such as asthma and chronic obstructive pulmonary disease (COPD). Comparison of upper and lower airway epithelial cells for the study of inflammatory diseases revealed that nasal epithelial cultures constitute an easily accessible surrogate for studying lower airway inflammation.17 The findings of altered gene expression in ethmoid polyps and consistent expression at other sites support use of the inferior turbinate as a proxy for tissue in the remainder of the sinonasal cavity, except for patients with polyp disease. Care must be taken when collecting specimens for study of the airway to differentiate polyp from nonpolyp tissue.
The validity of any study is dependent on the control group that is chosen for comparison. The strength of prior gene expression profiling experiments for sinus polyps rests in part on how well the control tissue represents the sinonasal cavity. The current study examined 5 distinct anatomic subsites within the sinonasal cavity of patients with sinus polyps and controls. Four of the 5 previously highlighted genes were again found to be differentially expressed in polyp tissue as compared to nonpolyp tissue from both polyp patients and controls. The finding of equivalent gene expression at the nonpolyp subsites further supports the conclusion that the expression pattern of these genes is unique to the polyp tissue itself and not simply related to chronic mucosal inflammation (as might be seen in nonpolyp subsites of patients with polyps) or to regional variation (as might be seen in subsites of controls without sinonasal disease).
The current study found similar expression of PPP1R9B in polyp tissue and nonpolyp tissue, which contradicts prior work6 showing differential expression of this gene. The discrepancy in PPP1R9B expression is most likely due to sample bias in the initial experiments. An alternative explanation is that additional unrecognized distinct polyp subgroups exist with varying gene expression of PPP1R9B. A limitation of polyp study is the inherent difficulty in recognizing a uniform phenotype in a complex, multifactorial disease.
Although differential expression of MET, periostin, PIP, and AZGP1 is characteristic of sinus polyps, the clinical significance of this finding remains undefined. The known functions of these genes certainly suggest that they may play a vital role in polyp development and/or maintenance.18–20 Periostin is a protein secreted by fibroblasts that promotes cell adhesion and autocrine activation and is recognized to be a potent regulator of fibrosis and collagen deposition.21 PIP is secreted by various apocrine glands and has been implicated in host defense mechanisms against infections and tumors.22 AZGP1 is a member of a distinct, heterogeneous lineage of major histocompatibility complex class I genes whose function has included anti-infectious and tumor immunity. If future studies prove these gene products to be critical to polyp development, then directed treatments could be efficacious. These genes currently have value as biomarkers of the polyp state and targets for future research.
Finally, an inherent limitation of this study relates to the use of sinonasal tissue from patients with Graves disease to serve as control specimens. Sinus mucosal thickening23 and alterations in gene expression24 have been reported in patients with this autoimmune disorder. Although none of the Graves patients in this study had evidence of mucosal thickening on CT scan or altered expression for the studied genes, the inclusion of such patients could affect study results.
Conclusion
Ethmoid polyps in chronic sinusitis appear to have a unique transcriptional pattern compared to other nonpolyp subsites in the sinonasal cavity of patients with and without sinusitis. The inferior turbinate mucosa is a viable representation of the sinonasal cavity for the selected genes in this study of airway disease and possibly other airway diseases; however, polyp tissue must be differentiated from nonpolyp mucosa. Further research is necessary to determine the significance of differential expression of MET, periostin, PIP, and AZGP1 in the development and/or maintenance of polyp tissue.
Acknowledgments
Funding source: This project was funded by the Massachusetts Eye and Ear Infirmary Research Foundation. KMS is supported by grants from NIH-NIDCD K08DC010419-01, Massachusetts Life Sciences Center, the Boston Foundation, and the Shore Fellowship at Harvard Medical School.
Footnotes
Reprints and permission: sagepub.com/journalsPermissions.nav
Author Contributions
Michael P. Platt, concept and design, acquisition of data, analysis and interpretation of data, drafting the article, final approval; Zachary M. Soler, analysis and interpretation of data, critical revision, final approval; Shyan-Yuan Kao, interpretation of data, critical revision, final approval; Ralph Metson, concept and design, acquisition of data, critical revision, final approval; Konstantina M. Stankovic, concept and design, analysis and interpretation of data, critical revision, final approval.
Disclosures
Competing interests: None.
Sponsorships: None.
References
- 1.Lund VJ. Impact of chronic rhinosinusitis on quality of life and health care expenditure. Clin Allergy Immunol. 2007;20:15–24. [PubMed] [Google Scholar]
- 2.Gliklich R, Metson R. Economic implications of chronic sinusitis. Otolaryngol Head Neck Surg. 1998;118:344–349. doi: 10.1016/S0194-59989870313-4. [DOI] [PubMed] [Google Scholar]
- 3.Andrews AE, Bryson JM, Rowe-Jones JM. Site of origin of nasal polyps: relevance to pathogenesis and management. Rhinology. 2005;43:180–184. [PubMed] [Google Scholar]
- 4.Larsen PL, Tos M. Site of origin of nasal polyps. Rhinology. 1995;33:185–188. [PubMed] [Google Scholar]
- 5.Fritz SB, Terrell JE, Conner ER, et al. Nasal mucosal gene expression in patients with allergic rhinitis with and without nasal polyps. J Allergy Clin Immunol. 2003;112:1057–1063. doi: 10.1016/j.jaci.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 6.Stankovic KM, Goldsztein H, Reh DD, Platt MP, Metson R. Gene expression profiling of nasal polyps associated with chronic sinusitis and aspirin-sensitive asthma. Laryngoscope. 2008;118:881–889. doi: 10.1097/MLG.0b013e31816b4b6f. [DOI] [PubMed] [Google Scholar]
- 7.Wang X, Dong Z, Zhu DD, Guan B. Expression profile of immune-associated genes in nasal polyps. Ann Otol Rhinol Laryngol. 2006;115:450–456. doi: 10.1177/000348940611500609. [DOI] [PubMed] [Google Scholar]
- 8.Reno C, Marchuk L, Sciore P, Frank CB, Hart DA. Rapid isolation of total RNA from small samples of hypocellular, dense connective tissues. Biotechniques. 1997;22:1082–1086. doi: 10.2144/97226bm16. [DOI] [PubMed] [Google Scholar]
- 9.Stankovic KM, Kristiansen AG, Bizaki A, Lister M, Adams JC, McKenna MJ. Studies of otic capsule morphology and gene expression in the Mov13 mouse—an animal model of type I osteogenesis imperfecta. Audiol Neurootol. 2007;12:334–343. doi: 10.1159/000104789. [DOI] [PubMed] [Google Scholar]
- 10.Grove D. Quantitative real-time polymerase chain reaction for the core facility using TaqMan and the Perkin-Elmer/Applied Biosystems Division 7700 Sequence Detector. J Biomol Tech. 1999;10:11–16. [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Krouse JH, Veling MC, Ryan MW, et al. Executive summary: asthma and the unified airway. Otolaryngol Head Neck Surg. 2007;136:699–706. doi: 10.1016/j.otohns.2007.02.023. [DOI] [PubMed] [Google Scholar]
- 13.Hao J, Pang YT, Wang DY. Diffuse mucosal inflammation in nasal polyps and adjacent middle turbinate. Otolaryngol Head Neck Surg. 2006;134:267–275. doi: 10.1016/j.otohns.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 14.Shaari J, Palmer JN, Chiu AG, et al. Regional analysis of sinonasal ciliary beat frequency. Am J Rhinol. 2006;20:150–154. [PubMed] [Google Scholar]
- 15.Devalia JL, Sapsford RJ, Wells CW, Richman P, Davies RJ. Culture and comparison of human bronchial and nasal epithelial cells in vitro. Respir Med. 1990;84:303–312. doi: 10.1016/s0954-6111(08)80058-3. [DOI] [PubMed] [Google Scholar]
- 16.Bridges MA, Walker DC, Davidson AG. Cystic fibrosis and control nasal epithelial cells harvested by a brushing procedure. In Vitro Cell Dev Biol. 1991;27A:684–686. doi: 10.1007/BF02633211. [DOI] [PubMed] [Google Scholar]
- 17.McDougall CM, Blaylock MG, Douglas JG, et al. Nasal epithelial cells as surrogates for bronchial epithelial cells in airway inflammation studies. Am J Respir Cell Mol Biol. 2008;39:560–568. doi: 10.1165/rcmb.2007-0325OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic program for cancer and stem cells. Nat Rev Cancer. 2006;6:637–645. doi: 10.1038/nrc1912. [DOI] [PubMed] [Google Scholar]
- 19.Gentile A, Trusolino L, Comoglio PM. The Met tyrosine kinase receptor in development and cancer. Cancer Metastasis Rev. 2008;27:85–94. doi: 10.1007/s10555-007-9107-6. [DOI] [PubMed] [Google Scholar]
- 20.Birchmeier C, Birchmeier W, Gherardi E, Vande Woude GF. Met, metastasis, motility and more. Nat Rev Mol Cell Biol. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- 21.Norris RA, Damon B, Mironov V, et al. Periostin regulates collagen fibrillogenesis and the biomechanical properties of connective tissues. J Cell Biochem. 2007;101:695–711. doi: 10.1002/jcb.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci U S A. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gouveris HT, Al-Homsi J, Gosepath J, Mann WJ. Histological and radiological signs indicative for chronic sinus mucosal inflammation in Graves’ ophthalmopathy. Rhinology. 2009;47:144–147. [PubMed] [Google Scholar]
- 24.Soler ZM, Platt MP, Leung MK, Mong S, Metson R. Sinonasal abnormalities in patients with Graves’ orbitopathy. Laryngoscope. 2010 Nov 18; doi: 10.1002/lary.21392. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]